Abstract
Objectives:
The accurate distinction of leiomyoma from leiomyosarcoma is essential for patient management. However, the distinction can be difficult to make, particularly in tissue biopsy samples. Immunohistochemistry has been established as a useful technique to aid in the diagnosis of malignancies. The advantages of immunohistochemical studies are their ease of use and interpretation. This study is the first to evaluate the utility of the promyelocytic leukemia zinc finger (PLZF) protein and the histone 1.5 (H1.5) protein as potential diagnostic immunohistochemical markers for distinguishing leiomyosarcoma from leiomyoma.
Methods:
Tissue samples from 21 leiomyosarcomas and 26 leiomyomas were studied. The student t-test and the Fisher exact test were used to calculate the differences in staining between the 2 groups.
Results:
Statistically significant differences were found in the staining indices of anti-PLZF and anti-H1.5 when comparing benign and malignant tumors (P < .0001 and P < .0001, respectively). The mean H1.5 staining score in leiomyosarcomas was 158.3, compared to 28.3 in leiomyomas. The mean PLZF score in leiomyosarcomas was 1.5 in contrast to 71.5 in leiomyomas. For H1.5 at a score ≥60, the sensitivity and specificity were 90.5% and 84.6%, respectively. For PLZF, a score ≤15 had a test sensitivity and specificity of 100% and 80.8%, respectively. This suggests that staining for H1.5 or PLZF can serve as a good screening test. Additionally, combining the 2 immunostains results in a sensitivity and specificity of 90.5% and 97.5%, respectively, in differentiating between leiomyoma and leiomyosarcoma.
Conclusions:
We describe immunostaining for PLZF and H1.5 in benign and malignant uterine smooth muscle tumors. Statistically significant differences in staining patterns were found, suggesting utility in distinguishing leiomyosarcomas from leiomyomas.
Keywords: leiomyosarcoma, leiomyoma, promyelocytic leukemia zinc finger (PLZF) protein, histone H1.5 protein
Introduction
Leiomyosarcoma is a rare malignant tumor that constitutes 2% to 7% of uterine malignancies.1 The frequency of leiomyosarcoma is 0.64/100 000 women per year, with a prevalence of 0.23% to1% in patients operated on for presumed leiomyoma.2–4 Although the incidence is low, uterine leiomyosarcoma is an aggressive malignancy with a poor prognosis; whereas leiomyoma is the most common benign uterine tumor. Hence, accurate diagnosis is essential. The clinical presentation of leiomyosarcoma is similar to that of leiomyoma, with presenting signs and symptoms of vaginal bleeding, infertility, and pelvic pain.5
Currently, leiomyosarcoma and leiomyoma are distinguished histopathologically by the mitotic index, the presence or absence of tumor cell necrosis, and the degree of nuclear pleomorphism in tissue samples.5–7 However, certain variants of leiomyoma—namely cellular, atypical, mitotically active, myxoid, or epithelioid subtypes—may exhibit atypical histologic features and unusual growth patterns.7–9 Conversely, some low-grade forms of leiomyosarcoma—particularly the myxoid or epithelioid variants—can be difficult to distinguish from myxoid or epithelioid leiomyomas.7,8 Such variants make the distinction of leiomyosarcomas from benign smooth muscle tumors challenging. For these cases, the discovery of a reliable cellular or tissue marker to unambiguously distinguish leiomyosarcoma from leiomyoma would be of considerable value. A number of proteins have been found to be expressed in leiomyosarcoma including angiogenic factors, vascular endothelial growth factor (VEGF), VEGF receptors, epidermal growth factor receptor,10,11 phosphatase and tensin homolog,12 Ki-ras,13 MDM2,13 Ki-67,5,8,14 p16,5,8,15 and p53.5,8,15 However, none of these markers have been found to be clinically useful because they either stain positively in both benign and malignant smooth muscle tumors or they show immunostaining that is difficult to interpret.16
The promyelocytic leukemia zinc finger (PLZF) protein is a DNA-binding transcriptional repressor.17 Immunohistochemistry (IHC) has revealed that PLZF is highly expressed in endometrial and myometrial cells during the secretory phase, with regulation by progesterone.18 Additionally, the protein has been shown to be downregulated in hematopoietic malignancies, mesothelioma, melanoma, and lung cancer.19–24
Histone 1.5 (H1.5) is a protein responsible for chromatin structure and folding that has been shown to play a role in transcriptional regulation.25,26 Immunohistochemical staining of the protein is strong in prostate cancer and in pulmonary neuroendocrine tumors, correlating with worsening disease.21,27
This is the first study to explore the utility of immunohistochemical staining of PLZF and H1.5 in the distinction of leiomyosarcoma from leiomyoma. We expect that PLZF is underexpressed and H1.5 is overexpressed in leiomyosarcoma based on the aforementioned studies of other malignancies.
Materials and Methods
Archival slides from the Mount Sinai Hospital Department of Pathology, comprising leiomyosarcomas and leiomyomas samples, were selected based on their respective pathology reports. Age and race were recorded. Histologic diagnoses were confirmed for all samples by 1 gynecologic pathologist (TK) who reviewed all of the hematoxylin and eosin (H&E) slides and selected 1 representative section from each case. The institutional review board of the Mount Sinai School of Medicine approved the study protocol.
Immunohistochemistry
Consecutive tissue sections were cut at 5-μm thickness from representative formalin-fixed and paraffin-embedded sections and placed on positively charged slides. A set of 3 consecutive sections from each sample was subjected to IHC with anti-PLZF and anti-H1.5, as well as H&E staining. For immunostaining, slides were deparaffinized with a xylene and alcohol series. Rehydration was followed by incubation with 3% hydrogen peroxide for 10 minutes to block endogenous peroxidase activity. Slides were then processed for antigen retrieval with 10 mmol/L citrate buffer (pH 6.0) using a pressure cooker (Pascal; Dako Cytomation, Glostrup, Denmark) for 4 minutes at 125°C, followed by slow cooling. The samples were washed in phosphate-buffered saline (PBS; 137 mmol/L sodium chloride, 2.7 mmol/L potassium chloride, 4.2 mmol/L sodium phosphate, and 1.5 mmol/L potassium phosphate). They were then incubated for 2 hours at room temperature with mouse monoclonal anti-PLZF (D-9; sc-28319, Santa Cruz Biotechnology, Santa Cruz, California) at 1:1000 dilution or with rabbit monoclonal anti-H1.5 (Abcam, ab18208, Cambridge, Massachusetts) at 1:800 dilution, along with 1% bovine serum albumin and 5% normal goat serum in PBS. The reaction was amplified using the UltraVision Quanto Detection System HRP (Thermo Fisher Scientific, Waltham, Massachusetts) according to the manufacturer’s protocol. Immunoreactivity was visualized with diaminobenzidine, and slides were counterstained with hematoxylin, dehydrated, and coverslipped.
Morphologic Evaluation
The diagnoses of leiomyoma and leiomyosarcoma were established by examination of H&E-stained sections. Leiomyosarcoma was diagnosed if at least 2 of the following features were present: diffuse moderate to severe cytologic atypia, a mitotic index of more than 10 mitotic figures per ten 400× power fields, and tumor cell necrosis. Immunohistochemical staining for both PLZF and H1.5 was scored in a semiquantitative fashion incorporating both the intensity and the extensiveness of staining. A staining score was generated by multiplying the percentage of positively stained tumor cells by the degree of staining intensity. Intensity was quantified as follows: 0 (no staining); 1+ (weak but detectable above control); 2+ (moderate, distinct); and 3+ (strong). The staining score ranged from 0 to 300. Only nuclear staining was considered specific. Control myometrium slides showed minimal background staining.
Statistical Analysis
The staining differences between leiomyoma and leiomyosarcoma were evaluated by the Student t test and Fisher exact test. A P value of <.05 was considered statistically significant. Based on the final diagnosis and the staining score of each sample, a receiver–operating characteristic (ROC) curve was generated to illustrate the relation between sensitivity and specificity. Threshold values for differentiating between leiomyoma and leiomyosarcoma were determined, according to the ROC curves. These cutoff scores were used to determine sensitivity and specificity. The statistical power of each immunohistochemical marker was evaluated using a post hoc 2-tailed, 2-sample power analysis. Statistical analysis was performed using SPSS19 and STATA10 software.
Results
A total of 47 cases, 26 leiomyomas and 21 leiomyosarcomas, were evaluated. Patients with leiomyoma were younger than those with leiomyosarcoma (P < .001). Table 1 shows the characteristics of the cases.
Table 1.
Characteristics of Women With Leiomyoma Versus Leiomyosarcoma.
| Leiomyoma (n = 26) | Leiomyosarcoma (n = 21) | P Value | |
|---|---|---|---|
| Age, mean (SD) | 40.1 (7.9) | 56.6 (10.3) | <.001 |
| Ethnicity | .04 | ||
| Caucasian | 7 (26.9%) | 17 (81.0%) | |
| African American | 6 (23.1%) | 3 (14.2%) | |
| Hispanic | 6 (23.1%) | 0 (0.0%) | |
| Asian | 3 (11.5%) | 0 (0.0%) | |
| Unknown | 4 (15.4%) | 1 (4.8%) |
Abbreviation: SD, standard deviation.
The expressions of PLZF and H1.5 were examined by IHC (Figures 1 and 2). There was a statistically significant difference in the staining with anti-PLZF and anti-H1.5 between both groups (Table 2). The mean H1.5 staining score was 158.3 (95% confidence interval [CI] 125-192) in the leiomyosarcoma group and 28.3 (95% CI 16-41) in the leiomyoma group (P < .0001). On the other hand, the mean PLZF staining score was 1.5 (95% CI −0.3-3.3) in the leiomyosarcoma group and 71.5 (95% CI 52-91) in the leiomyoma group (P < .0001).
Figure 1.
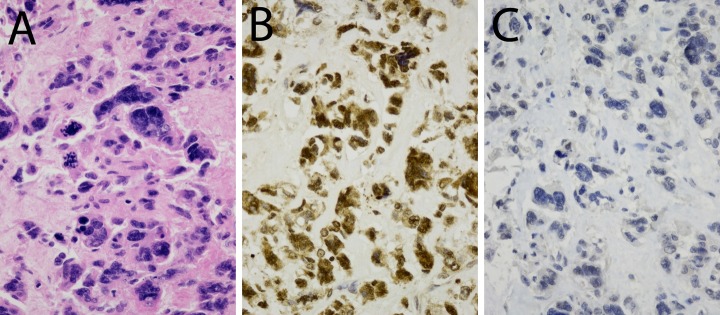
Expression of histone 1.5 (H1.5) and promyelocytic leukemia zinc finger (PLZF) in leiomyosarcoma. Diaminobenzidine (DAB). 400×. A, Hematoxylin and eosin (H&E) staining; (B) H1.5 staining (dilution 1/800) and; (C) PLZF staining (dilution 1/1000).
Figure 2.
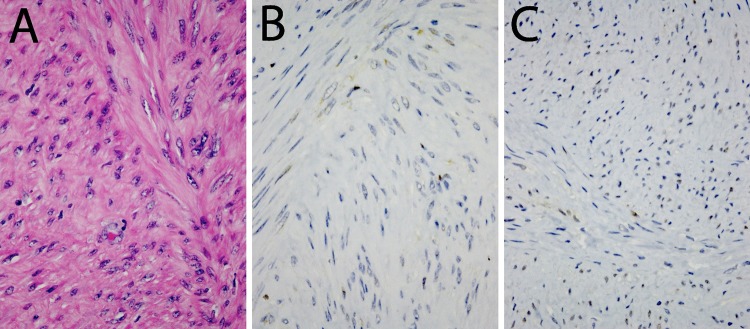
Expression of histone 1.5 (H1.5) and promyelocytic leukemia zinc finger (PLZF) in leiomyoma. Diaminobenzidine (DAB). 400×. A, Hematoxylin and eosin (H&E) staining; (B) H1.5 staining (dilution 1/800); and (C) PLZF staining (dilution 1/1000).
Table 2.
PLZF and H1.5 expression in both groups.
| N | Mean Staining Score | Standard Deviation | |
|---|---|---|---|
| PLZF | |||
| Leiomyoma | 26 | 71.5 | 47.8 |
| Leiomyosarcoma | 20 | 1.5 | 3.9 |
| H1.5 | |||
| Leiomyoma | 26 | 28.3 | 30.7 |
| Leiomyosarcoma | 21 | 158.3 | 73.9 |
Abbreviations: H1.5, histone 1.5; PLZF, promyelocytic leukemia zinc finger.
Although all of the leiomyosarcoma samples stained negatively or weakly for PLZF (14 of 20 [70%] had no staining), most of the leiomyoma samples displayed moderately positive staining. On the other hand, most of the leiomyosarcoma samples stained intensely for H1.5 (16 of 21 [76%]). In other words, PLZF and H1.5 showed inversely correlated staining in both leiomyoma and leiomyosarcoma.
A range of cutoff values is demonstrated in the ROC curve analysis (Figure 3 and Table 3). A staining score ≥60 for H1.5 was found to have a sensitivity and a specificity of 90.5% and 84.6%, respectively. A staining score ≤15 for PLZF had a specificity of 80.8% and a sensitivity of 100%. Additionally, the sensitivity and specificity were found to be 90.5% and 97.5%, respectively, if both tests were used together. This suggests that both of these markers are good screening tests, alone or combined.
Figure 3.
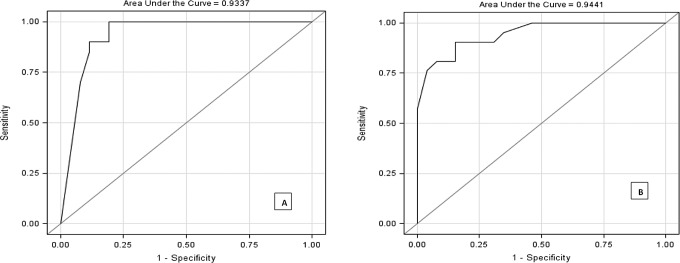
Promyelocytic leukemia zinc finger (PLZF) and histone 1.5 (H1.5), receiver–operating characteristic (ROC) curves A, ROC curve for PLZF and (B) ROC curve for H1.5.
Table 3.
Sensitivity and Specificity of H1.5 and PLZF Immunostaining for Detecting Leiomyosarcoma at Various Cutoff Values.a
| H1.5 Cutoff Value | Sensitivity, % | Specificity, % | Correctly Classified, % | PLZF Cutoff Value | Sensitivity, % | Specificity, % | Correctly Classified, % |
|---|---|---|---|---|---|---|---|
| ≥0 | 100.00 | 0.00 | 44.68 | =0 | 70.00 | 92.31 | 82.61 |
| ≥5 | 100.00 | 7.69 | 48.94 | ≤1 | 85.00 | 88.46 | 86.96 |
| ≥7.5 | 100.00 | 23.08 | 57.45 | ≤5 | 90.00 | 84.62 | 86.96 |
| ≥10 | 100.00 | 26.92 | 59.57 | ≤10 | 95.00 | 80.77 | 86.96 |
| ≥15 | 100.00 | 42.31 | 68.09 | ≤15 | 100.00 | 80.77 | 89.13 |
| ≥22.5 | 100.00 | 53.85 | 74.47 | ≤30 | 100.00 | 76.92 | 86.96 |
| ≥30 | 95.24 | 65.38 | 78.72 | ≤35 | 100.00 | 73.08 | 84.78 |
| ≥37.5 | 90.48 | 69.23 | 78.72 | ≤40 | 100.00 | 69.23 | 82.61 |
| ≥40 | 90.48 | 76.92 | 82.98 | ≤50 | 100.00 | 65.38 | 80.43 |
| ≥50 | 90.48 | 80.77 | 85.11 | ≤60 | 100.00 | 61.54 | 78.26 |
| ≥60 | 90.48 | 84.62 | 87.23 | ≤75 | 100.00 | 38.46 | 65.22 |
| ≥67.5 | 85.71 | 84.62 | 85.11 | ≤80 | 100.00 | 34.62 | 63.04 |
| ≥75 | 80.95 | 84.62 | 82.98 | ≤85 | 100.00 | 30.77 | 60.87 |
| ≥90 | 80.95 | 92.31 | 87.23 | ≤90 | 100.00 | 26.92 | 58.70 |
| ≥120 | 76.19 | 96.15 | 87.23 | ≤100 | 100.00 | 23.08 | 56.52 |
Abbreviations: H1.5, histone 1.5; PLZF, promyelocytic leukemia zinc finger.
a The bold values signify the optimal cutoffs.
The post hoc power analysis using the average staining scores and standard deviations of each PLZF and H1.5, with an α value of 1%, showed that the study was 100% powered.
Discussion
The identification of a molecular marker that can complement histologic evaluation in distinguishing leiomyoma from leiomyosarcoma would be of clinical value for both pathologic diagnosis and optimal patient management. However, a reliable positive biomarker or a biomarker panel that can unambiguously distinguish between these 2 entities is yet to be reported.
In this study, we demonstrate that PLZF and H1.5 are inversely correlated as immunohistochemical biomarkers for leiomyosarcoma and leiomyoma. Although PLZF staining is virtually negative in leiomyosarcoma, H1.5 strongly stains in this malignancy. Conversely, leiomyoma shows moderate staining for PLZF and weak staining for H1.5.
Promyelocytic leukemia zinc finger belongs to the family of Kruppel-like zinc finger proteins. The PLZF gene, Zfp145, was first identified in acute promyelocytic leukemia.28 The protein is localized to distinct nuclear foci and appears to interact with the promyelocytic leukemia and BCL6 proteins in large nuclear bodies.19,20,29 Overexpression of PLZF protein leads to suppression of cell growth.17 In embryogenesis, PLZF has been noted to be a repressor of HOX and BMP genes.30 Additionally, PLZF has been implicated in limb development, differentiation of myeloid cells, and spermatogenesis.17
Fahnenstich et al observed that PLZF is upregulated in a cyclical manner in female bovine reproductive tissues and subsequently investigated the protein’s presence in tissue samples from hysterectomies performed for uterine prolapse or leiomyomata. Using immunofluorescence, PLZF was shown to be highly expressed in the nuclei of endometrial stromal cells and myometrial smooth muscle cells during the secretory phase of the menstrual cycle.18 As expected, we found that PLZF is upregulated in leiomyoma. Both estrogen and progesterone are required for the growth of leiomyomas. Progesterone acts on the nuclear and the cytoplasmic progesterone receptors to inhibit cell death and to stimulate proliferation. The regulation of PLZF has been found to be progesterone-mediated, further supporting our findings.18 We demonstrated that 70% of the leiomyosarcoma samples did not have detectable PLZF staining, suggesting that this transcriptional repressor is downregulated in patients with leiomyosarcoma. Although the role of progesterone in patients with leiomyosarcoma is not well understood, patients with progesterone receptor (+) disease confined to the uterine body are thought to have better prognosis.31
Histone 1.5 is a complex family of linker proteins that are responsible for chromatin higher order structures. These isozymes localize to the base of nucleosomes near the DNA entry and exit sites to stabilize 2 full turns of DNA. They are also involved in packaging DNA into chromatin, regulating nucleosome spacing, and stabilizing the 10 nm chromatin fibers.25,26 It is not well known whether each subtype has a distinct role or whether each regulates specific promoters.32,33 Histone 1.5 proteins are also effective inhibitors of nucleosome sliding, consequently, blocking accessibility to transcription factors or RNA polymerase.25,34 A recent study showed that the subtype H1.5 cooperates with the transcription factor MSX1 for inhibition of specific target promoters in myogenesis.35 Histone 1.5 has also been suggested to maintain pluripotency/self-renewal in embryonic stem cells.36 In prostate cancer, H1.5 was found to have a direct correlation with the Gleason pattern.27 In pulmonary neuroendocrine tumors, H1.5 stains strongly in high-grade and weakly in low-grade malignancies.21 We found 76% of leiomyosarcomas in our study to have strong nuclear staining for H1.5. This finding suggests that H1.5 may play a role in the pathogenesis of leiomyosarcoma.
Although our results were statistically significant, the study was limited by the small sample size. Leiomyosarcoma is a rare malignancy; performing an analysis on a larger sample would require a multicenter approach. The medical center at which this study was conducted is a tertiary referral hospital located in Manhattan, where many of the patients are caucasians. This makes generalization to other races or ethnic backgrounds difficult. The control group was randomly chosen from patients who received surgery during the same time period as the patients with leiomyosarcoma. The patients were not matched according to age or tumor size. Unfortunately, the groups had a statistically significant difference in age. This could be explained by the fact that many of the control patients were still premenopausal and were therefore still symptomatic. On the other hand, the patients with leiomyosarcoma may have developed symptoms in the postmenopausal period. These issues might be more appropriately addressed in a larger study with a case-matched control group.
Straightforward cases of both leiomyoma and leiomyosarcoma were intentionally chosen for study. The true value of the chosen biomarker panel would be in the ability to accurately diagnose challenging variants in which morphology alone cannot clarify benign from malignant behavior, namely, in cellular, atypical, mitotically active, and epithelioid variants of leiomyoma as well as myxoid variants of leiomyosarcoma. Additionally, the diagnostic criteria used were stringent and did not include smooth muscle tumors of uncertain malignant potential. The combination of PLZF and H1.5 may be useful in diagnosis of difficult cases of leiomyosarcoma, especially the more uncommon variants.
We propose that when used with histologic criteria, PLZF and H1.5 immunostains can be useful adjuncts in distinguishing between malignant and benign smooth muscle tumors, increasing the pathologist’s level of confidence in establishing a definitive diagnosis. Using these 2 tests together, the sensitivity and specificity are 90.0% and 92.0%, respectively. Further research using ambiguous pathology samples would aid in determining the true value of the immunohistochemical panel.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: This research funded partially by a generous bequest to D.B. from the Leveen Family Trust.
References
- 1. Charles Z, Henry JN. Mesenchymal tumors of the uterus. In: Kurman RJ, Hedrick EL, Ronnett BM, eds. Blaustein's Pathology of the Female Genital Tract. New York: Springer Verlag; 2002:561 [Google Scholar]
- 2. Parker WH, Fu YS, Berek JS. Uterine sarcoma in patients operated on for presumed leiomyoma and rapidly growing leiomyoma. Obstetrics Gynecol. 1994;83(3):414–448 [PubMed] [Google Scholar]
- 3. Harlow BL, Weiss NS, Lofton S. The epidemiology of sarcomas of the uterus. J Natl Cancer Instit. 1986;76(3):399–402 [PubMed] [Google Scholar]
- 4. Leibsohn S, d'Ablaing G, Mishell DR, Jr, , Schlaerth JB. Leiomyosarcoma in a series of hysterectomies performed for presumed uterine leiomyomas. Am J Obstet Gynecol. 1990;162(4):968–974 [DOI] [PubMed] [Google Scholar]
- 5. D'Angelo E, Prat J. Uterine sarcomas: a review. Gynecol Oncol. 2010;116(1):131–139 [DOI] [PubMed] [Google Scholar]
- 6. Fattaneh AT, Peter D. Pathology and Genetics of Tumours of the Breast and Female Genital Organs (WHO/IARC Classification of Tumours). Lyon: IARC Press; 2003 [Google Scholar]
- 7. Toledo G, Oliva E. Smooth muscle tumors of the uterus: a practical approach. Arch Pathol Lab Med. 2008;132(4):595–605 [DOI] [PubMed] [Google Scholar]
- 8. O'Neill CJ, McBride HA, Connolly LE, McCluggage WG. Uterine leiomyosarcomas are characterized by high p16, p53 and MIB1 expression in comparison with usual leiomyomas, leiomyoma variants and smooth muscle tumours of uncertain malignant potential. Histopathology. 2007;50(7):851–858 [DOI] [PubMed] [Google Scholar]
- 9. Ip PP, Tse KY, Tam KF. Uterine smooth muscle tumors other than the ordinary leiomyomas and leiomyosarcomas: a review of selected variants with emphasis on recent advances and unusual morphology that may cause concern for malignancy. Adv Anat Pathol. 2010;17(2):91–112 [DOI] [PubMed] [Google Scholar]
- 10. Hong T, Shimada Y, Uchida S, et al. Expression of angiogenic factors and apoptotic factors in leiomyosarcoma and leiomyoma. Int J Mol Med. 2001;8(2):141–148 [DOI] [PubMed] [Google Scholar]
- 11. Moore AB, He H, Yoshida A, Rico PJ, Haseman JK, Dixon D. Transforming growth factor-alpha, epidermal growth factor receptor, and PCNA immunoexpression in uterine leiomyosarcomas and leiomyomas in B6C3F1 mice. Exp Toxicol Pathol. 2000;52(3):195–200 [DOI] [PubMed] [Google Scholar]
- 12. Gokaslan H, Turkeri L, Kavak ZN, et al. Differential diagnosis of smooth muscle tumors utilizing p53, pTEN and Ki-67 expression with estrogen and progesterone receptors. Gynecol Obstet Invest. 2005;59(1):36–40 [DOI] [PubMed] [Google Scholar]
- 13. Hall KL, Teneriello MG, Taylor RR, et al. Analysis of Ki-ras, p53, and MDM2 genes in uterine leiomyomas and leiomyosarcomas. Gynecol Oncol. 1997;65(2):330–335 [DOI] [PubMed] [Google Scholar]
- 14. Mittal KR, Chen F, Wei JJ, et al. Molecular and immunohistochemical evidence for the origin of uterine leiomyosarcomas from associated leiomyoma and symplastic leiomyoma-like areas. Mod Pathol. 2009;22(10):1303–1311 [DOI] [PubMed] [Google Scholar]
- 15. Chen L, Yang B. Immunohistochemical analysis of p16, p53, and Ki-67 expression in uterine smooth muscle tumors. Int J Gynecol Pathol. 2008;27(3):326–332 [DOI] [PubMed] [Google Scholar]
- 16. Lee CH, Turbin DA, Sung YC, et al. A panel of antibodies to determine site of origin and malignancy in smooth muscle tumors. Mod Pathol. 2009;22(12):1519–1531 [DOI] [PubMed] [Google Scholar]
- 17. Yeyati PL, Shaknovich R, Boterashvili S, et al. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene. 1999;18(4):925–934 [DOI] [PubMed] [Google Scholar]
- 18. Fahnenstich J, Nandy A, Milde-Langosch K, et al. Promyelocytic leukaemia zinc finger protein (PLZF) is a glucocorticoid- and progesterone-induced transcription factor in human endometrial stromal cells and myometrial smooth muscle cells. Mol Hum Reprod. 2003;9(10):611–623 [DOI] [PubMed] [Google Scholar]
- 19. Koken MH, Reid A, Quignon F, et al. Leukemia-associated retinoic acid receptor alpha fusion partners, PML and PLZF, heterodimerize and colocalize to nuclear bodies. Proc Natl Acad Sci U S A. 1997;94(19):10255–10260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Licht JD, Shaknovich R, English MA, et al. Reduced and altered DNA-binding and transcriptional properties of the PLZF-retinoic acid receptor-alpha chimera generated in t(11;17)-associated acute promyelocytic leukemia. Oncogene. 1996;12(2):323–336 [PubMed] [Google Scholar]
- 21. Hechtman JF, Beasley MB, Kinoshita Y, Ko HM, Hao K, Burstein DE. Promyelocytic leukemia zinc finger and histone H1.5 differentially stain low- and high-grade pulmonary neuroendocrine tumors: a pilot immunohistochemical study. Hum Pathol. 2013;44(7):1400–1405 [DOI] [PubMed] [Google Scholar]
- 22. Shaknovich R, Yeyati PL, Ivins S, et al. The promyelocytic leukemia zinc finger protein affects myeloid cell growth, differentiation, and apoptosis. Mol Cell Biol. 1998;18(9):5533–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheung M, Pei J, Pei Y, Jhanwar SC, Pass HI, Testa JR. The promyelocytic leukemia zinc-finger gene, PLZF, is frequently downregulated in malignant mesothelioma cells and contributes to cell survival. Oncogene. 2010;29(11):1633–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Felicetti F, Bottero L, Felli N, et al. Role of PLZF in melanoma progression. Oncogene. 2004;23(26):4567–4576 [DOI] [PubMed] [Google Scholar]
- 25. Hizume K, Yoshimura SH, Takeyasu K. Linker histone H1 per se can induce three-dimensional folding of chromatin fiber. Biochemistry. 2005;44(39):12978–1289 [DOI] [PubMed] [Google Scholar]
- 26. Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431(1-2):1–12 [DOI] [PubMed] [Google Scholar]
- 27. Sato S, Takahashi S, Asamoto M, et al. Histone H1 expression in human prostate cancer tissues and cell lines. Pathology. international 2012;62(2):84–92 [DOI] [PubMed] [Google Scholar]
- 28. Chen SJ, Zelent A, Tong JH, et al. Rearrangements of the retinoic acid receptor alpha and promyelocytic leukemia zinc finger genes resulting from t(11;17)(q23;q21) in a patient with acute promyelocytic leukemia. J Clin Invest. 1993;91(5):2260–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reid A, Gould A, Brand N, et al. Leukemia translocation gene, PLZF, is expressed with a speckled nuclear pattern in early hematopoietic progenitors. Blood. 1995;86(12):4544–4552 [PubMed] [Google Scholar]
- 30. Barna M, Hawe N, Niswander L, et al. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25(2):166–172 [DOI] [PubMed] [Google Scholar]
- 31. Leitao MM, Jr, , Hensley ML, Barakat RR, et al. Immunohistochemical expression of estrogen and progesterone receptors and outcomes in patients with newly diagnosed uterine leiomyosarcoma. Gynecol Oncol. 2012;124(3):558–562 [DOI] [PubMed] [Google Scholar]
- 32. Fan Y, Nikitina T, Zhao J, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123(7):1199–1212 [DOI] [PubMed] [Google Scholar]
- 33. Shen X, Gorovsky MA. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86(3):475–483 [DOI] [PubMed] [Google Scholar]
- 34. Hill DA. Influence of linker histone H1 on chromatin remodeling. Biochem Cell Biol. 2001;79(3):317–324 [PubMed] [Google Scholar]
- 35. Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304(5677):1675–1678 [DOI] [PubMed] [Google Scholar]
- 36. Terme JM, Sese B, Millan-Arino L, et al. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J Biol Chem. 2011;286(41):35347–35357 [DOI] [PMC free article] [PubMed] [Google Scholar]


