Abstract
Human epidermal growth factor receptor 2 (HER2) and topoisomerase II alpha (TOP2A) genes have been proposed as predictive biomarkers of sensitivity to anthracycline chemotherapy. Recently, chromosome 17 centromere enumeration probe (CEP17) duplication has also been associated with increased responsiveness to anthracyclines. However, reports are conflicting and none of these tumor markers can yet be considered a clinically reliable predictor of response to anthracyclines. We studied the association of TOP2A gene alterations, HER2 gene amplification, and CEP17 duplication with response to anthracycline-based neoadjuvant chemotherapy in 140 patients with operable or locally advanced breast cancer. HER2 was tested by fluorescence in situ hybridization and TOP2A and CEP17 by chromogenic in situ hybridization. Thirteen patients (9.3%) achieved pathologic complete response (pCR). HER2 amplification was present in 24 (17.5%) of the tumors. TOP2A amplification occurred in seven tumors (5.1%). CEP17 duplication was detected in 13 patients (9.5%). CEP17 duplication correlated with a higher rate of pCR [odds ratio (OR) 6.55, 95% confidence interval (95% CI) 1.25-34.29, P = .026], and analysis of TOP2A amplification showed a trend bordering on statistical significance (OR 6.97, 95% CI 0.96-50.12, P = .054). TOP2A amplification and CEP17 duplication combined were strongly associated with pCR (OR 6.71, 95% CI 1.66-27.01, P = .007). HER2 amplification did not correlate with pCR. Our results suggest that CEP17 duplication predicts pCR to primary anthracycline-based chemotherapy. CEP17 duplication, TOP2A amplifications, and HER2 amplifications were not associated with prognosis.
Abbreviations: CEP17, chromosome 17 centromere enumeration probe; CI, confidence interval; CISH, chromogenic in situ hybridization; DFS, disease-free survival; EC-D, epirubicin (90 mg/m2) and cyclophosphamide (600 mg/m2) followed by docetaxel (100 mg/m2); ER, estrogen receptor; FEC75, fluorouracil (600 mg/m2), epirubicin (75 mg/m2), and cyclophosphamide (600 mg/m2); FISH, fluorescence in situ hybridization; HR, hazard ratio; HER2, human epidermal growth factor receptor 2; OR, odds ratio; OS, overall survival; pCR, pathologic complete response; PR, progesterone receptor; TOP2A, topoisomerase II alpha
Introduction
Predicting response to anthracycline-based therapy is a central challenge in patients with breast cancer. Several randomized studies have shown that anthracycline-based adjuvant therapy produces a modest improvement in survival in patients with early-stage breast cancer [1]. In addition, it has been shown that incorporation of taxanes further improves pathologic complete response (pCR) in the neoadjuvant setting [2], [3], [4], [5], [6], [7]. However, the risk of serious side effects must be considered and predictive factors are needed to help clinicians select the most appropriate drug for each patient. To date, no specific biomarkers have been identified to predict tumor response to anthracycline-based chemotherapy.
The human epidermal growth factor receptor 2 (HER2) and topoisomerase II alpha (TOP2A) genes have been proposed as markers of sensitivity to anthracycline chemotherapy. Although several studies have reported an association between HER2 amplification with anthracycline sensitivity [8], [9], [10], [11], [12], [13], only two of these were statistically significant [10], [13]. In vitro and in vivo studies indicate that HER2 positivity alone does not alter anthracycline sensitivity [14] and the underlying mechanism remains elusive. The TOP2A gene, located at 17q12-q21, close to the HER2 gene, encodes TOP2A, a key enzyme in DNA replication and the molecular target of anthracyclines [15], [16]. When HER2 is amplified, genes situated around 17q21, such as TOP2A gene, may be either co-amplified or deleted [17]. Because of this physical proximity, some researchers have proposed that the link between HER2-positive disease and anthracycline sensitivity is the presence of TOP2A alterations (amplifications and deletions) rather than HER2 amplifications [18], [19]. However, the results of these studies have not always been consistent [20].
Given the location of HER2 and TOP2A on chromosome 17, recent reports have focused on the predictive role of other molecular alterations localized within the 17q21 region, including alterations in key genes such as HER1-3 [21], p53 [22], and BRCA1 [23], and variations in the copy number of subchromosomal regions, including the chromosome 17 centromere (CEP17) duplication [21]. On the basis of recent data from array comparative genomic hybridization, CEP17 duplication is defined as increased copy number of CEP17 [21] rather than polysomies of the whole chromosome 17 [24], [25]. CEP17 duplication has been described as a marker of genomic instability [26] and has received a great deal of attention as a potential predictor of anthracycline benefit [21], [27].
We hypothesized that CEP17 duplication is associated with a higher response to neoadjuvant chemotherapy. We explored the association of CEP17 duplication and TOP2A alterations with response to anthracycline-based neoadjuvant therapy. We studied a cohort of patients with early or locally advanced breast cancer treated with anthracycline-based primary chemotherapy, and we chose pCR as a surrogate marker of chemosensitivity.
Materials and Methods
Ethics Statement
This study was conducted according to the Declaration of Helsinki principles, with approval from the Clinical Research Ethics Committee at Institut d'Investigacions Biomèdiques Sant Pau. Written informed consent was obtained from all patients.
Study Design and Patients
We retrospectively studied 140 consecutive patients with stage II or III breast cancer who received anthracycline-based neoadjuvant chemotherapy in our hospital between 1993 and 2010. All patients had confirmed diagnosis based on histopathology of biopsy and none of them had prior treatment with surgery, chemotherapy, or radiation. Our study included patients with HER2-positive carcinomas treated before the approval of trastuzumab in 2006. The study excluded patients with bilateral or inflammatory tumors. Fifty-five patients received neoadjuvant treatment with anthracyclines alone [FEC75: fluorouracil (600 mg/m2), epirubicin (75 mg/m2), and cyclophosphamide (600 mg/m2) given every 3 weeks for four to six cycles, n = 40, or FAC60: fluorouracil (500 mg/m2), doxorubicin (60 mg/m2), and cyclophosphamide (500 mg/m2) given every 3 weeks for four to six cycles, n = 15] between 1993 and 2002. Eighty-five patients received neoadjuvant treatment with anthracyclines in combination with taxanes [EC-D: epirubicin (90 mg/m2) and cyclophosphamide (600 mg/m2) given every 3 weeks for four cycles followed by docetaxel (100 mg/m2) every 3 weeks for four cycles] from 2003 to 2010. Patients were staged according to the tumor-node-metastasis (TNM) system. Clinical response was assessed by palpation, breast ultrasound, mammography, and/or magnetic resonance imaging before systemic therapy and before curative surgery. Clinical responses were evaluated according to the response evaluation criteria in solid tumors (RECIST) criteria [28] every two cycles. Patients treated with FEC75 or FAC60 with partial response received surgical treatment after four cycles of chemotherapy and two additional cycles of FEC75 or FAC60 were administered after surgery. Patients treated with EC-D underwent breast surgery after completion of chemotherapy. Patients with positive hormone receptor tumors received radiotherapy and endocrine therapy. The extent of residual disease was measured in the surgical specimen. The primary endpoint for this study was pCR defined as the absence of invasive cancer in the breast and axillary lymph nodes at the time of definitive surgery. The secondary endpoints for the study were disease-free survival (DFS) and overall survival (OS). Patients were followed up according to the breast cancer guidelines.
Tumor Samples and Tissue Microarrays
We analyzed 140 representative formalin-fixed, paraffin-embedded tumor core biopsies obtained before neoadjuvant treatment. Paraffin blocks were stored at room temperature. Samples were identified only by an identification number assigned to each patient. A stained section of each tumor sample was prepared to confirm the diagnosis and to identify representative tumor areas. Tissue microarrays were prepared from formalin-fixed, paraffin-embedded tissue taken from three representative areas of each tumor. Serial 5-μm sections were obtained for immunohistochemical, fluorescence in situ hybridization (FISH), and chromogenic in situ hybridization (CISH) analyses.
Immunohistochemistry
Prediluted antibodies for estrogen receptor (ER; clone EP1), progesterone receptor (PR; clone 636) and Ki67 (clone MIB-I) were obtained from Dako (Glostrup, Denmark). Sections were processed in a PT Module using Dako high pH buffer (Dako) for deparaffinization and antigen retrieval. Sections for the Ki-67 study were processed with Dako low pH buffer. All immunohistochemical stains were performed in an Autostainer Link using the EnVision method (Dako). HER2 overexpression was analyzed using the HercepTest assay (Dako). Tumors were classified as ER or PR positive when at least 1% of the tumor cells showed staining in the nuclei cells [29]. HER2 was considered overexpressed when a uniform intense (3 +) membrane staining was present in > 30% of invasive tumor cells [30]. The percentage of Ki67-stained nuclei was evaluated independently of the intensity and its positivity cutoff value was ≥ 20% [31]. Two pathologists independently evaluated all immunostainings, and discordant results were reviewed to reach an agreement.
In situ Hybridization Analyses
HER2 gene status was confirmed for all patients by FISH. TOP2A gene status was evaluated by CISH and FISH in equivocal cases. A good correlation of the results for TOP2A obtained by FISH or CISH has been previously reported [32]. We used the HER2 FISH pharmDX (Dako) and the TOP2A FISH pharmDX (Dako) assays, respectively. The CISH assay was performed using Dako dual color assay (Dako). All tests were performed following the manufacturer's recommendations. Fluorescence signals were evaluated using an Olympus BX51 fluorescent microscope and appropriate filter sets. The assessment of FISH results for TOP2A and HER2 was performed in ≥ 60 nuclei per case. TOP2A gene status was assessed by CISH in ≥ 30 tumor cells per case. HER2 gene amplification was defined when the ratio HER2/CEP17 was ≥ 2 in accordance with the manufacturer’s recommendations. TOP2A amplification was considered when the ratio TOP2A/CEP17 was ≥ 2 and TOP2A deletion was considered when the ratio TOP2A/CEP17 was < 0.8. We use the term CEP17 duplication throughout the text as defined previously [21]. The cutoff value for CEP17 duplication was determined as > 1.86 observed CEP signals per cell [21], [26].
Statistical Analyses
DFS was defined as the time from initiation of treatment to date of first relapse (local, regional, contralateral or metastatic), second primary cancer, or death resulting from any cause (whichever occurred first). OS was defined as the time from sample collection to death resulting from any cause. Patients lost to follow-up were censored at the last contact. Quantitative variables between groups were compared using the Student’s t test, and categorical variables were compared using the Chi-square or Fisher exact test. The univariable association between variables and pCR was assessed using the Chi-square test. The multivariable logistic regression model for pCR was adjusted for age at diagnosis (< 50 vs ≥ 50 years), tumor stage (T2 vs T3 vs T4), nodal stage (negative vs positive), histologic grade (1 or 2 vs 3), neoadjuvant chemotherapy (FEC75/FAC60 vs EC-D), ER (negative vs positive), PR (negative vs positive), HER2 (normal vs amplified), TOP2A status (normal vs amplified), and CEP17 status (normal vs duplicated). Kaplan-Meier and log-rank analyses were used to compare DFS and OS. The Cox proportional hazard model with a single covariate was used to obtain the hazard ratios (HRs) for relapse or death and associated 95% confidence interval (95% CI). HER2 (normal vs amplified), TOP2A status (normal vs amplified), and CEP17 status (normal vs duplicated) as individual variables were entered in a final Cox model adjusted for traditional prognostic factors including age at diagnosis (< 50 vs ≥ 50 years), nodal stage (negative vs positive), tumor size (T2 vs T3 vs T4), histologic grade (1 or 2 vs 3), neoadjuvant chemotherapy (FEC75/FAC60 vs EC-D), ER (negative vs positive), PR (negative vs positive), and Ki67 (low vs high).The Cox proportional hazard model was used in an exploratory analysis of tumors to analyze tumors with either TOP2A amplification or CEP17 duplication; we refer to these tumors throughout the text as tumors with combined TOP2A or CEP17 duplication. A two-sided P value ≤ .05 was considered significant. Statistical analyses were performed using the SPSS 19.0 statistical software package (SPSS, Chicago, IL).
Patient selection, assay performance, and data analysis concurred to the reporting recommendations for tumor marker prognostic studies (REMARK) guidelines [33].
Results
Patient Characteristics
Patient and tumor characteristics are summarized in Table 1. A total of 101 of the 140 patients had T3 or T4 tumors (72.1%), and 93 had lymph node–positive tumors (66.4%); these factors explain the observed low rate of conservative treatment (37.1%). A pCR after neoadjuvant chemotherapy was achieved in 13 patients (9.3%). Four patients (7.3%) achieved pCR after FEC75/FAC60 and nine (10.6%) patients after EC-D treatment. No significant differences in pCR were observed according to type of neoadjuvant treatment or tumor stage (Supplementary Table 1).
Table 1.
Basic Patient and Tumor Characteristics.
| Variable | n (%) | |
|---|---|---|
| N | 140 | |
| Age, years | Mean ± SD | 53.7 ± 13.5 |
| Median (range) | 51 (25.5-86.1) | |
| Menopausal status | Premenopausal | 65 (46.4) |
| Postmenopausal | 75 (53.6) | |
| Tumor stage | II | 50 (35.7) |
| III | 90 (64.3) | |
| Tumor status | T2 | 39 (27.9) |
| T3 | 42 (30.0) | |
| T4 | 59 (42.1) | |
| Node status | N0 | 47 (33.6) |
| N1 | 63 (45.0) | |
| N2 | 26 (18.6) | |
| N3 | 4 (2.9) | |
| Tumor grade | 1 | 11 (7.9) |
| 2 | 63 (45.0) | |
| 3 | 66 (47.1) | |
| ER | Positive | 87 (62.1) |
| Negative | 53 (37.9) | |
| PR | Positive | 68 (48.6) |
| Negative | 72 (51.4) | |
| Ki67 status | < 20% | 74 (42.9) |
| ≥ 20% | 60 (52.9) | |
| Missing data | 6 (4.3) | |
| HER2 | Normal | 113 (81) |
| Amplification | 24 (17) | |
| Missing data | 3 (2) | |
| TOP2A | Normal | 124 (89) |
| Amplification | 7 (5) | |
| Deletion | 6 (4) | |
| Missing data | 3 (2) | |
| Neoadjuvant therapy | FEC75/FAC60 | 55 (39.3) |
| EC-D | 85 (60.7) | |
| Surgical treatment | Mastectomy | 88 (62.9) |
| Lumpectomy | 52 (37.1) | |
| Pathologic response | Complete | 13 (9.3%) |
| Residual disease | 127 (90.7) |
Association of CEP17 Duplication with Clinicopathologic Variables
Table 2 shows the associations of CEP17 status with patient characteristics and tumor-related variables. Status of HER2, TOP2A, and CEP17 duplication was assessable in 137 (97.9%) tumors. HER2 amplification was found in 24 samples (17.5%), TOP2A was amplified in 7 (5.1%) of the tumors, TOP2A deletion was found in 6 samples (4.4%), and CEP17 duplication was detected in 13 (9.5%) samples. Tumors with combined TOP2A amplification and CEP17 duplication alterations were present in 20 (14.6%) of the tumors. CEP17 duplication was not associated with HER2 gene amplification (P = .166), high grade (P = .499), ER negativity (P = .613), or a high Ki67 (P = .937). Tumors with HER2 amplification had TOP2A amplification in three cases (42.9%, P = .062), TOP2A deletion in three cases (50%, P = .028), and CEP17 aneusomy in two cases (40% P = .164). HER2 amplifications were significantly associated with TOP2A alterations (amplifications and deletions; P = .003) and with CEP17 aneusomy (P = .048; data not shown).
Table 2.
Correlation between Clinicopathologic Characteristics and CEP17 Status.
| Variable | n (%) | CEP17 Status |
P⁎ | ||
|---|---|---|---|---|---|
| Normal | Duplicated | ||||
| CEP17 | 137 | 124 (90.5) | 13 (9.5) | ||
| Age, years | ≤ 50 | 65 (47.5) | 61 (49) | 4 (30.8) | .206 |
| > 50 | 72 (52.5) | 63 (51) | 9 (69.2) | ||
| Menopausal status | Premenopausal | 63 (46) | 59 (47) | 4 (30.8) | .247 |
| Postmenopausal | 74 (54) | 65 (53) | 9 (69.2) | ||
| Tumor stage | II | 49 (35.8) | 44 (33.5) | 5 (38.5) | .830 |
| III | 88 (64.2) | 80 (64.5) | 8 (61.5) | ||
| Tumor status | T2 | 38 (27.7) | 33 (27) | 5 (38.6) | .606 |
| T3 | 42 (30.7) | 38 (30) | 4 (30.7) | ||
| T4 | 57 (41.6) | 53 (43) | 4 (30.7) | ||
| Node status | N0 | 46 (33.5) | 42 (34) | 4 (31) | .538 |
| N1 | 62 (45.5) | 54 (44) | 8 (61.5) | ||
| N2 | 25 (18) | 24 (19) | 1 (7.5) | ||
| N3 | 4 (3) | 4 (3) | 0 (0) | ||
| Tumor grade | 1 | 11 (8) | 11 (9) | 0 (0) | .499 |
| 2 | 62 (45) | 55 (44) | 7 (54) | ||
| 3 | 64 (47) | 58 (47) | 6 (46) | ||
| ER | Positive | 86 (62.8) | 77(62) | 9 (69.2) | .613 |
| Negative | 51 (37.2) | 47 (38) | 4 (30.8) | ||
| PR | Positive | 68 (49.6) | 62 (50) | 6 (46) | .792 |
| Negative | 69 (50.4) | 62 (50) | 7 (54) | ||
| Ki67 status | < 20% | 73 (53.3) | 66 (53) | 7 (54) | .937 |
| ≥ 20% | 60 (50.4) | 54 (43.5) | 6 (46) | ||
| Missing data | 4 (2.9) | 4 (3) | 0 (0) | ||
| HER2 | Normal | 112 (82) | 103 (83) | 9 (69) | .166 |
| Amplification | 23 (17) | 19 (15) | 4 (31) | ||
| Missing data | 2 (1) | 2 (2) | 0 (0) | ||
| TOP2A | Normal | 124 (91) | 111 (89.5) | 13 (100) | .220 |
| Amplification | 7 (5) | 7 (5.5) | 0 (0) | ||
| Deletion | 6 (4) | 6 (5) | 0 (0) | ||
| Neoadjuvant therapy | FEC75/FAC60 | 84 (61.3) | 76 (61) | 8 (61.5) | .986 |
| EC-D | 53 (38.7) | 48 (39) | 5 (38.5) | ||
| Surgical treatment | Mastectomy | 86 (62.9) | 78 (62.9) | 8 (61.5) | .923 |
| Lumpectomy | 51 (37.1) | 46 (37.1) | 5 (38.5) | ||
| Pathologic response | Complete | 12 (8.7) | 9 (7.3) | 3 (23) | .055 |
| Residual disease | 125 (91.3) | 115 (92.7) | 10 (77) | ||
P values were calculated using the Chi-square test.
Association of Clinicopathologic Parameters with pCR
In the multivariable analysis, CEP17 duplication associated with a high percentage of pCR [odds ratio (OR) 6.55, 95% CI 1.25-34.29, P = .026] and TOP2A gene amplification showed a benefit with borderline significance (OR 6.97, 95% CI 0.96-50.12, P = .054; Table 3). When TOP2A amplifications and CEP17 duplication were combined, we observed a significant association with pCR (OR 6.71, 95% CI 1.66-27.01, P = .007; Table 3). We did not find a significant difference in the benefit of treatment with anthracyclines when assessing deletion (P = .429) or combined TOP2A amplifications and deletions (P = .089) in univariable analysis. HER2 amplifications showed no direct association with pCR (P = .144). When pCR was assessed according to treatment received (Supplementary Table 1), CEP17 duplications were the only factor associated with better response to FEC70/FAC65 in both univariable (P = .004) and multivariable analyses (OR 14, 95% CI 1.427-137.324, P = .025). Meanwhile, the multivariable analysis showed that ER status (OR 12.68, 95% CI 1.427-137.324, P = .011) and TOP2A/CEP17 (OR 6.31, 95% CI 1.981-81.186, P = .007) were associated with better response to EC-D (data not shown).
Table 3.
Multivariable Analysis of Predictive Factors for pCR.
| Variable | Score | pCR |
|
|---|---|---|---|
| OR⁎ (95% CI) | P⁎ | ||
| ER | Positive | 1 | |
| Negative | 7.12 (1.69-30.03) | .007 | |
| TOP2A | Normal | 1 | |
| Amplification | 6.97 (0.96-50.12) | .054 | |
| CEP17 | Normal | 1 | |
| Duplication | 6.55 (1.25-34.29) | .026 | |
| HER2 | Normal | 1 | |
| Amplification | 0.804 (0.18-3.597) | .755 | |
| TOP2A/CEP17† | Normal | 1 | |
| Altered | 6.71 (1.66-27.01) | .007 | |
Logistic regression models adjusted for known prognostic factors (age, nodal status, tumor size, tumor grade, and hormone receptor status) and neoadjuvant therapy.
Tumors with combined TOP2A amplification and CEP17 duplication.
Prognostic Associations with DFS and OS
The univariable analysis showed that CEP17 duplication, HER2 amplification, and TOP2A amplification were not significantly associated with DFS or OS (Table 4). However, when TOP2A amplification and CEP17 duplication were combined, Kaplan-Meier and log-rank analyses showed a significant association with better DFS (P = .026) and a borderline non-significant trend with OS (P = .054; Figure 1). The univariable analysis showed a similar association of the combination of TOP2A amplification and CEP17 duplication with DFS (HR 4.34, 95% CI 1.05-17.92, P = .042) and OS (HR 3.70, 95% CI 0.89-15.34, P = .071; Table 4). However, after adjustment for the classic prognostic factors, age, and neoadjuvant therapy, the multivariable analysis showed that CEP17 duplication, TOP2A amplification, HER2 amplification, and the combination of TOP2A amplifications and CEP17 duplication failed to show an association with DFS or OS (Table 5).
Table 4.
Biomarker Analysis and Relation with DFS and OS.
| Variable | Score | n | DFS |
OS |
||
|---|---|---|---|---|---|---|
| HR⁎ (95% CI) | P⁎ | HR⁎ (95% CI) | P⁎ | |||
| CEP17 | Normal | 124 | 1 | 1 | ||
| Duplicated | 13 | 2.64 (0.64-10.9) | .179 | 2.42 (0.58-10.02) | .223 | |
| HER2 | Normal | 113 | 1 | 1 | ||
| Amplified | 24 | 0.97 (0.45-2.08) | .960 | 0.96 (0.44-2.06) | .907 | |
| TOP2A | Non-amplified | 130 | 1 | 1 | ||
| Amplified | 7 | 21.9 (0.92-5275) | .269 | 21.73 (0.04-11545) | .336 | |
| Ki67 | ≥ 20% | 60 | 1 | 1 | ||
| < 20% | 74 | 1.89 (1.04-3.46) | .036 | 1.91 (1.02-3.58) | .044 | |
| TOP2A/CEP17† | Normal | 117 | 1 | 1 | ||
| Altered | 20 | 4.35 (1.05-17.93) | .042 | 3.71 (0.89-15.34) | .071 | |
Univariable Cox proportional hazard models based on 136 patients, 44 recurrences, and 40 deaths.
Tumors with combined TOP2A amplification and CEP17 duplication.
Figure 1.
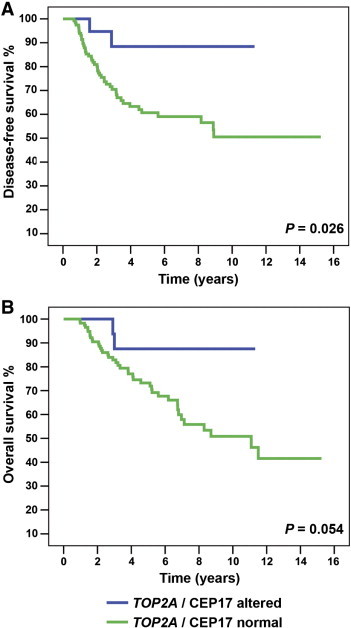
Association of combined TOP2A amplification and CEP17 duplication with patient outcome. DFS (A) and OS (B) Kaplan-Meier curves and log-rank tests according to the presence or absence of tumors with combined TOP2A amplification and CEP17 duplication.
Table 5.
Multivariable Analyses for All Prognosis Variables and Relation with DFS and OS.
| Variables | DFS |
OS |
||
|---|---|---|---|---|
| HR (95% CI) | P⁎ | HR (95% CI) | P⁎ | |
| Age (< 50 vs ≥ 50 years) | 1.03 (0.5-2.15) | .91 | 1.09 (0.5-2.41) | .81 |
| Lymph nodes (negative vs positive) | 2.95 (1.23-7.07) | .01 | 3.12 (1.21-8.03) | .01 |
| Tumor size (T2 vs T3 vs T4) | 1.78 (0.85-3.7) | .02 | 1.63 (0.68-3.91) | .9 |
| Tumor grade (1 vs 2 vs 3) | 1.53 (0.77-3.03) | .22 | 2.07 (1.06-4.05) | .03 |
| ER (positive vs negative) | 2.20 (1.24-4.01) | .01 | 1.95 (0.93-4.07) | .07 |
| pCR (positive vs negative) | 4.41 (0.54-30.65) | .08 | 4.49 (0.58-34.72) | .15 |
| Ki67 (< 20% vs ≥ 20%) | 1.29 (0.65-2.54) | .45 | 1.33 (0.65-2.71) | .43 |
| Treatment (EC-D vs FEC/FAC) | 1.17 (0.61-2.24) | .62 | 1.07 (0.54-2.12) | .83 |
| CEP17 (duplicated vs normal) | 2.46 (0.54-11.06) | .24 | 2.29 (0.51-10.35) | .27 |
| HER2 (amplified vs normal) | 1.23 (0.51-2.97) | .63 | 1.06 (0.43-2.62) | .89 |
| TOP2A/CEP17† (altered vs normal) | 3.91 (0.89-17.2) | .71 | 3.72 (0.84-16.43) | .08 |
Multivariable Cox proportional analysis based on 136 patients, 44 recurrences, and 40 deaths.
Tumors with combined TOP2A amplification and CEP17 duplication.
Discussion
The main finding in this study is that CEP17 duplication associates with pCR to anthracycline-based neoadjuvant therapy in patients with breast cancer. To our knowledge, this is the first study to analyze the association of CEP17 duplication with response to neoadjuvant chemotherapy using pCR as a surrogate marker of chemosensitivity. Our rationale in investigating the association of CEP17 duplication and response to chemotherapy was based on the recognition that CEP17 duplication could be a potential marker of genomic instability and DNA repair dysfunction [26], [34]. Chromosome 17 is the second highest density gene in the human genome, and it includes several key genes that have a role in breast cancer (HER2, TOP2A, BRCA1, and Tau) and DNA repair (P53, RAD51C, and RAD52B) [35], [36]. Analyses of rearrangement have shown that chromosome 17 harbors a region of high genomic instability centered around 17q12 and characterized by multiple gene amplifications and deletions [37], [38], [39]. This pattern has traditionally been associated with polysomies of chromosome 17. However, recent data using array comparative genomic hybridization suggest that chromosome 17 polysomies detected by FISH or CISH are in fact very rare in breast cancer; what is being detected are gains or amplification in the centromere region of chromosome 17 rather than a copy number gain of the whole chromosome 17 [24], [25]. An increase in the copy numbers of CEP17 might cause several key genetic changes, such as HER2 amplification, TOP2A amplification or deletion, P53 loss, and BRCA1 loss. All these chromosome abnormalities have been linked to mechanisms of breast tumorigenesis including cell proliferation, tumor angiogenesis, and reduced apoptosis [38], [39]. Together, these findings suggest that CEP17 duplication is an indicator of genetic instability around 17q12 locus that could be linked to aberrations in the copy number of DNA repair genes, such as TOP2A. Therefore, analysis of CEP17 duplication may allow for an improvement of treatment efficacy based on therapies targeting DNA repair genes.
Our results showed that CEP17 duplication was present in 9.3% of the tumors, a result that is somehow inferior to results reported by others. However, the interlaboratory reproducibility of CEP17 testing is unknown, and the prevalence differs among studies, ranging from 8% to 68% depending on the type of material examined, cell counting methods, and the threshold criteria for CEP17 duplication [36]. Moreover, there are conflicting results in earlier studies in relation to the correlation between CEP17 duplication and a more aggressive tumor phenotype [36]. Our results did not show an association between CEP17 duplication and high tumor grade, lymph node metastasis, ER negativity, HER2 amplification, or high Ki67.
Another finding of interest in our study is that TOP2A gene amplification correlated with pCR to anthracycline-based neoadjuvant therapy. However, the response was of borderline significance (OR 6.97, 95% CI 0.96-50.12, P = .054), but when combined with CEP17 duplication, the pCR response was significant (OR 6.71, 95% CI 1.66-27.01, P = .007). The small number of patients in our study might explain that TOP2A gene amplification alone (found in only 5% of tumors) was not associated with pCR. These observations suggest that patients with tumors that harbor gene alterations that may increase TOP2A copy number benefit more from anthracycline-based neoadjuvant therapy. We hypothesized that anthracycline sensitivity is not only the result of an increased number of genes secondary to gene amplification of TOP2A but also the result of concomitant gains in pericentromeric regions on chromosome 17 (CEP17 duplication). Thus, CEP17 duplication would be an indirect mechanism of increasing TOP2A gene copy number. However, we cannot yet identify which of these chromosomal abnormalities might be associated with the underlying mechanism of chemosensitivity.
As in previous studies [40], [41], we found that HER2 amplification failed to have a predictive value, supporting that patients with HER2-negative tumors may benefit from treatment with anthracyclines. We also observed that patients with TOP2A normal tumors might have some additional benefit from treatment with anthracyclines. Our results suggest that other mechanisms, such as the immune response, tumor-stromal interactions, and CEP17 duplication, may participate in the increased sensitivity to anthracyclines in these patients [42], [43], [44], [45]. Therefore, the use of anthracyclines should not be limited to patients with HER2 amplification or TOP2A amplification.
We were unable to identify any statistically significant relationship between CEP17 duplication and patient outcome. Our results are consistent with two previous studies [21], [27] that analyzed prognostic and predictive associations of CEP17 duplication in early-stage breast cancer patients treated with adjuvant chemotherapy regimens containing anthracyclines and non-anthracyclines. Bartlett et al. [21] assessed the role of CEP17 duplication in 1931 tumors from 2391 women enrolled onto the UK National Epirubicin Adjuvant Trial (NEAT/BR9601). Interestingly, although this study also failed to find a significant association of CEP17 duplication with patient prognosis (DFS HR 1.12, 95% CI 0.92-1.36, P = .24, OS HR 1.17, 95% CI 0.95-1.44, P = .13), it showed that the benefit on survival from the addition of epirubicin was confined to tumors with CEP17 duplication. Similar results were obtained by Pritchard et al. [27] in the randomized controlled mammary 5 (MA.5) adjuvant trial. The results did not identify a significant association of CEP17 with risk of recurrence (HR 1.15, 95% CI 0.86-1.54, P = .34) or risk of death (HR 1.39, 95% CI 0.90-2.15, P = .14) in multivariable analysis. However, in patients whose tumors exhibit CEP17 duplication and who received anthracyclines, there was a significant benefit for recurrence-free survival (HR 0.59, 95% CI 0.37-0.92, P = .02) and OS (HR 0.61, 95% CI 0.37-1.01, P = .05) in univariable analysis. This association disappeared after adjustment for other variables.
The potential limitations of our study are the restricted sample size and the retrospective design, which could bias patient selection. In addition, the absence of a study group receiving non-anthracycline-based chemotherapy meant we were unable to distinguish whether the pCR rate was attributed to higher sensitivity to anthracyclines or to chemotherapy with anthracyclines, taxanes, cyclophosphamide, and/or 5-fluorouracil. These results are therefore preliminary and require confirmation in larger prospective studies specifically designed to validate the predictive role of these biomarkers and to compare regimens with and without anthracyclines.
In conclusion, our results suggest that CEP17 duplication may be a useful biomarker of sensitivity to neoadjuvant anthracycline-based chemotherapy. A better understanding of CEP17 abnormalities might help to select patients who benefit most from anthracycline-based chemotherapy regimens and to identify patients in whom toxicity from these drugs could be avoided.
The following is the supplementary data related to this article.
Correlation between Clinicopathologic Characteristics and pCR
Acknowledgments
The authors thank Carolyn Newey for manuscript editing.
Footnotes
This article refers to supplementary material, which is designated by Supplementary Table 1 and is available online at www.neoplasia.com.
This study was supported by a grant from the Red Temática de Investigación Cooperativa de Cáncer (RTICC; RD12/0036/0076) to all authors, by grants from the Spanish Society of Clinical Oncology (SEOM; 2009 and 2012), the Molt Il lustríssima Administració (MIA) Foundation (2009), and Hospital de la Santa Creu i Sant Pau to A.T., by a grant from the Instituto de Salud Carlos III (ISCIII, PI10/0307) to D.E., and by a Pfizer award to A.B. The study sponsors had no involvement in the experimental work, data analysis, or manuscript preparation. Conflict of interests: The authors declare that they have no conflict of interest.
Contributor Information
Agustí Barnadas, Email: abarnadasm@santpau.cat.
Daniel Escuin, Email: descuin@santpau.cat.
References
- 1.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Gogas H, Fountzilas G. The role of taxanes as a component of neoadjuvant chemotherapy for breast cancer. Ann Oncol. 2003;14:667–674. doi: 10.1093/annonc/mdg210. [DOI] [PubMed] [Google Scholar]
- 3.Estevez LG, Gradishar WJ. Evidence-based use of neoadjuvant taxane in operable and inoperable breast cancer. Clin Cancer Res. 2004;10:3249–3261. doi: 10.1158/1078-0432.CCR-03-0133. [DOI] [PubMed] [Google Scholar]
- 4.Trudeau M, Sinclair SE, Clemons M. Neoadjuvant taxanes in the treatment of non-metastatic breast cancer: a systematic review. Cancer Treat Rev. 2005;31:283–302. doi: 10.1016/j.ctrv.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Schott AF, Hayes DF. Defining the benefits of neoadjuvant chemotherapy for breast cancer. J Clin Oncol. 2012;30:1747–1749. doi: 10.1200/JCO.2011.41.3161. [DOI] [PubMed] [Google Scholar]
- 6.Thompson AM, Moulder-Thompson SL. Neoadjuvant treatment of breast cancer. Ann Oncol. 2012;23(Suppl 10):x231–x236. doi: 10.1093/annonc/mds324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peto R, Davies C, Godwin J. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379:432–444. doi: 10.1016/S0140-6736(11)61625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Leo A, Gancberg D, Larsimont D. HER-2 amplification and topoisomerase IIalpha gene aberrations as predictive markers in node-positive breast cancer patients randomly treated either with an anthracycline-based therapy or with cyclophosphamide, methotrexate, and 5-fluorouracil. Clin Cancer Res. 2002;8:1107–1116. [PubMed] [Google Scholar]
- 9.Di Leo A, Larsimont D, Gancberg D. HER-2 and topo-isomerase IIalpha as predictive markers in a population of node-positive breast cancer patients randomly treated with adjuvant CMF or epirubicin plus cyclophosphamide. Ann Oncol. 2001;12:1081–1089. doi: 10.1023/a:1011669223035. [DOI] [PubMed] [Google Scholar]
- 10.Pritchard KI, Shepherd LE, O'Malley FP. HER2 and responsiveness of breast cancer to adjuvant chemotherapy. N Engl J Med. 2006;354:2103–2111. doi: 10.1056/NEJMoa054504. [DOI] [PubMed] [Google Scholar]
- 11.Muss HB, Thor AD, Berry DA. c-erbB-2 expression and response to adjuvant therapy in women with node-positive early breast cancer. N Engl J Med. 1994;330:1260–1266. doi: 10.1056/NEJM199405053301802. [DOI] [PubMed] [Google Scholar]
- 12.Paik S, Bryant J, Tan-Chiu E. HER2 and choice of adjuvant chemotherapy for invasive breast cancer: National Surgical Adjuvant Breast and Bowel Project Protocol B-15. J Natl Cancer Inst. 2000;92:1991–1998. doi: 10.1093/jnci/92.24.1991. [DOI] [PubMed] [Google Scholar]
- 13.Paik S, Bryant J, Park C. erbB-2 and response to doxorubicin in patients with axillary lymph node-positive, hormone receptor-negative breast cancer. J Natl Cancer Inst. 1998;90:1361–1370. doi: 10.1093/jnci/90.18.1361. [DOI] [PubMed] [Google Scholar]
- 14.Pegram MD, Finn RS, Arzoo K. The effect of HER-2/neu overexpression on chemotherapeutic drug sensitivity in human breast and ovarian cancer cells. Oncogene. 1997;15:537–547. doi: 10.1038/sj.onc.1201222. [DOI] [PubMed] [Google Scholar]
- 15.Jarvinen TA, Tanner M, Rantanen V. Amplification and deletion of topoisomerase IIalpha associate with ErbB-2 amplification and affect sensitivity to topoisomerase II inhibitor doxorubicin in breast cancer. Am J Pathol. 2000;156:839–847. doi: 10.1016/s0002-9440(10)64952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cummings J, Smyth JF. DNA topoisomerase I and II as targets for rational design of new anticancer drugs. Ann Oncol. 1993;4:533–543. doi: 10.1093/oxfordjournals.annonc.a058584. [DOI] [PubMed] [Google Scholar]
- 17.Jarvinen TA, Tanner M, Barlund M. Characterization of topoisomerase II alpha gene amplification and deletion in breast cancer. Genes Chromosomes Cancer. 1999;26:142–150. [PubMed] [Google Scholar]
- 18.Knoop AS, Knudsen H, Balslev E. Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J Clin Oncol. 2005;23:7483–7490. doi: 10.1200/JCO.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 19.O'Malley FP, Chia S, Tu D. Topoisomerase II alpha and responsiveness of breast cancer to adjuvant chemotherapy. J Natl Cancer Inst. 2009;101:644–650. doi: 10.1093/jnci/djp067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Leo A, Desmedt C, Bartlett JM. HER2 and TOP2A as predictive markers for anthracycline-containing chemotherapy regimens as adjuvant treatment of breast cancer: a meta-analysis of individual patient data. Lancet Oncol. 2011;12:1134–1142. doi: 10.1016/S1470-2045(11)70231-5. [DOI] [PubMed] [Google Scholar]
- 21.Bartlett JM, Munro AF, Dunn JA. Predictive markers of anthracycline benefit: a prospectively planned analysis of the UK National Epirubicin Adjuvant Trial (NEAT/BR9601) Lancet Oncol. 2010;11:266–274. doi: 10.1016/S1470-2045(10)70006-1. [DOI] [PubMed] [Google Scholar]
- 22.Bidard FC, Matthieu MC, Chollet P. p53 status and efficacy of primary anthracyclines/alkylating agent-based regimen according to breast cancer molecular classes. Ann Oncol. 2008;19:1261–1265. doi: 10.1093/annonc/mdn039. [DOI] [PubMed] [Google Scholar]
- 23.Fedier A, Steiner RA, Schwarz VA. The effect of loss of Brca1 on the sensitivity to anticancer agents in p53-deficient cells. Int J Oncol. 2003;22:1169–1173. [PubMed] [Google Scholar]
- 24.Marchiò C, Lambros MB, Gugliotta P. Does chromosome 17 centromere copy number predict polysomy in breast cancer? A fluorescence in situ hybridization and microarray-based CGH analysis. J Pathol. 2009;219:16–24. doi: 10.1002/path.2574. [DOI] [PubMed] [Google Scholar]
- 25.Yeh IT, Martin MA, Robetorye RS. Clinical validation of an array CGH test for HER2 status in breast cancer reveals that polysomy 17 is a rare event. Mod Pathol. 2009;22:1169–1175. doi: 10.1038/modpathol.2009.78. [DOI] [PubMed] [Google Scholar]
- 26.Watters AD, Going JJ, Cooke TG. Chromosome 17 aneusomy is associated with poor prognostic factors in invasive breast carcinoma. Breast Cancer Res Treat. 2003;77:109–114. doi: 10.1023/a:1021399923825. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard KI, Munro A, O'Malley FP. Chromosome 17 centromere (CEP17) duplication as a predictor of anthracycline response: evidence from the NCIC Clinical Trials Group (NCIC CTG) MA.5 Trial. Breast Cancer Res Treat. 2012;131:541–551. doi: 10.1007/s10549-011-1840-4. [DOI] [PubMed] [Google Scholar]
- 28.Jaffe CC. Measures of response: RECIST, WHO, and new alternatives. J Clin Oncol. 2006;24:3245–3251. doi: 10.1200/JCO.2006.06.5599. [DOI] [PubMed] [Google Scholar]
- 29.Hammond ME, Hayes DF, Dowsett M. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolff AC, Hammond ME, Schwartz JN. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 31.Goldhirsch A, Winer EP, Coates AS. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hannemann J, Kristel P, van Tinteren H. Molecular subtypes of breast cancer and amplification of topoisomerase II alpha: predictive role in dose intensive adjuvant chemotherapy. Br J Cancer. 2006;95:1334–1341. doi: 10.1038/sj.bjc.6603449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McShane LM, Altman DG, Sauerbrei W. REporting recommendations for tumor MARKer prognostic studies (REMARK) Breast Cancer Res Treat. 2006;100:229–235. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 34.Corzo C, Bellosillo B, Corominas JM. Does polysomy of chromosome 17 have a role in ERBB2 and topoisomerase IIalpha expression? Gene, mRNA and protein expression: a comprehensive analysis. Tumour Biol. 2007;28:221–228. doi: 10.1159/000107583. [DOI] [PubMed] [Google Scholar]
- 35.Zody MC, Garber M, Adams DJ. DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature. 2006;440:1045–1049. doi: 10.1038/nature04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinholz MM, Bruzek AK, Visscher DW. Breast cancer and aneusomy 17: implications for carcinogenesis and therapeutic response. Lancet Oncol. 2009;10:267–277. doi: 10.1016/S1470-2045(09)70063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orsetti B, Nugoli M, Cervera N. Genomic and expression profiling of chromosome 17 in breast cancer reveals complex patterns of alterations and novel candidate genes. Cancer Res. 2004;64:6453–6460. doi: 10.1158/0008-5472.CAN-04-0756. [DOI] [PubMed] [Google Scholar]
- 38.Chin SF, Wang Y, Thorne NP. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26:1959–1970. doi: 10.1038/sj.onc.1209985. [DOI] [PubMed] [Google Scholar]
- 39.Fridlyand J, Snijders AM, Ylstra B. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartlett JM, Munro A, Cameron DA. Type 1 receptor tyrosine kinase profiles identify patients with enhanced benefit from anthracyclines in the BR9601 adjuvant breast cancer chemotherapy trial. J Clin Oncol. 2008;26:5027–5035. doi: 10.1200/JCO.2007.14.6597. [DOI] [PubMed] [Google Scholar]
- 41.Cardoso F, Durbecq V, Larsimont D. Correlation between complete response to anthracycline-based chemotherapy and topoisomerase II-alpha gene amplification and protein overexpression in locally advanced/metastatic breast cancer. Int J Oncol. 2004;24:201–209. [PubMed] [Google Scholar]
- 42.Desmedt C, Di Leo A, de Azambuja E. Multifactorial approach to predicting resistance to anthracyclines. J Clin Oncol. 2011;29:1578–1586. doi: 10.1200/JCO.2010.31.2231. [DOI] [PubMed] [Google Scholar]
- 43.Farmer P, Bonnefoi H, Anderle P. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 44.Ejlertsen B, Jensen MB, Nielsen KV. HER2, TOP2A, and TIMP-1 and responsiveness to adjuvant anthracycline-containing chemotherapy in high-risk breast cancer patients. J Clin Oncol. 2010;28:984–990. doi: 10.1200/JCO.2009.24.1166. [DOI] [PubMed] [Google Scholar]
- 45.West NR, Milne K, Truong PT. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011;13:R126. doi: 10.1186/bcr3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between Clinicopathologic Characteristics and pCR


