Abstract
There is no consensus whether Sertoli cells express estrogen receptor 1 (Esr1). Reverse transcription-polymerase chain reaction, Western blot, and immunofluorescence demonstrated that mouse Sertoli cell lines, TM4, MSC-1, and 15P-1, and purified primary mouse Sertoli cells (PSCs) contained Esr1 messenger RNA and proteins. Incubation of Sertoli cells with 17β-estradiol (E2) or ESR1 agonist stimulated the expression of an estrogen responsive gene Greb1, which was prevented by ESR inhibitor or ESR1 antagonist. Overexpression of Esr1 in MSC-1 enhanced E2-induced Greb1 expression, while knockdown of Esr1 by small interfering RNA in TM4 attenuated the response. Furthermore, E2-induced Greb1 expression was abolished in the PSCs isolated from Amh-Cre/Esr1-floxed mice in which Esr1 in Sertoli cells were selectively deleted. Chromatin immunoprecipitation assays indicated that E2-induced Greb1 expression in Sertoli cells was mediated by binding of ESR1 to estrogen responsive elements. In summary, ligand-dependent nuclear ESR1 was present in mouse Sertoli cells and mediates a classical genomic action of estrogens.
Keywords: Esr1, estrogen, gene regulation, Sertoli cells, testis, mouse
Introduction
Several lines of evidence from cell-based and animal model studies and human genetic data have generally concluded that estrogen, once considered as a female hormone, also plays important roles in the male reproductive system.1–9 Either estrogen insufficiency or estrogen overexposure causes impaired spermatogenesis and infertility,10–15 suggesting that maintenance of a delicate balance between androgens and estrogens is critical for normal testicular function.15,16 The testis is 1 of the 2 major organs which produces estrogen in males. Here, the testosterone synthesized by Leydig cells is converted to estrogens by aromatase in almost all cell types in the testis.3 In addition to feedback regulation on the hypothalamus and pituitary gland that indirectly influences testicular functions,17,18 there is convincing evidence showing direct estrogen action within the testes that includes the modulation of testicular androgenesis and spermatogenesis.2,4,8,10,11,15,19–21
Estrogen is well established to elicit a myriad of biological processes in target tissues primarily through its binding to estrogen receptors (ESRs) ESR1 and ESR2 (also known as ERα and ERβ, respectively). Estrogen receptors belong to the steroid/thyroid hormone superfamily of nuclear transcription factors encoded by 2 different genes. There are 2 different but interrelated ESR-dependent estrogen signaling pathways commonly described as genomic and nongenomic mechanisms. The classic estrogen signaling pathway is through nuclear ESR-mediated genomic activity. The binding of estrogen to ESRs causes the dimerization of ESRs either as homodimers (eg, ESR1/ESR1 and ESR2/ESR2) or as heterodimers (eg, ESR1/ESR2 and ESR/other transcription factor) and interacts with the consensus DNA sequences called estrogen response elements (EREs) present in the promoter region of the target gene to alter the transcription activity. The nongenomic mechanism refers to membrane ESR-initiated rapid estrogen responses. This pathway is mediated mainly by nonclassical, membrane-associated ESRs through cross talk with other membrane receptors or direct activation of various kinase cascades to exert the biological effects.22–25
Testicular ESR1-mediated local estrogen actions have been demonstrated to be essential for male reproduction.2,26 Although both ESRs are present in the testes, deletion of Esr1 in mice causes male infertility, whereas an adverse testicular phenotype is not observed in mice with a targeted disruption of Esr2, suggesting that ESR2 has a very limited function in the testes.7,20,27 However, the mechanism by which ESR1-mediated estrogen activity affects male fertility is not completely understood. The fundamental question regarding which testicular cell types express functional ESR1 remains uncertain. Leydig cells are generally considered to express ESR1 in several species examined, such as fish, rodents, domestic animals, primates, and humans.7 Studies in Esr1 knockout mice reveal that the androgenesis in Leydig cells is enhanced in the absence of ESR1.28,29 This protein is also detected in the seminiferous tubules. Transplantation experiments in mice demonstrate that germ cells lacking Esr1 develop normally in the wild-type seminiferous tubules, and the mature sperm can fertilize wild-type oocytes to generate offspring.19,30 Hence, ESR1 has been postulated to play a role in testicular somatic cells that provide an environment for gametes to develop and mature.2 This notion is concurred by a recent study in mice that estrogen-dependent ESR1 action is required for germ cell survival and most likely involves the support of Sertoli cell functions.31 Sertoli cells are the somatic epithelial cells that line the seminiferous tubules of the testes in continuous contact with spermatogenic cells. It is known that these cells play critical roles in nursing and support of spermatogenic cell differentiation and maturation in response to a variety of hormone actions. However, there is no consensus regarding whether these cells express Esr1. In several earlier reports, no ESR1 protein was detected in Sertoli cells by immunohistochemistry in bank vole, bird, rat, mouse, dog, cat, goat, boar, monkey, and human (reviewed by Carreau and Hess7), whereas numerous more recent studies indicate that ESR1 is present in the Sertoli cells of multiple species including the hystricognath rodent, rat, cat, boar, pig, and human.31–40 The expression of ESRs in Sertoli cells from rat testes has recently been studied in more detail.31,35 The transcripts and proteins of both Esr1 and Esr2 are detected in premature and adult rat Sertoli cells. Furthermore, estrogen treatment of primary rat Sertoli cells reveals a membrane ESR-mediated rapid signal, involving the activation of the mitogen-activated protein kinases.34,35
Mice are one of the most common laboratory animals used in the studies of reproductive biology, but it is still debatable whether mouse Sertoli cells express Esr1. The results of the present study demonstrate the presence of both ESR1 and ESR2 in mouse Sertoli cell lines as well as primary Sertoli cells (PSCs). The ESR1 in mouse Sertoli cells mediates the classic genomic mechanism of estrogen action in the transactivation of its target gene Greb1 (gene regulated by estrogen in breast cancer protein 1) expression.
Materials and Methods
Animals
All animals were housed on 12-hour light–dark cycles with food and water provided ad libitum. All mice were maintained as required under the National Institutes of Health guidelines for the Care and Use of Laboratory Animals. The use of animals in this study has been approved by the Animal Care and Use Committee of the University of Louisville. All the mice were killed under ketamine anesthesia and all efforts were made to minimize their discomfort.
Primary Cell Culture and Cell Lines
Primary Sertoli cells were isolated from 30-day-old wild-type, Amh-Cre−/Esr1fl/fl and Amh-Cre+/Esr1fl/fl mice using a procedure described previously41 with a minor modification. Briefly, the testes were decapsulated and incubated with a collagenase type II solution (0.5 mg/mL; Sigma, St Louis, Missouri) to separate interstitial cells and seminiferous tubules. The dispersed seminiferous tubules were cut into small pieces and digested with a solution containing 1 mg/mL trypsin (Sigma) and 10 µg/mL DNase I (Sigma) at 32°C for 30 minutes. The reaction was stopped by adding trypsin inhibitor (Sigma) and Hanks balanced salt solution (HBSS; Invitrogen, Carlsbad, California). The supernatant that contained germ cells was discarded. The pellet was incubated with a collagenase type II solution at 32°C for 15 minutes and settled down by unit gravity sedimentation. The cell pellet, containing Sertoli cells, was rinsed with HBSS 3 times and plated with a 1:1 mixture of Dulbecco modified Eagle medium (DMEM) and F12 Ham medium supplemented with 10% fetal bovine serum (FBS; Invitrogen) overnight and the residual germ cells were hypotonically removed.
The purity of Sertoli cell preparations was verified by performing (1) reverse transcription-polymerase chain reaction (RT-PCR) analysis of the putative marker genes, (2) microscopic examination of their morphology following fixation with 10% formalin and stained with hematoxylin and eosin, and (3) immunostaining of a Sertoli cell-specific marker GATA-2 using an avidin–biotin immunoperoxidase method (Figure 1I-L). The transcripts of putative Sertoli cell marker genes (eg, Fshr [follicle-stimulating hormone receptor] and Pem [placenta and embryos oncofetal gene]) were readily detectable by RT-PCR, whereas Leydig cell marker genes (eg, Lhcgr [luteinizing hormone receptor] and Cyp17 [17α-hydroxylase]), myoid cell marker genes (eg, Alp [alkaline phosphatase] and Fn1 [fibronectin 1]), and germ cell maker genes (eg, Prm2 [protamine2] and Stra8 [stimulated by retinoic acid gene 8]) were not detected using the same RT-PCR conditions. Over 92% of isolated cells were positively immunostained for GATA-2. The mouse Sertoli cell lines, TM4 and 15P-1, were purchased from ATCC (Manassas, Virginia), and the MSC-1 cell line was kindly provided by Dr Griswold Washington State University (Pullman, Washington). MA10 cells (a mouse Leydig cell line) were a gift from Dr Ascoli (The University of Iowa, Iowa City, Iowa). All the cells were maintained in a mixture of DMEM and F12 medium supplemented with 10% FBS (Sigma) and antibiotic–antimycotic solution (Invitrogen). On the day of hormonal treatments, the medium was changed to phenol red- and serum-free DMEM and cells were then incubated with 17β-estradiol (E2, 10−10 mol/L; Sigma), ESR inhibitor ICI 182,780 (10−8 mol/L), ESR1 agonist PPT (4,4',4”-(4-Propyl-[1H]-pyrazole-1,3,5-triyl) trisphenol) (10−7 mol/L), ESR1 antagonist MPP (1,3-Bis(4-hydroxyphenyl)-4-methyl-5-[4-(2-piperidinylethoxy)phenol]-1H-pyrazole dihydrochloride) (10−7 mol/L), ESR2 agonist Diarylpropionitrile (DPN) (10−7 mol/L), ESR2 antagonist PHTPP (4-[2-Phenyl-5,7-bis(trifluoromethyl)pyrazolo[1,5-a]pyrimidin-3-yl]phenol) (10−7 mol/L; Tocris, Minneapolis, Minnesota), or vehicle dimethyl sulfoxide (Sigma) for 16 hours unless indicated elsewhere.
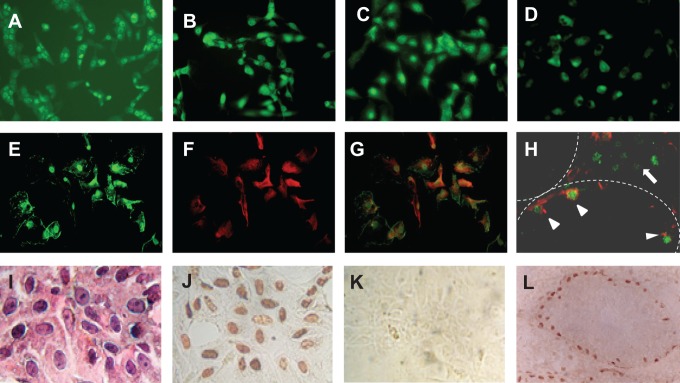
Figure 1.
Immunofluorescence showing membrane, cytoplasmic, and nuclear staining of ESR1 in MA10 (A), TM4 (B), 15P-1 (C), MSC-1 (D), and PSCs (E) cells. To differentiate ESR1-positive Sertoli cells from germ cells in isolated PSCs (G) and the testicular section (H), double labeling of ESR1 (E, green) and a Sertoli cell marker vimentin (F, red) was performed. Arrow heads in panel (H) indicate Sertoli cells in a somniferous tubule, and an arrow indicates Leydig cells in the interstitial space that is stained positively for ESR1. Panel (I) is isolated PSCs stained with hematoxylin and eosin. Panel (J) shows that the nuclei of isolated PSCs are immunostained for a Sertoli cell-specific marker GATA-2. Panel (K) is a negative control, in which GATA-2 antibody was omitted and panel (L) is a positive control, in which only the nuclei of Sertoli cells in the seminiferous tubules were immunostained for GATA-2. The immunostaining procedure was performed by an avidin–biotin immunoperoxidase method as described previously.42 Mag (A) to (H) and (L) = ×350 and (I) to (K) = ×500. ESR indicates estrogen receptor; mag, magnification; PSC, primary mouse Sertoli cell.
Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from the testes and the isolated testicular cells using Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. Total RNA was adjusted to a concentration of approximately 1 µg/µL. Total RNA of 1 µg was reverse transcribed into complementary DNA (cDNA) using random primers (Invitrogen) and avian myeloblastosis virus (AMV) reverse transcriptase (Promega Corporation, Madison, Wisconsin).43 The cDNA was amplified by PCR using the primer sets of the target gene and a housekeeping gene, ribosomal protein large subunit 19 (Rpl19), as an internal control for both cDNA quantity and quality. The PCR primers, as listed in Table 1, were designed according to the sequences obtained from GenBank using the Vector NTI 12.0 program (Invitrogen) and synthesized by Operon Technologies (Alameda, California). All primers were designed to amplify products that covered more than 1 exon. Each PCR cycle consisted of denaturation for 45 seconds at 94°C, annealing for 1 minute at 57°C, and extension for 1 minute at 72°C. Avian myeloblastosis virus was omitted in reactions used as procedure controls. The amplified products were separated by electrophoresis. For semiquantitative analysis, the intensity of specific bands was scanned using the TotalLab (Nonlinear USA Inc, Durham, North Carolina) image analysis software. The results were presented as the ratio of target gene to Rpl19.
Table 1.
Primer Sequences for Semiquantitative RT-PCR, ChIP, and Genotyping.
| Gene | Primer Sequence (5′-3′) | PCR Cycles |
|---|---|---|
| RT-PCR | ||
| Greb1 | F: TGCAGCATACAACACGTACCA R: GGGCTTTTGATGTGTTCATG | 27 for TM4, 34 for MSC-1 |
| Esr1 | F: CCGCAGCTGTCTCCTTTCCT R: CGGTTCTTGTCAATGGTGCA | 35 for all cell lines, 32 for TM4 |
| Esr2 | F: CGCTCAGGGACCGAGGAAAGTACGT R: GTCATGGCTGAGTATTCGTGACGG | 39 for all cell lines |
| Rpl19 | F: CTCAGGCTACAGAAGAGGCTT R: GGACAGAGTCTTGATGATCTC | Same as the coamplified target gene |
| ChIP | ||
| ERE1 | F: TTGGAAGATCCACCGCAAACT R: AGACAGGCTCGGGCATGTATC | 40 for TM4 |
| ERE2 | F: TCACCCACAGTGCTGCGAGA R: GCCCTTGACCGAGGAGATGA | 40 for TM4 |
| Genotyping | ||
| Amh-Cre | F: GCGGTCTGGCAGTAAAAACTATC R: GTGAAACAGCATTGCTGTCACTT | 30 cycles |
| Floxed and ▵Esr1 | F: TTGCCCGATAACAATAACAT R: GGCATTACCACTTCTCCTGGGAGTCT | 30 cycles |
| Floxed Esr1 | F: GTGTCAGAAAGAGACATT R: GGCATTACCACTTCTCCTGGGAGTCT | 30 cycles |
Abbreviations: ChIP, chromatin immunoprecipitation; ESR, estrogen receptor; ERE, estrogen response element; F, Forward primer; R, Reverse primer; RT-PCR, reverse transcription-polymerase chain reaction; Rpl19, ribosomal protein large subunit 19.
Western Blotting
Sertoli cells were homogenized by sonication in an ice-cold lysis buffer containing protease inhibitors (Roche, Indianapolis, Indiana). The protein concentrations were measured using the Bradford method (Bio-Rad laboratories, Hercules, California). An equal quantity of proteins was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. The membranes were then blocked with 3% nonfat milk and incubated overnight with rabbit anti-ESR1 (H184, 1:500), rabbit anti-ESR2 antibody (h-150, 1:500; Santa Cruz Biotech, California), and mouse anti-ESR1 (1:500; Abcam, Cambridge, Massachusetts), respectively, and detected by ECL (Amersham, Piscataway, New Jersey). Peroxidase-conjugated antirabbit or anti-mouse immunologublin G (IgG, 1:2000; Vector Laboratories, Burlingame, California) was used as the secondary antibody. Immunoblotting signals were detected using the Amersham ECL plus Western blotting detection system (GE Healthcare Biosciences, Pittsburgh, Pennsylvania). All the membranes were reprobed with anti-β actin or β-tubulin antibody (Sigma), which served as a loading control. The intensity of specific bands was scanned using the TotalLab (Nonlinear USA Inc) image analysis software. The results were presented as the ratio of target protein to β-actin or β-tubulin.
Immunofluorescence
The coverslips of cultured cells and frozen sections of the testes were fixed in freshly prepared 2% paraformaldehyde and permeabilized with 0.01% saponin. The coverslips and frozen sections were then incubated with goat antivimentin (1:150; Sigma) and rabbit anti-ESR1 (H184, 1:50) overnight at 4°C. Subsequently, ESR1 and ESR2 expression levels were detected with fluorescein isothiocyanate-labeled donkey antirabbit IgG and vimentin was detected by Texas red-labeled donkey antigoat IgG (1:100; Jackson, West Grove, Pennsylvania). The coverslips and frozen sections were covered with a 4',6-diamidino-2-phenylindole (DAPI)-containing mounting medium (Santa Cruz Biotech) to visualize the nuclei and then were viewed and photographed with an Olympus fluorescence microscope (B & B Microscopes, LTD, Pittsburgh, Pennsylvania). Replacement of the primary antibody with normal rabbit or mouse serum was used as a procedure control.
Chromatin Immunoprecipitation
The chromatin immunoprecipitation (ChIP) assays were performed using an Imprint Ultra ChIP kit (Sigma) according to the manufacturer’s instructions. Briefly, TM4 cells were cultured with DMEM medium supplemented with 10% FBS in 500 cm2 dishes. The chromatin was cross-linked by treating the cells with 1% formaldehyde for 10 minutes at room temperature. The cells were harvested and homogenized with a glass Dounce homogenizer (Fisher Scientific, Pittsburgh, Pennsylvania). The cross-linked chromatin was fragmented by sonication (3 × 10 minutes) with a sonic dismembrator model 150 (Fisher Scientific, Pittsburgh, Pennsylvania). The ESR1 (H-184) and ESR2 (H-150) antibodies (Santa Cruz Biotech) at a 1:50 dilution were used for immunoprecipitation, respectively. The DNA was then purified and used for PCR with the primer sets listed in Table 1 and the conditions as follows: 40 cycles of denaturation for 30 seconds at 95°C, annealing for 30 seconds at 60°C, and extension for 1 minute at 72°C.
Overexpression of Esr1 in MSC-1 Cells
MSC-1, the cell line expressing low amount of endogenous Esr1, was selected for overexpression of a human ESR1 cDNA. To generate stable ESR1 expression clones, the cells were cultured to approximately 60% confluence in a 6-well plate and were transfected with 2 µg of either pcDNA3-ESR1 or pcDNA3 vector (a control) using Lipofectamine 2000 (Invitrogen). The cells were then cultured in a medium containing DMEM, 10% FBS, and 600 µg/mL G418 (Cellgro Molecular Genetics, Manassas, Virginia) for 14 days. The G418-resistant clones were selected and subcultured for subsequent RT-PCR and Western blot analyses. The overexpression of ESR1 protein levels in these clones was determined by Western blot.
Knockdown of Esr1 in TM4 Cells
TM4, the cell line expressing high levels of endogenous Esr1, was selected for Esr1 small interfering RNA (siRNA) transfection using a procedure as described previously.43 The cells were used for transfection once they reached approximately 50% confluence. The Esr1 siRNA, purchased from Santa Cruz Biotech, consisted of a pool of 3 to 5 target-specific siRNAs against mouse Esr1 (Gene ID: 13982). Typically, a mixture of 0.25 μg Esr1 siRNA, 100 μL DMEM, and 4 μL Lipofectamine 2000 (Invitrogen) was added to the cells in a 12-well plate and incubated overnight. The transfection medium was replaced with 0.5 mL DMEM containing 10% FBS for an additional 24 hours and these cells were used for subsequent experiments. A control siRNA (Santa Cruz Biotech) consisting of a scrambled RNA sequence was transfected in parallel with Esr1 siRNA as a specificity control for the Esr1 knockdown. The RT-PCR and Western blotting were used to determine the effectiveness of Esr1 siRNA in suppressing Esr1 messenger RNA (mRNA) and protein levels in TM4 cells.
Sertoli Cell-Specific Esr1 Ablation in Mice
Floxed Esr1 male mice (provided by Drs Andree Krust and Pierre Chambon, Institute for Genetics and Cellular and Molecular Biology, Strasbourg, France)44 were mated with female Amh-Cre mice (purchased from Jackson Laboratories, Bar Harbor, Maine), where the Cre recombinase is specifically expressed in Sertoli cells under the control of the anti-Müllerian hormone promoter45 to obtain biogenic heterozygous females (ie, Amh-Cre+/Esr1flox/wt). These heterozygous females were bred with male Esr1flox/flox to generate Sertoli cell-specific Esr1 mutant mice (Amh-Cre+/Esr1flox/flox). Tail biopsies were obtained and used for genotyping as described previously.43 The primer sets listed in Table 1 were used to determine the deletion of the floxed Esr1 allele by PCR as described previously.46 The efficiency of the excised floxed Esr1 allele in the Sertoli cells was evaluated by performing semiquantitative RT-PCR with PSCs isolated from the testes of Amh-Cre+/Esr1flox/flox and Amh-Cre−/Esr1flox/flox mice.
Statistical Analysis
The data presented are the mean ± standard error of the mean. All results were analyzed by 1-way analysis of variance and the Tukey multiple comparison posttest using the version 3.06 Instat program (Graphpad Software, San Diego, California). A P value <.05 was considered as statistically significant.
Results
Mouse Sertoli cell Lines and PSCs Express Esr1 and Esr2
Reverse transcription PCR detected the expression of Esr1 and Esr2 mRNA in 3 established mouse Sertoli cell lines, TM4, 15P-1, and MSC-1 as well as PSCs isolated from 30-day-old mice (Figure 2A). A mouse Leydig cell line MA-10 was used as a positive control. No PCR-amplified products were observed when reverse transcriptase was omitted (data not shown).
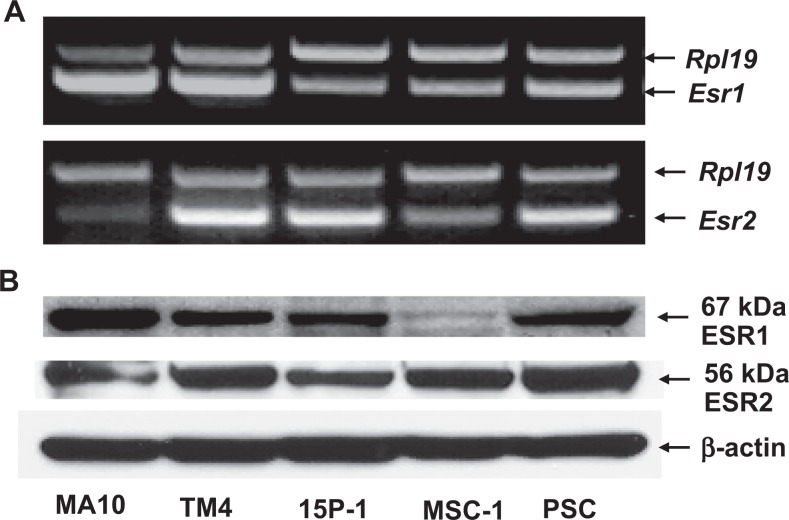
Figure 2.
The RT-PCR (A) and Western blot (B) analyses demonstrate that similar to mouse Leydig cells (MA10), all Sertoli cell lines (TM4, 15P-1, and MAC-1) and primary Sertoli cells (PSC) express both Esr1 and Esr2. MA10 serves as a positive control cell line. Ribosomal protein large subunit 19 (Rpl19) and β-actin serve as internal controls for RT-PCR and Western blotting, respectively. ESR indicates estrogen receptor; RT-PCR, reverse transcription-polymerase chain reaction.
Comparable with MA-10 cells, Western blot analysis revealed the presence of 67-kDa ESR1 and 56-kDa ESR2 proteins in all 3 Sertoli lines and PSCs (Figure 2B). Both monoclonal and polyclonal antibodies to ESR1 demonstrated the same results (data not shown). These proteins were absent when the primary antibodies were replaced by normal mouse or rabbit serum (data not shown). In agreement with the RT-PCR results, relatively high levels of ESR1 expression were observed in the TM4, 15P-1 cell lines, and PSCs but not in the MSC-1 cell line.
Immunofluorescent staining of the cultured Sertoli cells showed that the ESR1 protein was primarily localized in the nuclei of all 3 Sertoli cells lines and PSCs (Figure 1). To a lesser extent, ESR1 immunofluorescence was also observed in the membrane and cytoplasm of these cells (Figure 1). In the isolated PSCs (G) and the testicular sections (H), we performed double-labeling immunofluorescent staining for ESR1 (E, green) and a somatic cell marker vimentin (F, red) to differentiate ESR1-positive Sertoli cells from germ cells. Estrogen receptor 1 was localized to the nuclei of vimentin-positive Sertoli cells in frozen sections of the adult testes and isolated PSC (Figure 1H). Both monoclonal and polyclonal ESR1 antibodies showed the same immunostaining pattern. No specific immunofluorescence was detected when ESR1 antibodies were omitted (data not shown).
The Expression of Greb1 in Mouse Sertoli Cell Lines and PSCs
Estrogen-dependent modulation of target gene transcription is well established as the classic genomic function of ESR1 and ESR2. Reverse transcription PCR showed that all 3 Sertoli cell lines, PSCs, and MA-10 expressed mRNA for a well-characterized estrogen responsive gene Greb1 (Figure 3), which suggests that Greb1 can be used as an endogenous reporter gene to investigate ESR1-mediated gene expression in mouse Sertoli cells.
Figure 3.
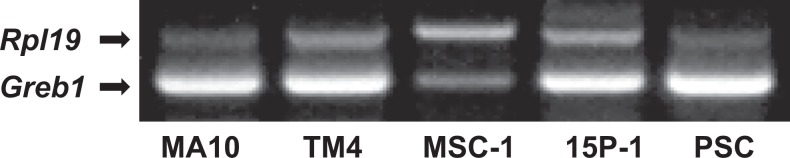
The RT-PCR analysis demonstrates that all Sertoli cell lines and primary Sertoli cells express Greb1 (A). Ribosomal protein large subunit 19 (Rpl19) serves as an internal control for RT-PCR. RT-PCR indicates reverse transcription-polymerase chain reaction.
Activation of ESR1 Induces Greb1 Expression in Mouse Sertoli Cells
To determine whether Esr1 expressed in mouse Sertoli cells is functional, we first challenged mouse TM4 Sertoli cells with an ESR ligand and selective synthetic agonists or antagonists and then measured the Greb1 mRNA levels. The expression of Greb1 mRNA was markedly increased in TM4 cells when the cells were incubated with 17β-estradiol (E2) or a synthetic selective ESR1 agonist PPT (Figure 4). However, a synthetic selective ESR2 agonist, DPN, had no effect on Greb1 mRNA levels.
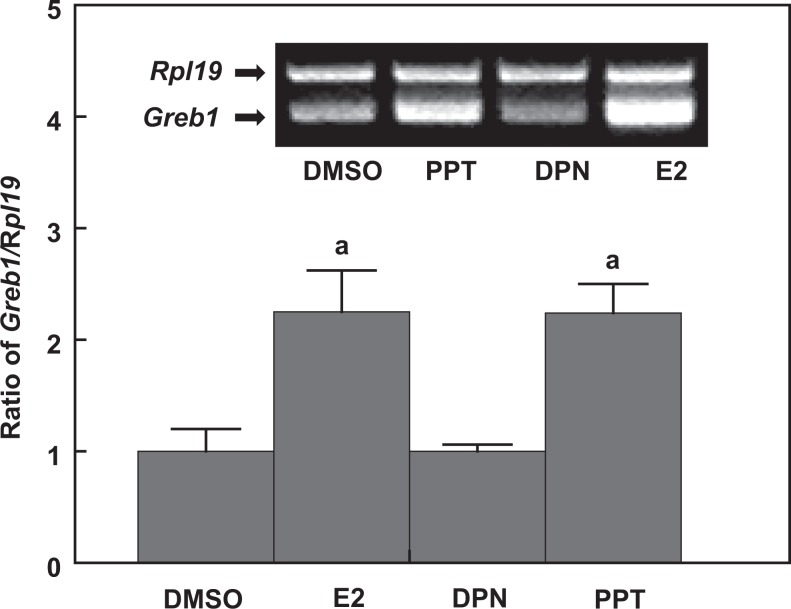
Figure 4.
The RT-PCR analysis shows that 17β-estradiol (E2) and PPT (selective ESR1 agonist) but not DPN (selective ESR2 agonist) induce Greb1 expression in TM4 cells. Ribosomal protein large subunit 19 (Rpl19) serves as an internal control for RT-PCR. Data are presented as mean ± standard error of the mean (SEM; n = 3). a P < .01 compared to the vehicle dimethyl sulfoxide (DMSO). ESR indicates estrogen receptor; RT-PCR, reverse transcription-polymerase chain reaction.
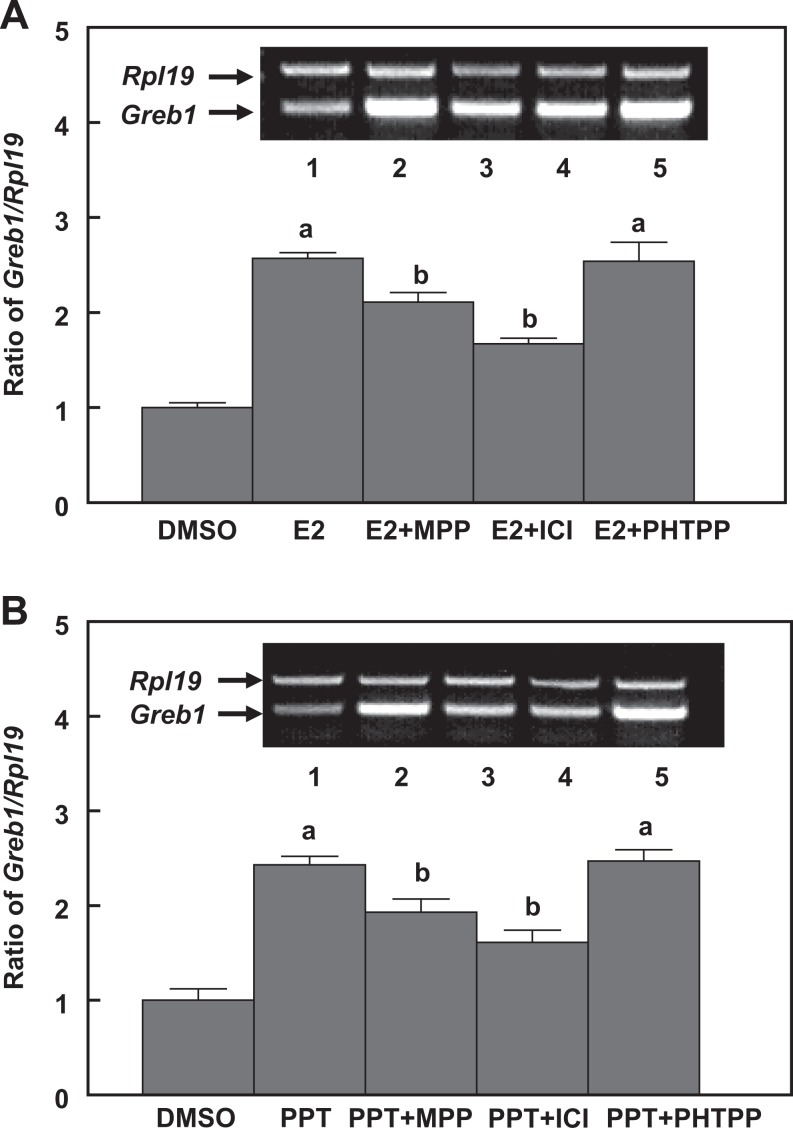
When TM4 cells were treated with E2 and PPT (a selective ESR1 agonist), Greb1 expression was increased in a dose- and time-dependent manner (Figure 5). However, when the cells coincubated with either ICI 182,780 (a nonselective ESR inhibitor) or MPP- (a selective ESR1 antagonist), E2-, and PPT-induced Greb1 expression was significantly attenuated. Meanwhile, a selective ERS2 antagonist PHTPP did not affect Greb1 mRNA levels (Figure 6). Together, these results suggest that regulation of Greb1 expression in mouse Sertoli TM4 cells is mediated by ESR1.
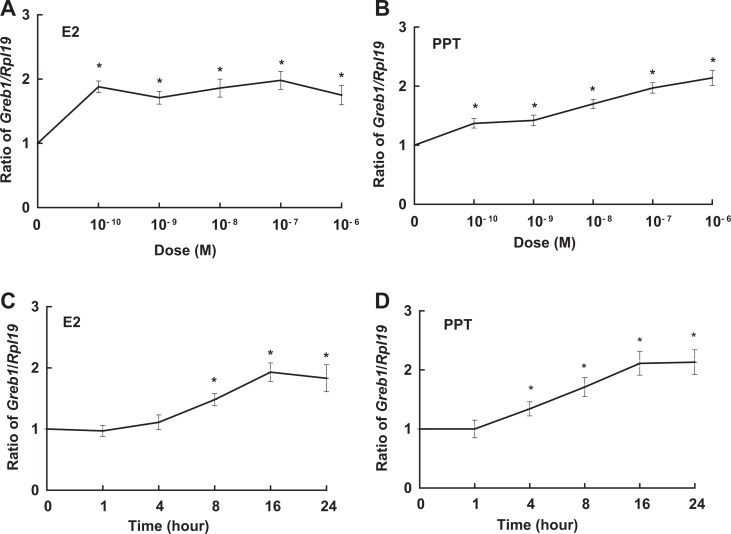
Figure 5.
Dose-dependent (A and B) and time-dependent (C and D) induction of Greb1 expression by 17β-estradiol (E2, A and C) and selective estrogen receptor 1 (ESR1) agonist PPT (B and D) in TM4 cells. Ribosomal protein large subunit 19 (Rpl19) serves as an internal control for reverse transcription-polymerase chain reaction (RT-PCR). Data are presented as mean ± standard error of the mean (SEM; n = 3). *P < .05 compared to dimethyl sulfoxide (DMSO) or time 0.
Figure 6.
Induction of Greb1 expression by 17β-estradiol (A) and the selective ESR1 agonist PPT in TM4 cells is attenuated by coincubation with either the selective ESR1 antagonist MPP or an ESR inhibitor ICI 182,780 but not selective ESR2 antagonist PHTPP. Ribosomal protein large subunit 19 (Rpl19) serves as an internal control for reverse transcription-polymerase chain reaction (RT-PCR). Data are presented as mean ± standard error of the mean (SEM; n = 3). a P < .01 compared to dimethyl sulfoxide (DMSO) and b P < .05 compared to PPT. ESR indicates estrogen receptor.
ESR1-Dependent Modulation of Greb1 Expression in Mouse Sertoli Cells
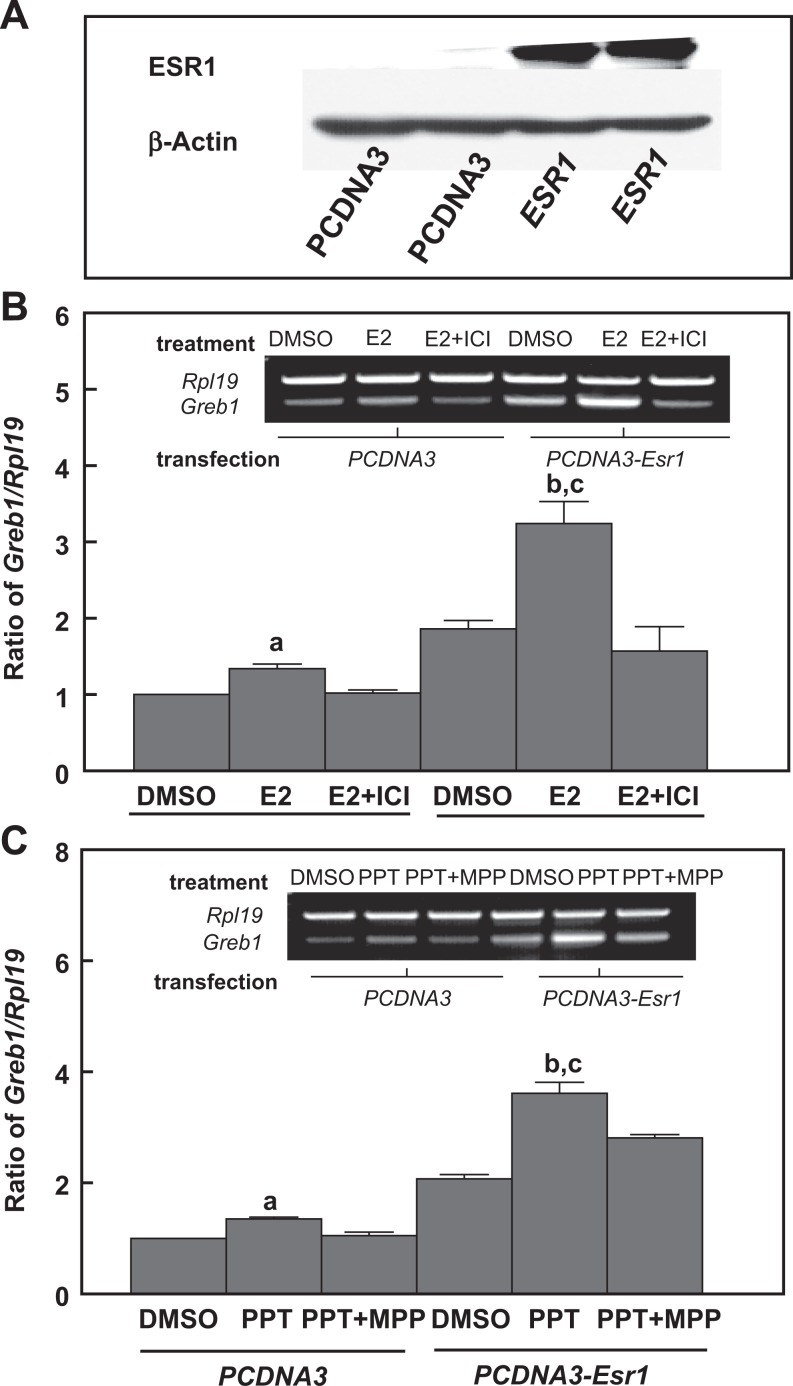
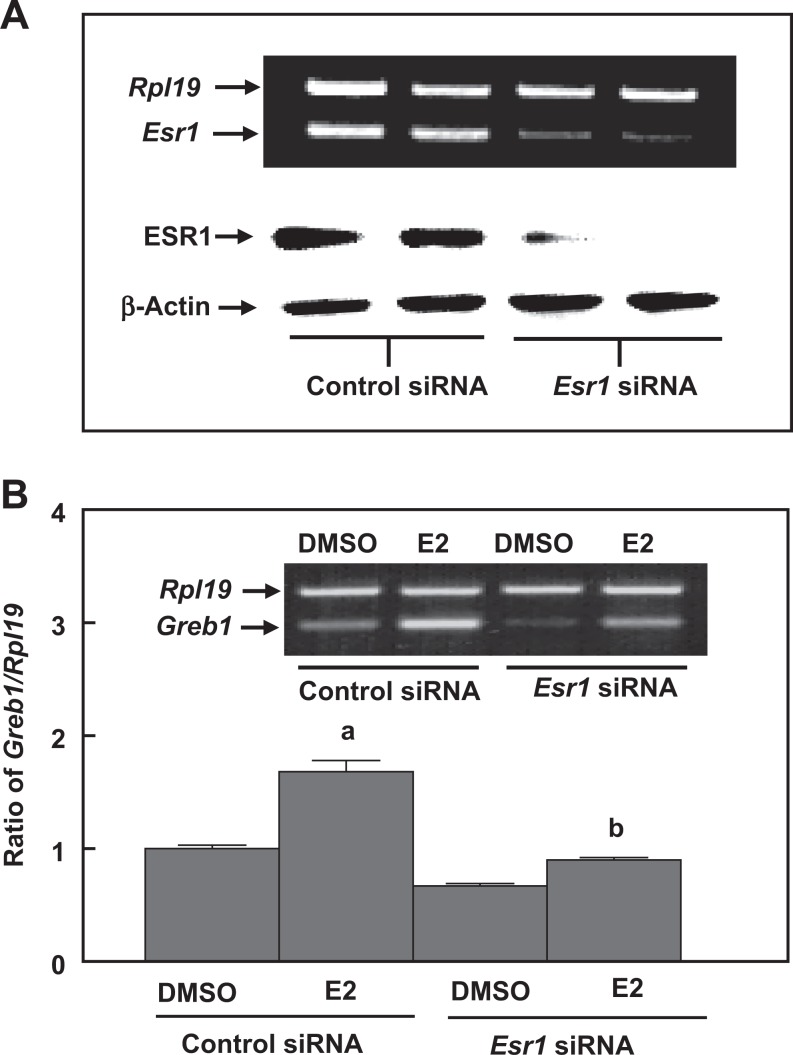
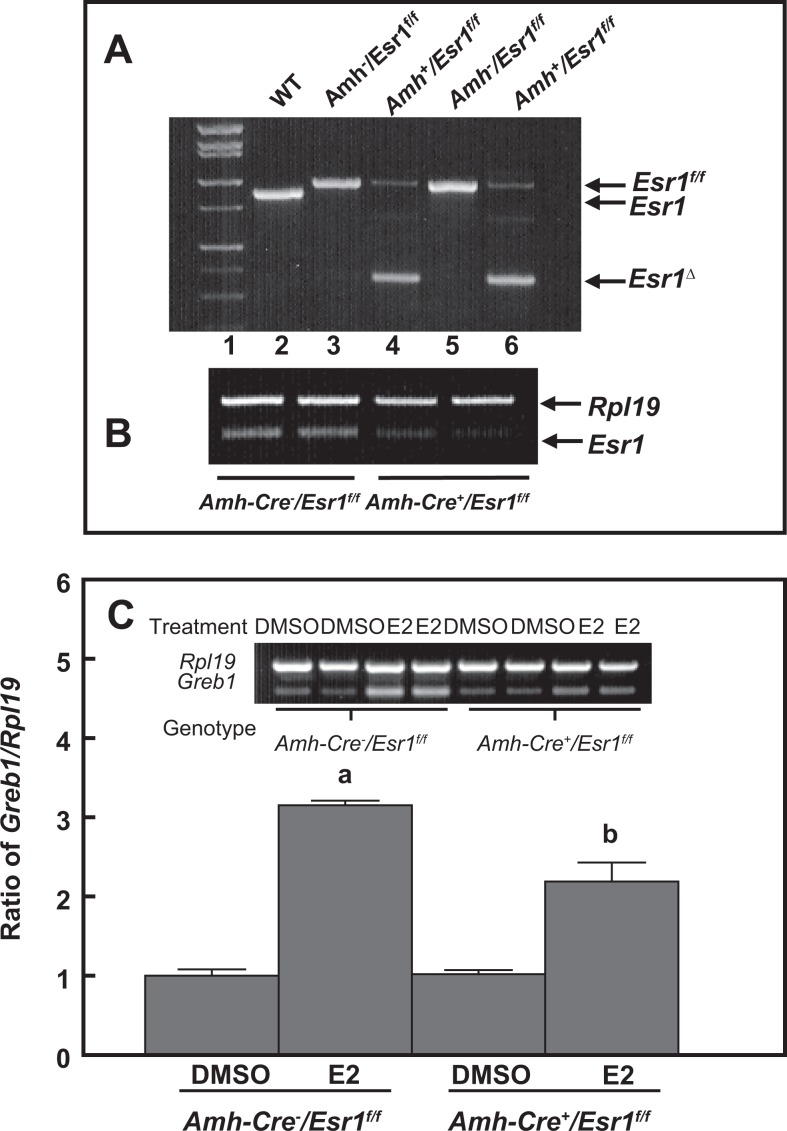
To characterize ESR1-dependent transactivation of Greb1 expression in mouse Sertoli cells, we used genomic approaches and performed both gain-of-function and loss-of-function experiments. Overexpression of human ESR1 in MSC-1, a mouse Sertoli cell line with a relatively low endogenous Esr1 expression (Figure 7A), dramatically increased the responses to E2- or PPT-induced Greb1 expression. This increase was curtailed by coincubation with either ICI 182,780 or MPP ESR inhibitors (Figure 7B and C). When Esr1 was knocked down by siRNA in a mouse Sertoli cell line with a relatively high expression of endogenous Esr1 TM4 cells, E2-induced Greb1 expression was significantly attenuated (Figure 8). Moreover, ESR1-dependent modulation of Greb1 expression was further examined in primary cultured mouse Sertoli cells. The Sertoli cells were isolated from the Amh-Cre+/Esr1fl/fl bigeneric mice, in which the exon 3 of the mouse Esr1 in Sertoli cells was selectively deleted by Amh-Cre (Figure 9A). The RT-PCR revealed that the mRNA levels of Esr1 in purified Sertoli cells from these mice were markedly reduced (Figure 9B). These purified Esr1-deficient Sertoli cells exhibited a significant decrease in E2-induced Greb1 expression (Figure 9C).
Figure 7.
Western blot demonstrating overexpression of ESR1 protein levels in MSC-1 cells (A). Overexpression of Esr1 in MSC-1 cells enhances 17β-estradiol (B) and the ESR1 agonist PPT-induced Greb1 expression, which can be counteracted by coincubation with an ESR inhibitor ICI 182,780 (B) or the selective ESR1 antagonist MPP (C), respectively. The empty vector pcDNA3 is transfected in parallel with Esr1 expression vector as a negative control for overexpression of ESR1. Ribosomal protein large subunit 19 (Rpl19) and β-actin serve as internal controls for reverse transcription-polymerase chain reaction (RT-PCR) and Western blotting, respectively. Data are presented as mean ± standard error of the mean (SEM; n = 3). a P < .05 and b P < .01 compared to dimethyl sulfoxide (DMSO), c P < .01 compared to E2 (B) and PPT (C) in PCDNA3. ESR indicates estrogen receptor.
Figure 8.
The RT-PCR and Western blot analyses show that Esr1 siRNA transfection markedly reduces Esr1 messenger RNA (mRNA) and protein levels in TM4 cells (A). Knockdown of Esr1 in TM4 cells abolishes 17β-estradiol (E2)-induced Greb1 expression (B). A scrambled RNA sequence (control siRNA) is transfected in parallel with the Esr1 siRNA as a specificity control for Esr1 knockdown. Ribosomal protein large subunit 19 (Rpl19) and β-actin serve as internal controls for RT-PCR and Western blotting, respectively. Data are presented as mean ± standard error of the mean (SEM; n = 3).a P < .01 compared to dimethyl sulfoxide (DMSO) and b P < .01 compared to E2 in control siRNA. ESR indicates estrogen receptor; RT-PCR, reverse transcription-polymerase chain reaction; siRNA, small interfering RNA.
Figure 9.
Panel (A) is a representative genotyping picture, which demonstrates that a deleted form of Esr1 (Esr1Δ) is only detected in Amh-Cre+/Esr1flox/flox mice (Lines 4 and 6) but not in Amh-Cre−/Esr1flox/flox mice (Lines 3 and 5). Panel (B) shows a dramatic decrease in Esr1 messenger RNA (mRNA) levels in primary Sertoli cells isolated from the Amh-Cre+/Esr1flox/flox testes. Panel (C) demonstrates that Sertoli cell-specific ablation of Esr1 significantly curtails 17β-estradiol-induced Greb1 expression. Ribosomal protein large subunit 19 (Rpl19) serves as an internal control for reverse transcription-polymerase chain reaction (RT-PCR). Data are presented as means ± standard error of the mean (SEM; n = 3). a P < .01 and b P < .05 compared to dimethyl sulfoxide (DMSO).
Estrogen Receptor 1 Binds to EREs in the Promoter Region of the Greb1 Gene
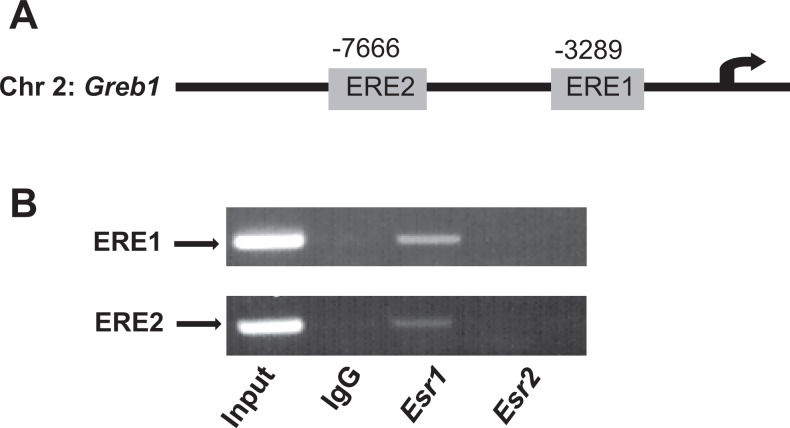
We finally determined whether the ESR1 or ESR2 physically interact with the promoter of Greb1 gene in TM4 cells. Two previously identified canonical EREs (ERE1 and ERE2) were present in the Greb1 gene at −3289 and −7666 bp upstream of its transcription start site (Figure 10A).47,48 The ChIP analysis showed that only ESR1 but not ESR2 bound to both the ERE1 and the ERE2 (Figure 10B). The physical interaction between ESR1 and EREs in the Greb1 promoter suggests that ESR1 contributes to the upregulation of Greb1 expression in TM4 cells through a genomic action.
Figure 10.
A schematic drawing illustrating the relative positions of 2 consensus estrogen responsive elements (ERE1 and ERE2) in the mouse Greb1 upstream regulatory regions (A). Chromatin immunoprecipitation (ChIP) assays reveal that ESR1 but not ESR2 binding to both EREs in the Greb1 promoter in mouse Sertoli cell line TM4 (B). Genomic DNA input and rabbit immunologublin G (IgG) serve as a positive and a negative control, respectively. ESR indicates estrogen receptor; ERE, estrogen response element.
Discussion
Increasing lines of evidence now demonstrated that the testis is a site of estrogen production and is also a target of estrogen action.2–4,15,16,26 Further evidence from the studies on Esr knockout mice suggests that ESR1 but not ESR2 expressed in the gonadal somatic cells, but not germ cells, plays a critical role in spermatogenesis.20,26 However, whether Sertoli cells express Esr1 is still controversial because although several studies showed its presence in these cells, other studies did not report any such expression.7,34 In the present study, we utilized multiple approaches to determine whether Esr1 is expressed in mouse Sertoli cells. The data demonstrate that mouse Sertoli cell lines and PSC express Esr1, and the protein is localized in nuclei, membrane, and cytoplasm. Our findings are consistent with several recent reports that Sertoli cells of the hystricognath rodent, rat, cat, boar, pig, and human express Esr1.31–40 Although it appears difficult to reconcile, the possibilities of these conflicting results in the literature with regard to the presence of ESR1 in Sertoli cells may be due to the differences in the origin, condition, and age of the samples examined and the techniques and reagents used. In rat and human testes, the controversial results are most likely attributed to the various tissue preservations and antibodies used as well as to the origin of the tissues, which have been extensively discussed in recent studies.35,38 The expression of Esr1 in Sertoli cells is affected by various conditions including developmental stage, hormonal milieu, and even cell density in culture. For instance, the expression of Esr1 in rat Sertoli cells is downregulated by estrogen49 whereas ESR1 in pig Sertoli cells generally decreases with age.32 Additionally, the expression levels of ESRs in mouse and rat Sertoli cells highly depend on cell density in vitro.50 If the cell or tissue preservation methods are not optimal or the techniques and reagents used for detection are not sensitive enough, it is perceivable that low expression levels of Esr1 mRNA or its protein in Sertoli cells may be difficult to detect.
The MSC-1 cell line, derived from a Sertoli cell tumor in the transgenic C57BL/6 X SJL-mixed hybrid adult mice, carries a transgene containing DNA encoding both small and large T antigen of the SV40 virus fused to the promoter for human Müllerian inhibiting substance. Several putative Sertoli cell marker genes are known to be expressed in this cell line, such as transferrin, clusterin, and inhibin βB.51,52 The 15P-1 cell line, established from testicular cells of transgenic adult mice, expresses the large T protein of polyoma virus (PyLT) in the seminiferous epithelium. This cell line is derived from a Sertoli cell origin by its expression of Wilm tumor and kit ligand.53 The TM4 cell line is the first and the most well-characterized mouse Sertoli cell line which were generated by Mather from primary cultures of Sertoli cells isolated from 11- to 13-day-old mice.54 This cell line possesses the most Sertoli cell characteristics, such as transferrin secretion, androgen receptor expression, response to FSH stimulation, and exhibition of phagocytotic activity.54–57 Our results demonstrate the presence of Esr1 mRNA and protein in all 3 cell lines. Although these immortalized Sertoli cell lines provide a pure population of cells for the study, they do not phenocopy all the properties of native Sertoli cells. For example, MSC-1 cells lack some of the immune privilege functions,58 and TM4 cells do not secrete androgen-binding protein associated with primary Sertoli cells.59 Therefore, we also include primary mouse Sertoli cells and testicular in situ immunolocalization of ESR1 in this study. These cell lines, freshly isolated primary Sertoli cells, and immunostained testicular tissues all demonstrate similar results, suggesting the presence of ESR1 in mouse Sertoli cells is unlikely to be an artifact.
To provide further evidence for the presence of ESR1 in mouse Sertoli cells, we examined its interaction with the promoter of Greb1, a known estrogen responsive gene. Greb1 was initially identified as an estrogen target gene in human breast tumor cells through a genome-wide screen.47,60 There are 3 consensus EREs consisting of a 15-bp palindromic sequence presence in the 5′ flanking region of the Greb1 gene, which are conserved in human and mouse.47,48 Our results show that all 3 mouse Sertoli cell lines and mouse PSC express Greb1 and its expression is estrogen inducible. More importantly, ChIP analysis indicated that ESR1 but not ESR2 binds to 2 proximal EREs in the mouse Greb1 promoter region, which is in consonance with previous studies.47,48,60 Therefore, Greb1 was chosen as the endogenous reporter gene for functional analysis of ESR1 in mediating estrogen action to regulate target gene expression in mouse Sertoli cells. Pharmacological and genetic approaches were used in this study to investigate the functional significance of ESR1 in mouse Sertoli cells. The Greb1 expression was induced in TM4 cells via ESR1 but not ESR2. The complementary genetic gain-of-function and loss-of-function experiments further confirmed the importance of ESR1 in the transactivation of Greb1 expression in TM4 Sertoli cells. Furthermore, primary Sertoli cells isolated from a Sertoli cell-specific knockout mouse line also displayed a significant attenuation of estrogen-induced Greb1 expression although Esr1 mRNA levels in these Sertoli cells are noticeably reduced but not completely deleted. Taken together, these data obtained using multiple approaches in the present study clearly establish a classic genomic function of ESR1 in mouse Sertoli cells.
Several mechanisms of ESR-mediated estrogen actions are well established.22–25 A rapid estrogen-signaling cascade mediated by nonclassical membrane-associated ESRs in rat Sertoli cells has been recently characterized.31,34,35 However, these studies do not differentiate whether the estrogen action is mediated by ERS1 or ESR2. Although our study focuses on the genomic action of ESR1 in mouse Sertoli cells, we do not exclude that a nonclassical membrane-associated ESR1 transduces rapid estrogen signals in mouse Sertoli cells. In fact, immunostaining of ESR1 in the membrane and cytoplasm in mouse Sertoli cell lines and PSCs was observed, suggesting the likelihood of existing membrane-associated ESR1 in these cells. Although a role of Sertoli cell ESR1 in the regulation of a target gene’s expression has been demonstrated in this study, it remains to be established how ESR1-mediated estrogen signaling affects Sertoli cell functions that ultimately influence the spermatogenic processes.
In summary, the results of the present work indicate that Sertoli cell lines derived from either immature or adult mice, as well as primary Sertoli cells isolated from peripubertal mice, express both Esr1 and Esr2. Importantly, ESR1 functions as a nuclear transcription activator to mediate the classic genomic action of estrogen to regulate Greb1 expression in mouse Sertoli cells. This study provides essential information for our understanding of ESR1-mediated estrogen actions in the regulation of Sertoli cell functions, which may consequently influence germ cell development and maturation in the seminiferous tubules.
Acknowledgments
We are grateful to Drs Andree Krust and Pierre Chambon (Institute for Genetics and Cellular and Molecular Biology, Strasbourg, France) for providing floxed Esr1 mice, Dr Griswold, MD (Washington State University, Pullman, Washington) for providing mouse Sertoli cell line MSC-1, and Dr M. Ascoli (The University of Iowa, Iowa City, Iowa) for providing mouse Leydig cell line MA10.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: This work was supported by Grant R01-HD057501 from the National Institutes.
References
- 1. Sharpe RM. Do males rely on female hormones? Nature. 1997;390(6659):447–448 [DOI] [PubMed] [Google Scholar]
- 2. Couse JF, Korach KS. Reproductive phenotypes in the estrogen receptor-alpha knockout mouse. Ann Endocrinol (Paris). 1999;60(2):143–148 [PubMed] [Google Scholar]
- 3. Carreau S, de Vienne C, Galeraud-Denis I. Aromatase and estrogens in man reproduction: a review and latest advances. Adv Med Sci. 2008;53(2):139–144 [DOI] [PubMed] [Google Scholar]
- 4. Akingbemi BT. Estrogen regulation of testicular function. Reprod Biol Endocrinol. 2005;3:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rochira V, Granata AR, Madeo B, Zirilli L, Rossi G, Carani C. Estrogens in males: what have we learned in the last 10 years? Asian J Androl. 2005;7(1):3–20 [DOI] [PubMed] [Google Scholar]
- 6. D'Souza R, Pathak S, Upadhyay R, et al. Disruption of tubulobulbar complex by high intratesticular estrogens leading to failed spermiation. Endocrinology. 2009;150(4):1861–1869 [DOI] [PubMed] [Google Scholar]
- 7. Carreau S, Hess RA. Oestrogens and spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365(1546):1517–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carreau S, Bois C, Zanatta L, Silva FR, Bouraima-Lelong H, Delalande C. Estrogen signaling in testicular cells. Life Sci. 2011;89(15-16):584–587 [DOI] [PubMed] [Google Scholar]
- 9. Upadhyay RD, Kumar AV, Sonawane S, Gaonkar R, Balasinor NH. Estrogen effects on actin cytoskeletal and endocytic proteins associated with tubulobulbar complex disruption in rat testes. Reprod Sci. 2013;20(10):1162–1174 [DOI] [PubMed] [Google Scholar]
- 10. Robertson KM, O'Donnell L, Jones ME, et al. Impairment of spermatogenesis in mice lacking a functional aromatase (cyp 19) gene. Proc Natl Acad Sci USA. 1999;96(14):7986–7991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robertson KM, Simpson ER, Lacham-Kaplan O, Jones ME. Characterization of the fertility of male aromatase knockout mice. J Androl. 2001;22(5):825–830 [PubMed] [Google Scholar]
- 12. Kaushik MC, Misro MM, Sehgal N, Nandan D. Effect of chronic oestrogen administration on androgen receptor expression in reproductive organs and pituitary of adult male rat. Andrologia. 2010;42(3):193–205 [DOI] [PubMed] [Google Scholar]
- 13. Toyama Y, Hosoi I, Ichikawa S, et al. Beta-estradiol 3-benzoate affects spermatogenesis in the adult mouse. Mol Cell Endocrinol. 2001;178(1-2):161–168 [DOI] [PubMed] [Google Scholar]
- 14. Balasinor NH, D'Souza R, Nanaware P, et al. Effect of high intratesticular estrogen on global gene expression and testicular cell number in rats. Reprod Biol Endocrinol. 2010;8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carreau S, Bouraima-Lelong H, Delalande C. Estrogens in male germ cells. Spermatogenesis. 2011;1(2):90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Q, Bai Q, Yuan Y, Liu P, Qiao J. Assessment of seminal estradiol and testosterone levels as predictors of human spermatogenesis. J Androl. 2010;31(2):215–220 [DOI] [PubMed] [Google Scholar]
- 17. O'Donnell L, Robertson KM, Jones ME, Simpson ER. Estrogen and spermatogenesis. Endocr Rev. 2001;22(3):289–318 [DOI] [PubMed] [Google Scholar]
- 18. Allan CM, Couse JF, Simanainen U, et al. Estradiol induction of spermatogenesis is mediated via an estrogen receptor-{alpha} mechanism involving neuroendocrine activation of follicle-stimulating hormone secretion. Endocrinology. 2010;151(6):2800–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Couse JE, Mahato D, Eddy EM, Korach KS. Molecular mechanism of estrogen action in the male: insights from the estrogen receptor null mice. Reproduction, fertility, and development. 2001;13(4):211–219 [DOI] [PubMed] [Google Scholar]
- 20. Couse JF, Hewitt SC, Bunch DO, et al. Postnatal sex reversal of the ovaries in mice lacking estrogen receptors alpha and beta. Science. 1999;286(5448):2328–2331 [DOI] [PubMed] [Google Scholar]
- 21. Couse JF, Korach KS. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20(3):358–417 [DOI] [PubMed] [Google Scholar]
- 22. Gruber CJ, Gruber DM, Gruber IM, Wieser F, Huber JC. Anatomy of the estrogen response element. Trends Endocrinol Metab. 2004;15(2):73–78 [DOI] [PubMed] [Google Scholar]
- 23. Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19(4):833–842 [DOI] [PubMed] [Google Scholar]
- 24. Levin ER. Plasma membrane estrogen receptors. Trends Endocrinol Metab. 2009;20(10):477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schultz-Norton JR, Ziegler YS, Nardulli AM. ERalpha-associated protein networks. Trends Endocrinol Metab. 2011;22(4):124–129 [DOI] [PubMed] [Google Scholar]
- 26. Walker VR, Korach KS. Estrogen receptor knockout mice as a model for endocrine research. ILAR journal/national research council, institute of laboratory animal resources. 2004;45(4):455–461 [DOI] [PubMed] [Google Scholar]
- 27. Antal MC, Krust A, Chambon P, Mark M. Sterility and absence of histopathological defects in nonreproductive organs of a mouse ERbeta-null mutant. Proc Natl Acad Sci USA. 2008;105(7):2433–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akingbemi BT, Ge R, Rosenfeld CS, et al. Estrogen receptor-alpha gene deficiency enhances androgen biosynthesis in the mouse Leydig cell. Endocrinology. 2003;144(1):84–93 [DOI] [PubMed] [Google Scholar]
- 29. Gould ML, Hurst PR, Nicholson HD. The effects of oestrogen receptors alpha and beta on testicular cell number and steroidogenesis in mice. Reproduction. 2007;134(2):271–279 [DOI] [PubMed] [Google Scholar]
- 30. Mahato D, Goulding EH, Korach KS, Eddy EM. Spermatogenic cells do not require estrogen receptor-alpha for development or function. Endocrinology. 2000;141(3):1273–1276 [DOI] [PubMed] [Google Scholar]
- 31. Royer C, Lucas TF, Lazari MF, Porto CS. 17Beta-estradiol signaling and regulation of proliferation and apoptosis of rat Sertoli cells. Biol Reprod. 2012;86(4):108. [DOI] [PubMed] [Google Scholar]
- 32. Ramesh R, Pearl CA, At-Taras E, Roser JF, Berger T. Ontogeny of androgen and estrogen receptor expression in porcine testis: effect of reducing testicular estrogen synthesis. Animal Reprod Sci. 2007;102(3-4):286–99 [DOI] [PubMed] [Google Scholar]
- 33. Cavaco JE, Laurentino SS, Barros A, Sousa M, Socorro S. Estrogen receptors alpha and beta in human testis: both isoforms are expressed. Syst Biol Reprod Med. 2009;55(4):137–144 [DOI] [PubMed] [Google Scholar]
- 34. Lucas TF, Pimenta MT, Pisolato R, Lazari MF, Porto CS. 17beta-estradiol signaling and regulation of Sertoli cell function. Spermatogenesis. 2011;1(4):318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lucas TF, Siu ER, Esteves CA, et al. 17Beta-estradiol induces the translocation of the estrogen receptors ESR1 and ESR2 to the cell membrane, MAPK3/1 phosphorylation and proliferation of cultured immature rat Sertoli cells. Biol Reprod. 2008;78(1):101–114 [DOI] [PubMed] [Google Scholar]
- 36. Izumi Y, Yamaguchi K, Ishikawa T, et al. Molecular changes induced by bisphenol-A in rat Sertoli cell culture. Syst Biol Reprod Med. 2011;57:228–332 [DOI] [PubMed] [Google Scholar]
- 37. Gunawan A, Kaewmala K, Uddin MJ, et al. Association study and expression analysis of porcine ESR1 as a candidate gene for boar fertility and sperm quality. Anim Reprod Sci. 2011;128(1-4):11–21 [DOI] [PubMed] [Google Scholar]
- 38. Filipiak E, Suliborska D, Laszczynska M, et al. Estrogen receptor alpha localization in the testes of men with normal spermatogenesis. Folia Histochem Cytobiol. 2012;50(3):340–345 [DOI] [PubMed] [Google Scholar]
- 39. Gonzalez CR, Muscarsel Isla ML, Leopardo NP, Willis MA, Dorfman VB, Vitullo AD. Expression of androgen receptor, estrogen receptors alpha and beta and aromatase in the fetal, perinatal, prepubertal and adult testes of the South American plains vizcacha, Lagostomus maximus (Mammalia, Rodentia). J Reprod Dev. 2012;58(6):629–635 [DOI] [PubMed] [Google Scholar]
- 40. Schoen J, Sharbati S, Ritter J, Jewgenow K. Feline gonads exhibit tissue specific alternative splicing of oestrogen receptor alpha (ESR1). Reprod Domest Anim. 2012;47(suppl 6):30–34 [DOI] [PubMed] [Google Scholar]
- 41. Boucheron C, Baxendale V. Isolation and purification of murine male germ cells. Methods Mol Biol. 2012;825:59–66 [DOI] [PubMed] [Google Scholar]
- 42. Lei ZM, Mishra S, Zou W, et al. Targeted disruption of luteinizing hormone/human chorionic gonadotropin receptor gene. Mol Endocrinol. 2001;15(1):184–200 [DOI] [PubMed] [Google Scholar]
- 43. Yuan FP, Lin DX, Rao CV, Lei ZM. Cryptorchidism in LhrKO animals and the effect of testosterone-replacement therapy. Hum Reprod. 2006;21(4):936–942 [DOI] [PubMed] [Google Scholar]
- 44. Dupont S, Krust A, Gansmuller A, Dierich A, Chambon P, Mark M. Effect of single and compound knockouts of estrogen receptors alpha (ERalpha) and beta (ERbeta) on mouse reproductive phenotypes. Development. 2000;127(19):4277–4291 [DOI] [PubMed] [Google Scholar]
- 45. Holdcraft RW, Braun RE. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development. 2004;131(2):459–467 [DOI] [PubMed] [Google Scholar]
- 46. Gieske MC, Kim HJ, Legan SJ, et al. Pituitary gonadotroph estrogen receptor-alpha is necessary for fertility in females. Endocrinology. 2008;149(1):20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bourdeau V, Deschenes J, Metivier R, et al. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol. 2004;18(6):1411–1427 [DOI] [PubMed] [Google Scholar]
- 48. Deschenes J, Bourdeau V, White JH, Mader S. Regulation of GREB1 transcription by estrogen receptor alpha through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J Biol Chem. 2007;282(24):17335–17339 [DOI] [PubMed] [Google Scholar]
- 49. Lucas TF, Royer C, Siu ER, Lazari MF, Porto CS. Expression and signaling of G protein-coupled estrogen receptor 1 (GPER) in rat Sertoli cells. Biol Reprod. 2010;83(2):307–317 [DOI] [PubMed] [Google Scholar]
- 50. Nakhla AM, Mather JP, Janne OA, Bardin CW. Estrogen and androgen receptors in Sertoli, Leydig, myoid, and epithelial cells: effects of time in culture and cell density. Endocrinology. 1984;115(1):121–128 [DOI] [PubMed] [Google Scholar]
- 51. Peschon JJ, Behringer RR, Cate RL, et al. Directed expression of an oncogene to Sertoli cells in transgenic mice using Müllerian inhibiting substance regulatory sequences. Mol Endocrinol. 1992;6(9):1403–1411 [DOI] [PubMed] [Google Scholar]
- 52. McGuinness MP, Linder CC, Morales CR, Heckert LL, Pikus J, Griswold MD. Relationship of a mouse Sertoli cell line (MSC-1) to normal Sertoli cells. Biol Reprod. 1994;51(1):116–124 [DOI] [PubMed] [Google Scholar]
- 53. Paquis-Flucklinger V, Michiels JF, Vidal F, et al. Expression in transgenic mice of the large T antigen of polyomavirus induces Sertoli cell tumours and allows the establishment of differentiated cell lines. Oncogene. 1993;8(8):2087–2094 [PubMed] [Google Scholar]
- 54. Mather JP. Establishment and characterization of two distinct mouse testicular epithelial cell lines. Biol Reprod. 1980;23(1):243–252 [DOI] [PubMed] [Google Scholar]
- 55. Mather JP, Zhuang LZ, Perez-Infante V, Phillips DM. Culture of testicular cells in hormone-supplemented serum-free medium. Ann N Y Acad Sci. 1982;383:44–68 [DOI] [PubMed] [Google Scholar]
- 56. Tokuda N, Mano T, Levy RB. Phagocytosis by the murine testicular TM4 Sertoli cell line in culture. J Urol. 1992;147(1):278–282 [DOI] [PubMed] [Google Scholar]
- 57. Jabado N, Canonne-Hergaux F, Gruenheid S, Picard V, Gros P. Iron transporter Nramp2/DMT-1 is associated with the membrane of phagosomes in macrophages and Sertoli cells. Blood. 2002;100(7):2617–2622 [DOI] [PubMed] [Google Scholar]
- 58. Doyle TJ, Kaur G, Putrevu SM, et al. Immunoprotective properties of primary Sertoli cells in mice: potential functional pathways that confer immune privilege. Biol Reprod. 2012;86(1):1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ducray A, Bloquel M, Hess K, Hammond GL, Gerard H, Gerard A. Establishment of a mouse Sertoli cell line producing rat androgen-binding protein (ABP). Steroids. 1998;63(5-6):285–287 [DOI] [PubMed] [Google Scholar]
- 60. Lin CY, Strom A, Vega VB, et al. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004;5(9):R66. [DOI] [PMC free article] [PubMed] [Google Scholar]