Abstract
IFNβ has been implicated as an effector of oviduct pathology resulting from genital chlamydial infection in the mouse model. In this study, we investigated the role of cytosolic DNA and engagement of DNA sensors in IFNβ expression during chlamydial infection. We determined that TREX-1, a host 3’to 5’ exonuclease, reduced IFNβ expression significantly during chlamydial infection using siRNA and gene knock out fibroblasts, implicating cytosolic DNA as a ligand for this response. The DNA sensor cGAS has been shown to bind cytosolic DNA to generate cGAMP, which binds to the signaling adaptor STING to induce IFNβ expression. We determined that cGAS is required for IFNβ expression during chlamydial infection in multiple cell types. Interestingly, although infected cells deficient for STING or cGAS alone failed to induce IFNβ, co-culture of cells depleted for either STING or cGAS rescued IFNβ expression. These data demonstrate that cGAMP produced in infected cGAS+STING− cells can migrate into adjacent cells via gap junctions to function in trans in cGAS−STING+ cells. Further, we observed cGAS localized in punctate regions on the cytosolic side of the chlamydial inclusion membrane in association with STING, indicating that chlamydial DNA is likely recognized outside the inclusion as infection progresses. These novel findings provide evidence that cGAS-mediated-DNA sensing directs IFNβ expression during C.trachomatis infection and suggests that effectors from infected cells can directly upregulate IFNβ expression in adjacent uninfected cells during in vivo infection, contributing to pathogenesis.
Keywords: IFN-β, Chlamydia, cyclic GMP-AMP synthase, STING
Introduction
Chlamydia trachomatis is the most common sexually transmitted bacterial pathogen in the world and infection can lead to pelvic inflammatory disease and infertility in women. Chlamydial infection of epithelial cells upregulates proinflammatory cytokines, chemokines, type I IFNs, and IFN stimulatory genes (1–3). We and others have shown that type I IFN (IFNα and IFNβ) signaling exacerbates host pathology during the course of genital (4) or pulmonary (5) C. muridarum infection in the mouse model. Further, IFNβ depletion protects mice from oviduct pathology during genital chlamydial infection (6), demonstrating a significant contribution of IFNβ to host pathology. A similar detrimental effect of IFNβ signaling has been reported during other bacterial infections as well [reviewed in (7)].
A consensus mechanism for IFNβ induction during intracellular bacterial infection is yet to be defined. However, multiple host pathogen recognition receptors that can induce IFNβ expression during viral infection [reviewed in (8)] have been identified. These include the RNA sensors, RIG-I (Retinoic acid-inducible gene) and MDA5 (Melanoma differentiation-associated protein 5) (9, 10), which recognize viral RNA and signal via the adaptor MAVS (Mitochondrial antiviral signaling) to induce IFNβ expression (11). In addition, several DNA sensors have been identified that recognize cytosolic DNA and induce IFNβ expression. These include RNA polymerase III (9, 10), DAI (DNA-dependent activator of IFN regulatory factors) (12), IFI16 (IFNγ inducible protein 16) (13), LRRFIP1 (Leucine rich repeat protein FLII interacting protein) (14), DDX41 (DEAD box polypeptide 41) (15), MRE11 (Meiotic recombination 11 homolog) (16), LSm14A (member of LSm protein family) (17) and DNA-PKcs (DNA-protein kinase catalytic subunit) (18). The large number of DNA sensors identified in the host suggests that they may play redundant roles during infection. On the other hand, STING (Stimulator of IFN genes), an ER resident transmembrane protein, has been reported to be crucial for independent recognition of cytosolic DNA during viral infection and induction of IFN-β (19). STING is not a direct sensor of DNA, but functions as an integral adaptor molecule in DNA recognition. STING binds to a novel second messenger, cyclic GAMP (cGAMP) generated by a host DNA sensor, cGAS (cyclic GMP-AMP synthase) (20) upon DNA-binding in the cytosol (21). This interaction of cGAMP with STING activates the signaling events that lead to IFNβ expression. Additionally, STING also directly binds bacterial second messengers, cyclic di-GMP and di-AMP to induce IFNβ (22), suggesting that it can also function as a direct sensor of intracellular pathogens. Indeed, cyclic di-AMP has been shown to be produced by Listeria monocytogenes (23) and Chlamydia trachomatis (24). However, the direct contribution of STING relative to its co-operation with DNA sensors in IFNβ expression during bacterial infection remains unclear.
We have shown that STING is required for IFNβ induction during chlamydial infection in HeLa cells and murine oviduct epithelial cells, while the cytosolic RNA sensing pathway is dispensable for this response (25). In this study, we demonstrate for the first time that cytosolic DNA is a trigger for IFNβ expression during C. trachomatis infection and that the DNA sensor cGAS plays an integral role in sensing this DNA to induce IFNβ expression. cGAS localized in close proximity to the chlamydial inclusion membrane and co-localized with STING, suggesting that chlamydial DNA is likely recognized on membrane compartments outside the chlamydial inclusion. We also provide indirect evidence for cGAS-mediated generation of cGAMP during infection, by demonstrating rescue of IFNβ expression during co-culture of cGAS+STING− cells with cGAS−STING+ cells, suggesting that cGAMP from infected cGAS+ cells can migrate into adjacent cells to induce STING-dependent IFNβ expression.
Materials and Methods
Cell culture, Primary cells, Reagents, cDNA constructs, chlamydial stocks and infection
HeLa cells, WT (wild-type) and TREX1 KO mouse embryonic fibroblasts (MEFs), and HEK293T were cultured in supplemented DMEM and mouse J774 cells cultured in complete media for macrophages, as described earlier (25). Mouse BM1.11 cells (26) and human OE-E6/E7 (oviduct epithelial cells) (27) were cultured in supplemented F12-DMEM as described (25). Poly dA:dT/LyoVec (100 µg/ml), Poly I:C/LyoVec (50 µg/ml) and 2’3’ cGAMP (1 mg/ml) were purchased from Invivogen. Carbenoxolone (working concentration 0.2 mM) was purchased from SIGMA. pBluscript vector (1–2.5 µg) was used as immunostimulatory DNA (ISD). cDNA constructs for human cGAS (pCMV-cGAS) and STING (pCMV-STING) were purchased from Origene. C. muridarum, C. trachomatis D and L2 were propagated in McCoy cells and infections performed at 1 MOI or as indicated, as previously described (25).
Small interfering RNA (siRNA)
siRNA targeting human cGAS (MB21D1) (s41746 and s453378), human DDX41 (s28120), human TRIM56 (s37816), human LSM14A (s25051), human STING (s50645), or corresponding non-targeting (NT; control, Cat #4390843) were obtained from Ambion Life Technologies, and were used for HeLa and OE cells. For BM1.11 cells, accell™ SMART pool siRNA duplexes targeting mouse STING (E-055528), mouse cGAS (E-055608) and corresponding accell™ non-targeting siRNA (D-001910-10) from Dharmacon, were used. For J774 cells, two mouse cGAS (MB21D1) siRNA oligos (Cat# 2675, SIGMA), described earlier (20) were used in parallel with the corresponding non-targeting siRNA from SIGMA (Cat# SIC001). For TREX-1 knock down siGenome SMART pool mouse TREX1 (M-042223-00) were used in BM1.11 cells, while siGenome SMART pool human TREX1 (M-013239-02) were used in HeLa cells with corresponding non-targeting pools.
siRNA, ISD transfection and cGAMP treatment
HeLa cells were plated at 1 × 105 cells /well in 24-well plates for 18 to 24 h before transfection. Thirty pmol of siRNA were transfected using Lipofectamine RNAiMAX reagent (Invitrogen). Forty-eight hours post-transfection, siRNA transfected cells were split into 4 wells in a 24 well dish for multiple treatments. siRNA (10 pmol) transfections were repeated on each well the next day to achieve maximal knock down in expression. Twenty-four hours after second siRNA transfection, one set of cells were infected with Chlamydia and harvested 18–24 h post infection. In parallel, cells were transfected with ISD (positive control) or with poly I:C (negative control), 6–8 h prior to harvest, so that all cells, including untreated controls were harvested at the same time. BM1.11 cells were transfected directly using accell™ siRNA (100 pmol/well) in a 24 well dish for multiple treatments. OE-E6/E7 cells and J774 cells were transfected with siRNA (100 pmol/well and 30 µM, respectively) in a 24 well dish for multiple treatments using INTERFERin (Polyplus transfection™). Seventy-two hours after siRNA transfection, cells were infected with C. trachomatis or transfected with ISD (positive control) 8 h before harvest, as described previously. The effect of specific siRNAs on target gene mRNA was assessed by qRT-PCR. For ISD transfection, 1 µg of ISD were transfected into HeLa, and 2.5 µg of ISD was transfected into OE-E6/E7 or BM1.11 cells using FuGene HD transfection reagent (Promega). For cGAMP treatment, cGAMP (1 µg/ml) was added to cells at 37°C in a permeabilization buffer (50 mM HEPES, 100 mM KCl, 3 mM MgCl2, 85 mM Sucrose, 0.1 mM DTT, 0.2% BSA, 1mM ATP, 10 µg/ml Digitonin) described earlier (23) and replaced with media 30 min after treatment. Cells were harvested 6 h post treatment.
RNA extraction, quantitative RT-PCR analysis and ELISA
The ISD transfected cells, infected cells, or un-treated cells (UT) were processed at the same time for RNA extraction using the RNeasy kit (Qiagen). The RNA were then processed for reverse transcription and quantitative PCR using an SsoAdvanced SYBR mix (Bio-Rad) using a CFX iCycler (Bio-Rad) as described previously (25) . Primers sequences for IFNβ, IL-8, 16S rRNA were described previously (25). Additional primers include, mcGAS-F: TAGCGGTCTCAACTCAAG, mcGAS-R: TGGTGTCTGTTCATAGCA, hcGAS-F: CCTGCTGTAACACTTCTTAT, hcGAS-R; TTAGTCGTAGTTGCTTCCT, hTREX1-F: TGCCTTCTGTGTGGATAG, hTREX1-R: AGTGTAGATGCTGCCTAG, mTREX1-F: CAATAGCCACTCTGTATG, and mTREX1-R: TGACCGCTATGACTTTCC. All primers were designed using Beacon Design software (Bio-Rad). Culture supernatants were collected at 24 h post infection (p.i) for IFNβ and CXCL10 by ELISA (R&D).
HeLa cell co-culture following siRNA transfection to demonstrate cGAMP transfer
Twenty-four hours after second individual siRNA transfection, the cells were trypsinized and counted. cGAS siRNA and STING siRNA cells were re-plated individually or mixed at different ratio (1:3, 1:1, and 3:1). Equal numbers of the mixed or individual siRNA transfected cells were plated into a new 24-well plate (2 × 105cell /well) and were infected with C. muridarum (3 moi) 18 h later. ISD transfection (1 µg/well) was done on parallel wells 6 h before harvest, so that all cells were harvested at the same time for RNA extraction. In an independent experiment, cGAS KD and STING KD cells were co-cultured in a transwell separated by permeable support, to determine the requirement of cell-cell contact.
HEK293T transfection and Immunoblots
HEK293T cells were transfected with pCMV-cGAS (Origene), pCMV-STING (Origene) or pcDNA3.1 vector (60 ng DNA). Cells were trypsinized 24 post transfection and re-plated individually or mixed at 1:1 ratio, such that each well contained a total of 4 × 105 cells/well in a 24 well dish. Cells were infected, or permeabilized with positive control ligands cGAMP (1µg/ml) or transfected with ISD (1µg), 6 hours after plating and harvested at indicated times and processed for RNA and qRT-PCR. Immunoblots for cGAS or STING protein in HEK293, HEK293T and HeLa cells were carried out using cell lysate prepared using RIPA buffer (Pierce-Thermo Scientific) and protease inhibitor cocktails (Sigma). Anti cGAS Ab (Cat# AP10510c, Abgent), anti-STING poly clonal Ab (Cat# PA5-26751, Thermo Scientific) and anti-actin Ab (Cat# A2228, Sigma) were used at the 1:500, 1:500 and 1:1000 concentration respectively.
Confocal microscopy
HeLa cells were grown on 12 mm glass coverslips (#1, 0.17 mm thickness) at 50% density in 24-well plate, one day before infection with 1 MOI of C. muridarum or C. trachomatis. Twenty four hours post-infection (pi), cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for immunostaining as described previously (25). Rabbit anti cGAS was used at 1:200 dilution. Chlamydiae were stained with C. muridarum antiserum obtained from convalescent mice post infection diluted 1:300, while Alexa Fluor (AF) 488 conjugated anti-mouse and AF568 anti-rabbit (Invitrogen) were used at 1:1000 dilution as secondary Abs for detection. AF488 conjugated mouse mAb for STING (R & D) was used at 1:100 dilution in conjunction with anti-cGAS polyclonal antibody for co-localization studies. AF647 anti GM130 (BD BioScience) was used for Golgi staining at 1:100 dilution in some experiments. Transfected cells were stained using mouse anti-FLAG (Origene). Cells were washed and mounted using Prolong anti-fade containing DAPI (Invitrogen). Confocal images were acquired with the 63X oil 0.8 numerical aperture objective using Zeiss confocal microscope (LSM 510 META) and images analyzed using AxioVision software (Thornwood, NY).
Statistical analysis
At least three independent repeats were performed for each siRNA experiment and a representative experiment shown. Error bar indicates the standard error for technical replicates for qRT-PCR. To determine statistical significance in siRNA experiments, percent decrease/increase in expression levels of IFNβ relative to non-targeting (NT) siRNA (100%) were averaged from multiple experiments and significance determined by paired T test or one-way ANOVA with Holm-Sidak multiple comparison test using Graphpad Prism™. For transfection experiments in HEK293T cells, fold changes from 3 independent experiments were averaged, represented with SD, and significance determined by one way ANOVA with Holm-Sidak multiple comparison test.
Results
Cytosolic nucleic acid is a potential ligand for IFNβ expression during chlamydial infection
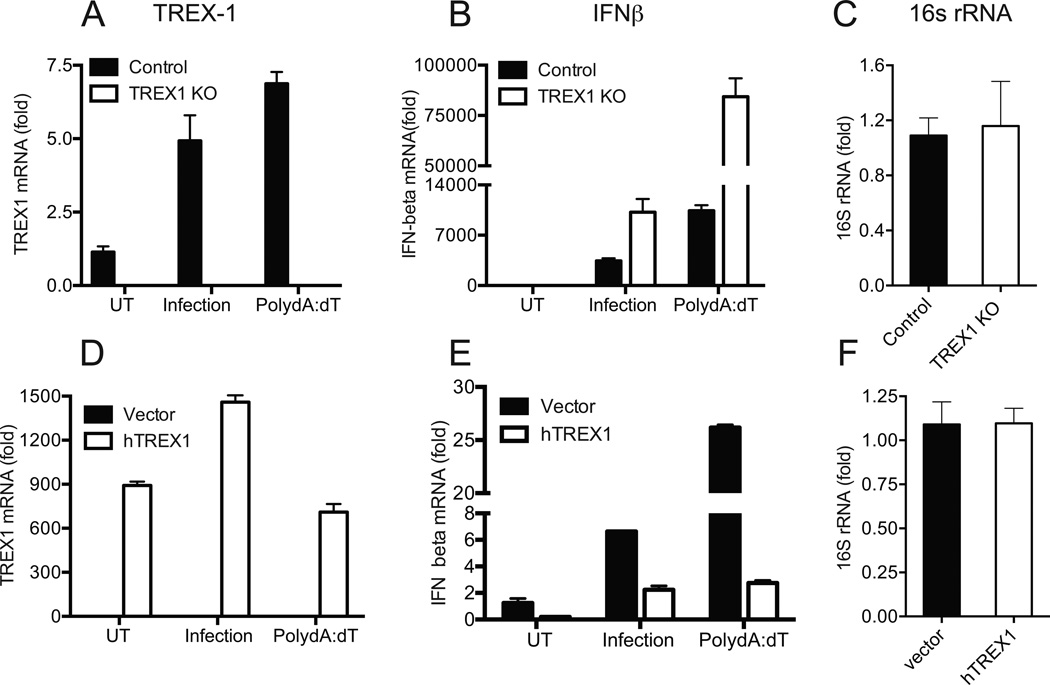
During intracellular bacterial or viral infection, nucleic acids released into the cytosol can result in IFNβ expression. Host exonucleases, such as Three prime Repair EXonuclease-1 (TREX1), cleave intracellular DNA and have been shown to regulate IFNβ expression during viral (28) or bacterial infection (29). To determine if cytosolic DNA contributes to IFNβ expression during chlamydial infection, TREX1 KO mouse embryonic fibroblast (MEF) and their corresponding WT controls (Fig 1A) were infected with C. muridarum. TREX1 KO cells showed a significant 2–3 fold increase in IFNβ expression compared to wild type (WT) MEF during C. muridarum infection (Fig 1B), without altering chlamydial growth, as measured by comparable levels of chlamydial 16S rRNA (Fig 1C). TREX1 KO cells were also hyper-responsive to the DNA analog poly (dA-dT), used as a positive control (Fig 1B). Further, complementation of TREX1 KO cells with a cDNA construct expressing human TREX1 (Fig 1D), resulted in 50–70% reduction in IFNβ expression during chlamydial infection or poly (dA-dT) treatment (Fig 1E) relative to their respective vector controls, without altering chlamydial replication (Fig 1F).
Figure 1. IFNβ expression during chlamydial infection is elevated in TREX1 KO MEFs and reduced by TREX-1 over-expression.
(A–C) Control MEFs and TREX1 KO MEFs were infected with C. muridarum at 1 MOI or transfected with poly dA:dT for 6 h, before harvest. Infected cells were harvested at 24 h p.i. TREX1 mRNA (A), IFNβ mRNA (B) and chlamydial 16S rRNA (C) levels were measured by qRT-PCR. (D–F) TREX1 KO cells were transfected with human TREX1 or vector control. Twenty four hour post transfection, cells were infected with C.muridarum or transfected with poly dA:dT. Relative expression levels of TREX1 (D), IFNβ (E) and chlamydial 16S rRNA (F) normalized to GAPDH are shown. Panels are representative of three independent experiments and error bars represent the range in technical replicates. Statistical significance for qPCR data between multiple experiments was determined by using paired T tests on percent change in IFNβ levels between WT and TREX1 KO infected cells (P=0.01). In experiment involving transfection of TREX KO cells, percent change in IFNβ between vector and TREX-1 cDNA from multiple experiments were used to calculate significance (Infection; P=0.001, poly dA:dT; P=0.009). UT = Untreated.
To further establish the contribution of cytosolic DNA to IFNβ expression in chlamydial infection, siRNA knock down of TREX1 was carried out in mouse oviduct epithelial cells BM1.11 (Fig 2A). A significant increase in IFNβ expression was observed in cells transfected with TREX1 siRNA compared to non-targeting (NT) siRNA, during poly (dA-dT) treatment (P=0.04) or C. muridarum infection (P=0.01) (Fig 2B and Fig S1), again without affecting chlamydial replication (Fig 2C). The role of TREX-1 was further confirmed in HeLa cells, where siRNA transfection can be performed with high efficiency. Knock down of TREX-1 (Fig 2D) resulted in significant increase (P=0.045) in chlamydia-induced IFNβ expression relative to NT siRNA (Fig 2E and Fig S1), without altering chlamydial replication (Fig 2F). The effect of TREX-1 siRNA was specific for immunostimulatory DNA (ISD)- but not RNA-induced IFNβ expression as evidenced in cells transfected with the RNA–analog poly I:C (Fig 2E and Fig S1). Overall, these data suggest that cytosolic DNA is a ligand required for IFNβ expression during chlamydial infection.
Figure 2. siRNA knockdown of TREX-1 in epithelial cells increases IFNβ expression, during chlamydial infection.
(A–C) Mouse BM1.11 were transfected with TREX1 siRNA or non-targeting (NT) siRNA (A) and infected with C. muridarum at 1 MOI, 72 h post transfection. TREX-1 (A) and IFNβ (B) mRNA were measured at 24 h p.i. In parallel, cells were transfected with dsDNA analog poly dA:dT, 6 h before harvest. Chlamydial replication was monitored by comparing 16S rRNA levels (C) in infected cells between treatments. (D–F) HeLa cells were transfected with siRNA for human TREX1 or NT siRNA. Cells were infected with C.muridarum at 1 MOI for 24 h or transfected 6 h before harvest with ISD or RNA analog poly I:C. TREX1 (D), IFN beta (E) and chlamydial 16S rRNA levels (F) were measured by qRT-PCR. Panels are representative of three independent experiments and error bars represent the mean ± error of technical replicates. Statistical significance for qPCR data between multiple experiments was determined by using paired T tests on percent change in IFNβ levels between TREX1 siRNA relative to NT siRNA in each experiment. For BM1.11 cells, infection; P=0.014 and poly dA-dT; P=0.047. For HeLa, infection; P=0.04, ISD; P=0.04, poly IC; P=NS. UT=Un-treated.
The DNA sensor, cyclic GMP-AMP synthase (cGAS) is essential for IFNβ expression during C. muridarum and C. trachomatis infection
That DNA serves as a ligand for IFNβ expression during chlamydial infection suggested that host DNA sensors detect cytosolic DNA during infection. We have previously shown that the adaptor molecule STING is required for IFNβ expression during chlamydial infection in HeLa cells (25). Using these cells, we investigated potential DNA sensors involved in recognition of this ligand during infection. siRNA knockdown of several DNA sensors, specifically DAI, IFI16, LRRFIP1, DDX41, Lsm14A, and TRIM56 in HeLa cells had no effect on IFNβ expression during infection (data not shown). Recently, the host enzyme cGAS was shown to catalyze the generation of the STING ligand, cyclic GAMP, after binding cytosolic DNA (20, 21). We tested the contribution of cGAS to IFNβ expression during chlamydial infection via knock down using two independent siRNAs directed against cGAS in comparison to a non-targeting (NT) siRNA from the same provider. siRNA-mediated knock down for cGAS and STING resulted in >90% and >99% reduction in mRNA expression of cGAS and STING, respectively (data not shown). cGAS knock down with cGAS1 siRNA and cGAS2 siRNA decreased the expression of IFNβ by > 75% (p<0.001) and >97% (p<0.001) relative to NT controls during C. muridarum infection. (Fig 3A and Fig 3K). Similar results were observed with human C. trachomatis serovar D, and C. trachomatis L2 (Fig 3A). Cells transfected with ISD, where induction of IFNβ was significantly compromised in STING and cGAS knock down cells, served as positive controls for functional siRNA knockdown, showing >95% decrease (P<0.001) in IFNβ expression relative to NT controls (Fig 3B and Fig 3K). To demonstrate the specificity of the siRNAs used, cells were transfected with poly I:C in parallel. cGAS and STING siRNA did not decrease IFNβ induction in poly IC transfected cells suggesting that RNA-sensing pathways or downstream effectors for IFNβ expression were not targeted (Fig 3C and 3K), although some increase in IFNβ expression was observed in cells transfected with one of the 2 cGAS siRNA tested (P=0.024). cGAS and STING siRNA significantly reduced the expression of another IRF3-dependent gene IFNλ during infection and ISD transfection (Fig 3D–F), but did not reduce the expression of IL-8, an IRF3-independent (Fig 3G), demonstrating a specific effect on IRF3 pathway. Chlamydial replication remained unaltered in response to cGAS or STING knockdown as evidenced by 16S rRNA levels (Fig 3H). IFNβ protein was not detectable in supernatants because it is likely rapidly endocytosed by its receptor IFNAR. Therefore, protein levels of an interferon response gene, CXCL10, which serves as a functional surrogate for biologically active IFNβ, was measured in the culture supernatants by ELISA and was found to correspond to IFNβ mRNA levels in the cells (Fig 3I). siRNA knock down of STING and cGAS resulted in undetectable or low levels of the respective proteins in cell lysates detected by western blot (Fig 3J). Further, cGAS knock down did not alter STING levels and vice versa (Fig 3J), confirming the effect of cGAS on IFNβ expression was independent of STING protein levels.
Figure 3. cGAS is required for IFNβ expression in HeLa cells infected with C. muridarum or C. trachomatis (D and L2).
HeLa cells were transfected with non-targeting siRNA (NT), 2 different siRNA for cGAS (cGAS1 and cGAS2), or a siRNA for STING as described in Methods. Significant knock down of STING (99%) and cGAS (90%) mRNA was achieved. Cells were infected with 1 MOI of C. muridarum (C.M) or 5 MOI of C.trachomatis -serovar D (C.T-D) or C. trachomatis L2 (C.T-L2). Cells were harvested at 24 h p.i and analyzed for expression of IFNβ (A), IFN λ (D), IL-8 (G), and chlamydial 16S rRNA (H). In parallel, cells were transfected with ISD (positive control) or poly IC-LyoVec (negative control) and harvested at 6 h post transfection and analyzed for expression of IFNβ (B, C), IFNλ (E, F). Culture supernatants from infected or transfected cells were collected at 24 h and CXCL10 protein levels assayed by ELISA (I). Mean ± SD of samples from 3 independent experiments are shown for ELISA. A representative western blot showing the levels of STING and cGAS following siRNA knock down in uninfected HeLa cells (J). A representative of five independent experiments is presented in A–H for qRT-PCR data and error bars represent range in technical replicates. Statistical significance for qPCR data between multiple experiments was determined by one way ANOVA with multiple comparison tests on the percent decrease in IFNβ levels for the siRNA used relative to NT siRNA in each experiment (K). UT=Un-treated.
The role of cGAS and STING was further assessed in mouse and human oviduct epithelial cells infected with C. muridarum or C. trachomatis, respectively (Fig 4). About 50% and 90% reduction in IFNβ expression was observed in mouse oviduct epithelial cells (BM1.11) with cGAS and STING knock down, respectively during C. muridarum infection (Fig 4A and 4D). These results paralleled a 50% and 80% decrease of cGAS and STING expression, respectively (Fig 4B and 4C) while chlamydial growth remained unaffected by cGAS and STING knock down (data not shown). IFNβ expression during ISD transfection in BM1.11 cells was also significantly reduced in cells transfected with cGAS and STING siRNA relative to NT siRNA (Fig 4A and 4D). In OE-E6/E7 cells (27), a transformed human oviduct cell line, a similar effect of cGAS and STING knock down was observed (Fig 4E and 4H), although these cells only induced low levels of IFNβ during C.trachomatis infection. The reduction in IFNβ expression paralleled about 50% knock down achieved for both cGAS and STING expression in these cells (Fig 4F and 4G). cGAS was also found to be essential for IFNβ expression during chlamydial infection in the mouse macrophage line J774 (data not shown), which supports the growth of C. muridarum. These data indicate that DNA sensing by cGAS during chlamydial infection occurs in multiple cells types and with both mouse and human C. trachomatis strains.
Figure 4. cGAS is required for IFNβ expression during Chlamydia spp. infection of mouse oviduct epithelial cells (BM1.11 cells) and human oviduct epithelial cells (OE-E6/E7).
siRNA knock down in mouse BM1.11 cells were carried out using accell™ NT, mouse cGAS or STING siRNA pools. Seventy-two hours after transfection, cells were infected with C. muridarum or transfected with ISD (DNA) 6 h before harvest, and analyzed simultaneously at 24 h p.i for expression of mouse IFNβ (A), cGAS (B) and STING (C). Human oviduct epithelial cells (OE-E6/E7) were transfected with non-targeting (NT), human cGAS (cGAS siRNA 1 and 2) or human STING siRNA. Seventy-two hours after transfection, cells were infected with C. trachomatis (serovar D) at 5 MOI or transfected with ISD (DNA) 6 h before harvest and analyzed concurrently at 24 h p.i for expression of human IFNβ (E), cGAS (F) and STING (G). A representative of three experiments for BM1.11 cells and OE cells is presented for qRT-PCR data. Statistical significance for qPCR data between multiple experiments was determined by one way ANOVA with multiple comparison tests on the percent decrease in IFNβ levels for the siRNA used relative to NT siRNA in each experiment (D and H).
cGAS is recruited to the inclusion membrane during chlamydial infection
We have previously shown that STING and the ER protein Sec16α localized in close proximity to the chlamydial inclusion (25). cGAS has been shown to colocalize with transfected DNA in the host cytosol (20). To determine the intracellular niche(s) where cGAS recognizes DNA during chlamydial infection, immunostaining of endogenous cGAS and STING was performed in infected cells. Antibody specificity was confirmed by transfecting cGAS or STING into HEK293T cells (data not shown), which do not express either protein at high levels (20). Endogenous cGAS was found to localize in close proximity to chlamydial inclusion membrane during both C. muridarum (Fig 5A) and C. trachomatis (Fig 5B) infection. In uninfected cells, cGAS was found distributed in the cytosol (Fig 5C), while in infected cells cGAS was found enriched at certain points around the chlamydial inclusion membrane (Fig 5D). Co-localization of STING and cGAS was observed at several contact points around the inclusion (Fig 5D), but no co-localization of STING or cGAS was observed with the Golgi marker GM130 (data not shown). To confirm the data obtained by endogenous staining of cGAS, we examined trafficking and localization of cGAS in HeLa cells transfected with FLAG-cGAS. A distinct enrichment of FLAG-cGAS was observed as punctate staining around the inclusion in infected cells (Fig 5E). These data are reminiscent of what was previously observed with STING (25). cGAS is a cytosolic protein and not associated with the membrane component of the cells. Therefore, these data suggest that cGAS is trafficked to the inclusion membrane during chlamydial infection.
Figure 5. cGAS localizes in punctate regions around the chlamydial inclusion membrane.

HeLa cells infected with C. muridarum (A) or C. trachomatis, serovar D (B) were fixed and stained for endogenous cGAS (red) and Chlamydia (green). Cells were fixed with Prolong gold™ with DAPI (blue) and analyzed by confocal microscopy. Uninfected HeLa cells (C) and cells infected with C. muridarum (D) for 18 h were fixed and stained for endogenous cGAS (red) and STING (green). In an independent experiment HeLa cells were transfected with FLAG-cGAS (E) and infected with C. muridarum 24 h later. Infected cells were fixed at 18 h p.i and stained using mouse monoclonal Ab for FLAG. Chlamydial inclusions and cell nucleus are marked with an “I” and “N” respectively on the DAPI staining.
Evidence for cGAMP generation during chlamydial infection
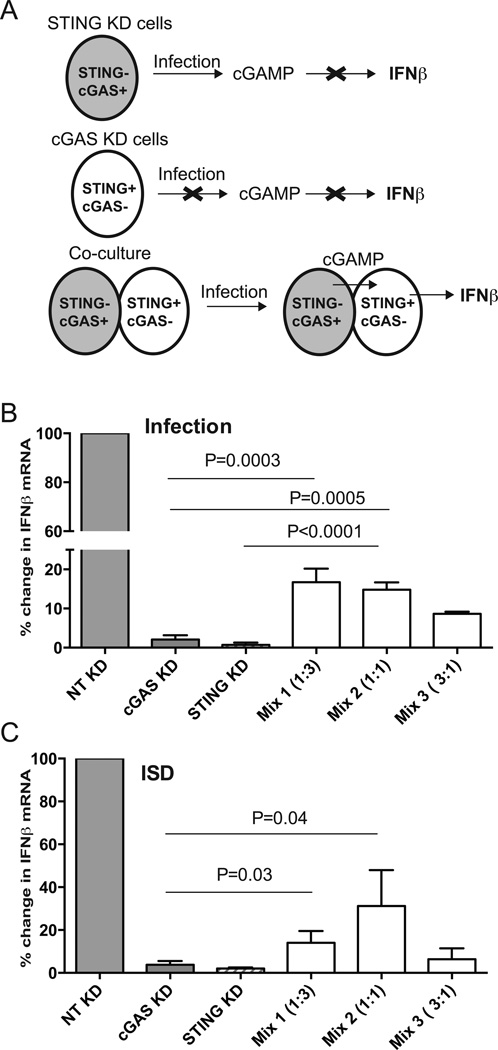
Recognition of DNA by cGAS results in the generation of the STING ligand, cGAMP (20, 21). To determine if cGAMP is generated during chlamydial infection, we used an indirect approach based on the recent finding that cGAMP can cross gap junctions between epithelial cells (30). HeLa cells knocked down for STING expression should retain the capacity to generate cGAMP, since they express cGAS, but are incapable of inducing IFNβ expression in the absence of STING. Similarly, cells knocked down for cGAS cannot generate cGAMP, but retain their ability to activate IFNβ signaling because STING can respond to cGAMP provided in trans. Therefore, we hypothesized that IFNβ expression would be rescued if the cells knocked down for cGAS or STING were mixed prior to infection (Fig 6A). siRNA knock down of these proteins (cGAS or STING) was carried out in HeLa cells as before and cells were mixed at varying ratios with equal cell number in all wells. IFNβ expression was rescued by >10–20 fold in infected cells that were co-cultured relative to cells with cGAS or STING knock downs, respectively (Fig 6B). Chlamydial growth was similar in co-cultured cells relative to individually plated cells as evidenced by 16s rRNA levels (data not shown). Cells transfected with ISD served as a positive control and showed a similar trend of rescue of IFNβ expression resulting from cGAMP transfer (Fig 6C). GAP junction inhibitors such as Carbenoxolone, were able to abrogate cGAMP transfer in co-cultured cells transfected with ISD (data not shown) as previously reported (30), but could not be used in conjunction with infected cells as they also function as Pannexin inhibitor and abrogated chlamydial growth (31). Cell-cell contact was essential for IFNβ rescue to occur during co-culture, since IFNβ rescue was compromised when the STING KD and cGAS KD cells were cultured separately on a transwell (Fig S2). Together, these results provide indirect evidence for generation of cGAMP during infection and its transfer from infected cell to adjacent cell through gap junctions.
Figure 6. Evidence of cGAMP transfer from infected cells to adjacent cells.
Schematic representation of the experimental plan (A). STING−cGAS+ can produce cGAMP but cannot induce IFNβ since they lack STING, while STING+cGAS− cells cannot produce cGAMP upon infection. Co-culture can rescue IFNβ expression during infection if cGAMP from cGAS+ cells can migrate into STING+ cells. HeLa cells knocked down for STING (>99% KD) or cGAS (>95% KD) were cultured individually or co-cultured at different ratios 18 h before infection in 24 well dishes (2 × 105 cells/well). The mix numbers (1, 2, and 3) represent cGAS KD:STING KD cells in the indicated ratios. Cells were infected with C. muridarum at 3 MOI and IFNβ mRNA measured at 24 h post infection (B). A parallel set of cells were transfected with ISD as a positive control, 6 h before harvesting all the cells for RNA (C). Data is represented as mean of percent decrease relative to NT control from three experiments with SD. Significance determined by one way ANOVA with multiple comparison tests and indicated. Differences between Mix 1:3 and 1:1 co-culture were not significant.
DNA is the predominant ligand inducing IFNβ expression during chlamydial infection
Recently it was reported that chlamydial EBs produce the bacterial second messenger cyclic di-AMP, which can directly bind to STING to induce IFNβ expression (24). This was shown by infecting HEK293T cells overexpressing STING, and transfected with IFNβ promoter-driven luciferase reporter construct. In light of our finding that cGAS is required for chlamydia-induced IFNβ, we investigated the contribution of STING in the presence or absence of cGAS, to determine the relative contributions of DNA and chlamydial cyclic di-AMP to IFNβ expression. HEK293 cells (without T antigen) express STING but not cGAS, while HEK293T (with T antigen) have undetectable cGAS and STING protein, as reported earlier (20) contrasting with HeLa cells, which express high levels of both STING and cGAS (Fig 7A). The STING protein in HEK293 cells was fully functional as evidenced by high levels of IFNβ expression in response to the commercially available STING ligand 2’3’cGAMP (Fig 7B), but these cells expressed very low levels of endogenous IFNβ expression (2-fold increase) in response to chlamydial infection (Fig 7C). These data suggested that STING expression alone was insufficient to drive biologically relevant endogenous IFNβ expression. HEK293T cells were then used to examine the independent and combined role of cGAS and STING in IFNβ expression during infection. Co-transfection of cGAS and STING cDNA into HEK293T cells resulted in significant background IFNβ expression- a consequence of recognition of transfected DNA by cGAS protein in the presence of STING (20). Therefore, the alternative approach of examining the contribution of cGAS in trans was employed. HEK293T cells were transfected with STING or cGAS, which resulted in equivalent levels of STING or cGAS mRNA (data not shown). HEK293T cells transfected with STING were responsive to exogenous cGAMP but did not induce IFNβ expression to transfected DNA. Conversely, cGAS transfected cells were unresponsive to either cGAMP or ISD transfection in the absence of STING signaling (Fig 7D). When cGAS-expressing cells were co-cultured with STING-expressing cells, 6 hours before infection at a 1:1 ratio, IFNβ expression was rescued 40-fold, relative to infected cells expressing STING or cGAS alone (Fig. 7E). Chlamydial growth was similar in co-cultured cells to individually plated cells as evidenced by 16s rRNA levels (data not shown). Cells transfected with ISD served as positive control (Fig. 7E). These data suggest that cGAMP produced in cGAS-expressing cells migrated into STING-expressing cells to induce IFNβ expression during infection, and DNA transfection. Together these data demonstrate that STING alone is insufficient to rescue significant IFNβ expression in the absence of cGAS, and that both cGAS and STING function cooperatively to induce maximal IFNβ expression during chlamydial infection.
Figure 7. Both cGAS and STING are required for maximal IFNβ expression during chlamydial infection.
Protein levels of cGAS and STING in HeLa, HEK293T, and HEK293 cell lysates (A). Actin blots were carried out using 1/ 10th of cell lysates used for cGAS and STING western blots. HEK293 cells were permeabilized with cGAMP and IFNβ expression measured 6 h post treatment (B). HEK293 cells were infected with C. muridarum or transfected with ISD and IFNβ expression measured 24 h post infection or 6 h post treatment, respectively (C). HEK293T cells were transiently transfected with pcDNA3.1 (vector), STING, or cGAS expression constructs. Twenty-four hours post transfection, cells were trypsinized and plated individually or mixed at indicated ratio. Six hours after plating, individual cells were transfected with ISD or permeabilized with cGAMP (D). Individual or mixed cells were transfected with ISD or infected with C. muridarum (E). Expression of IFNβ measured at 24 h p.i or 6 h after ISD/cGAMP transfection. Data represents average of three experiments and error bars indicate SD. Significance was determined by one way ANOVA with multiple comparison test. For (D), p=0.004 for STING transfected-untreated (UT) vs cGAMP treated cells.
Discussion
A previous study from our laboratory showed that STING, the central adaptor molecule involved in cytosolic DNA recognition, is essential for IFNβ induction during chlamydial infection (25). Recent identification of the DNA sensor, cGAS that catalyzes the formation of cGAMP, a STING ligand (20), led us to investigate if this was the major pathway leading to IFNβ expression during chlamydial infection. Our results show that cGAS is required for IFNβ expression during infection with multiple Chlamydia strains in HeLa, epithelial cell lines that were derived from human Fallopian tube or mouse oviducts, mouse macrophages and HEK293T cells. cGAS meets the requirements for the DNA sensor during chlamydial infection, because it is specifically required for IRF3-mediated IFNβ expression and it is localized adjacent to inclusion membrane. This is the first report on a role for cGAS during intracellular bacterial infection, although its role as a DNA sensor during viral infection has been described (32).
The first evidence that DNA is a potential ligand for IFNβ expression during infection came from TREX1 KO cells. TREX1, a member of TREX-family of proteins, is essential for degradation of cytosolic DNA (33) and could function to reduce autoimmune response to host DNA. During HIV infection, host TREX1 targets HIV DNA to abrogate IFNβ expression, a classic example of viral hijacking of host machinery (28). Using siRNA knock down of TREX1 in mouse epithelial cells and HeLa cells and TREX1 KO cells, we determined that cytosolic DNA is targeted by these nucleases during chlamydial infection. Subsequent screening of multiple host DNA sensors did not identify a Chlamydia-specific sensor until we tested the role for cGAS in IFNβ expression and correlated its function to its cellular localization during chlamydial infection. In addition to demonstrating a role for cGAS in chlamydia-induced IFNβ expression, we also provide indirect evidence for generation of cGAMP during chlamydial infection and its transfer from infected cell to adjacent cells. These experiments were built on the recent finding that cGAMP can cross gap-junctions and activate STING in neighboring cells (30). These data suggest that during in vivo infection, IFNβ induced in uninfected cells adjacent to infected epithelial cells could either make them refractory to infection, as during viral infection or induce cell death pathways to affect tissue pathology.
It has been shown that the expression level of cGAS in multiple cell types is correlated to IFNβ expression in response to ISD (20). Therefore, the contribution of cGAS to IFNβ expression during chlamydial infection, relative to other receptors is likely dependent on their expression levels in multiple cell types. We have shown that cGAS is active in multiple cell types during chlamydial infection, including those where other candidate receptors e.g. TLR3 (34) have been associated with IFNβ expression. Interestingly, cGAS is a bonafide IFN-inducible protein (35) and we have observed a significant increase in cGAS expression following infection or ISD transfection. Therefore, during in vivo infection, both IFNβ and IFNγ can induce cGAS expression in epithelial cells. Analysis of cGAS and STING gene knock out mice should provide a more definitive answer on the relative contribution of cGAS over other receptors on IFNβ expression in the genital tract during in vivo genital infection.
Barker et al recently showed that Chlamydia trachomatis strain L2 generates the bacterial second messenger cyclic di-AMP which induced a IFNβ promoter-driven luciferase-reporter in HEK293T cells over expressing STING (24). Further the authors concluded that cyclic di-AMP is the predominant inducer of IFNβ during chlamydial infection using a mouse STING mutant (mR231A) defective for cyclic di-AMP binding (22, 36), in STING KO mouse fibroblasts. Our findings conflict with the conclusions of Barker et al (24) because our data supports a model where DNA sensing via cGAS rather than cyclic di-AMP is the predominant pathway of IFNβ induction during chlamydial infection of human epithelial cells, but without disregarding the findings that chlamydial EBs generate cyclic di-AMP. Our model is supported by the following observations. Firstly, if recognition of cyclic di-AMP by STING was sufficient to drive IFNβ expression during chlamydial infection, cGAS knockdown should not have altered IFNβ expression significantly in multiple cell types that express abundant and functional STING. Secondly, STING expression was insufficient to rescue endogenous IFNβ expression during infection in cell lines expressing a functional STING but not cGAS, such as HEK293 cells. Finally, HEK293T transfected with cGAS or STING did not induce significant endogenous IFNβ expression during chlamydial infection. However co-culturing the cells rescued expression, further confirming the requirement of cGAS-mediated cGAMP generation and transfer during infection. cGAMP binds to the same pocket in STING as cyclic di-AMP/di-GMP, but at much lower concentration with higher affinity (21). Indeed, the cGAS product, 2’3’cGAMP, is a much more potent ligand of STING than all other bacterial cyclic di-nucleotides described (37). Further, human STING is responsive only to cGAMP and unresponsive to the STING ligands CMA (38) and cyclic di-AMP/cyc di-GMP (39), unlike mouse STING which is responsive to both cyclic dinucleotides and cGAMP (40). These studies combined with our findings would significantly shift the importance of cGAMP over bacterial cyclic dinucleotides during Chlamydia trachomatis infection in human cells.
An important question that remains unclear at present is what is the source of the cytosolic DNA during infection? We speculate that the source of the DNA is chlamydial. Mitochondrial damage has not been observed at 24 h p.i (data not shown). Further, addition of chloramphenicol to block chlamydial growth after inclusion formation abrogates IFNβ expression, confirming the consistent requirement of bacterial growth (41). The localization of cGAS in punctate regions around the inclusion is also suggestive of a chlamydial source for the DNA. Manzanillo et al. have shown that during Mycobacterium tuberculosis infection, phagosomal permeabilization mediated by the bacterial ESX-1 secretion system allows cytosolic recognition pathways access to DNA (29). Numerous studies have linked IFNβ expression to bacterial secretion systems (42–45) and we have shown that IFNβ expression is abrogated in C. muridarum infected cells exposed to drugs that inhibit type III secretion system (T3SS) (41), suggesting a similar role for chlamydial T3SS in permeabilization of inclusion membrane. Previous studies (46) have shown that chlamydial reticulate bodies (RB) make direct contact with the inclusion membrane, likely through T3SS. These could be potential permeabilization points where nucleic acids could leak into cytosol and made available for host recognition. It is important to note that only viable chlamydiae induce IFNβ expression. This would suggest that condensed DNA from UV-killed chlamydial EBs do not provide an acceptable form of DNA for recognition by cGAS. While EB are a known source of cyclic di-AMP (24), when these developmental forms transition into RB, and RB replicate, this may initiate and accelerate DNA recognition, significantly amplifying IFNβ expression levels. Lateral gene transfer has been shown to occur between chlamydial RBs (47), consistent with a model where extra-chlamydial DNA is available for sensing and supporting the possibility of DNA transfer into cytosol, although DNA could be also released as a passive process resulting from non-viable RBs inside the inclusion. It has been shown that Chlamydia hijacks the host ER and several ER proteins were found localized on inclusion membrane (48, 49). The localization of the ER protein STING (25) and cytosolic cGAS in close proximity to the inclusion membrane suggest that STING could serve as a membrane scaffold for the interactions between DNA-cGAS to take place. Activation of STING would then result in phosphorylation and nuclear translocation of the critical transcription factor IRF3 for IFNβ expression (Fig 8).
Figure 8. A schematic model of cGAS recognizing DNA during Chlamydia infection, in human epithelial cells.

Left panel shows an electron micrograph of an inclusion, where chlamydial RBs are in close contact with the inclusion membrane and rough ER (arrows). Right panel shows a hypothetical model of DNA recognition by cGAS leading to cGAMP generation, STING activation and IRF3 phosphorylation, resulting in IFNβ expression during infection. Contribution of TREX-1 in diminishing this response is also shown.
An IFNβ response occurs during a vast number of intracellular infections, arising from bacteria that can occupy diverse niches in the cell (cytosolic, lysosomal, or vacuolar). In all cases cytosolic DNA could be available for sensing, thus we predict that cGAS may be a common recognition mechanism for sensing other intracellular pathogens. Further, the intensity of IFNβ expression could be directly correlated with the availability of foreign DNA in the cytosol, with cytosolic pathogens inducing a stronger response relative to those sequestered in membrane organelles. Collectively, our data demonstrates that cGAS is a novel pathogen recognition receptor involved in recognition of chlamydial infection and implicate cytosolic DNA-recognition during infection as an inducer of this response. The requirement of cGAS for chlamydia-induced IFNβ expression also provides a novel therapeutic target to block this response using a cGAMP antagonist in order to protect against oviduct disease during genital chlamydial infection.
Supplementary Material
Acknowledgements
A significant portion of the work was carried out at Department of Pediatrics, University of Pittsburgh. TREX1 KO and WT mouse embryonic fibroblasts were provided by Cancer Research (UK). BM1.11 cells were provided by Dr. Raymond Johnson (Indiana University). We thank Dr. Toni Darville and Dr. Catherine O’ Connell (University of North Carolina, Chapel Hill) for helpful suggestions, and Theodor Danciu (University of Pittsburgh) for laboratory assistance.
Abbreviations used in this paper
- cGAS
cyclic GMP-AMP synthase
- CXCL10
C-X-C-motif chemokine 10
- cGAMP
cyclic GMP-AMP
- ER
Endoplasmic reticulum
- IRF3
IFN regulatory factor 3
- MOI
multiplicity of infection
- siRNA
small interfering RNA
- STING
stimulator of interferon genes
- TEM
Transmission electron microscopy
- TREX-1
Three prime repair exonuclease-1
Footnotes
The project described was supported largely by PHS (NIAID) grant AI067678 (UN) and partly by GM083144 (VL).
References
- 1.Xia M, Bumgarner RE, Lampe MF, Stamm WE. Chlamydia trachomatis infection alters host cell transcription in diverse cellular pathways. J Infect Dis. 2003;187:424–434. doi: 10.1086/367962. [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen SJ, Eckmann L, Quayle AJ, Shen L, Zhang YX, Anderson DJ, Fierer J, Stephens RS, Kagnoff MF. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J Clin Invest. 1997;99:77–87. doi: 10.1172/JCI119136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lad SP, Fukuda EY, Li J, de la Maza LM, Li E. Up-regulation of the JAK/STAT1 signal pathway during Chlamydia trachomatis infection. J Immunol. 2005;174:7186–7193. doi: 10.4049/jimmunol.174.11.7186. [DOI] [PubMed] [Google Scholar]
- 4.Nagarajan UM, Prantner D, Sikes JD, Andrews CW, Jr, Goodwin AM, Nagarajan S, Darville T. Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect Immun. 2008;76:4642–4648. doi: 10.1128/IAI.00629-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu H, Fan Y, Joyee AG, Wang S, Han X, Bai H, Jiao L, Rooijen NVan, Yang X. Type I IFNs enhance susceptibility to Chlamydia muridarum lung infection by enhancing apoptosis of local macrophages. J Immunol. 2008;181:2092–2102. doi: 10.4049/jimmunol.181.3.2092. [DOI] [PubMed] [Google Scholar]
- 6.Prantner D, Sikes JD, Hennings L, Savenka AV, Basnakian AG, Nagarajan UM. Interferon regulatory transcription factor 3 protects mice from uterine horn pathology during Chlamydia muridarum genital infection. Infection and Immunity. 2011;79:3922–3933. doi: 10.1128/IAI.00140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagarajan U. Induction and function of IFNbeta during viral and bacterial infection. Crit Rev Immunol. 2011;31:459–474. doi: 10.1615/critrevimmunol.v31.i6.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF (3) Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 13.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature immunology. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Chen R, Zhou Q, Xu Z, Li C, Wang S, Mao A, Zhang X, He W, Shu HB. LSm14A is a processing body-associated sensor of viral nucleic acids that initiates cellular antiviral response in the early phase of viral infection. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11770–11775. doi: 10.1073/pnas.1203405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. STING-Dependent Recognition of Cyclic di-AMP Mediates Type I Interferon Responses during. Chlamydia trachomatis Infection. MBio. 2013:4. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prantner D, Darville T, Nagarajan UM. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J Immunol. 2010;184:2551–2560. doi: 10.4049/jimmunol.0903704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson RM. Murine oviduct epithelial cell cytokine responses to Chlamydia muridarum infection include interleukin-12-p70 secretion. Infect Immun. 2004;72:3951–3960. doi: 10.1128/IAI.72.7.3951-3960.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YL, Lee KF, Xu JS, Wang YL, Tsao SW, Yeung WS. Establishment and characterization of an immortalized human oviductal cell line. Mol Reprod Dev. 2001;59:400–409. doi: 10.1002/mrd.1046. [DOI] [PubMed] [Google Scholar]
- 28.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type (1) Nature immunology. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium Tuberculosis Activates the DNA-Dependent Cytosolic Surveillance Pathway within Macrophages. Cell host & microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKuen MJ, Dahl G, Fields KA. Assessing a potential role of host Pannexin 1 during Chlamydia trachomatis infection. PLoS One. 2013;8:e63732. doi: 10.1371/journal.pone.0063732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Molecular cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Derbigny WA, Johnson RM, Toomey KS, Ofner S, Jayarapu K. The Chlamydia muridarum-induced IFN-beta response is TLR3-dependent in murine oviduct epithelial cells. J Immunol. 2010;185:6689–6697. doi: 10.4049/jimmunol.1001548. [DOI] [PubMed] [Google Scholar]
- 35.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Shi H, Wu J, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Molecular cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conlon J, Burdette DL, Sharma S, Bhat N, Thompson M, Jiang Z, Rathinam VA, Monks B, Jin T, Xiao TS, Vogel SN, Vance RE, Fitzgerald KA. Mouse, but not Human STING, Binds and Signals in Response to the Vascular Disrupting Agent 5,6-Dimethylxanthenone-4-Acetic Acid. Journal of immunology. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, Hartmann G, Barchet W, Tuschl T, Patel DJ. Structure-function analysis of STING activation by c[G(2',5')pA(3',5')p] and targeting by antiviral DMXAA. Cell. 2013;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prantner D, Nagarajan UM. Role for the chlamydial type III secretion apparatus in host cytokine expression. Infect Immun. 2009;77:76–84. doi: 10.1128/IAI.00963-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Roux CM, Rolan HG, Santos RL, Beremand PD, Thomas TL, Adams LG, Tsolis RM. Brucella requires a functional Type IV secretion system to elicit innate immune responses in mice. Cell Microbiol. 2007;9:1851–1869. doi: 10.1111/j.1462-5822.2007.00922.x. [DOI] [PubMed] [Google Scholar]
- 44.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 45.Crimmins GT, Herskovits AA, Rehder K, Sivick KE, Lauer P, Dubensky TW, Jr, Portnoy DA. Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc Natl Acad Sci U S A. 2008;105:10191–10196. doi: 10.1073/pnas.0804170105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson DP, Timms P, McElwain DL, Bavoil PM. Type III secretion, contact-dependent model for the intracellular development of Chlamydia . Bull Math Biol. 2006;68:161–178. doi: 10.1007/s11538-005-9024-1. [DOI] [PubMed] [Google Scholar]
- 47.DeMars R, Weinfurter J. Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance. J Bacteriol. 2008;190:1605–1614. doi: 10.1128/JB.01592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dumoux M, Clare DK, Saibil HR, Hayward RD. Chlamydiae assemble a pathogen synapse to hijack the host endoplasmic reticulum. Traffic. 2012;13:1612–1627. doi: 10.1111/tra.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giles DK, Wyrick PB. Trafficking of chlamydial antigens to the endoplasmic reticulum of infected epithelial cells. Microbes Infect. 2008 doi: 10.1016/j.micinf.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.