Abstract
The challenging task of studying and modeling complex dynamics of biological systems in order to describe various human diseases has gathered great interest in recent years. Major biological processes are mediated through protein interactions, hence there is a need to understand the chaotic network that forms these processes in pursuance of understanding human diseases. The applications of protein interaction networks to disease datasets allow the identification of genes and proteins associated with diseases, the study of network properties, identification of subnetworks, and network-based disease gene classification. Although various protein interaction network analysis strategies have been employed, grand challenges are still existing. Global understanding of protein interaction networks via integration of high-throughput functional genomics data from different levels will allow researchers to examine the disease pathways and identify strategies to control them. As a result, it seems likely that more personalized, more accurate and more rapid disease gene diagnostic techniques will be devised in the future, as well as novel strategies that are more personalized. This mini-review summarizes the current practice of protein interaction networks in medical research as well as challenges to be overcome.
Keywords: Interactome, Network, Disease genes, Gene expression, Human diseases
1. Introduction
We have come a long way from “one-gene/one-enzyme/one-function” concept originally framed by Beadle and Tatum [1]. They provided a basic explanation of how genes work at the molecular level, however, we now know that the picture is more complex. Biological processes inside our body are governed by the well-defined organization of proteins into complexes, which perform different functions acting as molecular machines [2]. Major biological processes such as immunity (antigen–antibody interaction), metabolism (enzyme–substrate interaction), signaling (interaction of messenger molecules, hormones, neurotransmitters with their cognate receptors), and gene expression (DNA–protein interactions), as well as the building of supramolecular assemblies (collagens, elastic fibers, actin filaments) and molecular machines (molecular motors, ribosomes, proteasome) were mediated through protein interactions. Studying the interactome, which is the whole set of molecular physical interactions between biological entities in cells and organisms, is essential in understanding how gene functions and regulations are integrated at the level of an organism [3].
The notion that, a disease is rarely a consequence of an abnormality on a single gene, but it is usually the result of complex interactions and perturbations involving large sets of genes and their relationships with several cellular components, has led to the development of the network based approaches to understand human disease [4]. Theoretical advances in network science and paralleling advances in high-throughput efforts to map biological networks have provided a conceptual framework with which we interpret large interactome network maps. Protein–protein interaction (PPI) networks are increasingly serving as tools to decipher the molecular basis of diseases. Furthermore, the sequencing of the genome and advances in proteomics leads to the identification of proteins of unknown functions. Interaction networks might give clues on the functions of these newly discovered proteins or on new functions of already identified proteins [5–9].
The promising applications of PPI networks to disease datasets are concentrated on four major areas: (i) the identification of genes and proteins associated with diseases, (ii) the study of network properties and their relation to disease states, (iii) the identification of disease-related subnetworks, and (iv) network-based disease classification [10]. Fig. 1 gives an example for the schematic representation of understanding disease–PPI network relationship using systems biomedicine approach.
Fig. 1.
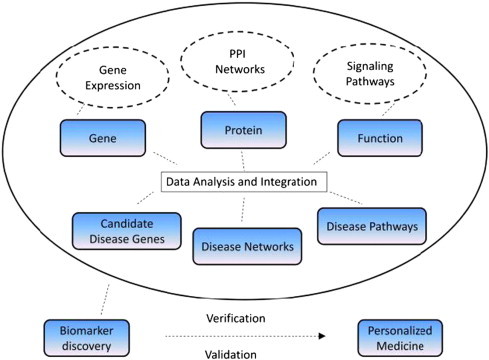
A systems biomedicine based approach to understanding PPI network–disease relationship.
Global understanding of networks will allow researchers to examine the disease pathways and identify strategies to control them. The integration of functional genomic and proteomic data to obtain dynamic network analysis will further improve the success of medical research.
2. The Role of Networks in Medicine
Networks provide a systems-level understanding of the mechanisms underlying diseases by serving as a model for data integration and analysis. They have been used to gain insight into disease mechanisms [11,12], study comorbidities [13,14], analyze therapeutic drugs and their targets [15–17], and discover novel network-based biomarkers [18,19].
Network science deals with complexity by “simplifying” complex systems to components (nodes) and interactions (edges) between them (Fig. 2). These simplifications help researchers make useful discoveries. Networks can be constructed purely based on gene expression information, including transcriptional regulatory networks [20] and co-expression networks [21], or can also be built upon prior knowledge of protein–protein interactions [22]. The nodes in a network representation are metabolites or macromolecules such as proteins, RNA molecules and gene sequences, while the edges are physical, biochemical or functional interactions. The resulting “interactome” network can serve as scaffold information to extract global or local graph theory properties, which lead to a better understanding of biological processes. Since cellular networks consist of various types of interaction and regulation, networks reflecting this complex scenario will provide better insight into the problem in hand.
Fig. 2.
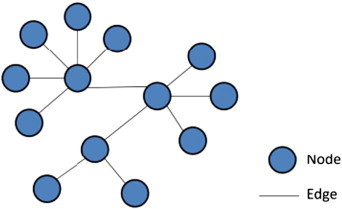
Simple representation of a network, nodes representing components and edges representing interactions.
Regulatory interaction networks, metabolic networks, signaling networks, and protein–protein interaction networks cannot be considered in isolation or as independent entities. Rather, we have to incorporate their intricate interwoven structure. Proteins might act alone or in combination: as transcription factors and regulators of protein abundances, as enzymes they catalyze and coordinate the basic cellular metabolic processes, and they react to external and internal stimuli activating other proteins in signaling cascades. All of these processes in turn provide cues that may lead to the formation or termination of protein interactions and complexes. PPI networks in particular have become a valuable resource in this context [23,24]. An example for a PPI network can be seen in Fig. 3 for psoriasis disease.
Fig. 3.
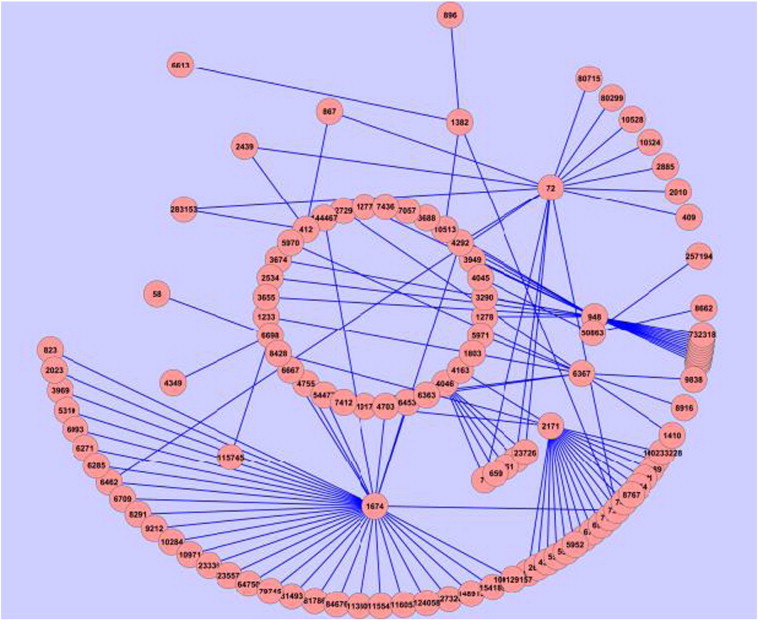
A simplified PPI network of psoriasis disease visualized using Cytoscape (proteins are represented by Entrez ID's).
The current estimates suggest that the human interactome comprises approximately 130,000–650,000 protein interactions [25,26]; however, only a subset of these has been experimentally identified [24].
There are currently two widely used high throughput methodologies for large scale mapping of PPI. The yeast two-hybrid (Y2H) system [27] maps binary interactions. Mapping of direct interactions of protein complexes is carried out by affinity or immune-purification, followed by mass spectrometry (AP/MS) to identify protein constituents of these complexes as well as other protein–protein interactions [28]. It should be noted that these high-throughput methods are prone to a high rate of false-positives and false-negatives, i.e. protein interactions which are identified by the experiment do not take place in the cell or interacting protein pairs cannot be identified by current experiment technology [29]. Sprinzak et al. [30] have shown that the false positive (FP) rate of high-throughput yeast two-hybrid assays is ∼ 50%. The rate of false negative (FN) is also very high [31]. Another downside of these technologies is that they are expensive and time consuming [32].
The reconstruction of interaction networks has been performed via three distinct approaches: (1) the manual curation of already existing data available in literature, usually obtained from one or just a few types of physical or biochemical interactions [33], (2) computational predictions based on available “orthogonal” information apart from physical or biochemical interactions, such as sequence similarities, gene-order conservation, co-presence and co-absence of genes in completely sequenced genomes and protein structural information [34], and (3) systematic and high throughput whole genome or proteome mapping strategies [35].
Studies employing these reconstructions ended up with more advances in network biology such as, studies of global relationships between human disorders, associated genes and interactome networks [36,37], predictions of new human disease-associated genes [38], analyses of network perturbations by pathogens [39], and emergence of node removal versus edge specific or “edgetic” models [40] to explain genotype–phenotype relationships [41]. The global topological analysis of PPI networks is increasingly being replaced by more detailed analyses of binary interactions, their determinants, characteristics, and effects [42,43]. In the first stage such analyses will inform us about the functional role of particular interactions, and later they will hopefully enable us to predict PPI and their roles in silico.
PPI networks also include small interwoven modules inside them. These modules contain functional information on complex biological networks and interaction between these proteins comprises information related to biological processes of the interactants. Using networking approaches to study biological problems can provide an intuitive picture or useful insights to help analyze complicated relations in these systems [2]. Because PPI networks are large and complex, it becomes necessary to develop efficient and biologically meaningful algorithms for their modular analysis. Graphlets (small induced subgraphs of large networks) can be applied to analyze the modular characteristics of PPI Networks [44–46].
We also have to keep in mind that molecular networks exhibit dynamic responses to both internal states and external signals. Ultimately, health or disease states emerge from an individual's integration of these internal and external signals [47]. PPI networks are also dynamic. They change over seconds as cellular processes require protein complexes to assemble and disassemble, and also evolve over millions of years as interactions and genes are gained and lost. It is thus crucial to understand how these dynamic and evolutionary changes operate, in order to grasp how cellular machineries function and have been shaped during evolution. In recent years such properties of PPI's and networks have been amenable to study, thanks to the ever-increasing body of data on interactions, atomic structures and mRNA expression levels [48].
3. Strategies to Identify Disease Genes
High-throughput gene expression profiling technologies have allowed researchers to characterize molecular differences between healthy and disease states which has led to the identification of an increasing amount of disease related genes. PPI network based approaches have proved to be useful in enlightening relevant disease related biological processes, and identifying candidate disease genes. Using a detailed interaction map of a given disease can help in elucidating its mechanism and can suggest potential points for biomarkers or drug targets [49].
The analysis of disease networks has also permitted the discovery of novel classes of therapeutic targets not amenable to classical drug-like compounds but reachable through novel bioactive compounds [50].
Disease genes tend to be highly expressed and have tissue-specific expression patterns and a higher mutation rate over evolutionary time [51]. With this information at hand different studies have been performed on omics platforms at various levels to identify, predict or prioritize disease genes.
Wu et al. [52] have used gene classification strategies to detect whether a gene is disease-associated or not. They have explored gene classification with topological features by using k-shell decomposition and the average distance to the center (essential genes) to analyze the hierarchy structure of their PPI network. Assuming essential genes as the center of the network, the disease genes appeared closer to the center than other genes. Their findings concluded that compared with other genes, both disease genes and non-essential disease genes are topologically more important. Nguyen and Ho [53] enriched the disease gene prediction by incorporating known disease genes with neighbors of disease genes and integrating multiple data sources in their semi-supervised Learning scheme. It is also feasible to use gene networks to prioritize positional candidate genes in various heritable disorders with multiple associated genes. Franke et al. [54] proposed methods to integrate gene networks with various genetic studies which can be useful in identifying disease genes. Köhler et al. [55] have investigated the hypothesis that global network-similarity measures are better suited to capture relationships between disease proteins than are algorithms based on direct interactions or shortest paths between disease genes. They have presented a novel method for candidate-gene prioritization based on the random walk method, which they used to calculate a score reflecting the global similarity of candidate genes to known members of a disease-gene family. Schlicker et al. [56] introduced a novel approach for ranking candidate genes for a particular disease based on functional comparisons involving the Gene Ontology. Lee et al. [57] analyzed a large-scale, human gene functional interaction network (dubbed HumanNet). They showed that candidate disease genes can be effectively identified by GBA (guilt by association) in cross-validated tests using label propagation algorithms related to Google's PageRank. They resolved the issue of GBA working poorly in genome-wide association studies by explicitly modeling the uncertainty of the associations and incorporating the uncertainty for the seed set into the GBA framework. Re and Valentini [58] proposed a novel semi-supervised method to rank genes with respect to cancer modules using networks constructed from different sources of functional information, not limited to gene expression data. Aerts et al. [59] described a bioinformatics approach which generates distinct prioritizations for multiple heterogeneous data sources, which are then integrated, or fused, into a global ranking using order statistics. Navlakha and Kingsford [60] have found that random-walk approaches individually outperform clustering and neighborhood approaches by examining the performance of seven recently developed computational methods.
4. Challenges and Limitations of PPI Networks
There are still challenges regarding PPI network based approaches stemming from the huge body of information that seems to be getting bigger by the day.
Providing maps of PPI networks using systematic and standardized approaches and assays that are as unbiased as possible is a grand challenge of network biology.
There is an impressive amount of data on sequence alterations and biomolecular profiles (mRNA expression, miRNA and noncoding RNA profiling, proteomics, and metabolomic measurements) for many human diseases, which can be accessed from specialized databases and publications. However, we still have not succeeded in translating this wealth of information into actionable knowledge about disease pathogenesis for the development of better strategies for disease prevention, diagnosis, and treatment. Progress is limited by the difficulties in assessing the functional consequences of disease-associated sequence variants and understanding how phenotype is affected by the combined effect of environmental and genomic variation [24].
Though high-throughput gene expression profiling has permitted the characterization of molecular differences between healthy and disease states, a clear limitation of these approaches is that they often deal with data about single players (i.e. changes in the expression of individual genes). Novel strategies should integrate systemic information to contextualize the differential expression patterns observed [51].
Another limitation of network biology is the coverage and quality of interactome data. The data incompleteness of the human PPI network poses limitations to any study of network properties of disease genes. Also, the availability of condition-specific interactomes that are more representative of the interactions of the proteins in a given tissue or under certain conditions will improve the significance of such analysis. Differential network mapping should offer more insight into the network rewiring that occurs during disease. This approach will enable monitoring changes in the role of the individual nodes (hubs, bottlenecks, etc.) and changes in the global topology of the PPI network, as well as how all these alterations correlate with cell function. By fully characterizing network rewiring in disease, a deeper understanding of how sequence variation shapes cellular networks and leads to observed phenotypic changes would be gained [24].
Although the amount of protein interactome data obtained by high-throughput protein interaction techniques is increasing rapidly, they contain a significant proportion of false positives and false negatives, which exhibits the need to prioritize the interactome reported in such assays for further validation. Computational analysis techniques for assessing and ranking the reliability of binary PPI are highly desirable [61]. Though many methods have emerged for ranking the reliability of protein interactions reported by high-throughput assays, they are mostly successful in detecting false positives. However, these assays are also known to produce a large number of false negatives. There is an immediate need for methods that detect false negatives. The identification of false negatives is equivalent to the problem of predicting new protein interactions [62].
Another challenge in integrating physical and genetic maps is to reconcile the variety of interaction types (i.e. genetic and physical interactions) that are currently available [63]. Complex diseases stem from interplay of genetic and environmental factors, which complicate the study of human diseases, since it is difficult to create a controlled environment that enables scientists to study environmental effects on disease development [64].
Researchers are increasingly using model organisms such as fruit flies, mice, and zebrafish to examine human diseases because they are easy to grow, dissect, and genetically manipulate in the laboratory. The issue of the transfer of phenotypic information from animal models to human is also a challenge [65,66].
5. Concluding Remarks and Future Work
We are still far from unraveling the molecular mechanisms of most diseases, thus developing effective methods to uncover disease genes remains a great challenge.
First and foremost, the construction of a more reliable PPI network for human genes is a must. This might be achieved by assembling a large PPI dataset with the inclusion of all or most of the major and minor PPI databases and ranking the interactions according to a well established scoring system. This also means less false positives and false negatives.
In conjunction with the use of PPI networks, studies evaluating the correlation between human disease genes for a specific disease and cross-comparison of those disease genes to other human diseases are needed for a deeper understanding of the relationship of disease genes within a disease and between diseases.
Understanding specific molecular pathways unique to a specific disease and elucidation of differences in molecular pathways for different diseases will help us recognize the similarities and differences in those pathways which will lead to a better understanding of the relationship between human diseases.
Constructing a ‘linkage network’ between diseases that are linked to each other by one or more than one gene is also another great way to associate human diseases by a simplified network.
It is also important to identify the ‘triggers’, key disease genes that initiate a specific disease, and constructing a ‘triggers network’ to further the unveiling of the disease mechanisms. This will aid us in the optimization of primary prevention of preventable human diseases and also lead to the discovery of biomarker or biomarker groups for early diagnosis and intervention of human diseases. Studies to evaluate individualized therapy that could lead to personalized medicine for patients is also needed.
The impact of outside effects to the progress of human diseases and the risk factors for the initiation and advancement of human diseases also need to be analyzed for a better evaluation of each disease.
The never ending information flow will revolutionize how we look at human diseases and disease networks for many years to come. The prospects of predictive and personalized medicine are enormous. The future holds the cures and the medicine.
Acknowledgments
The financial support by the Marmara University Research Fund through project FEN-B-090414-0089 is greatly acknowledged.
References
- 1.Beadle G.W., Tatum E.L. Genetic control of biochemical reactions, in Neurospora. Proc Natl Acad Sci U S A. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma A., Costantini S., Colonn G. The protein–protein interaction network of the human Sirtuin family. Biochim Biophys Acta. 2013;1834:1998–2009. doi: 10.1016/j.bbapap.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Albert R., Barabasi A.L. Statistical mechanics of complex networks. Rev Mod Phys. 2002;74:47–97. [Google Scholar]
- 4.Valentini G., Paccanaro A., Caniza H., Romero A.E., Matteo R. An extensive analysis of disease-gene associations using network integration and fast kernel-based gene prioritization methods. Artif Intell Med. 2014 doi: 10.1016/j.artmed.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arga K.Y., Önsan Z.I., Kırdar B., Ülgen K.Ö., Nielsen J. Understanding signaling in yeast: insights from network analysis. Biotechnol Bioeng. 2007;97(5):1246–1258. doi: 10.1002/bit.21317. [DOI] [PubMed] [Google Scholar]
- 6.Sharan R., Ulitsky I., Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Re M., Mesiti M., Valentini G. A fast ranking algorithm for predicting gene functions in biomolecular networks. IEEE ACM Trans Comput Biol Bioinf. 2012;9(6):1812–1818. doi: 10.1109/TCBB.2012.114. [DOI] [PubMed] [Google Scholar]
- 8.Mostafavi S., Ray D., Warde-Farley D., Grouios C., Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(S4) doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mesiti M., Re M., Valentini G. Think globally and solve locally: secondary memory-based network learning for automated multi-species function prediction. GigaScience. 2014;3:5. doi: 10.1186/2047-217X-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chautard E., Thierry-Mieg N., Ricard-Blum S. Interaction networks: from protein functions to drug discovery: a review. Pathol Biol. 2009;57:324–333. doi: 10.1016/j.patbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Goh K., Cusick M.E., Valle D., Childs B., Vidal M., Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X., Deignan J.L., Qi H., Zhu J., Qian S., Torosyan G. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee D.S., Park J., Kay K.A., Christakis N.A., Oltvai Z.N. The implications of human metabolic network topology for disease comorbidity. Proc Natl Acad Sci U S A. 2008;105:9880–9885. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park J., Lee D.S., Christakis N.A., Barabási A.L. The impact of cellular networks on disease comorbidity. Mol Syst Biol. 2009;5:262. doi: 10.1038/msb.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yildirim M.A., Goh K.I., Cusick M.E., Barabási A.L., Vidal M. Drug–target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 16.Winter C., Kristiansen G., Kersting S., Roy J., Aust D., Knösel T. Google goes cancer: improving outcome prediction for cancer patients by network-based ranking of marker genes. PLoS Comput Biol. 2012;8(5) doi: 10.1371/journal.pcbi.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottlieb A., Stein G.Y., Ruppin E., Sharan R. PREDICT, a method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol. 2011;7:496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor I.W., Linding R., Warde-Farley D., Liu Y., Pesquita C., Faria D. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 19.Re M., Valentini G. Network-based drug ranking and repositioning with respect to DrugBank therapeutic categories. IEEE ACM Trans Comput Biol Bioinf. 2013;10(6):1359–1371. doi: 10.1109/TCBB.2013.62. [DOI] [PubMed] [Google Scholar]
- 20.Chen X., Chen M., Ning K. BNArray: an R package for constructing gene regulatory networks from microarray data by using Bayesian network. Bioinformatics. 2006;22:2952–2954. doi: 10.1093/bioinformatics/btl491. [DOI] [PubMed] [Google Scholar]
- 21.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin A., Ochagavia M.E., Rabasa L.C., Miranda J., Fernandez-de Cossio J., Bringas R. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11:91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly W.P., Stumpf M.P.H. Protein–protein interactions: from global to local analyses. Curr Opin Biotechnol. 2008;19:396–403. doi: 10.1016/j.copbio.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Furlong L. Human diseases through the lens of network biology. Trends Genet. 2013;29:3. doi: 10.1016/j.tig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Stumpf M.P.H., Thorne T., de Silva E., Stewart R., An H.J., Lappe M. Estimating the size of the human interactome. Proc Natl Acad Sci U S A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkatesan K., Rual J.-F., Vazquez A., Stelzl U., Lemmens I., Hirozane-Kishikawa T. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fields S., Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 28.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;10:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 29.You Z.-H., Lei Y.-K., Gui J., Huang D.-S., Zhou X. Using manifold embedding for assessing and predicting protein interactions from high-throughput experimental data. Bioinformatics. 2010;26(21):2744–2751. doi: 10.1093/bioinformatics/btq510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sprinzak E., Sattath S., Margalit H. How reliable are experimental protein–protein interaction data? J Mol Biol. 2003;327:919–923. doi: 10.1016/s0022-2836(03)00239-0. [DOI] [PubMed] [Google Scholar]
- 31.Chua H.N., Sung W.-K., Wong L. Exploiting indirect neighbours and topological weight to predict protein function from protein–protein interactions. Bioinformatics. 2006;22:1623–1630. doi: 10.1093/bioinformatics/btl145. [DOI] [PubMed] [Google Scholar]
- 32.Deane C.M., Salwiński Ł., Xenarios I., Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics. 2002;1(5):349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 33.Roberts P.M. Mining literature for systems biology. Brief Bioinform. 2006;7:399–406. doi: 10.1093/bib/bbl037. [DOI] [PubMed] [Google Scholar]
- 34.Marcotte E.M., Date S.V. Exploiting big biology: integrating large-scale biological data for functional inference. Brief Bioinform. 2001;2(4):363–374. doi: 10.1093/bib/2.4.363. [DOI] [PubMed] [Google Scholar]
- 35.Walhout A.J., Vidal M. Protein interaction maps for model organisms. Nat Rev Mol Cell Biol. 2001;2:55–62. doi: 10.1038/35048107. [DOI] [PubMed] [Google Scholar]
- 36.Rende D., Baysal N., Kirdar B. A novel integrative network approach to understand the interplay between cardiovascular disease and other complex disorders. Mol Biosyst. 2011;7:2205–2219. doi: 10.1039/c1mb05064h. [DOI] [PubMed] [Google Scholar]
- 37.Rende D., Baysal N., Kirdar B. Complex disease interventions from a network model for type 2 diabetes. PLoS ONE. 2013 doi: 10.1371/journal.pone.0065854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu C., Wu C., Aronow B.J., Jegga A.G. Computational approaches for human disease gene prediction and ranking. Adv Exp Med Biol. 2014;799:69–84. doi: 10.1007/978-1-4614-8778-4_4. [DOI] [PubMed] [Google Scholar]
- 39.Gulbahce N., Yan H., Dricot A., Padi M., Byrdsong D., Franchi R. Viral perturbations of host networks reflect disease etiology. PLoS Comput Biol. 2012;8:6. doi: 10.1371/journal.pcbi.1002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong Q., Simonis N., Li Q.R., Charloteaux B., Heuze F., Klitgord N. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidal M., Cusick M.E., Barabasi A.-L. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dreze M., Monachello D., Lurin C., Cusick M.E., Hill D.E., Vidal M. High-quality binary interactome mapping. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- 43.Yu H., Braun P., Yıldırım M.A., Lemmens I., Venkatesan K., Sahalie J. High–Quality Binary Protein Interaction Map of the Yeast Interactome Network. Science. 2008;322(5898):104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Przulj N. Biological network comparison using graphlet degree distribution. Bioinformatics. 2007;23:177–183. doi: 10.1093/bioinformatics/btl301. [DOI] [PubMed] [Google Scholar]
- 45.Przulj N., Corneil D.G., Jurisica I. Modeling interactome: scale-free or geometric? Bioinformatics. 2004;20:3508–3515. doi: 10.1093/bioinformatics/bth436. [DOI] [PubMed] [Google Scholar]
- 46.Hayes W., Sun K., Przulj N. Graphlet-based measures are suitable for biological network comparison. Bioinformatics. 2013;29(4):483–491. doi: 10.1093/bioinformatics/bts729. [DOI] [PubMed] [Google Scholar]
- 47.Pe'er D., Hacohen N. Principles and strategies for developing network models in cancer. Cell. 2011;144:864–873. doi: 10.1016/j.cell.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levy E.D., Pereira-Leal J.B. Evolution and dynamics of protein interactions and Networks. Curr Opin Struct Biol. 2008;18:349–357. doi: 10.1016/j.sbi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Tekir S.D., Arga K.Y., Ülgen K.Ö. Drug targets for tumorigenesis: insights from structural analysis of EGFR signaling network. J Biomed Inform. 2009;42:228–236. doi: 10.1016/j.jbi.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Zanzoni A., Soler-López M., Aloy P. A network medicine approach to human disease. FEBS Lett. 2009;583:1759–1765. doi: 10.1016/j.febslet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Oti M., Brunner H.G. The modular nature of genetic diseases. Clin Genet. 2007;71:1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- 52.Wu S-Y, Shaoa F-J, Sun R-C, Sui Y., Wang Y., Wang J-L. Analysis of human genes with protein–protein interaction network for detecting disease genes. Physica A. 2014;398:217–228. [Google Scholar]
- 53.Nguyen T.P., Ho T.B. Detecting disease genes based on semi-supervised learning and protein–protein interaction networks. Artif Intell Med. 2012;54:63–71. doi: 10.1016/j.artmed.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 54.Franke L., van Bakel H., Fokkens L., de Jong E.D., Egmont-Petersen M., Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet. 2006;78 doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Köhler S., Bauer S., Horn D., Robinson P.N. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlicker A., Lengauer T., Albrecht M. Improving disease gene prioritization using the semantic similarity of gene ontology terms. Bioinformatics. 2010;26(18):561–567. doi: 10.1093/bioinformatics/btq384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee I., Blom U., Wang P., Shim J., Marcotte E. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Re M., Valentini G. Cancer module genes ranking using kernelized score functions. BMC Bioinformatics. 2012;13(S14):S3. doi: 10.1186/1471-2105-13-S14-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aerts S., Lambrechts D., Maity S., Van Loo P., Coessens B., De Smet F. Gene prioritization through genomic data fusion. Nat Biotechnol. 2006;24(5):537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 60.Navlakha S., Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics. 2010;26(8):1057–1063. doi: 10.1093/bioinformatics/btq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karagöz K., Arga K.Y. Assessment of high-confidence protein–protein interactome in yeast. J Comput Biol Chem. 2013;45:1–8. doi: 10.1016/j.compbiolchem.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Chua H.N., Wong L. Increasing the reliability of protein interactomes. Drug Discov Today. 2008;13:15–16. doi: 10.1016/j.drudis.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 63.Beyer A., Bandyopadhyay S., Ideker T. Integrating physical and genetic maps: from genomes to interaction networks. Nat. Rev. Genet. 2007;8:699–710. doi: 10.1038/nrg2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nabhan A.R., Sarkar I.N. Structural network analysis of biological networks for assessment of potential disease model organisms. J Biomed Inform. 2014;47:178–191. doi: 10.1016/j.jbi.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 65.Washington N.L., Haendel M.A., Mungall C.J., Ashburner M., Westerfield M., Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotations. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smedley D., Oellrich A., Kohler S., Ruef Barbara, Sanger Mouse Genetics Project, Westerfield M. PhenoDigm: analyzing curated annotations to associate animal models with human diseases. Database. 2013;2013 doi: 10.1093/database/bat025. [Article ID bat025] [DOI] [PMC free article] [PubMed] [Google Scholar]


