Abstract
Human chorionic mesenchymal stem/stromal cells (CMSCs) derived from the placenta are similar to adult tissue-derived MSCs. The aim of this study was to investigate the role of these cells in normal placental development. Transcription factors, particularly members of the homeobox gene family, play crucial roles in maintaining stem cell proliferation and lineage specification in embryonic tissues. In adult tissues and organs, stem cells proliferate at low levels in their niche until they receive cues from the microenvironment to differentiate. The homeobox genes that are expressed in the CMSC niche in placental tissues have not been identified. We used the novel strategy of laser capture microdissection to isolate the stromal component of first trimester villi and excluded the cytotrophoblast and syncytiotrophoblast layers that comprise the outer layer of the chorionic villi. Microarray analysis was then used to screen for homeobox genes in the microdissected tissue. Candidate homeobox genes were selected for further RNA analysis. Immunohistochemistry of candidate genes in first trimester placental villous stromal tissue revealed homeobox genes Meis1, myeloid ectropic viral integration site 1 homolog 2 (MEIS2), H2.0-like Drosophila (HLX), transforming growth factor β-induced factor (TGIF), and distal-less homeobox 5 (DLX5) were expressed in the vascular niche where CMSCs have been shown to reside. Expression of MEIS2, HLX, TGIF, and DLX5 was also detected in scattered stromal cells. Real-time polymerase chain reaction and immunocytochemistry verified expression of MEIS2, HLX, TGIF, and DLX5 homeobox genes in first trimester and term CMSCs. These data suggest a combination of regulatory homeobox genes is expressed in CMSCs from early placental development to term, which may be required for stem cell proliferation and differentiation.
Keywords: placenta, mesenchymal, stem cells, homeobox, chorion
Introduction
The human placenta has emerged as a valuable source of transplantable cells of mesenchymal origin for multiple cytotherapeutic purposes, including enhanced engraftment of hematopoietic cells, modulation of inflammation, bone repair, and cancer.1–3 Human chorionic mesenchymal stem/stromal cells (CMSCs, also known as placental mesenchymal stem cells) are isolated from the chorion of the postpartum human placenta and display stem cell-like properties such as high proliferative capacity, self-renewal, multipotent differentiation, and migration potential.4–8 In culture, CMSCs can be readily induced to commit to mesodermal lineage pathways including adipogenesis, chondrogenesis, osteogenesis, and myogenesis.6–10 Although the ability of CMSCs to differentiate into multiple lineages is important in regenerative medicine, with the exception of myogenesis and angiogenesis, the other lineages are not relevant to normal placental development. Our knowledge of how CMSC differentiation is controlled in the human placenta is rudimentary.
Transcription factors play crucial roles in the development of the placenta, in particular the regulation of cell differentiation and proliferation in specific placental cell types.11–16 Microarray analysis of human iliac mesenchymal stem cells (MSCs) revealed a large number of transcription factors are expressed in MSCs that are potentially important in MSC proliferation and differentiation.17 Since CMSCs have been identified relatively recently, there is scant knowledge of transcription factor control of CMSC proliferation and differentiation.
Homeobox genes comprise a large family of transcription factors, and several are key regulators in stem cell proliferation and differentiation, which is consistent with their role as master regulators of cell fate.15 Homeobox genes nanog, Oct3/4, Pax3/4/6, Nkx2.5, PDX1, LHX2, and HOXB4 are critical for maintaining stem cell proliferation and regulation of differentiation.15,18,19 The formation of trophoblast cells, the key placental cell lineage, involves downregulation of the homeobox genes Oct3/4 (POU5F1) and nanog in stem cells, which indicates that these genes are required for stem cell maintenance. Conversely, upregulation of the homeobox gene CDX2 is required for trophoblast differentiation.20–22
MSCs from various organs of the mouse (eg, lung, thymus, sternum, forelimb, femur, and tibia) express a distinct combination of homeobox genes.23 In humans, genomic profiling studies on cultured MSCs from various sources (ie, bone marrow, adipose tissue, and umbilical cord) reveal that the homeobox genes HOXA5 and HOXB6 are highly expressed and that ZEB is increased in differentiating MSCs.24 Similar studies to identify the homeobox expression profile of placental CMSCs have not yet been carried out.
We used MSC surface markers to provide evidence for a vascular microenvironment or “niche” for CMSCs.7 The vascular niche, as defined by Nikolova et al,25 is a microenvironment that is generated by endothelial cells and/or mural cells (pericytes or smooth muscle cells), which affects the behavior of adjacent cells. According to this definition of the vascular niche, an influence of nonvascular cells in the niche is not excluded. We also demonstrated that stem cell surface marker expression in the vascular niche does not change significantly from first trimester to term as would be expected of stem cells that reside in a niche. We also provided evidence of a vascular niche for MSCs in the decidua parietalis associated with the fetal membranes.26 Normally, the niche maintains MSCs in a quiescent state but when triggered by developmental or environmental stimuli, the surrounding microenvironment signals to MSCs to either promote self-renewal or differentiate to form new tissue.27,28 Thus, identification of homeobox genes expressed in the stem cell niche through normal placental development should reveal candidate homeobox genes required for stem cell proliferation and/or differentiation, which is the aim of this study.
As described earlier, screens for homeobox genes in cultured MSCs from various tissue sources have been carried out. Nevertheless, the process of in vitro culturing, which includes additives such as serum, changes global gene expression patterns in human bone MSCs.29 Variation in the expression of some “stemness” markers is also observed with passaging of CMSCs.30 Thus, in vitro CMSC gene expression patterns do not necessarily reflect the in vivo patterns. Our novel strategy was to use laser capture microdissection to isolate the stromal component of first trimester villi, which includes the CMSC niche and then identify expressed homeobox genes by genome-wide microarray analysis. The homeobox genes identified were subsequently screened for expression in first trimester placental tissue, as well as in unpassaged and early passage CMSCs, which were derived from early pregnancy tissue (early CMSCs) and term CMSCs. This allowed us to identify candidate homeobox genes, which are potentially important for proliferation and differentiation of CMSCs throughout placental development.
Materials and Methods
Tissue Collection
First trimester placental tissue for laser capture microdissection was obtained by vacuum suction, following termination of uncomplicated pregnancies for psychosocial reasons (7 to 9 weeks of gestation). The study was approved by the ethics committee of the Medical University of Vienna (Vienna, Austria). For immunohistochemical and immunofluorescence studies, first trimester placentae (8-12 weeks of gestation) were obtained by author UM following elective termination of pregnancy and with ethics approval (Southern Health Human Research Ethics Committee) and written informed patient consent. For preparation of early CMSCs, early pregnancy placental chorionic villous samples (12-14 weeks of gestation) were provided by M. Pertile of the Victorian Clinical Genetics Services and were obtained with informed patient consent and ethics approval from The Royal Women’s Hospital Human Research and Ethics Committee and The Royal Children’s Hospital Melbourne Research and Ethics Committee.
For the preparation of term CMSCs, placentae were obtained from uncomplicated, term pregnancies by vaginal delivery or cesarean section, with informed patient consent and with the approval of The Royal Women’s Hospital Human Research and Ethics Committee.
Laser Capture Microdissection, RNA Processing, and Microarray Analysis
First trimester placentae (n = 3; 7-8 weeks, 8 weeks and 9 weeks of gestation) were fixed overnight in a 30% sucrose solution and embedded in Optimum cutting temperature (O.C.T.) compound (Tissue Tek, Australia). Approximately 12 hours elapsed between tissue collection and final embedding. Serial cryosections (7 µm) were prepared and mounted on polyethylene napthalate-membrane slides (P.A.L.M. Microlaser Technologies AG, Bernried, Germany; Carl Zeiss MicroImaging GmbH, Göttingen, Germany). Subsequently, slides were quickly stained with hematoxylin and total villous stroma was dissected from chorionic villi in each slide, using a laser capture microdissection unit as described by the manufacturer (P.A.L.M. Microlaser Technologies AG; Carl Zeiss MicroImaging GmbH, Göttingen, Germany). The dissected material of 20 cryosections of each placental sample was pooled together before RNA isolation. RNA extraction of microdissected tissue yielded approximately 150 ng of total RNA after isolation with peqGOLD TriFast reagent (Peqlab, Erlangen, Germany). RNA integrity was checked using the Agilent RNA 6000 Pico Kit and the Agilent 2100 Bioanalyzer (Agilent; Palo Alto, California). The quality criterion for the isolated RNA was the presence of both 28S and 18S ribosomal RNA (rRNA) peaks. The range of the RNA ratio 28S/18S was from 0.3 to 0.5. RNA was amplified to 8 to 9 µg per sample and biotin-labeled using a Two-Cycle Target Labeling kit as described by the supplier (Affymetrix, Santa Clara, California). Labeled RNA of 5 μg was used to prepare complementary DNA (cDNA) and this was subsequently used for GeneChip analysis. Hybridization to human U133A, B GeneChips (Affymetrix, California) and scanning of the arrays were carried out according to the manufacturer’s protocols (https://www.affymetrix.com) as recently described by Bilban et al.31
Immunohistochemistry
Placental tissue was dissected from random areas of central cotyledons, placed into O.C.T. freezing medium, snap frozen in liquid nitrogen, and then stored at −80°C. Sections of 5-μm thickness were cut and placed on SuperFrost coated slides (Menzel-Glaser, Germany). Tissue sections were treated with a blocking reagent (Histostain-Plus Broad Spectrum kit, Zymed Laboratories, California) and subsequently washed in phosphate-buffered saline (PBS). The sections were incubated overnight at 4°C with either one of the following primary mouse monoclonal or rabbit polyclonal antibodies at 1 μg/mL concentration: Meis1, myeloid ectropic viral integration site 1 homolog 2 (MEIS2), H2.0-like Drosophila (HLX), Msh homeobox 2 (MSX2), hematopoietically expressed homeobox (HHEX), transforming growth factor β-induced factor (TGIF), HOXB7, distal-less homeobox 5 (DLX5), or distal-less homeobox 4 (DLX4). Negative controls included omission of primary antibodies and incubation with the appropriate mouse or rabbit immune preimmune serum control. Sections were incubated with the appropriate biotinylated secondary antibody for 15 minutes followed by the addition of streptavidin horseradish peroxidase conjugate for 15 minutes at room temperature. Detection was by 3-amino-9-ethylcarbazole (AEC; Zymed, California). Morphological assessment was carried out on sections counterstained with 0.5% methyl green. All slides were mounted in 80% glycerol, visualized using a Zeiss Axioscop microscope and analyzed with Axiovision software (Carl Zeiss Inc, Germany).
Multilabel Immunofluorescence
Frozen first trimester placental blocks were cut to a 5-µm thickness and placed onto Superfrost Plus slides (Menzel-Glaser, Germany). Mouse antihuman frizzled family receptor 9 (FZD9/CD349) (1µg/mL, Biolegend, California) was added together with the following rabbit antihuman primary antibodies: HLX (3.2 µg/mL, Sigma-Aldrich, Australia), DLX5 (1/200 dilution, Abcam, Cambridge, United Kingdom), and α-1integrin (ITGA1/CD49a/VLA-1) (1/250 dilution, Millipore, Australia). Conjugated secondary antibodies used were goat antirabbit Alexa Fluor 568 (8 µg/mL; Life Technologies, Australia) and donkey antimouse Alexa Fluor 488 (4 µg/mL, Life Technologies, Australia). Nonimmune rabbit serum (1 µg/mL, Flinders Technologies, Australia) and control mouse monoclonal antibody X63 (see subsequently), which has no known cross-reactivity with human proteins, were used as negative controls. Nuclei were counterstained with Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, California). Sections were mounted and coverslipped using Dako fluorescence mounting medium (Dako, Australia). Fluorescent staining was visualized on an Olympus AX70 fluorescence microscope with the appropriate filters and the resulting multicolor image was produced by Axiovision 4 software (Carl Zeiss, Jena, Germany).
Isolation of First Trimester (Early) CMSCs
The method was based on the study of Poloni,32 which describes the isolation of MSCs from first trimester chorionic villi of human placenta. Chorionic villus samples were obtained from pregnancies between 12 and 14 weeks of gestation by ultrasound-guided transabdominal chorionic villus sampling. Samples were collected into flasks containing 1× PBS without calcium and magnesium (Gibco, California), supplemented with 50 IU/mL heparin (Pfizer, New York) and 100U/mL penicillin and 100µg/mL streptomycin (Roche, Germany). The aspirated chorionic villi were transferred to a 55-mm Petri dish and washed free of maternal blood using prewarmed (37°C) PBS. Washed villi were examined under a stereomicroscope (×16 magnification) and fine sterile forceps were used to dissect away any contaminating maternal decidua. The cleaned villi were removed to a second Petri dish and rinsed again using PBS. After removing the PBS, the cleaned chorionic villi (10-20 mg) were finely minced in 2 to 3 drops of 0.25% trypsin (Gibco, California) using a sterile, size 22 surgical scalpel blade (Swann-Morton, Sheffield, United Kingdom). Minced villi were transferred into a conical centrifuge tube containing 2 mL 0.25% trypsin, agitated thoroughly, and incubated for 30 minutes at 37°C. The digested tissue was then neutralized by adding 5 mL complete medium (Roswell Park Memorial Institute supplemented with 20% fetal bovine serum) and pelleted by centrifugation at 560g at room temperature for 6 minutes. The supernatant was removed and the cells were seeded onto glass coverslips in Petri dishes with Amniomax C100 medium (Life Technologies, Australia). Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2 until ready for routine diagnostic karyotype analysis. Once karyotype analysis was complete, cells from back-up cultures were used to prepare early CMSCs by subculturing in Amniomax medium. Only samples with normal fetal ultrasound findings and normal diagnostic karyotype were included for subculture. Typically, these subcultures of early CMSCs were at passages P1 or P2 prior to release for subculturing early CMSCs. Following subculture, early CMSCs from P2 or P3 were assessed for their proliferative potential, expression of cell surface markers, and differentiation potential as described previously.7,26 Early CMSCs up to P5 were used for experiments. Preparations of early CMSCs from male placentae were routinely tested for the presence of maternal cell contamination.
Isolation of Term CMSCs
Term placentae were used to prepare CMSCs using a method adapted from Fukuchi et al10 and described in more detail elsewhere.7 Term placentae (n = 5) from uncomplicated pregnancies were used to prepare cultures of CMSCs.
The maternal side of the placenta was used for MSC preparation following careful removal of decidual tissue that was attached to the maternal surface. Placental tissue from central cotyledons of about 10 g (2.5 cm cubes) was dissected, extensively minced, and then centrifuged at 550g for 5 minutes. The pellet was hemolyzed in red blood cell lysis buffer (155 mmol/L NH4Cl, 10 mmol/L KHCO3, 0.1 mmol/L EDTA, pH 8.0) and then centrifuged again at 550g for 5 minutes at room temperature. The pellet was incubated at 37°C in 0.05% Trypsin-EDTA solution (Invitrogen, Australia) for 10 minutes, resuspended twice in 50% α-Minimum Essential Medium (α-MEM)/50% fetal bovine serum (FBS), centrifuged at 550g for 10 minutes at room temperature, and then plated in α-MEM medium supplemented with 20% FCS, 0.02% ascorbate-2-phosphate, 1% l-glutamine, 0.5% penicillin/streptomycin, 0.5% fungizone, and 1% tylosin. Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2. After incubation for 3 days, cells had adhered to the plate and the digested tissue pieces were removed by washing in Hank Balanced Salt Solution (HBSS). Cultures were maintained in α-MEM complete medium until 80% confluent and then passaged (P) up to 5 times.
Flow Cytometry Analysis of CMSC Preparations
For flow cytometry, P2 to P4 early CMSCs or term CMSCs were detached with TrypLE Express (Invitrogen, Australia) and resuspended in HBSS(Invitrogen, Australia) with 2% FBS. Approximately 105 cells per sample were stained with CD73-PE (BD Biosciences, Australia), CD105-APC (Caltag Laboratories, Buckingham, United Kingdom), and CD45APC-Cy7 (BD Biosciences, Australia) for 30 minutes each at 4°C. Mouse IgGκPE (BD Biosciences, Australia) isotype control was used to detect nonspecific fluorescence. Cell preparations were assayed on a BD LSRII flow cytometer and analyzed using FACSDiva software (BD Biosciences, Australia).
Immunocytochemistry
The cultured CMSCs were placed onto 8 well-chamber slides (BD Biosciences) and fixed in 10% formalin for 10 minutes at room temperature and rehydrated twice separately in graded ethanol. Endogenous peroxide activity was quenched with 0.3% hydrogen peroxide (Merck, Australia) solution. The cells were blocked with blocking solution (solution A from Histostain-Plus Broad Spectrum Kit, Life Technologies, Australia) at room temperature for 20 minutes. Slides were then incubated with rabbit polyclonal antihuman MEIS2, HLX, MSX2, HHEX, TGIF, HOXB7, DLX5, and DLX4 at 1 μg/mL concentration. Negative controls included the omission of primary antibody (ie, using antibody diluent alone) and the X63 antibody (1 μg/mL, immunoglobulin 1; mouse monoclonal antibody P3X63.Ag8, supplied by the Department of Clinical Immunology, Flinders Medical Centre, Bedford Park, South Australia). X63 antibody is a negative control antibody with no known cross-reactivity to human tissue.33 The immunoreactivity was visualized using biotinylated- and HRP-conjugated secondary antibodies and the chromogen AEC (Zymed Laboratories, California). The control was counterstained with 0.5% methyl green to assess chorionic villous morphology.
RNA Isolation for Real-Time Polymerase Chain Reaction Analysis
Confluent cultures of early CMSCs or term CMSCs were homogenized using a QIAshredder spin column (Qiagen, Australia), and total RNA extracted using the RNeasy Micro Kit (Qiagen, Australia) as described.29 Relative quantitation of homeobox genes in early and term CMSCs was performed as described by Murthi et al30 on an ABI Prism 7700 Sequence Detector (Perkin-Elmer Applied Biosystems, Australia) using Inventoried gene expression assays consisting of a 20× mix of unlabeled polymerase chain reaction (PCR) primers and TaqMan MGB FAM dye-labeled probe ZHX1(Hs00232545_m1), MEIS2 (Hs00542636_m1), HLX (Hs00172035_m1), MSX2 (Hs00741177_m1), LIM6 (Hs00232660_m1), MOX2 (Hs00232248_m1), HHEX (Hs00242160_m1), TGIF (Hs00820148_g1), HOXA9 (Hs00365956_m1), PHOX1 (HsHs00246567), HOXB7 (Hs00270131_m1), VSX1 (Hs00232724_m1), NKX3-1(Hs00171834_m1), DLX5 (Hs00193291_m1), DLX4 (Hs00231080_m1), SIX6 (Hs00231050_m1), and PITX1 (Hs00267528_m1). All probes were obtained from Applied Biosystems, Australia. Relative quantitation of homeobox gene expression was normalized to 18S rRNA (TaqMan MGB VIC dye labeled probe, Applied Biosystems, Australia) in a duplex reaction and calculated according to the 2−ΔΔCT method of Livak and Schmittgen.34
Data Analysis
Differences in homeobox gene messenger RNA (mRNA) expression normalized to 18S rRNA in CMSCs were determined using the Mann-Whitney U test. Statistical calculations were performed on the GraphPad Prism software. Results were considered significant at P < .05.
Results
Laser Capture Microdissection and RNA Analysis of Homeobox Genes
RNA analysis of laser capture microdissected first trimester placental chorionic villi was used to detect mRNAs encoding homeobox genes. DNA microarrays contained about 20 000 transcripts and more than 100 of the 235 probable functional human homeobox genes35 were represented. Array analysis was carried out on 3 cDNA preparations from independent laser capture stroma preparations from 3 different patients. Where multiple probes for individual homeobox genes were present on the microarray, the average was used, and the cutoff for average signal intensity was set at 100. The list of all genes in the array was cited in Bilban et al.31 Eighteen homeobox genes were detected in the laser capture microdissected first trimester placental chorionic villous stroma, with signal intensities above the arbitrary cutoff of 100 (Table 1, footnotes a-c). The highest average signal intensity was observed for zinc finger homeobox gene 1 (ZHX1), MEIS2, and HLX homeobox gene. The majority of homeobox genes showed signal intensities of less than 100.
Table 1.
Homeobox Gene Expression Profile in First-Trimester Chorionic Villous Stroma and in Isolated Term CMSCs.
| First Trimester Chorionic Villous Stroma. Average Signal Intensitya | Gene Symbolb | Gene Namec | Term-CMSCs. Relative Quantitation by Real-Time PCRd |
|---|---|---|---|
| 3800.35 | ZHX1 | Zinc finger and homeoboxes 1 | 1.85 |
| 2248.10 | MEIS2* | Meis1, myeloid ectropic viral integration site 1 homolog 2 | 1.05 |
| 728.60 | HLX* | H2.0-like Drosophila | 0.44 |
| 482.20 | MSX2* | Msh homeobox 2 | 0.52 |
| 415.10 | LIM6 | LIM-domain binding 6 | 0.70 |
| 400.53 | MOX2 | Mesenchyme homeobox 2 | 0.61 |
| 387.83 | HHEX* | Hematopoietically expressed homeobox | 1.29 |
| 363.83 | TGIF* | TGFB-induced factor | 1.24 |
| 339.90 | HOXA9 | Homeobox A9 | 1.1 |
| 258.80 | PHOX2A | Paired-like (aristaless) homeobox 2a | 0.70 |
| 254.27 | HOXB7* | Homeobox B7 | 0.56 |
| 229.40 | VSX1 | Visual system homeobox 1 homolog, CHX10-like | 0.80 |
| 223.33 | NKX3-1 | NK3 transcription factor related, locus 1 | 0.52 |
| 157.70 | DLX5* | Distal-less homeobox 5 | 0.57 |
| 154.80 | DLX4* | Distal-less homeobox 4 | 0.61 |
| 128.70 | PRRX1 | Pared related homeobox 1 | 0.39 |
| 124.00 | SIX6 | Sine oculis homeobox homolog 6 | 0.41 |
| 104.20 | PITX1 | Paried-like homeodomain transcription factor 1 | 0.66 |
Abbreviations: CMSCs, chorionic mesenchymal stem/stromal cells; PCR, polymerase chain reaction; TGFB, transforming growth factor β; mRNA, messenger RNA; rRNA, ribosomal RNA.
a Average signal intensity above the arbitrary cutoff of 100.
b Gene symbol with asterisk represents a gene chosen for further mRNA/protein analysis.
c Gene name.
d Real-time PCR was used to detect homeobox gene mRNA relative to 18S rRNA in term CMSC.
Candidate Protein Expression in First Trimester Placentae
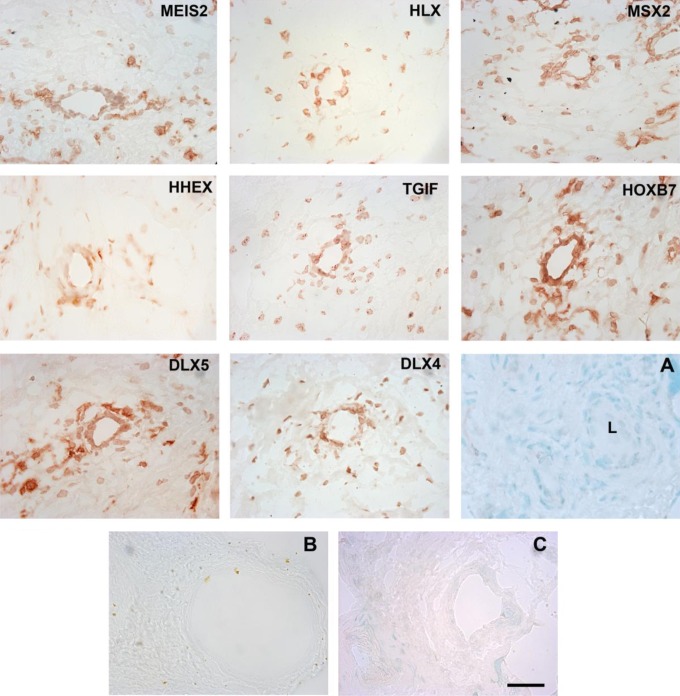
Candidate homeobox genes were selected based on the range of average signal intensity scores obtained from laser capture microdissection and RNA analysis. Zinc finger homeobox gene 1, which had the highest score (Table 1) in both categories, was not included because a well-characterized antibody was not available. Figure 1 reveals that the antibodies to MEIS2, HLX, MSX2, HHEX, TGIF, HOXB7, DLX5, and DLX4 were immunoreactive to vascular cells within the chorionic villi of first trimester placental tissue. Immunoreactivity was predominantly nuclear. All antibodies showed immunoreactivity in scattered stromal cells.
Figure 1.
Immunohistochemical staining of first trimester human placental tissue (n = 3) with antibodies to MEIS2, HLX, MSX2, HHEX, TGIF, HOXB7, DLX5, and DLX4. Panels are labeled with the antibodies to homeobox gene protein products, which were used to generate the staining pattern. Panel A is a methyl green nuclear stain showing a ∼50 μm vessel and surrounding stromal morphology. L indicates lumen. Panels B and C show representative controls. Panel B shows an IgG1 control for mouse monoclonal antibodies (MEIS2, MSX2, and TGIF), the IgG2 control for mouse monoclonal antibodies (DLX5) showed a similar low background level of staining (data not shown). Panel C shows a preimmune serum control for rabbit polyclonal antibodies (HLX, HHEX, and DLX4). Detection was with AEC. Magnification was ×400 and the scale bar is 50 μm. IgG indicates immunoglobulin. MEIS2 indicates Meis1, myeloid ectropic viral integration site 1 homolog 2; HLX, H2.0-like Drosophila; MSX2, Msh homeobox 2; HHEX, hematopoietically expressed homeobox; TGIF, transforming growth factor β-induced factor; HOXB7, homeobox B7; DLX5, distal-less homeobox 5; DLX4, distal-less homeobox 4. (The color version of this figure is available in the online version at http://rs.sagepub.com/.)
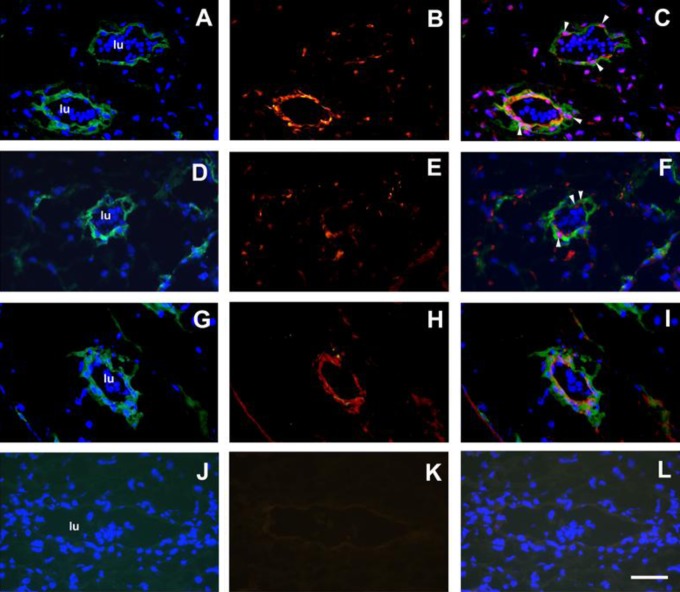
Immunofluorescence staining with a mouse monoclonal antibody to CMSC cell surface marker FZD936 and rabbit polyclonal antibodies to DLX5 or HLX was carried out on first trimester placental sections (n = 3). The FZD9 antigen was detected with a donkey antimouse Alexafluor 488 secondary antibody in the cytoplasm of cells around vessels in DAPI counterstained sections (Figure 2A, D, and G). The DLX5 nuclear antigen was detected with a goat antirabbit Alexafluor568 secondary antibody in nuclei of cells around vessels and scattered cells in the stroma (Figure 2B, red). The combined image (Figure 2c) shows cells with DLX5 and DAPI nuclear staining (purple) with FZD9 cytoplasmic staining around vessels (Figure 2C, white arrows). Similarly, HLX nuclear antigen was detected in nuclei of vascular cells and scattered cells in the stroma (Figure 2E, red). The combined image (Figure 2F) shows cells with HLX and DAPI nuclear staining (purple) with FZD9 cytoplasmic staining of vascular cells (Figure 2C, white arrows). Figure 2G and H shows FZD9 and another MSC cell surface marker, ITGA1, staining, respectively. These 2 MSC cell surface markers colocalize to vascular cells (Figure 2I). Nonimmune rabbit serum and X63 negative controls showed no significant staining.
Figure 2.
Multilabel immunofluorescence staining of first trimester human placental stroma and blood vessels with DLX5 and HLX antibodies (n = 3). Panel A, FZD9 staining (donkey antimouse Alexafluor488, green) and DAPI staining (blue). Panel B, DLX5 staining (goat antirabbit Alexafluor568, red). Panel C, Combined FZD9/DAPI/DLX5 image. Panel D, FZD9 staining and DAPI staining. Panel E, HLX staining (red). Panel F, combined FZD9/DAPI/HLX image. White arrowheads show representative cells that stain positively with DLX5, DAPI (purple nuclei), and FZD9 (Panel C), or with HLX, DAPI (purple nuclei), and FZD9 (Panel F). Panel G, FZD9 staining and DAPI staining. Panel H, ITGA1 staining (red). Panel I, Combined FZD9/DAPI/ITGA1 image. Panel J, Mouse monoclonal X63 (green) and DAPI. Panel K, nonimmune rabbit serum (NIRS; red). Panel I, Combined X63/DAPI/NIRS image. Nucleated erythrocytes were detected within the vessel lumen (lu) by DAPI staining (Panels A, D, G, and J). Magnification was at ×600 and scale bar is 50 μm. DAPI indicates 4′,6-diamidino-2-phenylindole. HLX indicates H2.0-like Drosophila; DLX5, distal-less homeobox 5.
Preparation and Characterization of Early CMSCs and Term CMSCs
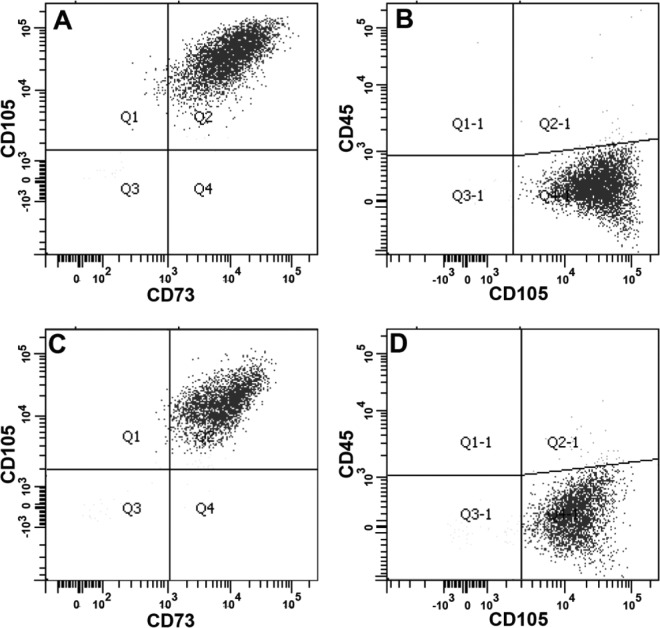
Chorionic villus sampling cell preparations obtained postdiagnosis were used to prepare early CMSCs, while term CMSCs were prepared from term placentae following delivery. We previously published flow cytometry analysis for term CMSCs and decidua parietalis MSCs.7,26 Our preparations of term CMSCs for the present study were of similar quality. Figure 3 shows a representative triple antibody staining flow cytometry profile for both early CMSCs and term CMSCs, which stained positively with MSC markers CD105 and CD73 (>98%) and negatively for the leukocyte common antigen CD45 (<2%). Both early CMSCs and term CMSCs were characterized using methods described in detail elsewhere7 and included colony-forming unit-fibroblast assays and differentiation potential (data not shown).
Figure 3.
Representative flow cytometry analyses for CMSCs. Early CMSCs (panels A and B) and term CMSCs (panels C and D) were CD105+, CD73+ (>98%), and CD45− (<2%). Antibodies used are described on each axis. CMSCs indicate chorionic mesenchymal stem/stromal cells.
In addition, all term CMSC preparations were assessed by immunocytochemistry using the MSC marker ITGA1 (CD49a)37 to which >90% of the population were positive (>200 cells per slide assessed, data not shown).
Real-Time PCR Analysis of Homeobox Gene Expression in Early and Term CMSCs
Table 1 (see footnote d) shows that all homeobox genes identified in laser capture microdissected first trimester chorionic villous stroma, from which early CMSCs are derived, were also detected by real-time PCR in term CMSCs. Zinc finger homeobox gene 1 showed the highest relative expression of the 18 homeobox genes in term CMSCs as was the case in the first trimester tissue (Table 1, see footnote a).
Real-time PCR analyses of cultured term CMSC preparations from passages P2 to P4 detected mRNA for selected homeobox genes (MEIS2, HLX, HHEX, TGIF, and DLX5). There were no significant differences in mRNA levels in passages between P2 and P3 (n = 3 samples in each passage, P > .05).
Relative to P2, there was no significant difference in relative mRNA levels for any of the homeobox genes in both freshly isolated, unpassaged term CMSCs and in cultured term CMSCs at passages P2 to P4 (n = 5 for each P > .05).
Figure 4 shows that cDNA products of homeobox genes MEIS2, HLX, HHEX, TGIF, and DLX5 were detected relative to 18S rRNA for both early CMSCs (n = 5) and term CMSCs (n = 5). There was no significant difference in expression of MEIS2 (P = .30), HLX (P = .12), HHEX (P = .19), TGIF (P = .42), and DLX5 (P = .17) between early and term CMSCs.
Figure 4.
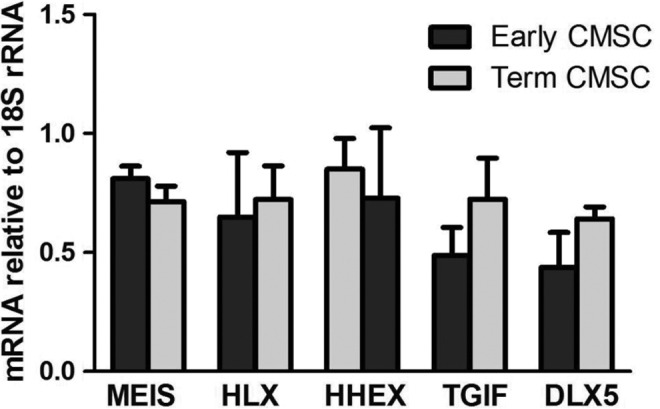
Real-time PCR analysis in early and term CMSCs. Homeobox gene mRNA levels relative to 18S rRNA in passages P2 to P4 in early and term CMSCs. CMSC indicates chorionic mesenchymal stem/stromal cells; mRNA, messenger RNA; rRNA, ribosomal RNA.
Expression of Homeobox Genes in Early- and Term CMSCs
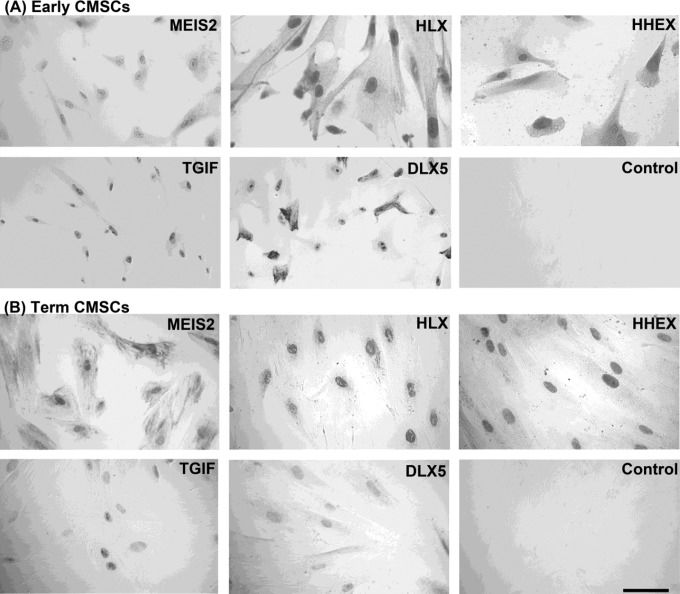
Figure 5 shows immunocytochemical analyses for early and term CMSCs. MEIS2, HLX, HHEX, TGIF, and DLX5 protein expressions were detected in both early and term CMSCs. Expression was predominantly nuclear for all antibodies tested but there was some variation in the intensity of staining and in the degree of the cytoplasmic staining.
Figure 5.
Immunocytochemical staining of (A) first trimester and (B) term-human placental CMSCs with antibodies to MEIS2, HLX, HHEX, TGIF, and DLX5. A representative control with IgG2 or rabbit nonimmune serum is shown. Detection was with AEC and differential interference contrast microscopy. Magnification was ×400 and the scale bar is 50 μm. IgG indicates immunoglobulin. MEIS2 indicates Meis1, myeloid ectropic viral integration site 1 homolog 2; HLX, H2.0-like Drosophila; HHEX, Hematopoietically expressed homeobox; TGIF, transforming growth factor β-induced factor; DLX5, distal-less homeobox 5.
Discussion
Laser Capture Microdissection and RNA Analysis of Homeobox Genes
Microarray screening of cDNA prepared from laser capture microdissected first trimester chorionic villous stromal mRNA detected 18 candidate homeobox genes. Laser capture microdissection and mRNA analysis allow us to conclude that these homeobox genes were expressed in vivo. Zinc finger homeobox gene 1, MEIS2, and HLX homeobox genes had the highest average signal intensity scores (Table 1). The ZHX proteins are major transcriptional regulators of podocyte gene expression during the development of nephrotic syndrome.38 In addition, real-time PCR analysis showed ZHX1 had the highest mRNA expression relative to 18S rRNA in term CMSCs (Table 1). The ZHX1 expression has not been previously described in MSCs or the human placenta. However, since there is no well-characterized antibody for human ZHX1, we did not pursue further analysis of this gene in this study but it clearly warrants further investigation.
Candidate Gene Expression in First Trimester Placentae
Selected candidate homeobox genes were assessed for expression in first trimester placental tissue. Antibodies to MEIS2, HLX, MSX2, HHEX, TGIF, HOXB7, DLX5, and DLX4 were immunoreactive predominantly in the nuclei of cells as would be expected of nuclear transcription factors. Immunoreactivity for all antibodies to homeobox gene protein products in the villous stroma was predominantly vascular but there were also some positive scattered cells in the villous stroma.
In multilabel immunofluorescence analysis, MSC cell surface markers FZD9 and ITGA1 were used. FZD9 has been used in several studies to identify CMSCs in the human placenta. FZD9 and ITGA1 (CD49a) are used to enrich and identify MSCs with high clonogenic potential.36,37,39 Immunoreactivity of FZD9 and ITGA1 was detected in vascular cells. Vascular cells with immunoreactivity for both FZD9 and DLX5 or FZD9 and HLX were detected. Not all vascular cells were immunoreactive for both FZD9 and DLX5 or FZD9 and HLX suggesting that CMSCs represent a subset of the vascular cells.
The vascular expression patterns of MEIS2, HLX, MSX2, HHEX, TGIF, HOXB7, DLX5, and DLX4 were consistent with a vascular niche for CMSCs, which was previously shown using stem cell surface marker antibodies in chorionic villi in both first trimester and term placentae.7 Vascular and perivascular niches have been identified for MSCs in many tissues and organs.40
We previously showed HLX protein expression in vascular cell types in first trimester and term placental tissues41 and more recently we found a similar protein expression pattern for TGIF.42 HLX, MSX2, HHEX, TGIF, HOXB7, and DLX4 mRNAs are expressed in isolated and enriched vascular (endothelial) cells from term placental vessels.43,44 This study is the first to demonstrate MSX2, HOXB7, DLX5, and DLX4 protein expression in vascular cells in first trimester placental tissue.
The vascular niche comprises a number of cell types including MSCs, mural cells (pericytes or smooth muscle cells), and endothelial cells which also express homeobox genes (see earlier). Therefore, it was important to demonstrate homeobox gene expression in isolated, enriched, and well-characterized early and term CMSCs.
Preparation and Characterization of Early and Term CMSCs
Early and term CMSCs showed a typical MSC cell surface marker phenotype (CD105+, CD73+, and CD45−). Thus, our preparations of early and term CMSCs showed characteristics reported in the literature for early CMSCs32 and term CMSCs.5–7
Real-Time PCR Analysis in Early and Term CMSCs
A subset of the candidate homeobox genes (MEIS2, HLX, HHEX, TGIF, and DLX5) was assessed in more detail for mRNA expression in early and term CMSCs. To determine whether homeobox gene expression varies significantly during passaging, we determined homeobox gene mRNA levels relative to 18S rRNA from early passages (P2-P4) in term CMSCs and found no significant changes with passaging up to P4 (ie, about 20-25 population doublings).
Relative to P2, there was no significant difference in mRNA levels for any of the homeobox genes in either freshly isolated (n = 5), unpassaged term CMSCs (n = 5), or in cultured term CMSCs at passages P2 to P4 (n = 3 each passage). There were no significant differences between freshly isolated and cultured term CMSCs P2 to P4, or unpassaged term CMSCs and cultured term CMSCs P2 to P4 cells. Thus, passaging of CMSCs up to P4 in vitro did not significantly change the levels of MEIS2, HLX, HHEX, TGIF, and DLX5 mRNA from the levels detected in unpassaged CMSCs isolated directly from tissue.
Expression of MEIS2, HLX, HHEX, TGIF, and DLX5 homeobox gene relative to 18S rRNA was not significantly different between early and term CMSCs. These data are consistent with this combination of homeobox genes being expressed in CMSCs from early placental development through to term. Others have analyzed MSCs from mouse bone marrow, thymus, and lung and showed they express specific combinations of Hox cluster homeobox genes depending on the anatomical location of the MSCs.23 Previous studies that have compared early to term CMSCs concluded that with respect to cell surface phenotype, differentiation potential, and expression of certain “stemness” markers, there is no significant difference between them,45,46 whereas other functions such as growth kinetics, ability to form embryoid bodies, and tissue repair potential are significantly different.8,45 Our data indicate that a particular combination of homeobox genes is expressed in CMSCs throughout placental development.
Immunocytochemistry
As expected for nuclear transcription factors, immunocytochemistry on CMSCs revealed that MEIS2, HLX, HHEX, TGIF, and DLX5 antibodies were immunoreactive with cell nuclei in both early and term CMSCs. These data further support the notion that a particular combination of homeobox gene nuclear transcription factors is expressed in CMSCs through placental development.
What is the Function of Homeobox Genes in CMSCs?
The function of these homeobox genes in CMSCs is not known but there is evidence that they may play a role in the control of stem cell proliferation and differentiation. In this study, we showed that MEIS2, HLX, HHEX, TGIF, and DLX5 are expressed in the vascular niche and stroma but HLX and TGIF are also expressed in villous cytotrophoblast (CTB) cells, which are the stem cell population for the trophoblast cell lineage. In previous studies, we showed that HLX is expressed in CTB41 and is a regulator of cell proliferation.41,47,48 We have shown that TGIF is a regulator of CTB differentiation.42 Therefore, based on the function of homeobox genes in the CTB stem cells, there is a potential role for HLX and TGIF in the regulation of CMSC proliferation and differentiation.
Some evidence for the potential function of other homeobox genes we detected in CMSCs comes from published studies of their roles in nonplacental stem cells. For example, MEIS2 is critical for the differentiation in human embryonic stem cells into the cardiac cell lineage.49 Expression of Meis2 in chick and mouse embryos plays an evolutionary conserved role in maintaining proliferating retinal progenitor cells.50 Hex, the murine homolog of human HHEX, is important for murine liver development, specifically for hemangioblast differentiation to definitive hematopoietic progenitors.51
TGIF was first identified as a competitor of the retinoic acid receptor for binding to retinoic acid response elements and shows restricted expression to only a few tissues where it regulates proliferating and differentiating cell lineages.52,53 Dlx5, the murine homolog of human DLX5, plays a critical role in the lineage commitment of bone marrow MSCs into adipocytes and osteoblasts.54
Thus, there is supporting evidence from various animal models for the potential role of MEIS2, HLX, HHEX, TGIF, and DLX5 in the regulation of CMSC proliferation and differentiation. Future work will elucidate the functional role of these homeobox genes in CMSCs.
Acknowledgments
The authors gratefully acknowledge specimen collection by Clinical Research Midwives Susan Nisbet and Sue Duggan, Pregnancy Research Centre, and Department of Perinatal Medicine at the The Royal Women’s Hospital. We thank Dr Neil M. Gude for assistance with manuscript preparation.
Footnotes
Authors’ Note: Haiying Liu and Padma Murthi are equal first authors.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: We acknowledge the financial support of KAIMRC Grant No. RC08/114, KACST Grant No. ARP-29-186, and NHMRC Grant No. 509178.
References
- 1. Abumaree M, Al Jumah M, Pace RA, Kalionis B. Immunosuppressive properties of mesenchymal stem cells. Stem Cell Rev. 2012;8(2):375–392 [DOI] [PubMed] [Google Scholar]
- 2. Longo UG, Loppini M, Berton A, La Verde L, Khan WS, Denaro V. Stem cells from umbilical cord and placenta for musculoskeletal tissue engineering. Curr Stem Cell Res Ther. 2012;7(4):272–281 [DOI] [PubMed] [Google Scholar]
- 3. Manuelpillai U, Moodley Y, Borlongan CV, Parolini O. Amniotic membrane and amniotic cells: potential therapeutic tools to combat tissue inflammation and fibrosis? Placenta. 2011;32(suppl 4):S320–S325 [DOI] [PubMed] [Google Scholar]
- 4. Parolini O, Alviano F, Bagnara GP, et al. Concise review: isolation and characterization of cells from human term placenta: outcome of the first international workshop on placenta derived stem cells. Stem Cells. 2008;26(2):300–311 [DOI] [PubMed] [Google Scholar]
- 5. In ’t Anker PS, Scherjon SA, Kleijburg-van der Keur C, et al. Isolation of mesenchymal stem cells of fetal or maternal origin from human placenta. Stem Cells. 2004;22(7):1338–1345 [DOI] [PubMed] [Google Scholar]
- 6. Abumaree MH, Al Jumah MA, Kalionis B, et al. Phenotypic and functional characterization of mesenchymal stem cells from chorionic villi of human term placenta. Stem Cell Rev. 2013;9(1):6. [DOI] [PubMed] [Google Scholar]
- 7. Castrechini NM, Murthi P, Gude NM, et al. Mesenchymal stem cells in human placental chorionic villi reside in a vascular Niche. Placenta. 2010;31(3):203–212 [DOI] [PubMed] [Google Scholar]
- 8. Jones GN, Moschidou D, Puga-Iglesias TI, et al. Ontological differences in first compared to third trimester human fetal placental chorionic stem cells. PLoS One. 2012;7(9):e43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arakawa R, Aoki R, Arakawa M, Saito K. Human first-trimester chorionic villi have a myogenic potential. Cell Tissue Res. 2012;348(1):189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fukuchi Y, Nakajima H, Sugiyama D, Hirose I, Kitamura T, Tsuji K. Human placenta-derived cells have mesenchymal stem/progenitor cell potential. Stem Cells. 2004;22(5):649–658 [DOI] [PubMed] [Google Scholar]
- 11. Maltepe E, Bakardjiev AI, Fisher SJ. The placenta: transcriptional, epigenetic, and physiological integration during development. J Clin Invest. 2010;120(4):1016–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cross JC. How to make a placenta: mechanisms of trophoblast cell differentiation in mice--a review. Placenta. 2005;26(suppl A):S3–S9 [DOI] [PubMed] [Google Scholar]
- 13. Cross JC, Baczyk D, Dobric N, et al. Genes, development and evolution of the placenta. Placenta. 2003;24(2-3):123–130 [DOI] [PubMed] [Google Scholar]
- 14. Hemberger M, Cross JC. Genes governing placental development. Trends Endocrinol Metab. 2001;12(4):162–168 [DOI] [PubMed] [Google Scholar]
- 15. Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17(1):42–49 [DOI] [PubMed] [Google Scholar]
- 16. Loregger T, Pollheimer J, Knofler M. Regulatory transcription factors controlling function and differentiation of human trophoblast--a review. Placenta. 2003;24(suppl A):S104–S110 [DOI] [PubMed] [Google Scholar]
- 17. Kubo H, Shimizu M, Taya Y, et al. Identification of mesenchymal stem cell (MSC)-transcription factors by microarray and knockdown analyses, and signature molecule-marked MSC in bone marrow by immunohistochemistry. Genes Cells. 2009;14(3):407–424 [DOI] [PubMed] [Google Scholar]
- 18. Klump H, Schiedlmeier B, Baum C. Control of self-renewal and differentiation of hematopoietic stem cells: HOXB4 on the threshold. Ann N Y Acad Sci. 2005;1044:6–15 [DOI] [PubMed] [Google Scholar]
- 19. Trosko JE. From adult stem cells to cancer stem cells: Oct-4 Gene, cell-cell communication, and hormones during tumor promotion. Ann N Y Acad Sci. 2006;1089:36–58 [DOI] [PubMed] [Google Scholar]
- 20. Tolkunova E, Cavaleri F, Eckardt S, et al. The caudal-related protein cdx2 promotes trophoblast differentiation of mouse embryonic stem cells. Stem Cells. 2006;24(1):139–144 [DOI] [PubMed] [Google Scholar]
- 21. Niwa H, Toyooka Y, Shimosato D, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123(5):917–929 [DOI] [PubMed] [Google Scholar]
- 22. Chen L, Yabuuchi A, Eminli S, et al. Cross-regulation of the Nanog and Cdx2 promoters. Cell Res. 2009;19(9):1052–1061 [DOI] [PubMed] [Google Scholar]
- 23. Ackema KB, Charite J. Mesenchymal stem cells from different organs are characterized by distinct topographic Hox codes. Stem Cells Dev. 2008;17(5):979–991 [DOI] [PubMed] [Google Scholar]
- 24. Menicanin D, Bartold PM, Zannettino AC, Gronthos S. Genomic profiling of mesenchymal stem cells. Stem Cell Rev. 2009;5(1):36–50 [DOI] [PubMed] [Google Scholar]
- 25. Nikolova G, Strilic B, Lammert E. The vascular niche and its basement membrane. Trends Cell Biol. 2007;17(1):19–25 [DOI] [PubMed] [Google Scholar]
- 26. Castrechini NM, Murthi P, Qin S, et al. Decidua parietalis-derived mesenchymal stromal cells reside in a vascular niche within the choriodecidua. Reprod Sci. 2012;19(12):1302–1314 [DOI] [PubMed] [Google Scholar]
- 27. Martini MM, Jeremias Tda S, Kohler MC, Marostica LL, Trentin AG, Alvarez-Silva M. Human placenta-derived mesenchymal stem cells acquire neural phenotype under the appropriate niche conditions. DNA Cell Biol. 2013;32(2):58–65 [DOI] [PubMed] [Google Scholar]
- 28. Gomez-Gaviro MV, Lovell-Badge R, Fernandez-Aviles F, Lara-Pezzi E. The vascular stem cell niche. J Cardiovasc Transl Res. 2012;5(5):618–630 [DOI] [PubMed] [Google Scholar]
- 29. Shahdadfar A, Fronsdal K, Haug T, Reinholt FP, Brinchmann JE. In vitro expansion of human mesenchymal stem cells: choice of serum is a determinant of cell proliferation, differentiation, gene expression, and transcriptome stability. Stem Cells. 2005;23(9):1357–1366 [DOI] [PubMed] [Google Scholar]
- 30. Nur Fariha MM, Chua KH, Tan GC, Tan AE, Hayati AR. Human chorion-derived stem cells: changes in stem cell properties during serial passage. Cytotherapy. 2011;13(5):582–593 [DOI] [PubMed] [Google Scholar]
- 31. Bilban M, Tauber S, Haslinger P, et al. Trophoblast invasion: assessment of cellular models using gene expression signatures. Placenta. 2010;31(11):989–996 [DOI] [PubMed] [Google Scholar]
- 32. Poloni A, Rosini V, Mondini E, et al. Characterization and expansion of mesenchymal progenitor cells from first-trimester chorionic villi of human placenta. Cytotherapy. 2008;10(7):690–697 [DOI] [PubMed] [Google Scholar]
- 33. Hawes CS, Petropoulos A, Lopata A, Kalionis B, Jones WR. Reactivity of human trophoblast monoclonal antibodies with marmoset monkey trophoblast cultures. Hum Reprod. 1998;13(5):1169–1174 [DOI] [PubMed] [Google Scholar]
- 34. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408 [DOI] [PubMed] [Google Scholar]
- 35. Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Battula VL, Treml S, Abele H, Buhring HJ. Prospective isolation and characterization of mesenchymal stem cells from human placenta using a frizzled-9-specific monoclonal antibody. Differentiation. 2008;76(4):326–336 [DOI] [PubMed] [Google Scholar]
- 37. Rider DA, Nalathamby T, Nurcombe V, Cool SM. Selection using the alpha-1 integrin (CD49a) enhances the multipotentiality of the mesenchymal stem cell population from heterogeneous bone marrow stromal cells. J Mol Histol. 2007;38(5):449–458 [DOI] [PubMed] [Google Scholar]
- 38. Liu G, Clement LC, Kanwar YS, Avila-Casado C, Chugh SS. ZHX proteins regulate podocyte gene expression during the development of nephrotic syndrome. J Biol Chem. 2006;281(51):39681–39692 [DOI] [PubMed] [Google Scholar]
- 39. Buhring HJ, Battula VL, Treml S, et al. Novel markers for the prospective isolation of human MSC. Ann N Y Acad Sci. 2007;1106:262–271 [DOI] [PubMed] [Google Scholar]
- 40. da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119(pt 11):2204–2213 [DOI] [PubMed] [Google Scholar]
- 41. Rajaraman G, Murthi P, Quinn L, Brennecke SP, Kalionis B. Homeodomain protein HLX is expressed primarily in cytotrophoblast cell types in the early pregnancy human placenta. Reprod Fertil Dev. 2008;20(3):357–367 [DOI] [PubMed] [Google Scholar]
- 42. Pathirage NA, Cocquebert M, Sadovsky Y, et al. Homeobox gene transforming growth factor beta-induced factor-1 (TGIF-1) is a regulator of villous trophoblast differentiation and its expression is increased in human idiopathic fetal growth restriction. Mol Hum Reprod. 2013;19(10):665–675 [DOI] [PubMed] [Google Scholar]
- 43. Murthi P, So M, Gude NM, Doherty VL, Brennecke SP, Kalionis B. Homeobox genes are differentially expressed in macrovascular human umbilical vein endothelial cells and microvascular placental endothelial cells. Placenta. 2007;28(2-3):219–223 [DOI] [PubMed] [Google Scholar]
- 44. Murthi P, Hiden U, Rajaraman G, et al. Novel homeobox genes are differentially expressed in placental microvascular endothelial cells compared with macrovascular cells. Placenta. 2008;29(7):624–630 [DOI] [PubMed] [Google Scholar]
- 45. Sung HJ, Hong SC, Yoo JH, et al. Stemness evaluation of mesenchymal stem cells from placentas according to developmental stage: comparison to those from adult bone marrow. J Korean Med Sci. 2010;25(10):1418–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Portmann-Lanz CB, Baumann MU, Mueller M, et al. Neurogenic characteristics of placental stem cells in preeclampsia. Am J Obstet Gynecol. 2010;203(4):399.e391–397 [DOI] [PubMed] [Google Scholar]
- 47. Rajaraman G, Murthi P, Pathirage N, Brennecke SP, Kalionis B. Downstream targets of homeobox gene HLX show altered expression in human idiopathic fetal growth restriction. Am J Pathol. 2010;176(1):278–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rajaraman G, Murthi P, Leo B, Brennecke SP, Kalionis B. Homeobox gene HLX1 is a regulator of colony stimulating factor-1 dependent trophoblast cell proliferation. Placenta. 2007;28(10):991–998 [DOI] [PubMed] [Google Scholar]
- 49. Paige SL, Thomas S, Stoick-Cooper CL, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151(1):221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Heine P, Dohle E, Bumsted-O’Brien K, Engelkamp D, Schulte D. Evidence for an evolutionary conserved role of homothorax/Meis1/2 during vertebrate retina development. Development. 2008;135(5):805–811 [DOI] [PubMed] [Google Scholar]
- 51. Chan RJ, Hromas R, Yoder MC. The role of Hex in hemangioblast and hematopoietic development. Methods Mol Biol. 2006;330:123–133 [DOI] [PubMed] [Google Scholar]
- 52. Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem. 1995;270(52):31178–31188 [DOI] [PubMed] [Google Scholar]
- 53. Bertolino E, Wildt S, Richards G, Clerc RG. Expression of a novel murine homeobox gene in the developing cerebellar external granular layer during its proliferation. Dev Dyn. 1996;205(4):410–420 [DOI] [PubMed] [Google Scholar]
- 54. Baek K, Baek JH. The transcription factors myeloid elf-1-like factor (MEF) and distal-less homeobox 5 (Dlx5) inversely regulate the differentiation of osteoblasts and adipocytes in bone marrow. Adipocyte. 2013;2(1):50–54 [DOI] [PMC free article] [PubMed] [Google Scholar]