Abstract
Studies investigating mechanisms that control gene regulation frequently examine the accessibility of specific DNA sequences to nuclease cleavage. In general, sequences that are sensitive to nuclease cleavage are considered to be in an “open” chromatin conformation that is associated with regulatory factor binding, while sequences resistant to nuclease cleavage are considered to be in a “closed” conformation commonly associated with chromatin that is neither poised for transcription nor being actively transcribed. Changes in nuclease accessibility at specific genomic sequences reflect changes in the local chromatin structure that can occur as a result of signaling cues in the extracellular environment. These changes in chromatin structure usually precede or are coincident with changes in gene expression patterns and are therefore a useful marker of regulatory events controlling transcription.
We describe a method to perform restriction enzyme accessibility assays (REAA) that utilizes ligation-mediated polymerase chain reaction (LM-PCR) technology and that permits assessment of samples from any source containing as few as 1,000 cells. Use of this modified REAA protocol will enhance analysis of chromatin structural changes at specific DNA sequences of interest by making it possible to analyze samples where unrestricted amounts of sample are not readily available.
Keywords: Restriction enzyme accessibility assay, Chromatin accessibility, Chromatin remodeling enzymes, Skeletal muscle, Satellite cells
1. Introduction
Eukaryotic genomic DNA is packaged around histone octamers to form nucleosomes. Nucleosomes form arrays on the DNA and fold and condense to form filaments and fibers of chromatin, which is the natural state of the genomic DNA in a eukaryotic cell nucleus. However, chromatin structure is not uniform across the nucleus; indeed it is dynamic and can change as a function of input from a multitude of signaling pathways regulating nearly every biological process and response. In particular, it was recognized many years ago that the structure of those portions of the genome encoding actively transcribed genes was inherently different from genomic regions devoid of genes or containing transcriptionally silent genes. This conclusion was based on the observation that different genomic regions exhibited differential sensitivity to digestion with DNAse I. Specifically, this study demonstrated that the globin genes in nuclei isolated from chicken erythrocytes were more accessible to cleavage by DNAse I than were the globin sequences in nuclei isolated from other cell types in which globin is not expressed (1). Subsequent studies expanded the concept of differential DNAse I accessibility by demonstrating that specific sequences within DNAse I accessible chromatin at active genes were hypersensitive to enzyme cleavage (2, 3) and that these sequences likely corresponded to regulatory regions controlling the expression of the gene being analyzed (4). Work from many laboratories over the following years clearly established the idea that DNAse I hypersensitivity frequently correlated with the location of cis-acting regulatory sequences (reviewed in refs. (5–7)).
Restriction endonuclease enzymes have also been used to probe accessibility of genomic DNA, first to analyze satellite DNA sequences (8), and later to also demonstrate that actively transcribed genes are more accessible than nontranscribed genes (9). A significant advantage to the use of restriction enzymes as opposed to DNAse I is that the sequence specificity with which they cleave DNA clearly delineates the site of DNA cleavage. However, the sequence specificity also limits use of the assay to genomic regions near where the restriction enzyme recognition sequence exists. Typically, if the restriction enzyme cleavage site is incorporated into a nucleosome, cleavage is inhibited. In contrast, if nucleosome positioning is altered or nucleosomes are remodeled or otherwise structurally altered, the restriction enzyme cleavage site usually becomes more accessible to the enzyme and results in an increased frequency of cleavage. For these reasons, restriction enzyme accessibility is a useful assay to generally measure changes in chromatin structure at specific gene loci and has been particularly useful in monitoring structural changes caused by chromatin remodeling enzymes. It is important to note that changes in nuclease accessibility do not structurally define the changes that are occurring to genomic chromatin. Instead, they simply indicate that a structural change has occurred.
Although some of the initial studies utilized solution hybridization to detect cleaved genomic DNA, the field quickly turned to detection of nuclease cleavage by Southern blot. In this technique, the nuclease-digested chromatin is deproteinated and purified, then subjected to restriction enzyme cleavage at sites flanking the sequence of interest to create a restriction fragment of known size. If cleavage of genomic DNA at the sequence of interest occurred, some fraction of the specific restriction fragment will be reduced in size. When the sample is separated on an agarose gel, transferred to a membrane and then hybridized with a suitably labeled probe to the sequences of interest; detection of the cleaved restriction fragment reflects the extent of nuclease accessibility, and by extension, the extent of change to the local chromatin structure.
The advent of PCR-based technologies led to the development of ligation-mediated polymerase chain reaction (LM-PCR) as a means to greatly increase the sensitivity of detecting cleaved genomic DNA (10). Genomic DNA cleaved by enzymatic or chemical methods is deproteinated and processed as necessary to prepare double-stranded fragments suitable for ligating to a defined linker DNA (11). Amplification of the cleaved genomic fragment can then be accomplished by PCR using one primer to the linker sequence and a second primer that hybridizes to the genomic DNA of interest near the cleavage site. Because of the ability to PCR amplify from small amounts of starting material, this method greatly increases the sensitivity of detection. Thus, it is ideal for analyzing chromatin accessibility when starting with limiting amounts of material. More detailed explanation of LM-PCR use to measure regions of genomic DNA cleaved by restriction endonucleases is presented below.
2. Materials
2.1. Isolation of Nuclei
Phosphate-Buffered Saline (PBS) pH 7.4: For 1 L of 1 × PBS, add 8 g sodium chloride, 0.2 g potassium chloride, 1.44 g disodium hydrogen phosphate, and 0.24 g potassium dihydrogen phosphate to 800 mL deionized water. Adjust pH to 7.4 with hydrochloric acid and make up the volume to 1 L with deionized water.
Rubber policeman or cell scraper.
1 M Piperazine-N,N'-bis(2-ethane-sulfonic acid) (PIPES) pH 8.0 stock solution.
1 M Potassium chloride (KCl) stock solution.
1 M Calcium chloride (CaCl2) stock solution.
10% Nonidet P-40 (NP-40) stock solution.
1 mg/mL Leupeptin stock solution: Dissolve 10 mg of leupeptin into 10 mL of deionized water and store in aliquots at −20°C (stable up to 6 months).
0.1 M Phenylmethanesulfonylfluoride (PMSF) stock solution: Add 174 mg of PMSF to 10 mL of isopropanol and dissolve by vortexing or rotation on a rotating platform.
Tissue culture cell (TCC) lysis buffer (10 mM PIPES pH 8, 85 mM KCl, 1 mM CaCl2, 5% sucrose, 0.5% NP-40, plus protease inhibitors added immediately before use, including 1 µg/mL leupeptin, and 1 mM PMSF): Prepare 500 mL of tcc buffer by dissolving 25 g sucrose in 400 mL of deionized water using a magnetic stirrer. Then add 5 mL of 1 M PIPES pH 8.0 stock solution, 42.5 mL of 1 M KCl stock solution, 0.5 mL of 1 M CaCl2 stock solution, and 25 mL of 10% NP-40 stock solution. Bring the volume to 500 mL. Immediately before use, aliquot the desired amount and add the protease inhibitors.
10 mL Dounce homogenizer with Pestle A.
Hoechst 33258 dye.
2.2. Restriction Enzyme Digestion and Ligation of Linker to Cleaved DNA
Restriction enzyme.
Appropriate 10× restriction enzyme reaction buffer.
DNeasy purification kit (Qiagen).
2.3. Preparation of Linker and Ligation of Linker to Cleaved DNA
1 M Tris–hydrochloride (Tris–HCl), pH 7.5 stock solution: For 1 L, add 121.1 g Tris to 800 mL of deionized water and dissolve using a magnetic stirrer. Adjust the pH to 7.5 with HCl and make up the volume to 1,000 mL using deionized water. Autoclave and store at room temperature.
0.5 M Ethylenediaminetetraacetic acid (EDTA) stock solution: Add 186.12 g EDTA to 800 mL of deionized water. Stir with a magnetic stirrer to begin to dissolve. Add NaOH pellets slowly to adjust the pH to 8.0. Note that EDTA will not fully dissolve unless the pH is near 8.0. Make up the volume to 1,000 mL using deionized water, autoclave and store at room temperature.
10× Annealing buffer (100 mM Tris, pH 7.5, 500 mM NaCl, 10 mM EDTA): For 50 mL, dissolve 0.58 g NaCl in 40 mL of deionized water. Add 5 mL of 1 M Tris–HCl pH 7.5 stock solution and 1 mL of 0.5 M EDTA stock solution. Bring the volume to 50 mL with deionized water. Filter sterilize through a 0.22 µm filter.
Mighty Mix Ligation kit (Takara).
RNAse/DNAse-free water.
3. Methods
3.1. Isolation of Nuclei
A detailed method for the isolation of nuclei from primary tissues is presented in the accompanying report (see Chapter 31). The same protocol can be used for isolation of nuclei from tissue culture cells, but we have found that gradient purification of the nuclei is not generally necessary. A slightly different method to isolate nuclei from a 100 mm plate of adherent tissue culture cells follows (see Notes 1 and 2).
Place the cell culture dishes containing the cells to be isolated on a tray of ice.
Aspirate or remove media covering the cells.
Wash the cells twice by slowly pouring or pipeting 5–10 mL of cold PBS onto the cells (see Note 3).
Add 1 mL of cold PBS to the cells.
Scrape cells off the plate with a rubber policeman or with a cell scraper. Hold the plate on an angle and use the scraper to push the cells and media to the bottom of the dish to facilitate collection.
Use a pipet or Pipetman to collect the cells in PBS and transfer to a prechilled 1.5 mL microcentrifuge tube.
Pellet by centrifugation at 300 × g for 2 min at 4°C.
Resuspend pellet in 500 µL of tcc lysis buffer.
Transfer suspension to a prechilled 10 mL Dounce homogenizer.
Lyse the cells by using a Dounce homogenizer with pestle A for seven to eight slow strokes on ice, taking care not to cause foaming. The optimum number of strokes may need to be determined empirically.
Check the release and integrity of nuclei under a light microscope after staining of a small aliquot (1–2 µL) with an equal volume of Hoechst 33258 dye; nuclei will stain blue while any remaining unlysed cells will clear the dye and appear unstained. If lysis is insufficient, repeat step 10.
Collect the resulting nuclei solution in a fresh, prechilled microcentrifuge tube.
Use 1–2 µL to determine the A260 reading using a spectrophotometer or spectrofluorometer.
3.2. Restriction Enzyme Digestion
The choice of restriction enzyme will be specific to the sequences of interest. Generally, a site at or within a few hundred base pairs of the sequence of interest should provide an accurate reflection of changes in local chromatin accessibility. Restriction enzymes inhibited by CpG methylation should be avoided. If multiple genomic sequences are of interest but do not share common restriction enzyme sites, multiple enzymes can be combined for genomic DNA digestion. The same considerations that exist for combining restriction enzymes to cleave naked DNA apply to genomic DNA digestion, including reaction components and total amount of enzyme added to the reaction. In practice, we have had success combining different restriction enzymes that will work in similar reaction conditions. Sequential digests are an alternate option.
Aliquot 100 µg of nuclei into a 1.5 mL microcentrifuge tube (see Notes 4 and 5).
Add the appropriate restriction enzyme 10× buffer, water, and the restriction enzyme of choice at 0.5 Units/µg nuclei. The reaction size should be approximately 100 µL and should not exceed 150 µL.
Digest for no more than 1 h at 37°C or the suggested optimum temperature for the restriction enzyme of choice (see Note 6).
Stop the digestion using the conditions recommended by the restriction enzyme manufacturer (see Note 7).
Purify the DNA using the DNeasy purification kit (Qiagen) in accordance with the manufacturer’s instructions. Elute the samples in 40 µL of elution buffer (supplied as part of the kit).
3.3. Preparation of Linker and Ligation of Linker to Cleaved DNA
We have frequently utilized the linkers LM-PCR1 and LM-PCR2 (11), which when annealed provide a linker suitable for ligation to genomic DNA cleaved with a blunt end cutter (see Fig. 1a; includes linker sequence). If the genomic DNA was cleaved with an enzyme leaving a 5' or 3' recessed end, the linker design can be modified to incorporate the bases complimentary to the existing overhang (see Fig. 1b). This strategy permits use of restriction enzymes that leave either a 3' or 5' recessed end.
To prepare the linker, mix 20 µL of each single stranded oligonucleotide from 100 pmol/µL stock in 1× annealing buffer in a 50 µL reaction volume, heat to 95°C for 5 min, incubate at 75°C for 15 min, followed by incubation at 35°C for 45 min, and cooling to 4°C to obtain 40 µM of double-stranded linker (see Notes 8 and 9).
Ligate 1 µg of purified digested DNA to 2.5 µL of 40 µM double-stranded linker oligonucleotide using the Mighty Mix Ligation kit (Takara) reagents as instructed by the manufacturer. The ligation reaction volume ideally should be 10 µL and should not exceed 20 µL (see Note 10).
Dilute the ligation reaction to 40–60 µL using RNAse/DNAse-free water. The sample may be stored at −20°C.
Fig. 1.
Schematic of alternate possibilities for linker ligation to restriction enzyme-cleaved genomic DNA. (a) ligation to blunt-ended genomic DNA. (b) ligation to genomic DNA with 3'- or 5'-recessed ends.
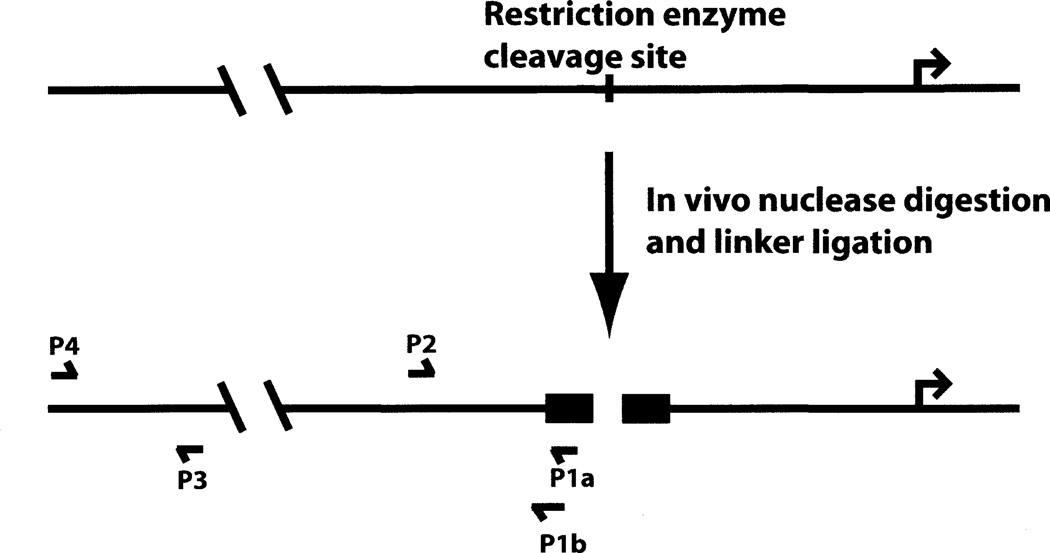
3.4 PCR Detection of Cleaved DNA
The standard amplification protocol utilizes a primer to the linker and a second primer upstream or downstream of the anticipated cleavage site (see Fig. 2; P1a and P2). This permits detection of the cleaved product, thereby providing direct measurement of increased enzyme accessibility at the genomic site examined. However, because the linker has been ligated onto every cleaved genomic DNA fragment, there is potential for increased background signal in the PCR reaction. One method to decrease background and improve specificity is to design the linker primer to span the linker sequence and the genomic sequence that is predicted to adjoin the linker (see Fig. 2; P1b and P2). Theoretically, this decreases the number of genomic fragments to which the primer can hybridize to the single fragment of interest. We have found this to be particularly helpful when performing experiments with limited amounts of starting material. Real-time PCR as a detection method also greatly improves the sensitivity of the assay as compared to conventional PCR, since this method allows quantification of the PCR product during the amplification stages of the reaction.
Fig. 2.
Schematic of ligation-mediated polymerase chain reaction (LM-PCR) protocol. P1a, P1b, P2, P3, and P4 represent primers that are described in the text.
Typically, a sequence upstream or downstream of the cleavage site of interest is amplified as a control for quantitative comparison of different samples in a particular experiment (see Fig. 2; P3 and P4). Alternatively, the quantity of a cleaved genomic fragment can be compared to input by using primers that flank the cleavage site of interest for amplification of undigested genomic DNA.
- Prepare the following reaction mixture on ice for real-time q-PCR using an ABI StepOne Plus (see Note 11). For a 20 µL reaction volume add;
- 2 µL of Ligated DNA from Subheading 3.3, step 3.
- 1 µL of Forward primer from 4 pmol/µL stock (200 nM final concentration).
- 1 µL of Reverse primer from 4 pmol/µL stock (200 nM final concentration) (see Note 12).
- 10 µL of ABI Fast SYBR green master mix.
- 6 µL of Sterile deionized water.
Analyze the data using delta delta Ct method (12). Most real-time PCR machine manuals will also have detailed explanations for the analysis of raw data by this method. For normalization, use the amplification values obtained from the upstream primer pairs, P3 and P4 (see Fig. 2).
3.5. Detection of Changes In Restriction Enzyme Accessibility in as Few as 1,000 Cells
An example of this methodology is presented in Fig. 3. Our laboratories have a longstanding interest in the mammalian SWI/SNF complexes, which are ATP-dependent chromatin remodeling enzymes that break histone:DNA contacts that facilitate both nucleosome mobility as well as regulatory factor interactions with chromatin (13, 14). Mammalian SWI/SNF complexes contain one of two highly related ATPase subunits called Brahma (Brm) or Brahma-related gene 1 (Brgl) (15). To identify biological roles for SWI/SNF enzymes, we created fibroblast cell lines that express ATPase-deficient, dominant negative Brm or ATPase-deficient, dominant negative Brgl in a tetracycline repressible manner (16). The dominant negative form of the protein interacts with the other subunits of the complex, but is functionally inactive (16,17). Thus, the expression of genes that require functional SWI/SNF enzymes is inhibited in cells expressing the mutant ATPase.
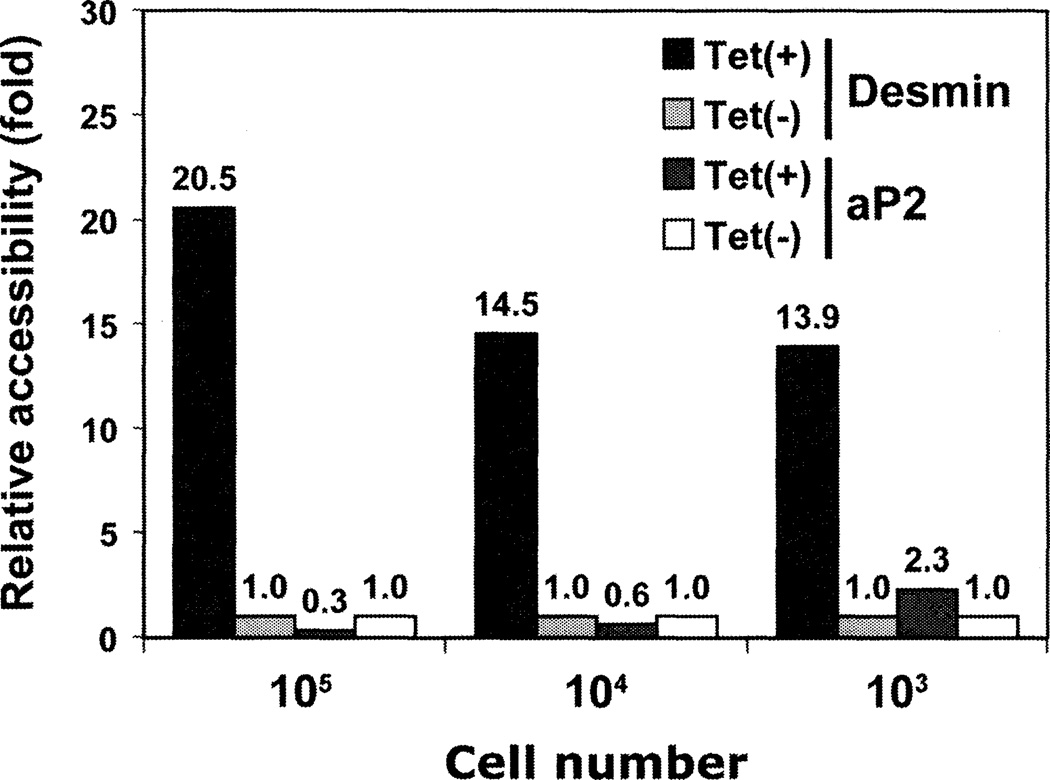
Fig. 3.
Restriction enzyme accessibility assays (REM) using as few as 1,000 cells. Restriction enzyme accessibility of Pvu II at the promoter of a muscle-specific (desmin) or fat-specific (aP2) gene in differentiated muscle cells in the presence (−tet) or absence (+tet) of a dominant negative version of Brahma-related gene 1 (Brg1) was measured using tenfold serial dilutions of isolated nuclei. Accessibility was observed in the muscle-, but not the fat-specific promoter in a Brg1 dependent manner, as expected in a muscle cell. The amount of cleavage at each promoter site in the cells expressing dominant negative Brg1 was set to 1; the amount of cleavage in cells not expressing dominant negative Brg1 is expressed relative to that number. Primers to amplify desmin regulatory sequences were described (19). Primers for the aP2 promoter were 5' caaaatgtgtgatgcctttgtgg 3' and 5' tccatgcgggaagcagcattttc 3'.
Fibroblasts can be reprogrammed along numerous different lineages, including skeletal muscle. Introduction of the MyoD regulator followed by serum withdrawal promotes skeletal muscle differentiation (18). We previously used this experimental system to show that expression of dominant negative Brm or dominant negative Brgl inhibits skeletal muscle-specific gene expression and differentiation because the chromatin at muscle-specific gene promoters is not remodeled (19–22). In the experiment presented in Fig. 3, cells were cultured in the presence or absence of tetracycline, infected with a retrovirus expressing MyoD, and subsequently differentiated. Nuclei were isolated from these cells, and side-by-side enzymatic digestion reactions containing 105,104, or 103 nuclei were performed. We used Pvu II for digestion because its recognition sequence matches the consensus sequence for E boxes, which are binding sites for MyoD- and MyoD-related myogenic factors. Thus, this experiment measures accessibility at muscle-specific genes in the regions where MyoD may be binding.
As shown in Fig. 3, cells maintained in tetracycline, which do not express dominant negative Brgl, show a robust increase in Pvu II accessibility in the promoter of the muscle-specific desmin gene. In contrast, in cells cultured in the absence of tetracycline, where the dominant negative Brgl enzyme was expressed, there was much less accessibility, resulting in greatly reduced levels of cleavage. This implicates functional SWI/SNF enzyme in the creation of the open chromatin structure that could be cleaved by the restriction enzyme. As a control, there were no changes in chromatin accessibility at the adipogenic aP2 gene, which contains a Pvu II site in its promoter but is not expressed in the MyoD differentiated cells. Importantly, the same results were obtained whether the reaction contained 100,000 nuclei or 1,000 nuclei. This demonstrates the exquisite sensitivity of this assay and validates use of this approach with samples where large quantities of starting material are not available. We have made use of this approach for measuring gene-specific changes in chromatin structure in embryonic mouse limb buds and embryonic mouse limb skeletal muscle (19, 23). This technique should be applicable to any tissue from which the nuclei can be isolated.
Acknowledgments
We thank C. Baron for assistance with the figures. This work was supported by NIH grant GM56244 to ANI and by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan to YO.
Footnotes
Volumes should be scaled proportionally for larger or smaller size samples.
For tissue culture cells grown in suspension, the first step is to pellet the cells by centrifugation. Cells should be poured or pipeted into 15 mL conical tubes and centrifuged at 300×g. Modifications to the centrifugation may be necessary based on the cell type being used.
For cell types that do not adhere strongly to the culture dish surface, do not pour the PBS onto the cells. Instead, tilt the dish with one hand, and with the other, slowly expel 5–10 mL of cold PBS from a pipet along the side of the culture dish, not directly on top of the cells.
When cell lines or commonly available primary tissue are used as starting materials, 100 µg of nuclei is a reasonable starting point for restriction enzyme digestion. However, if sample size is limiting, 10–20 µg; of nuclei can be utilized.
We recommend starting with trial experiments using commonly available nuclei and making dilutions of this material prior to initiating work with samples where availability and/or amount are restricted.
Limited digestion to optimize differences between accessible and inaccessible chromatin is essential; overdigestion will minimize accessibility differences. Some empirical determination of enzyme concentration and/or time of digestion may be necessary.
Although the digestion reactions can be stopped using conditions recommended by the enzyme supplier for inactivation, we do not find this to be a necessary step. Instead, we proceed directly to DNA purification as described in Subheading 3.2, step 5.
Preparation of the double-stranded linker should occur well in advance of the procedure. Large quantities can be prepared, aliquoted, and stored at −20°C.
An alternative annealing method is to place the linker mixture on the bench following the 95°C incubation and allow to cool to room temperature.
We have successfully utilized different ligation times at different temperatures. Ligations can be performed in as little as 30 min at 25°C. Alternatively, 4 h at 16°C or overnight at 4°C can be used.
The suggested protocol is optimized for the ABI StepOne Plus real-time q-PCR machine. Any real-time q-PCR machine is adequate; the reader will need to modify the protocol and reagents in accordance with the requirements of the machine to be used.
One of the primers should amplify the linker sequence or the linker sequence and adjacent sequence as discussed in Subheading 3.4.
References
- 1.Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 2.Wu C, Wong YC, Elgin SC. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979;16:807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- 3.Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SC. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979;16:797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- 4.Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 5.Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984;782:343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- 6.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 7.Elgin SC. DNAase I-hypersensitive sites of chromatin. Cell. 1981;27:413–15. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- 8.Lipchitz L, Axel R. Restriction endonuclease cleavage of satellite DNA in intact bovine nuclei. Cell. 1976;9:355–364. doi: 10.1016/0092-8674(76)90125-2. [DOI] [PubMed] [Google Scholar]
- 9.Pfeiffer W, Zachau HG. Accessibility of expressed and non-expressed genes to a restriction nuclease. Nucleic Acids Res. 1980;8:4621–4638. doi: 10.1093/nar/8.20.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller PR, Salser SJ, Wold B. Constitutive and metal-inducible protein:DNA interactions at the mouse metallothionein I promoter examined by in vivo and in vitro foot-printing. Genes Dev. 1988;2:412–427. doi: 10.1101/gad.2.4.412. [DOI] [PubMed] [Google Scholar]
- 11.Carey M, Smale ST. Transcriptional Regulation in Eukaryotes. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 14.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 16.de La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20:2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brgl and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 19.Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brgl. EMBO J. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 22.Mallappa C, Nasipak BT, Etheridge L, Androphy EJ, Jones SN, Sagerstrom CG, Ohkawa Y, Imbalzano AN. Myogenic microRNA expression requires ATP-dependent chromatin remodeling enzyme function. Mol Cell Biol. 2010;30:3176–3186. doi: 10.1128/MCB.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohkawa Y, Yoshimura S, Higashi C, Marfella CG, Dacwag CS, Tachibana T, Imbalzano AN. Myogenin and the SWI/SNF ATPase Brgl maintain myogenic gene expression at different stages of skeletal myogenesis. J Biol Chem. 2007;282:6564–6570. doi: 10.1074/jbc.M608898200. [DOI] [PubMed] [Google Scholar]