Abstract
Cytokines are soluble proteins that exert their functions by binding specific receptors. Many cytokines play essential roles in carcinogenesis and have been developed for the treatment of cancer. In this study, we identified a novel potential cytokine using immunogenomics designated colon-derived SUSD2 binding factor (CSBF), also known as chromosome 10 open reading frame 99 (C10orf99). CSBF/C10orf99 is a classical secreted protein with predicted molecular mass of 6.5 kDa, and a functional ligand of Sushi Domain Containing 2 (SUSD2). CSBF/C10orf99 has the highest expression level in colon tissue. Both CSBF/C10orf99 and SUSD2 are down-regulated in colon cancer tissues and cell lines with different regulation mechanisms. CSBF/C10orf99 interacts with SUSD2 to inhibit colon cancer cell growth and induce G1 cell cycle arrest by down-regulating cyclin D and cyclin-dependent kinase 6 (CDK6). CSBF/C10orf99 displays a bell-shaped activity curve with the optimal effect at ~10 ng/ml. Its growth inhibitory effects can be blocked by sSUSD2-Fc soluble protein. Our results suggest that CSBF/C10orf99 is a novel potential cytokine with tumor suppressor functions.
Cytokines are small secreted proteins (~5–20 kDa) that bind specific receptors to exert their effects in a paracrine or autocrine manner1,2. They are produced by immune cells, such as dendritic cells3, NK cells4 and T lymphocytes5,6, and non-immune cells7 including endothelial cells8, fibroblasts9,10, stromal cells11,12 et al. Cytokines play important roles in immune responses, inflammation and carcinogenesis13,14,15,16. EGF (epidermal growth factor)17, VEGF (vascular endothelial growth factor)18 and HGF (hepatocyte growth factor)19 have significant effects on oncogenesis, and they have been developed as targets for tumor therapy. On the other hand, some cytokines function as tumor suppressors, preventing and suppressing the development of cancer20, such as oncostatin M (OSM)21 and interleukin-24 (IL-24)22. OSM is a multifunctional interleukin 6-related cytokine, which inhibits the proliferation of various solid tumor cell lines. IL-24 is a cytokine tumor suppressor, which was identified initially as a melanoma differentiation-associated molecule. IL-24 is one of the IL-10 family cytokines and inhibits tumor cell growth, invasion, and migration by inducing tumor cell apoptosis and G1 cell cycle arrest with down-regulation of Cyclin D. IL-24 is significantly down-regulated in colorectal cancer (CRC) tissues compared with that in the adjacent normal mucosa with post-transcriptional modifications23.
CRC represents the third most common cancer in the world24. Its development is a multistage process characterized by a complex interaction between genetic alterations and host immune system25,26. Proto-oncogenes, including K-ras and c-myc27, play important roles in colorectal carcinogenesis. On the other hand, tumor suppressor genes (TSGs)28,29, such as p16, p21, p27, p53, APC, DCC, and MMR, are also of great importance during this process. TSGs inhibit unrestrained cell division in normal cells and limit tumor cell growth by decelerating cell division, repairing DNA mistakes and inducing programmed cell death. However, loss of function or decreased expression of TSGs, which is caused by genetic mutation, allele deletion, promoter methylation, or other mechanisms, may contribute to the development of uncontrolled cell growth of cancer. CRC can be treated with surgery in its early stages30, but it usually metastasizes at the time of diagnosis, which requires chemotherapy. However, chemotherapy is not very effective in the presence of metastasis, with survival rates lower than 10%. Taking together, failure of early diagnosis and therapy resistance are the major causes of death from CRC31,32,33.
So far many cytokines and their related proteins have been developed for treatment of cancers34. For example, tumor specific therapies toward CRC typically use Avastin (bevacizumab)35 or Erbitux (cetuximab)36, combined to traditional chemotherapy. Avastin inhibits growth of blood vessels by blocking VEGF, and Erbitux reduces growth of cancer by binding EGFR. A phase I clinical trial using a recombinant adenovirus vector expressing IL-24 (Ad-IL24/INGN241) reported that Ad-IL24 treatment had measurable tumor-killing effects in over 40% of patients37. It will therefore be of great value to identify and characterize novel cytokines for both basic research and clinical application.
To isolate novel potential cytokines, we have established a platform using immunogenomics as previously reported38 and found several secreted proteins with important functions. CSBF/C10orf99 is one of them. Here we show that CSBF/C10orf99 is a classical secreted protein with small molecular size. It has the highest expression level in normal colon tissue and decreases in colon cancer tissues and cell lines. CSBF/C10orf99 interacts with SUSD2 to inhibit colon cancer cell growth and induce G1 cell cycle arrest by down-regulating cyclin D and cyclin-dependent kinase 6 (CDK6). It shows a bell-shaped activity curve with the maximal effect at ~10 ng/ml and sSUSD2-Fc soluble protein can block its function. Our results demonstrate that CSBF/C10orf99 is a novel potential cytokine with tumor suppressor activities.
Results
CSBF/C10orf99 is a classical secreted protein
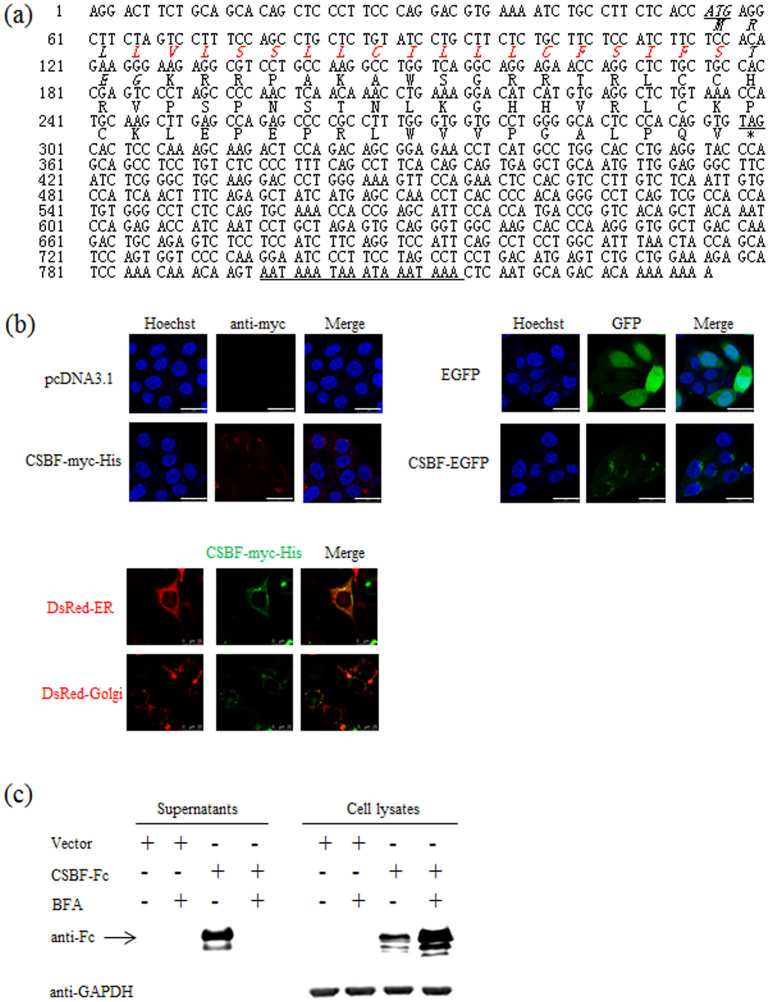
The complete cDNA and deduced amino acid sequences of CSBF/C10orf99 are shown in Figure 1a. The cDNA of CSBF/C10orf99 contains typical polyadenylation signal (AATAAA) within the 3' UTR and Kozak sequence (CACCATG)39 within the 5' UTR. CSBF/C10orf99 is composed of 81 amino acids, containing a signal peptide predicted by SignalP 4.0 Server (http://www.cbs.dtu.dk/services/SignalP/) and a transmembrane helices domain in the N-terminal predicted by TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The transmembrane helices domain is located in the signal peptide.
Figure 1. CSBF/C10orf99 is a classical secreted protein.
(a) The cDNA and deduced amino acid sequences of CSBF/C10orf99 are shown. The predicted signal peptide at the 5' end is typed in italic. The transmembrane helices domain is indicated by red color. Stop signal is replaced by asterisk. The putative signal for polyadenylation at the 3' end is underlined. (b) The subcellular localization of CSBF/C10orf99 was determined by microscopy. Up, Transfected HEK 293T cells were fixed, and stained with anti-myc and Hoechst 33258. EGFP is shown in green, whereas anti-myc staining is shown in red. The localization of CSBF-myc-His and the indicated GFP-fusion protein were observed by confocal laser scanning microscopy (LSM). The data shown are representative of at least three independent experiments. Scale bar = 25 μm. Down, Cells expressing DsRed-ER, DsRed-Golgi and CSBF-myc-His were fixed, stained and observed by LSM. (c) Western blot assay of CSBF/C10orf99 was using mouse anti-human IgG (Fc specific) antibody. HEK 293T cells transfected with pwYD11-CSBF-Fc (pwYD11, a modified vector, gained from National Research Council Biotechnology Research Institute, Canada.) or control vector were cultured in the absence or presence of 10 μg/ml of BFA. The supernatants and cell lysates were prepared 48 hours post-transfection. The GAPDH was detected as loading control. The data shown are representative of at least three independent experiments.
After molecular cloning of CSBF/C10orf99, we performed immunofluorescence microscopy to determine the subcellular localization of CSBF/C10orf99 in pcDNA3.1-CSBF-myc-His transfected HEK 293T cells. As illustrated in Figure 1b Up, the red fluorescence represented CSBF-myc-His was located in cytosol. This was confirmed by transfection of another GFP-tagged CSBF/C10orf99 plasmid. The green fluorescence signals revealed that CSBF-EGFP also had a cytoplasmic localization. There was no signal presented on cell membrane or in the nucleus. The DsRed-tagged organelle markers (DsRed-ER and DsRed-Golgi) were used to precisely determine the localization of CSBF-myc-His. The results showed that CSBF-myc-His was in the lumen of intracellular compartment along the classic secretion pathway (Figure 1b, Down).
Soluble proteins containing N-terminal signal peptides are usually secreted through the classical ER/Golgi dependent pathway, which can be blocked by brefeldin A (BFA)40. To confirm whether CSBF/C10orf99 is secreted via the classical pathway, we detected CSBF-Fc protein in the cell culture supernatant transfected with pwYD11-CSBF-Fc plasmid and then performed a BFA inhibition assay. As illustrated in Figure 1c, BFA treatment decreased the secretion of CSBF-Fc, confirming that CSBF/C10orf99 is a classical secretory protein. Purified CSBF-Fc protein was further sequenced by Edman degradation reaction41,42. As shown in Supplementary Figure S1, the N-terminal sequence KRRPAKAWSG is in accordance with the SignalP prediction.
CSBF/C10orf99 is a ligand of SUSD2
Collectively, the above findings verified that CSBF/C10orf99 is a classical secreted protein. Previous studies deduced high-confidence interactions between CSBF/C10orf99 and SUSD2, as well as Gal1 and SUSD243. SUSD2 encodes an 822-amino acid type I membrane protein containing extracellular domains of SO, AMOP, VWD, and CCP (Supplementary Figure S2a)44. In order to determine whether these four domains are indispensable for the location and function of SUSD2, we constructed truncated mutants of these four extracellular domains and checked the expression of these constructs (Supplementary Figure S2b). Then we examined subcellular localization of these mutants in LoVo cells. Supplementary Figure S2c showed different expression patterns of full length SUSD2 and all these mutants. Compared with the wild type of SUSD2, all these mutants are abundant in cytoplasm, suggesting that absence of any of these four domains would change the localization of this protein, which may further affect its function. So the following studies only included the function of intact SUSD2 protein.
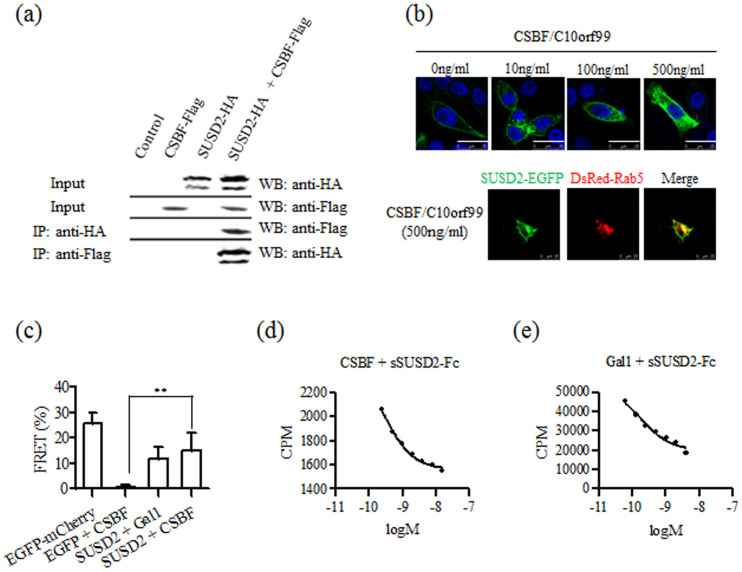
In order to verify the interaction between SUSD2 and CSBF/C10orf99, we constructed a dicistronic mammalian expression vector20, pcDNA3-SUSD2-HA::IRES-CSBF-Flag, which exploited the virus-derived IRES element for the co-expression of SUSD2-HA and CSBF-Flag. Then we conducted co-immunoprecipitation (co-IP) assay. As shown in Figure 2a, we found that CSBF/C10orf99 co-immunoprecipitated with SUSD2 and vice versa, indicating that the two proteins were in the same complex.
Figure 2. CSBF/C10orf99 is a ligand of SUSD2.
(a) Co-IP of the CSBF-SUSD2 complex. Co-IP studies were carried out with lysates prepared from HEK 293T cells. Cells were transfected with CSBF-Flag, SUSD2-HA or control vector alone for control. SUSD2-containing proteins were immunoprecipitated with a monoclonal anti-HA antibody and then blotted using an anti-Flag monoclonal antibody. In a reciprocal fashion, CSBF-containing proteins were immunoprecipitated with the anti-Flag antibody and then blotted using the anti-HA antibody. The data shown are representative of at least three independent experiments. (b) Up, Internalization of SUSD2 was induced by CSBF/C10orf99. LoVo cells transiently expressing SUSD2-EGFP were treated with CSBF/C10orf99 (0, 10, 100 and 500 ng/ml) at 37°C for 30 minutes, fixed and stained with Hoechst 33258. SUSD2-EGFP observed by confocal LSM is shown in green. The data shown are representative of at least three independent experiments. Scale bar = 25 μm. Down, Cells expressing SUSD2-EGFP or/and DsRed-Rab5 were treated with CSBF/C10orf99 (500 ng/ml). (c) FRET was taken out between CSBF-mCherry and SUSD2-EGFP by confocal LSM. FRET efficiency E = 1 − FD′/FD, where FD′ and FD are the donor fluorescence intensities with and without an acceptor respectively. Gal1 is a positive ligand control. Histogram represents mean ± SD. (d, e) Curve fitting showed the affinity of radioligand binding to sSUSD2-Fc. The radioligand competition binding assay was taken out between CSBF and sSUSD2-Fc. Gal-1 is a positive ligand control.
As ligand-dependent internalization is a characteristic of receptor45, we next managed to confirm whether SUSD2 goes through this process. Firstly, the localization of SUSD2-EGFP was detected on the plasma membrane of pEGFP-N1-SUSD2 transfected LoVo cells (Supplementary Figure S2c). Then we found out that CSBF/C10orf99 induced internalization of SUSD2-EGFP in a dose-dependent manner (Figure 2b Up). To precisely determine the internalization of SUSD2-EGFP, the endosome marker (DsRed-Rab5) was used. As illustrated in Figure 2b Down, the internalized SUSD2-EGFP was located in the endosome. Thses indicated that CSBF/C10orf99 can bind SUSD2, and trigger receptor internalization.
Furthermore, to determine whether CSBF/C10orf99 can directly bind SUSD2, we used in situ fluorescence resonance energy transfer (FRET) to confirm the co-localization between CSBF-mCherry and SUSD2-EGFP. The fluorescence energy could transfer from EGFP to RFP when the distance between them was less than 10 nm. Human galectin-1 (Gal1) was used as a positive ligand control. Figure 2c summarized the result of the FRET assay. FRET efficiency of CSBF/SUSD2 was 15.2%, slightly higher than that of positive ligand control Gal1/SUSD2 (11.6%), with fused EGFP-mCherry46 as a positive FRET control (25.9%).
To further elucidate the biochemical nature of the CSBF/SUSD2 interaction, we measured affinity of CSBF/C10orf99 to purified extracellular region of SUSD2, which is a soluble form of SUSD2 generated by fusing extracellular moiety of SUSD2 to the Fc region of human IgG1 (sSUSD2-Fc) (Supplementary Figure S3). Competition binding assay were taken to detect the affinity between CSBF/C10orf99 and sSUSD2-Fc after CSBF/C10orf99 labeled with 125I. The IC50-value was determined by nonlinear regression of competition binding. As shown in Figure 2e, curve fitting showed that unlabeled CSBF/C10orf99 displaced 125I-labeled CSBF/C10orf99 from sSUSD2-Fc with an IC50 of 0.28 nM, while Gal1 with an IC50 of 0.19 nM. Together, these data strongly confirmed that CSBF/C10orf99 is a ligand of SUSD2.
CSBF/C10orf99 and SUSD2 are decreased in colon cancer tissues
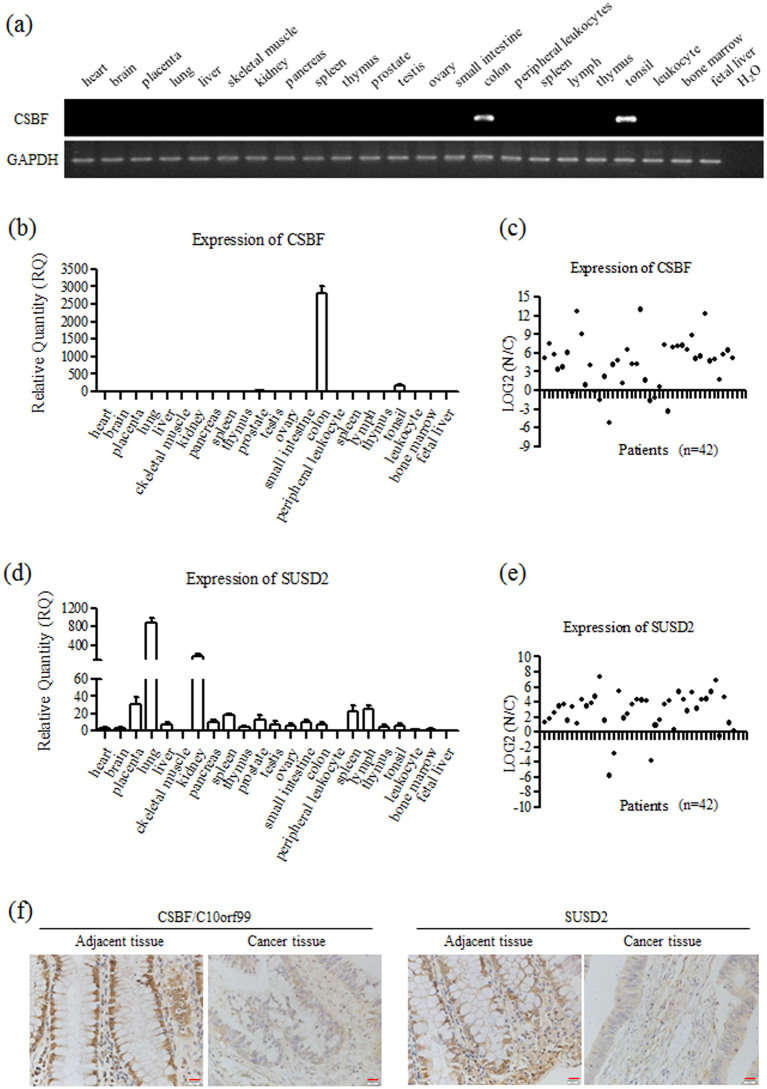
Since CSBF/C10orf99 is a novel secreted protein, to discover more about its function, we first searched the expression profile from known databases. GEO profile analysis (GDS3113) showed that CSBF/C10orf99 is highly expressed in colon tissue, and moderately expressed in tonsil tissue among 96 samples. The SAGE database (http://cgap.nci.nih.gov/SAGE/AnatomicViewer) and Human Protein Atlas (www.proteinatlas.org) reported that CSBF/C10orf99 is down-regulated in colorectal cancer tissues.
We then investigated the expression pattern of CSBF/C10orf99 in normal human tissues with RT-PCR and real-time PCR, which was detected in three cDNA panels from Clontech, including two Multiple Tissues cDNA libraries (16 samples) and one Immune System Tissues cDNA library (7 samples). As shown in Figure 3a and 3b, CSBF/C10orf99 mRNA was found to be expressed in specific tissues: at the highest level in colon, at moderate level in tonsil, and almost undetectable in other tissues.
Figure 3. CSBF/C10orf99 and SUSD2 are decreased in colorectal cancer tissues.
(a) Expression of CSBF/C10orf99 was carried out using RT-PCR. Three cDNA panels from Clontech were analyzed, including two Multiple Tissues cDNA libraries and one Immune System Tissues cDNA library. GAPDH was detected as a reference gene. (b, d) Expression of CSBF/C10orf99 and SUSD2 was carried out using real-time PCR. GAPDH was detected as a reference gene. Real-time PCR were performed in triplicate, and mean ± SD are shown. (c, e) Down-regulation of CSBF/C10orf99 or SUSD2 was detected in primary colorectal cancers by real-time PCR. N/C, paired adjacent tissues/cancer tissues. GAPDH was detected as a reference gene. (f) Representatives of immunohistochemical staining for CSBF/C10orf99 and SUSD2 in one pair of colorectal cancer tissue and adjacent tissue. Magnification: 400 times.
Next, we detected the expression of CSBF/C10orf99 on a cDNA panel derived from 42 pairs of colon cancer and adjacent tissues. As shown in Figure 3c, the expression of CSBF/C10orf99 was dramatically reduced in most of the primary cancer tissues compared with that in the adjacent tissues. To determine whether CSBF/C10orf99 was down-regulated on the protein level, we raised a rabbit polyclonal antibody against CSBF/C10orf99 with GST-CSBF fusion protein as immunogen and purified by CSBF/C10orf99 affinity chromatography. We analyzed CSBF/C10orf99 in representative samples by immunohistochemistry. As shown in Figure 3f, CSBF/C10orf99 was down-regulated in most of colon cancer tissues, which was consistent with mRNA level.
As our results indicated that SUSD2 is the receptor of CSBF/C10orf99, we checked the expression profile of SUSD2 further. Previous study showed that SUSD2 was highly expressed in lung and kidney, and moderately expressed in mammary gland by RT-PCR44. GEO profile analysis (GDS3113) showed that SUSD2 was highly expressed in spinal cord, skin and lung, and moderately expressed in colon and mammary gland tissues. The SAGE database and Human Protein Atlas reported that SUSD2 is down-regulated in colorectal cancer tissues. We investigated the expression pattern of SUSD2 in normal human tissues with real-time PCR. SUSD2 was highly expressed in lung and kidney, and detectable in colon tissue (Figure 3d). As shown in Figures 3e and 3f, SUSD2 was also down-regulated both on mRNA and protein levels in most of colon cancer tissues.
CSBF/C10orf99 and SUSD2 are decreased in colon cancer cell lines with different patterns of gene regulation
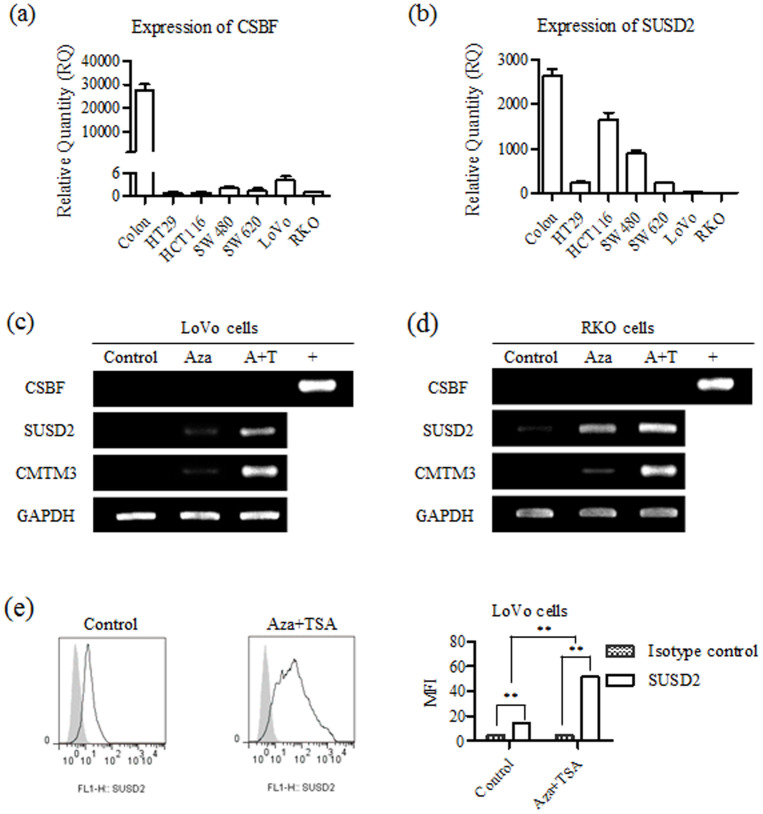
The above results confirmed that CSBF/C10orf99 and SUSD2 were significantly reduced in colon cancer tissues. Then we tried to investigate the expression of CSBF/C10orf99 and SUSD2 in colon cancer cell lines. As shown in Figures 4a and b, both CSBF/C10orf99 and SUSD2 were down-regulated in six colon cancer cell lines.
Figure 4. CSBF/C10orf99 and SUSD2 are decreased in colorectal cancer cell lines with different patterns of gene regulation.
(a, b) Expression of CSBF/C10orf99 and SUSD2 in six cell lines was carried out using real-time PCR. cDNA of colon tissue from Clontech was used as positive control. GAPDH was detected as a reference gene. Real-time PCR were performed in triplicate, and mean ± SD are shown. (c, d) Demethylation with Aza or combined with TSA (A+T) restored genes expression in LoVo or RKO cells. (e) Aza combined with TSA restored SUSD2 expression was analyzed in LoVo cells by FACS assay. FACS was performed in triplicate.
The putative CSBF/C10orf99 and SUSD2 promoters, identified by bioinformatics promoter analysis (http://www.genomatix.de), both contain non-typical CpG islands. To investigate whether the down regulation of these two genes were caused by promoter methylation, we treated LoVo and RKO cells with demethylating agent Aza, or combined with histone deacetylase inhibitor trichostatin A (TSA). The tumor suppressor CMTM3 was checked as positive control47. Indeed, SUSD2 expression was restored by Aza or combined with TSA both on mRNA (Figures 4c and 4d) and protein levels (Figure 4e), while the expression of CSBF/C10orf99 was not restored. These results clearly show that down-regulation of these two genes possesses different mechanisms.
CSBF/C10orf99 interacts with SUSD2 to inhibit colon cancer cell growth
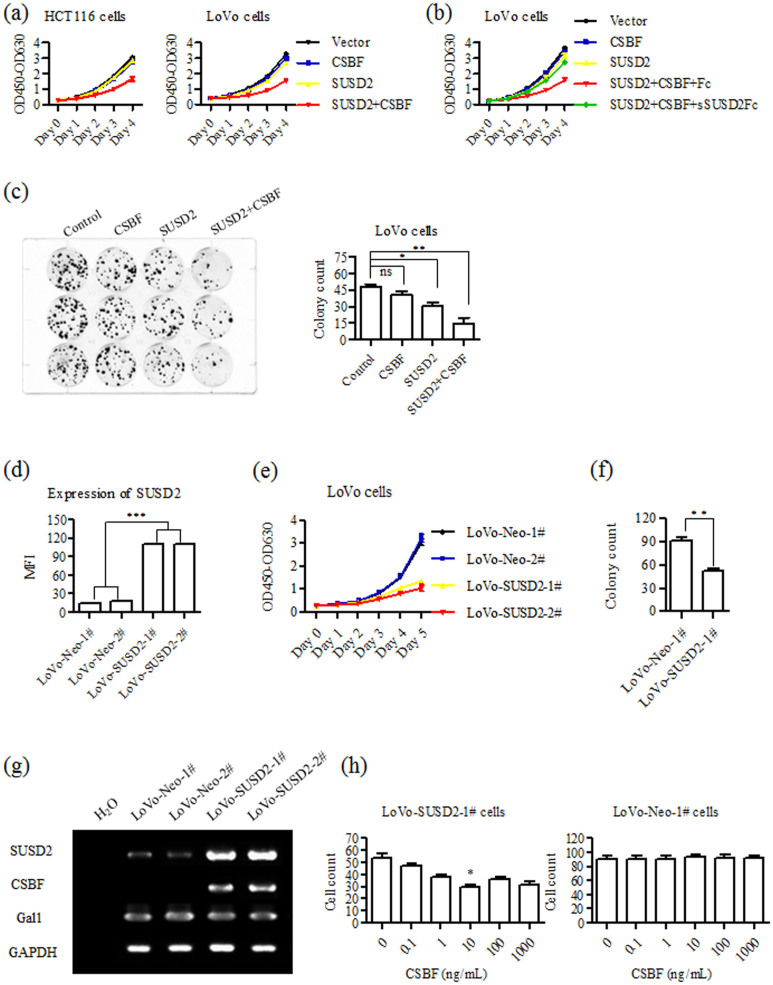
As CSBF/C10orf99 and SUSD2 are both down-regulated in colon cancer tissues and cell lines, we assumed whether CSBF/C10orf99 and SUSD2 have effects on growth of colon cancer cells. As shown in Figure 5a, proliferation of HCT116 or LoVo cells became significantly slower in CSBF/C10orf99 and SUSD2 co-expressed group compared to other groups. These results indicated that the growth inhibition has no cell specificity, and CSBF/C10orf99 or SUSD2 did not affect cell proliferation alone in the transiently transfected system. To further confirm if the inhibitory effect of CSBF/C10orf99 is dependent on the existence of SUSD2, we used purified recombinant sSUSD2-Fc protein in blocking experiments and purified recombinant Fc was used as negative control. As shown in Figure 5b, blockade of CSBF/SUSD2 with sSUSD2-Fc increased LoVo cell proliferation.
Figure 5. CSBF/C10orf99 interacts with SUSD2 to inhibit colon cancer cell growth.
(a) CCK-8 assay showed that cell growth was inhibited by co-expression of CSBF/C10orf99 and SUSD2 in HCT116 and LoVo cells. The data shown are representative of at least three independent experiments. (b) CCK-8 assay showed that LoVo cell growth inhibition was blocked by adding sSUSD2-Fc protein in experiment A. The data shown are representative of at least three independent experiments. (c) Co-expression of CSBF/C10orf99 and SUSD2 inhibited colony formation in LoVo cells. Assays were performed in triplicate, and mean ± SD are shown. (d) Expression of SUSD2 in LoVo-SUSD2 was analyzed by FACS assay. FACS was performed in triplicate. (e) CCK-8 assay showed that LoVo-SUSD2 cells had lower cell growth rate. The data shown are representative of at least three independent experiments. (f) Colony formation assay showed that LoVo-SUSD2-1# cells had fewer colonies. Assays were performed in triplicate, and mean ± SD are shown. (g) RT-PCR assay showed long term ectopic expression of SUSD2 restored the expression of CSBF/C10orf99 but not Gal1. The data shown are representative of at least three independent experiments. (h) Colony formation assay showed that the inhibitory effects of SUSD2 can be increased by adding CSBF/C10orf99. Assays were performed in triplicate, and mean ± SD are shown.
We further conducted colony formation assay to validate that the interaction between CSBF/C10orf99 and SUSD2 affects the growth of colon cancer cells. As shown in Figure 5c, transfected LoVo cells were cultured in the existence of G418 for two weeks. A significant reduction in colony was observed in CSBF/C10orf99 and SUSD2 co-expressed LoVo cells. The colony of SUSD2 single-expressed cells was also less than that of vector control cells, while there was no obvious change in CSBF/C10orf99-expressed cells. This result indicates that long-term expressed SUSD2 decreased LoVo cell growth.
To further confirm the inhibitory effects of long-term expression of SUSD2 on colon cancer cell growth, stable SUSD2-expressing cell lines were generated. As illustrated in Figure 5d, after screening with antibiotic selection, we obtained two cell strains of LoVo-SUSD2 (LoVo-SUSD2-1#, LoVo-SUSD2-2#) and two strains of LoVo-Neo cells (LoVo-Neo-1#, LoVo-Neo-2#). As illustrated in Fig. 5E, LoVo-SUSD2 cells showed lower growth rates in CCK-8 assay. Then we conducted colony formation assay with LoVo-SUSD2-1# and LoVo-Neo-1# cells. As shown in Figure 5f, the colony of LoVo-SUSD2-1# cells was also much less than that of LoVo-Neo-1# cells.
To elucidate why long-term expression of SUSD2 inhibits cell growth, we performed RT-PCR to investigate whether CSBF/C10orf99 was increased. As shown in Figure 5g, CSBF/C10orf99 was indeed up-regulated by restored expression of SUSD2. Furthermore, to clarify the effect of CSBF/C10orf99 on LoVo-SUSD2 cells, we added CSBF/C10orf99 in the colony formation assay system. As shown in Figure 5h, CSBF/C10orf99 could significantly increase the growth inhibitory effect of SUSD2, but not in LoVo-Neo cells.
CSBF/C10orf99 interacts with SUSD2 to induce G1 cell cycle arrest
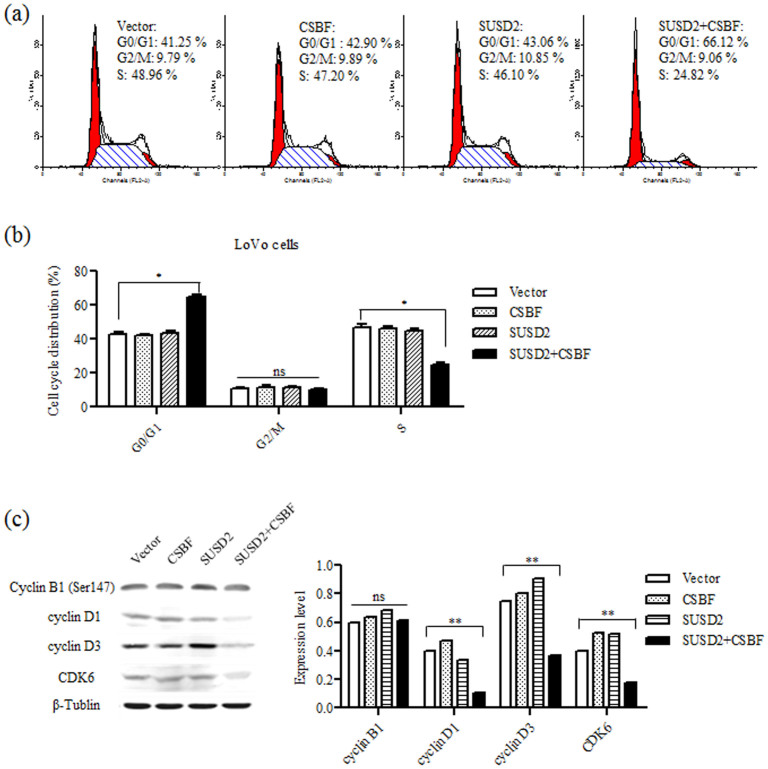
Usually, inhibition of cell growth is induced by apoptosis or cell cycle arrest48. Our results showed that there was no significant increase in the percentage of apoptotic cells (Supplementary Figure S4). Then we further detected cell cycle of transfected LoVo cells by FACS. As shown in Figures 6a and b, when CSBF/C10orf99 was co-expressed with SUSD2 in LoVo cells, G0/G1 phase cells were significantly increased and S phase cells were decreased, but no obvious change was observed in other groups. These results suggest that co-expressing of CSBF/C10orf99 and SUSD2 might induce G1 cell cycle arrest. We further examined several key cell cycle regulators by Western blotting. As shown in Figure 6c, cyclin D1, cyclin D3 and CDK6 were down-regulated in CSBF/C10orf99 and SUSD2 co-expressing LoVo cells, consistent with the phenotypes of G1 cell cycle arrest. While the expression of cyclin B1 was not changed, consisting with the undetectable change of G2/M phase. Overall, these results confirmed that CSBF/C10orf99 interacts with SUSD2 to induce G1 cell cycle arrest.
Figure 6. CSBF/C10orf99 interacts with SUSD2 to induce G1 cell cycle arrest.
(a) Flow cytometry was used to compare the DNA content between mock, CSBF-expressing, SUSD2-expressing and co-expressing cells. Summary of cell proportions in different phases of cell cycle (b). The results were expressed as Mean ± S.D. of three independent experiments. (c) Protein expression of cyclin B1, cyclin D1, cyclin D3 and CDK6 were detected by western blot (left). β-tublin was used as an internal control. And from three independent experiments, the levels of the cell cycle related proteins were normalized to β-tublin (right).
Discussion
Cytokines are usually small secreted proteins with optimal activity at quite low concentrations and their functions are dependent on the binding of specific receptors49. In the present study, we identified a novel potential cytokine CSBF/C10orf99 using immunogenomics. CSBF/C10orf99 is a classical secreted protein with a regular N-terminal signal peptide of 24 amino acids. SUSD2 is indispensable for the growth inhibitory effect of CSBF/C10orf99 on colon cancer cells and recombinant sSUSD2-Fc can block its function. CSBF/C10orf99 displays a bell-shaped activity curve and its optimal effect is about 10 ng/ml, which is in accordance with the characteristics of cytokines.
To our knowledge, this is the first systemic study of CSBF/C10orf99. GEO profile analysis showed that 2610528A11Rik, the mouse homolog of CSBF/C10orf99, was significantly up-regulated in psoriasis mouse models (GDS3907). Moreover, CSBF/C10orf99 was also up-regulated in psoriasis patients (GDS3539). Meanwhile CSBF/C10orf99 was speculated to be involved in the pathogenesis of psoriasis50, which is a chronic immunological skin and joint disease, traditionally believed to be involved in the Th1 pathway. By challenging of ovalbumin, Tang et al. reported that the homozygous 2610528A11Rik knockout mice exhibited an increased mean serum IgG2a response51, which is an immunoglobulin isotype marker for T helper 1 (Th1) lymphocytes associated function, indicating that 2610528A11Rik might inhibit Th1 cells to regulate adaptive immune responses. Further studies are necessary to elucidate the function of CSBF/C10orf99 in Th1 cell immune responses.
The expressional and functional characteristics of CSBF/C10orf99 indicate it may be a tumor suppressor. Its gene is located on chromosome 10q23.1 beside the genomic region of tumor suppressor PTEN (10q23.3)52. Inactivation of TSGs through promoter methylation, gene mutation, or loss of heterozygosity is important for carcinogenesis. In human colon cancer cell lines, the expression of CSBF/C10orf99 cannot be restored by Aza or combined with TSA, which indicates that it is not regulated by promoter methylation. The mechanism underlying the down-regulation of CSBF/C10orf99 remains to be studied further. Possible roles for genetic and/or other epigenetic controls must be considered. The promoter of SUSD2 contains non-typical CpG islands, but it can be restored by Aza or combined with TSA, indicating that promoter methylation manipulates its expression directly or transcription factors regulating its expression are epigenetically regulated.
Intriguingly, higher expression of CSBF/C10orf99 and SUSD2 has also been detected in a few colon cancer samples (7/42 and 4/42), which is similar to the increased expression of tumor suppressor p16 in many malignant tumors. There are some possible mechanisms to elucidate why p16 overexpression occurs. On one hand, partial loss of p16 function due to missense mutations can be compensated by elevated expression as observed in some tumor specimens. On the other hand, in the presence of wild-type p16, other molecular events, such as over-expression of CDC6 and cyclin D1, or deregulation of Rb in tumor cells and cancer tissues have the potential to positively feedback p16 expression53,54. Given these available mechanisms and our results, there might be some mutations of CSBF/C10orf99 and SUSD2, or other downstream molecules changing in the mentioned samples.
Sugahara and colleagues have demonstrated that the ectopic expression of Susd2, the mouse homolog of SUSD2, can inhibit growth and reverse tumorigenic phenotypes of HT1080 cells and HeLa cells in vitro55,56. Their results indicated that Susd2 is a possible tumor suppressor, which is similar as our results. Watson et al. reported that SUSD2 interacts with Gal1 and the cell surface localization of Gal1 is dependent on the existence of SUSD244. The interaction of SUSD2 with Gal1 increased the invasion of breast cancer cells and might play a significant role in modulating the body's immune response. GEO profile analysis showed that SUSD2 was significantly down-regulated in estrogen receptor (ER) alpha-silenced breast cancer cells (GDS4061), and it was up-regulated in ER positive cells (GDS4067) and some ovarian adenocarcinoma cells (GDS3592). Meanwhile, Gal1 is the reported ligand of SUSD2 and involved in tumor transformation, angiogenesis, cell-cycle regulation and apoptosis57. Our results showed that the affinity between CSBF/C10orf99 and SUSD2 is similar to that of Gal-1 and SUSD2, but the functions are different. Collectively, these results suggest that the expression and function of SUSD2 might be influenced by estrogen, ligands and microenvironment.
This study shows that CSBF/C10orf99 inhibits G1-S phase transition through down-regulating cyclin D and CDK6. G1-S phase transition is known to be a major checkpoint for cell cycle progression58. It is necessary to identify the intracellular interacting proteins of SUSD2 and elucidate its mechanism on the modulation of cyclin D and CDK6 in the future study, which will be helpful to understand the pathogenesis of colon cancer.
Methods
Cell lines, reagents and cancer samples
HEK 293T, HCT116, HT29, LoVo, SW480, SW620 and RKO cells were obtained from our collaborators and maintained in DMEM (GIBCO) supplemented with 10% FBS (HyClone). CSBF/C10orf99 was synthesized in Chinese Peptide Company (Hangzhou, China) with a purity > 97%. Gal1 was purchased from R&D Systems (1152-GA-050). Antibodies used in this paper were: mouse anti-myc (9E10) (# SAB4700447, Sigma-Aldrich), rabbit anti-HA (C29F4) (# 3724, Cell Signaling Technology), goat anti-Human IgG (Fc specific) (# I2136, Sigma-Aldrich), anti-cyclin D1 (# 2926, Cell Signaling Technology), anti-cyclin D3 (# 2936, Cell Signaling Technology), anti-CDK4 (# 2906, Cell Signaling Technology), anti-CDK6 (# 3136, Cell Signaling Technology), anti-phospho-Cyclin B1 (Ser147) (# 4131, Cell Signaling Technology), anti-SUSD2 rabbit polyclonal antibody (# HPA004117, Sigma-Aldrich), anti-β-tublin (# T8328, Sigma-Aldrich) and anti-GAPDH (# 2118, Cell Signaling Technology). Primary cancer tissues and paired adjacent tissues were obtained from patients under primary surgery at Peking University Cancer Hospital (Beijing, China), with patients' consent and institutional ethics approval. Fresh human tissues were fixed with 10% formalin in PBS for immunohistochemistry, or frozen in liquid nitrogen for RNA extraction. This investigation was carried out after approval by the Ethics Committee of Peking University Cancer Hospital.
Molecular cloning and sub-cloning of CSBF/C10orf99 and SUSD2
CSBF/C10orf99 was amplified from PBMC by PCR with primers: 5′-CAgaattcCACCATGAGGCTTCTAGTCCTTTC-3′ and 5′-GCggtaccCCCACCTGTGGGAGTGCCCCAG-3′, digested with EcoRI and KpnI and ligated into pEGFP-N1 (Clontech). This plasmid was then digested with EcoRI and BamHI and ligated into pcDNA3.1(-)-myc-His, pCMV-3×Flag-C and pwYD11 (containing fused human IgG1 Fc tag) to obtain pcDNA3.1-CSBF-myc-His, pCMV-CSBF-Flag and pwYD11-CSBF-Fc, respectively. SUSD2 was amplified from HCT116 cDNA library with primers: 5'-CCgaattcGCCACCATGAAGCCAGCCCTCCTGC-3' and 5'-CCgaattcGGGCTGTGCACCCCAGACG-3', digested with EcoRI and cloned into pCMV-C-HA. The EGFP-tagged SUSD2 plasmid was constructed using the full-length SUSD2 as a template for PCR with a pair of primers: 5'-CCgaattcGCCACCATGAAGCCAGCCCTCCTGC-3' and 5'-GGaccggtCCGGGCTGTGCACCCCAGACGT G-3', digested with EcoRI and AgeI and cloned into the pEGFP-N1 plasmid. The truncated plasmids of SUSD2-HA were constructed by standard method of site-directed mutagenesis by overlapping extension using the polymerase chain reaction (PCR) with different PCR primer sets presented in Supplementary Table S1.
Immunofluorescence microscopy
The cells were seeded on coverslips for 24 hours. At 24 hours post-transfection, the cells were washed and fixed for 20 minutes with prepared 4% PFA, rinsed in serum-free medium, and permeabilized for 30 min in 0.1% Triton X-100 and 3% BSA in PBS. Cells were incubated in succession with intervening washing, with primary antibody overnight at 4°C, and secondary antibody for 60 minutes at room temperature. Primary antibodies were the mouse anti-myc (9E10) and rabbit anti-HA (C29F4) diluted 1:1000. Rhodamine-conjugated secondary antibodies were applied for detection. To stain the nuclei, the cells were incubated for 60 minutes with 0.05 μg/ml Hoechst33342 dye (Sigma-Aldrich). Coverslips were mounted, sealed, examined and photographed under a confocal laser scanning microscope (Leica TCS SP5 Microsystems, Germany).
BFA inhibition assay
At 24 hours post-transfection, HEK 293T cells were addition of either 10 μg/ml of BFA (# B7651, Sigma-Aldrich) or ethanol (vehicle control), and cultured for another 24 h. Finally, the total cell lysates and the culture supernatants were harvested for Western blot analysis.
Western blot analysis
Details have been described previously59. Briefly, samples were separated and transferred onto nitrocellulose membranes, blocked and incubated in succession with primary antibody overnight at 4°C and then with horseradish peroxidase-coupled secondary antibody at a dilution of 1:2000. After washing, immunoreactive bands were detected by Odyssey Infrared Imager (LICOR Bioscience, Lincoln, NE, USA).
Antibodies used were: anti-myc (9E10), anti-Human IgG (Fc specific), anti-HA (C29F4), anti-cyclin D1, anti-cyclin D3, anti-CDK4, anti-CDK6, anti-phospho-Cyclin B1 (Ser147), anti-SUSD2 rabbit polyclonal antibody, anti-β-tublin, anti-GAPDH and IRDye secondary antibodies against mouse, rabbit or goat immunoglobulin G (Li-Cor Biosciences). The anti-CSBF/C10orf99 rabbit polyclonal antibody and were made in our lab.
Co-IP assays
The transfected HEK 293T cells were lysed in buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EDTA (pH 8.0) and protease inhibitor cocktail. Cell extracts were clarified by centrifugation, and the resulting supernatant was incubated overnight at 4°C with anti-HA or anti-Flag and then with protein G plus agarose beads. After intensive washing and centrifugation, immune complexes were analyzed by Western blotting.
Ligand-dependent internalization of SUSD2
LoVo cells were seeded on coverslips for 24 hours. After transfected with pEGFP-N1-SUSD2 (SUSD2-EGFP) or vector control, cells were cultured for another 24 hours. Then different doses of CSBF/C10orf99 were added to the medium for 30 minutes. BSA was added as reagent control. Coverslips were mounted, sealed, examined and photographed under a confocal laser scanning microscope.
FRET microscopy and image analysis
HEK 293T cells were seeded on coverslips for 24 hours. After transfected with EGFP-mCherry, EGFP, mCherry, CSBF-mCherry, Gal1-mCherry, SUSD2-EGFP, EGFP+CSBF-mCherry, EGFP+Gal1-mCherry, SUSD2-EGFP+mCherry, SUSD2-EGFP+CSBF-mCherry, SUSD2-EGFP+Gal1-mCherry, cells were cultured for another 24 hours. Gal1 was used as ligand control. Coverslips were mounted, sealed, examined and photographed under a confocal laser scanning microscope. Images were acquired under SP5-FRET acquisition module by interline scanning for EGFP (488 nm excitation; 510 nm emission) and mCherry (514 nm excitation; 600 nm emission), respectively. FRET efficiency was analyzed under SP5-FRET analysis module. Over 10 cells were gained in each group, and 10 ROIs were gained on each cell.
Radioligand binding to SUSD2
CSBF/C10orf99 and Gal1 were labeled with 125I by the chloramine-T (ch-T) method. Recombinant sSUSD2-Fc was used as potential receptor in the binding reaction. 125I-Gal1 was added to the binding reaction with unlabeled Gal1 of 7 appropriate concentrations (60, 30, 15, 7.5, 3.75, 1.875, and 0.9375 pg/μl). After an incubation time of 24 hrs at 4°C, the unbinding 125I-Gal1 was separated of the reaction mixture by precipitating the bound 125I-Gal1 with polyethylene glycol (PEG6000). After centrifugation the supernatant was removed and the bound radioligand was counted in a liquid scintillation gamma-counter. Results obtained from the standards were used to construct a standard dose-response curve. Nonspecific binding was determined within each experimental series. Meanwhile 125I-CSBF was added to the binding reaction with unlabeled CSBF of 7 appropriate concentrations (100, 50, 25, 12.5, 6.25, 3.125, and 1.5625 pg/μl).
Semi-quantitative RT–PCR and real-time PCR analysis
Human multiple tissue cDNA libraries and immune system tissues cDNA library were purchased from Clontech. Total RNA from colon cancer cells was isolated using TRIzol reagent (Invitrogen) and used as a template for cDNA synthesis. CSBF/C10orf99 PCR amplification was carried out using the following primers: 5'-CTCCACAGAAGGGAAGAGGCG-3' and 5'-GGACCACTGGATGCTGGTAG-3'; SUSD2 PCR amplification: 5'-CTTCTTCACGGACTACGGCT-3' and 5'-GACACACTCAGGAACGGG-3'; Gal1 PCR amplification: 5'-ATGGCTTGTGGTCTGGT-3' and 5'-CAGAGGGAGCAGAGGCA-3'; and GAPDH PCR amplification: 5'-CAAGGTCATCCATGACAACTTTG-3' and 5'-GTCCACCACCCTGTTGCTGTAG-3'. Quantitative PCR was run on ABI PRISM 7000 Sequence Detection System (Applied Biosystems). The amplification conditions were 10 min initial denaturation at 95°C, followed by 40 cycles of each 15 s at 95°C and 1 min at 60°C. CSBF/C10orf99 amplification was carried out using the following primers: 5'-GGTCAGGCAGGAGAACCAG-3' and 5'-AGCTTGCATGGTTTACAGAGC-3'. Primers for SUSD2 amplification were using 5'-AGAGCTGGATGGACCTGAAA-3' and 5'-ATGCCAGCATGATGGAGAC-3'. Samples were run in triplicate. All samples were normalized against GAPDH using comparative Ct method (ddCt).
Immunohistochemistry
Human tissue slides were deparaffinized, rehydrated and blocked in 10% normal goat serum for 30 min. The slides were then incubated at 37°C for 1 h with rabbit anti-CSBF/C10orf99 or anti-SUSD2 polyclonal antibody. Antibody against CSBF/C10orf99 was prepared and affinity-purified in our laboratory. After thorough washing, Dakocytomation Envision System HRP (DakoCytomation, USA) was applied for 30 min. After rinsing in PBS, all slides were visualized with 0.05% (w/v) 3,3′-diaminobenzidine.
Cell proliferation assay
Cells were harvested, plated in 96-well plates at a density of 2000 cells per well, and incubated at 37°C. Cell proliferation was analyzed using the Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Japan). At indicated time points, 10 μl of the CCK-8 solution was added into each well and incubated for 2 h. Absorbance at 450 nm and 630 nm was measured to calculate the number of viable cells.
Colony formation assay
Colony formation was analyzed by plating 200 cells in 12-well culture plates (in triplicate). After 8 days, colonies were fixed and stained with 0.5% crystal violet. Colonies were defined as a minimum of 50 cells in a group and were counted using image analysis software (IPP6).
Flow cytometry
For SUSD2 expression analysis, fixed and permeabilized cells were stained with mouse monoclonal W5C5 antibody (Catalog #327401, BioLegend) or the appropriate isotype control (mouse IgG1, kappa) (Catalog # 401401, BioLegend). Labeled cells were used for flow cytometric analysis by FACSCalibur (BD Biosciences). Data was analyzed with using FlowJo software (Tree Star).
Cell cycle analysis
Cells were harvested and fixed in 70% ethanol, treated with RNase A and stained with propidium iodide, and DNA content was analyzed by FACSCalibur. The results were analyzed by ModFit software.
Statistical analysis
Data were expressed as mean ± SD and tested for statistical significance by the two-tailed Student's t-test or ANOVAs with the appropriate between- and within-subjects factors for each experiment. Values of p < 0.05 were considered statistically significant, where * = p < 0.05, ** = p < 0.01, and *** = p < 0.001.
Author Contributions
W.P. designed and performed experiments. Y.C., H.Z. and B.L. prepared figures 1 and 3. X.C. and L.L. prepared the cancer samples used in this work. W.H. and W.P. discussed the results and wrote the main manuscript text. W.H., J.J., L.Z., P.W., T.L., X.C. and X.M. supervised the project. All authors reviewed the manuscript.
Supplementary Material
CSBF/C10orf99, a novel potential cytokine, inhibits colon cancer cell growth through inducing G1 arrest
Acknowledgments
This work was supported by the Peking University–National Taiwan University Cooperation Fund (BMU20120316) and the Program for Innovation of New Drugs (2013ZX09103003-023). The authors declare that they do not have any conflicts of interest (financial or otherwise) related to the data presented in this manuscript.
References
- Smith-Garvin J. E. & Sigal L. J. Immunology: Memory cells sound the alarm. Nature 497, 194–6 (2013). [DOI] [PubMed] [Google Scholar]
- Hsu S. M. & Hsu P. L. Autocrine and paracrine functions of cytokines in malignant lymphomas. Biomed Pharmacother 48, 433–44 (1994). [DOI] [PubMed] [Google Scholar]
- Clement M. et al. CD31 is a key coinhibitory receptor in the development of immunogenic dendritic cells. Proc Natl Acad Sci U S A 111, E1101–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki F., Sitnicka E. & Bryceson Y. T. NK cell development and function - Plasticity and redundancy unleashed. Semin Immunol 26, 114–26 (2014). [DOI] [PubMed] [Google Scholar]
- Shen P. et al. IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature 507, 366–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. J. et al. Insights into the Role of Follicular Helper T Cells in Autoimmunity. Immune Netw 14, 21–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Wu L. & Li X. IL-17 family: cytokines, receptors and signaling. Cytokine 64, 477–85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel H. L. & Mercurio A. M. VEGF targets the tumour cell. Nat Rev Cancer 13, 871–82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullens D. M., Decraene A., Seys S. & Dupont L. J. IL-17A in human respiratory diseases: innate or adaptive immunity? Clinical implications. Clin Dev Immunol 2013, 840315 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham B. W., Kavanaugh A. & Reich K. Interleukin-17A: a unique pathway in immune-mediated diseases: psoriasis, psoriatic arthritis and rheumatoid arthritis. Immunology 141, 133–42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oranger A., Carbone C., Izzo M. & Grano M. Cellular mechanisms of multiple myeloma bone disease. Clin Dev Immunol 2013, 289458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L. et al. Stromal cell-derived factor-1 and its receptor CXCR4 in adult neurogenesis after cerebral ischemia. Restor Neurol Neurosci 31, 239–51 (2013). [DOI] [PubMed] [Google Scholar]
- Bai F. et al. Genetically-engineered Newcastle disease virus expressing interleukin 2 is a potential drug candidate for cancer immunotherapy. Immunol Lett 159, 36–46 (2014). [DOI] [PubMed] [Google Scholar]
- Churlaud G. et al. Sustained stimulation and expansion of Tregs by IL2 control autoimmunity without impairing immune responses to infection, vaccination and cancer. Clin Immunol 151, 114–126 (2014). [DOI] [PubMed] [Google Scholar]
- Watanabe S. & Inoue J. Intracellular delivery of lipopolysaccharide induces effective Th1-immune responses independent of IL-12. PLoS One 8, e68671 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W. et al. Molecular cloning and characterization of chemokine-like factor 1 (CKLF1), a novel human cytokine with unique structure and potential chemotactic activity. Biochem J 357, 127–35 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T. et al. Cytohesins/ARNO: The Function in Colorectal Cancer Cells. PLoS One 9, e90997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P. & Jain R. K. Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q. et al. Overexpression of HGF Promotes HBV-Induced Hepatocellular Carcinoma Progression and Is an Effective Indicator for Met-Targeting Therapy. Genes Cancer 4, 247–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. et al. Enhanced in-vitro and in-vivo suppression of A375 melanoma by combined IL-24/OSM adenoviral-mediated gene therapy. Melanoma Res 24, 20–31 (2014). [DOI] [PubMed] [Google Scholar]
- Wang M. L. et al. Oncostatin m modulates the mesenchymal-epithelial transition of lung adenocarcinoma cells by a mesenchymal stem cell-mediated paracrine effect. Cancer Res 72, 6051–64 (2012). [DOI] [PubMed] [Google Scholar]
- Mao Z. et al. Adenovirus-mediated IL-24 expression enhances the chemosensitivity of multidrug-resistantgastric cancer cells to cisplatin. Oncol Rep 30, 2288–96 (2013). [DOI] [PubMed] [Google Scholar]
- Xu S. et al. Stabilization of MDA-7/IL-24 for colon cancer therapy. Cancer Lett 335, 421–30 (2013). [DOI] [PubMed] [Google Scholar]
- Bertorelle R., Rampazzo E., Pucciarelli S., Nitti D. & De Rossi A. Telomeres, telomerase and colorectal cancer. World J Gastroenterol 20, 1940–1950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasool S., Rasool V., Naqvi T., Ganai B. A. & Shah B. A. Genetic unraveling of colorectal cancer. Tumour Biol 35, 5067–82 (2014). [DOI] [PubMed] [Google Scholar]
- Bardhan K. & Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers (Basel) 5, 676–713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard J. Y., Rong A. & Sidhu R. RAS mutations in colorectal cancer. N Engl J Med 369, 2159–60 (2013). [DOI] [PubMed] [Google Scholar]
- Arends M. J. Pathways of colorectal carcinogenesis. Appl Immunohistochem Mol Morphol 21, 97–102 (2013). [DOI] [PubMed] [Google Scholar]
- Hammoud S. S., Cairns B. R. & Jones D. A. Epigenetic regulation of colon cancer and intestinal stem cells. Curr Opin Cell Biol 25, 177–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin J. J. et al. Chemoprevention, chemotherapy, and chemoresistance in colorectal cancer. Drug Metab Rev 44, 148–72 (2012). [DOI] [PubMed] [Google Scholar]
- Coppede F., Lopomo A., Spisni R. & Migliore L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J Gastroenterol 20, 943–956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett D. L. & Chu E. Can metastatic colorectal cancer be cured? Oncology (Williston Park) 26, 266–75 (2012). [PubMed] [Google Scholar]
- Kohne C. H., Folprecht G., Goldberg R. M., Mitry E. & Rougier P. Chemotherapy in elderly patients with colorectal cancer. Oncologist 13, 390–402 (2008). [DOI] [PubMed] [Google Scholar]
- Foster D., Parrish-Novak J., Fox B. & Xu W. Cytokine-receptor pairing: accelerating discovery of cytokine function. Nat Rev Drug Discov 3, 160–70 (2004). [DOI] [PubMed] [Google Scholar]
- Kim S. T., Park K. H., Shin S. W. & Kim Y. H. Dose KRAS Mutation Status Affect on the Effect of VEGF Therapy in Metastatic Colon Cancer Patients? Cancer Res Treat 46, 48–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razis E. et al. EGFR gene gain and PTEN protein expression are favorable prognostic factors in patients with KRAS wild-type metastatic colorectal cancer treated with cetuximab. J Cancer Res Clin Oncol 140, 737–48 (2014). [DOI] [PubMed] [Google Scholar]
- Ramesh R., Ioannides C. G., Roth J. A. & Chada S. Adenovirus-mediated interleukin (IL)-24 immunotherapy for cancer. Methods Mol Biol 651, 241–70 (2010). [DOI] [PubMed] [Google Scholar]
- Guo X. et al. VSTM1-v2, a novel soluble glycoprotein, promotes the differentiation and activation of Th17 cells. Cell Immunol 278, 136–42 (2012). [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15, 8125–48 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. G., Carnell L. & Moore H. H. Post-Golgi membrane traffic: brefeldin A inhibits export from distal Golgi compartments to the cell surface but not recycling. J Cell Biol 118, 267–83 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Bayer S., Cowley J. A. & Gorbalenya A. E. The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J Virol 77, 1415–26 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdomo-Morales R. et al. The trypsin inhibitor panulirin regulates the prophenoloxidase-activating system in the spiny lobster Panulirus argus. J Biol Chem 288, 31867–79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing R. M. et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3, 89 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A. P., Evans R. L. & Egland K. A. Multiple functions of Sushi Domain Containing 2 (SUSD2) in breast tumorigenesis. Mol Cancer Res 11, 74–85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland C. T., Salanga C. L., Kawamura T., Trejo J. & Handel T. M. The chemokine receptor CCR1 is constitutively active, which leads to G protein-independent, beta-arrestin-mediated internalization. J Biol Chem 288, 32194–210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertazzi L., Arosio D., Marchetti L., Ricci F. & Beltram F. Quantitative FRET analysis with the EGFP-mCherry fluorescent protein pair. Photochem Photobiol 85, 287–97 (2009). [DOI] [PubMed] [Google Scholar]
- Su Y. et al. CMTM3 inhibits cell migration and invasion and correlates with favorable prognosis in gastric cancer. Cancer Sci 105, 26–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. et al. Gli1 inhibition suppressed cell growth and cell cycle progression and induced apoptosis as well as autophagy depending on ERK1/2 activity in human chondrosarcoma cells. Cell Death Dis 5, e979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. Janeway's immunobiology. London, England: Garland Science, Taylor and Francis Group; 2007.
- Guo P. et al. Gene expression profile based classification models of psoriasis. Genomics 103, 48–55 (2014). [DOI] [PubMed] [Google Scholar]
- Tang T. et al. A mouse knockout library for secreted and transmembrane proteins. Nat Biotechnol 28, 749–55 (2010). [DOI] [PubMed] [Google Scholar]
- Molinari F. & Frattini M. Functions and Regulation of the PTEN Gene in Colorectal Cancer. Front Oncol 3, 326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M. The tumor suppressor protein p16INK4a. Exp Cell Res 237, 7–13 (1997). [DOI] [PubMed] [Google Scholar]
- Li J., Poi M. J. & Tsai M. D. Regulatory mechanisms of tumor suppressor P16(INK4A) and their relevance to cancer. Biochemistry 50, 5566–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara T. et al. Isolation of a novel mouse gene, mSVS-1/SUSD2, reversing tumorigenic phenotypes of cancer cells in vitro. Cancer Sci 98, 900–8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara T. et al. von Willebrand factor type D domain mutant of SVS-1/SUSD2, vWD(m), induces apoptosis in HeLa cells. Cancer Sci 98, 909–15 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier F., Bourin P., Bladier D., Joubert-Caron R. & Caron M. Effect of 5-azacytidine and galectin-1 on growth and differentiation of the human b lymphoma cell line bl36. Cancer Cell Int 1, 2 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. G1 cell-cycle control and cancer. Nature 432, 298–306 (2004). [DOI] [PubMed] [Google Scholar]
- Li H. et al. A novel 3p22.3 gene CMTM7 represses oncogenic EGFR signaling and inhibits cancer cell growth. Oncogene 33, 3109–3118 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CSBF/C10orf99, a novel potential cytokine, inhibits colon cancer cell growth through inducing G1 arrest