Abstract
Cesium-137 is a radionuclide of concern in fallout from reactor accidents or nuclear detonations. When ingested or inhaled, it can expose the entire body for an extended period of time, potentially contributing to serious health consequences ranging from acute radiation syndrome to increased cancer risks. To identify changes in gene expression that may be informative for detecting such exposure, and to begin examining the molecular responses involved, we have profiled global gene expression in blood of male C57BL/6 mice injected with 137CsCl. We extracted RNA from the blood of control or 137CsCl-injected mice at 2, 3, 5, 20 or 30 days after exposure. Gene expression was measured using Agilent Whole Mouse Genome Microarrays, and the data was analyzed using BRB-ArrayTools. Between 466–6,213 genes were differentially expressed, depending on the time after 137Cs administration. At early times (2–3 days), the majority of responsive genes were expressed above control levels, while at later times (20–30 days) most responding genes were expressed below control levels. Numerous genes were overexpressed by day 2 or 3, and then underexpressed by day 20 or 30, including many Tp53-regulated genes. The same pattern was seen among significantly enriched gene ontology categories, including those related to nucleotide binding, protein localization and modification, actin and the cytoskeleton, and in the integrin signaling canonical pathway. We compared the expression of several genes three days after 137CsCl injection and three days after an acute external gamma-ray exposure, and found that the internal exposure appeared to produce a more sustained response. Many common radiation-responsive genes are altered by internally administered 137Cs, but the gene expression pattern resulting from continued irradiation at a decreasing dose rate is extremely complex, and appears to involve a late reversal of much of the initial response.
INTRODUCTION
The fission product 137Cs is an environmental contaminant of concern following nuclear accidents such as those at Chernobyl and Fukushima-Daiichi and is also an important component of fallout from nuclear detonations, such as an improvised nuclear device. As 137Cs is also commonly used in medical and research irradiators, it has recently become of concern as a possible component of a terrorist “dirty bomb,” which could result in widespread radioactive contamination. With a 30 year physical half-life, 137Cs persists long-term in soil or water. However, when inhaled or ingested 137Cs has a shorter biological half-life due to elimination from the body in urine. For instance, the highly soluble chemical form, 137CsCl, provides a fairly uniform total-body exposure to beta particles and gamma rays over an extended period of time, but at a decreasing dose rate as it is eliminated from the body. As an example, Melo et al. (1) used data from 137Cs-exposed humans and dogs to develop a biokinetic model. Their results showed for male adult humans that 15% of the intake was cleared with a 3 day half-life, and the remainder was cleared with a 90 day half-life.
Exposure to ionizing radiation, including internal emitters acquired from fallout or other environmental contamination, can have deleterious health consequences, ranging from acute radiation syndrome to increased risks of late effects such as cancer. In light of growing concerns over the possibility of radiological events such as industrial accidents or terrorist attacks, much recent effort has gone into improving methods of using biological indicators for rapid detection of radiation-exposed individuals within large potentially exposed populations, and for estimating exposure doses (2). We have pioneered the application of gene expression profiling in peripheral blood to develop biodosimetric signatures for acute external radiation exposures, using both ex vivo models (3–7) and patients undergoing total-body irradiation for bone marrow transplantation (8–10). Although the signatures appear to be useful across a range of after exposure times, the gene expression responses vary with time since exposure. Radiation exposures at different dose rates have also been shown to result in different gene expression patterns (11–15), further complicating the interpretation of results from such studies.
Contamination by internal emitters is likely to be a factor in a large-scale radiological incident. Measurement of radioactivity by whole-body counting would not be practical for rapid screening of large populations, although counting of spot urines could be considered as an initial screen. As part of the development of gene expression signatures to provide general radiation biodosimetry, it would be advantageous to determine if this approach might be extended to indicate cases of contamination by internal emitters. As a first step toward investigating the potential usefulness of gene expression in this regard, we have used cohorts of mice injected with a single 8.0 MBq dose of 137CsCl as an initial model of internal emitter effects, and monitored global gene expression in peripheral blood at a range of times after injection. C57BL/6 mice were selected for this study as a mouse strain with intermediate radiation sensitivity and no known DNA repair defects. This widely used inbred mouse strain is a standard for basic radiation biodosimetry studies (16–24). The injection dose used in this preliminary study was calculated to produce accumulated total-body doses of up to 10 Gy within 30 days of first exposure. This injection dose was chosen to allow us to cover the range of doses of most interest for triage (2–10 Gy) without inducing massive toxicity in the animals, while providing doses in the range of interest within the first week, the target timing for triage. The results from this preliminary study may provide groundwork useful for designing more complex future studies.
METHODS AND MATERIALS
Animals and Exposure
All animal experiments were conducted in accordance with applicable federal and state guidelines and were approved by the Institutional Animal Care and Use Committee of the Lovelace Biomedical and Environmental Research Institute (LBERI). Male mice were used for this study to avoid potentially confounding gene expression changes that may result from metabolic fluctuations due to the estrous cycle in female mice. Male C57BL/6 mice (approximately 10–12 weeks old, 25–30 g) were received from Charles River Laboratories (Frederick, MD) and were quarantined for 14 days prior to group assignment by body weight stratification for randomization into the study. Each mouse was individually weighed just prior to group assignment and 137Cs injection.
Animals were injected intraperitoneally with 8.0 ± 0.3 MBq 137CsCl solution in a volume of 50 µL. Nearly identical biokinetics have been found after inhalation, ingestion, intraperitoneal or intravenous administration of 137Cs in multiple species including mice, rats, dogs, sheep and cattle [summarized in refs. (25, 26)]. It also has been concluded that for homeotherms, the retention and distribution of soluble Cs in vivo is a function of metabolic rate and body size (27). The large body of interspecies data revealing nearly identical biokinetics regardless of the means of exposure suggests that intraperitoneal injection will provide biokinetics similar to exposure routes such as inhalation or ingestion. In addition, unpublished work in our laboratory has demonstrated that the biokinetics of 137Cs in C57BL/6 mice are similar whether 137CsCl is delivered intravenously or intraperitoneally. Given the apparent equivalence of exposure modes, intraperitoneal administration was used in the current study, rather than inhalation or ingestion, as this greatly simplifies the delivery of a consistent and accurate dose. After dose administration, mice were housed individually in microisolator cages with lead shielding used to minimize cross-irradiation from adjacent mice. All animals had unlimited access to Teklad Certified Global Rodent Diet 2016 (Harlan Teklad, Madison, WI) and water except during dose administration and whole-body in vivo counting. No deaths or other adverse effects occurred during the study.
Biokinetics and Dosimetry of 137Cs in Mice
Animals were radioassayed for 137Cs whole-body content using the LBERI in vivo photon counting system, consisting of a single 5” diameter Phoswich [dual NaI(Tl) – CsI(Tl) detector and associated pulse height analysis electronics]. Animals were placed in small containers, with breathing holes, and measured to determine the amount of radioactivity present in each animal daily on days 0–7, then on days 10, 14, 17, 20, 23, 27 and 30 after 137Cs administration (until the time of necropsy). There were a total of 40 mice treated with 137Cs in the full study, with 8 mice being sacrificed on days 2, 3, 5, 20 and 30, and each mouse being assayed for 137Cs levels up to and including their day of sacrifice. This means that 40 mice were assayed for 137Cs levels on days 0–2, 32 mice on day 3, 24 mice on days 4 and 5, 16 mice on days 6, 7, 10, 14, 17 and 20, and 8 mice were assayed for 137Cs levels on days 23, 27 and 30.
The measurement system was calibrated each day prior to sample analysis. For calibration, phantoms representing the animal body and samples were developed using a 137Cs NIST-traceable standard solution. Each animal or sample was measured for 3 min. The 137Cs whole-body retention profile for each individual mouse was derived from its whole-body 137Cs measurements. The whole-body retention data from each mouse was fitted individually to negative exponential functions. The average whole-body retention equation was determined to be:
R(t) = 21e−1.0t + 79e−0.096t
Where R(t) represents the whole-body content at time (t), expressed as percentage of the injected 137Cs activity; and t is time in days. The decay constants (λ) of the two terms of the retention equation were 1 per day and 0.096 per day, respectively. The biological half-time values (T1/2) were calculated from these decay constants using the equation:
T1/2 = ln(2)/λ
Thus the biological half time related to the first term of the whole-body retention equation was 0.7 day, and that related to the second term was 7.21 days.
To convert the empirical 137Cs retention data into radiation absorbed dose, specific absorbed fractions for electrons and photons published by Stabin et al. (28), and which were developed specifically for young adult mice and rats, were converted into a dose coefficient (dose in Gy per unit decay of 137Cs in equilibrium with 137mBa), and integrated over the experimental lifespan from 137Cs injection to sacrifice for each individual mouse. Since 137CsCl is highly soluble and results in a relatively uniform biological distribution of 137Cs throughout the body, only the whole-body dose was calculated and used for this study. The current study was designed to deliver whole-body absorbed doses of 2, 3, 4, 9 and 10 Gy to the animals. The same 137Cs activity was injected into each animal, and the euthanasia time points after 137Cs administration were selected to result in the delivery of these doses to the whole body of the mice.
On scheduled necropsy days, animals were euthanatized by i.p. injection of Euthasol [>150 mg/kg (390 mg/mL pentobarbital and 50 mg/mL phenytoin in sterile saline)] and each mouse was weighed individually. Whole blood was collected by cardiac puncture in a sterile hood. For each animal 0.4 mL of blood was collected into 2 mL of PAXgene Blood RNA stabilization and lysis solution (Becton, Dickinson Company, Franklin Lakes, NJ), mixed thoroughly and shipped on ice.
For external gamma-ray exposure, 5 mice were exposed to 2.8 Gy gamma rays at the Center for Radiological Research at a dose rate of 0.82 Gy/min using a Gammacell-40 137Cs irradiator (AECL, Ontario, Canada), and 5 mice were mock irradiated as controls. Mice were sacrificed 3 days after exposure. Blood was collected by cardiac puncture and placed in PAXgene solution as above.
Microarray Hybridization and Data Extraction
RNA was purified following the directions in the PAXgene Blood RNA kit (Becton, Dickinson Company), scaling for the input volumes. Prior to microarray processing, globin transcripts were reduced using the Ambion GLOBINclear-Mouse/Rat kit (Life Technologies, Grand Island, NY). RNA yields were quantified using the NanoDrop ND1000 Spectrophotometer (Thermo Scientific, Wilmington, DE) and quality was checked using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA used for microarray hybridization had an average RNA Integrity Number (29) of 8.9.
Cyanine-3 (Cy3) labeled cRNA was prepared with the One-Color Low Input Quick Amp Labeling Kit (Agilent Technologies) according to the manufacturer’s instructions, followed by RNAeasy column purification (QIAGEN, Valencia, CA). Dye incorporation and cRNA yield were checked with the NanoDrop ND1000 Spectrophotometer; 1.5 µg of cRNA with incorporation of >15 pmol Cy3 per µg cRNA was fragmented and hybridized (17 h with rotation at 65°C) to Agilent Whole Mouse Genome Microarrays 4x44K v2 (G4846A), and then washed using the Gene Expression Hybridization Kit and GE Wash Buffers as recommended by Agilent. Slides were then scanned with the Agilent DNA Microarray Scanner (G2505B), and the images were analyzed (Agilent Feature Extraction Software v. 9.1) with default parameters for background correction and flagging nonuniform features.
Analyses with BRB-ArrayTools
Background-corrected hybridization intensities were imported into BRB-ArrayTools, Version 4.2.1 (30) log2-transformed and median normalized. Application of standard filtering parameters (9) yielded a set of 11,434 features that were used in subsequent analyses. The microarray data is available through the Gene Expression Omnibus with accession number GSE52690.
Due to marked time-dependent changes observed among controls at the later times, mice sacrificed at times up to day 5 were compared against the controls from days 3 and 5, while the day 20 and day 30 treated mice were compared only to their own controls. RNA from six individual mice was hybridized to a separate microarray for each point tested. Class comparisons were conducted using BRB-ArrayTools to identify genes that were differentially expressed between controls and 137Cs-exposed animals at each of the five sacrifice times using a random-variance t test (31). Genes with P values less than 0.001 were considered statistically significant. The false discovery rate (FDR) was also estimated for each gene using the method of Benjamini and Hochberg (32), to control for false positives. (33).
Gene Ontology Network Analyses
Lists of genes significantly over- or under-expressed relative to controls were imported separately into the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 to identify enriched biological themes and gene ontology (GO) terms using the functional annotation tool (33, 34). Benjamini corrected P values <0.05 were considered significant.
The significantly differentially expressed gene lists from each sacrifice time, along with their relative expression levels, were also imported into Ingenuity Pathways Analysis (IPA) (Ingenuity® Systems, http://www.ingenuity.com) and analyzed with the IPA Core Analysis and the Transcription Factor Analysis Tool. These analyses both use curated information on the published relationships between gene products to predict network information. The transcription factor analysis specifically uses information about the relationship between the activity of potential upstream regulatory factors and the expression changes of the measured genes to make predictions of which regulatory factors may be activated or inhibited. IPA generates a z score for each factor and uses a cutoff of z > 2 to predict activation and z < −2 to predict inhibition.
qRT-PCR
Total RNA was transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) following the manufacturer’s recommendations. Primers and probe specific for Phlda3 (forward primer: 5′-GGGCCTGGTCAAGTTCAAGA-3′; Reverse primer: 5′-CACATGTAGCCAGGTCCCAA-3′; Probe: 5′-ATCCAGACTGTGCGGGCCCG-3′) were designed using online TaqMan Primer Design software (GeneScript Corporation) and Primer Express® (Applied Biosystems). Probes were synthesized with 6-carboxyfluorescein (FAM) at the 5′ end and BHQ1quencer at the 3′ end by Operon Biotech, Inc. (Huntsville, AL). For all other genes, predesigned assays were purchased from Life Technologies: Rn18s (Mm03928990_g1), Gapdh (Mm99999915_g1), Aen (Mm00471554_m1), Bbc3 (Mm00519268_m1), Cdkn1a (Mm04205640_g1), Itga6 (Mm00434375_m1), Itgb5 (Mm00439825_m1), Nckap1 (Mm00550539_m1), Pls1 (Mm01223664_m1) and Unc93b1 (Mm00457643_m1). Standard curves were generated to optimize the input amount of cDNA used for each gene (5 or 10 ng). Real-time PCR reactions were carried out in duplicate using the ABI 7900 Real Time PCR System with Universal PCR Master Mix (Applied Biosystems) following the manufacturer’s protocols. Relative fold inductions were calculated using the ΔΔCT method as described previously (8) with Rn18s or Gapdh used for normalization. To test the significance of differential expression, unpaired two-tailed t tests were performed comparing the normalized CT values for each gene for control and treated samples at each time point using the GraphPad Software web tool at http://graphpad.com/quickcalcs/ttest1/. P values below 0.05 were considered significant.
RESULTS
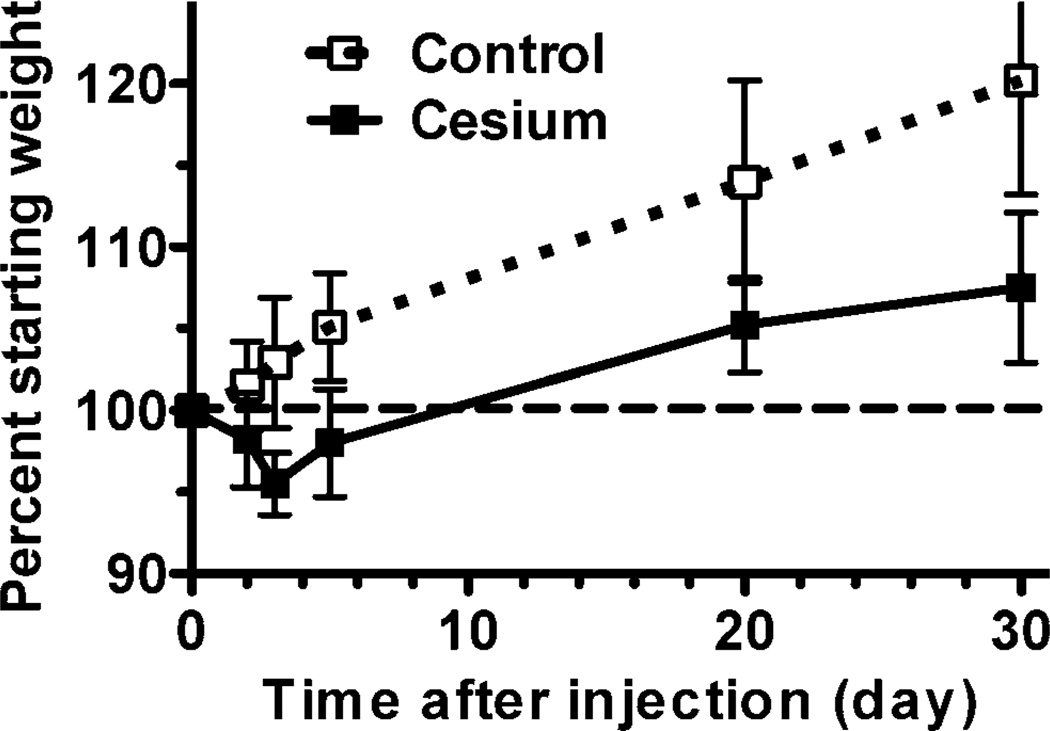
All animals remained in apparent good health, with no adverse events noted during the course of the study. Control mice gained weight steadily throughout the study. Mice injected with 137CsCl initially lost weight, losing up to an average of 4.5% of their starting weight by day 3 after injection, then resumed weight gain at approximately the same rate as the unexposed controls from day 3 onward (Fig. 1).
FIG. 1.
Changes in weight of mice during the study. Each mouse was weighed prior to the beginning of the study and again at the time of necropsy. The percentage of starting weight was calculated for each mouse relative to its own starting weight. The plot shows the mean percent starting weight for each treatment group and time (n = 8). Error bars are standard deviation. The dashed line at 100% indicates starting weight percentage.
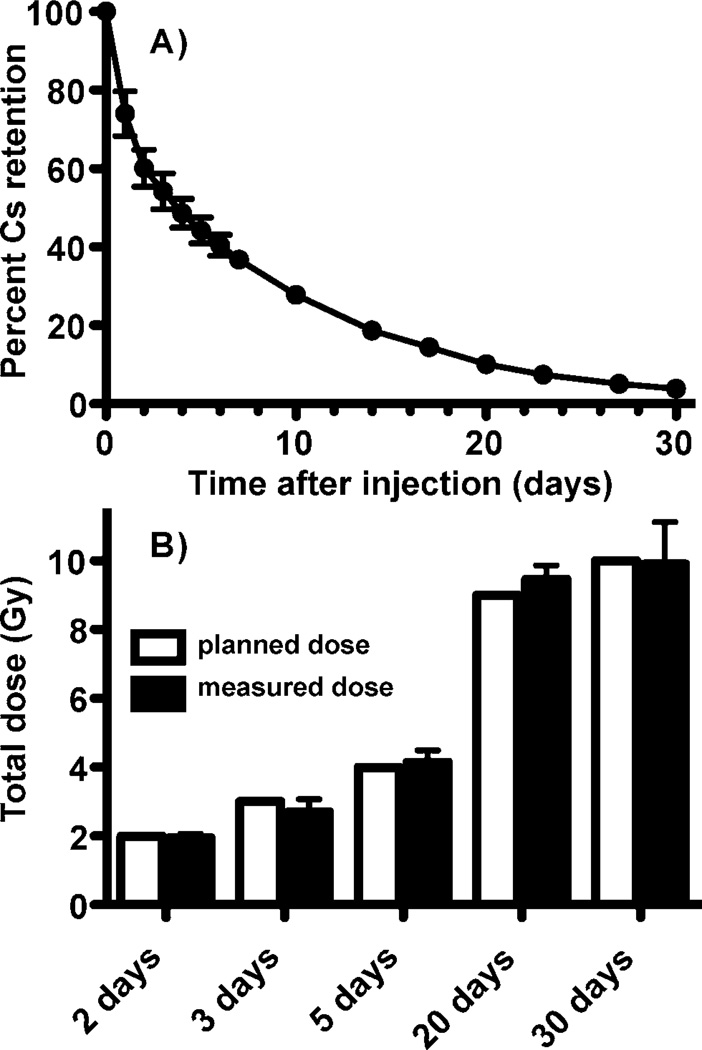
The relative retention of 137Cs over the course of the experiment is shown in Fig. 2A, where the 137Cs content in the animals on each day of measurement is represented as a fraction of the radioactivity present immediately following 137Cs administration. After necropsy, this information was used to reconstruct the actual dose to each individual mouse. The mean absorbed dose, calculated from the whole-body retention profiles, was very close to the intended absorbed dose for each day of sacrifice, as calculated in the experimental plan, with little variation among individual mice (Fig. 2B).
FIG. 2.
Panel A: Retention of 137Cs over the course of the study. The percentage of the original injected activity remaining at each time is plotted as the average for all animals on the study at each assay time (days 0–2, n = 40; day 3, n = 32; days 4–5, n = 24; days 6–7, 10, 14, 17 and 20, n = 16; days 23, 27 and 30, n = 8). Error bars are standard deviation. Panel B: The measured dose for each animal at the time of sacrifice was determined from whole-body counts and is shown averaged across animals for each day of sacrifice (n = 8; filled bars). Error bars are standard deviation. The doses planned in our experimental design are shown for comparison (open bars).
Microarray Experiments
Global gene expression was measured in the blood of mice sacrificed at 2, 3, 5, 20 and 30 days after injection of 137Cs, and in time-matched controls. Agilent whole genome microarrays were hybridized for six mice per point using the Agilent one-color workflow. Filtering for quality and minimum change yielded a set of 11,434 features that were used in subsequent analyses. The class comparison feature of BRB-ArrayTools was used to identify genes with significantly different expression levels between controls and 137Cs-exposed animals at each sacrifice time (summarized in Table 1; Supplementary Table S1: http://dx.doi.org/10.1667/RR13466.1.S1). Despite the large numbers of genes responding at the various times, only five genes were identified as significantly different from controls at all times assayed (Rnase6, March1, Bank1, Unc93b1 and Mmd).
TABLE 1.
Summary of Significantly Different Gene Expression
| Time (days) |
Accrued dose (Gy) |
No. features significant at P < 0.001 |
FDRa | Percentage over-expressed |
Percentage overlap with following time |
|---|---|---|---|---|---|
| 2 | 2 | 619 | <2% | 72.5 | 71 |
| 3 | 2.7 | 1493 | <1% | 81.7 | 29 |
| 5 | 4.1 | 466 | <2.5% | 7.5 | 90 |
| 20 | 9.5 | 6375 | <1% | 28.9 | 80 |
| 30 | 9.9 | 6213 | <1% | 30.4 | NAb |
False discovery rate.
Not applicable.
Gene Ontology Analysis
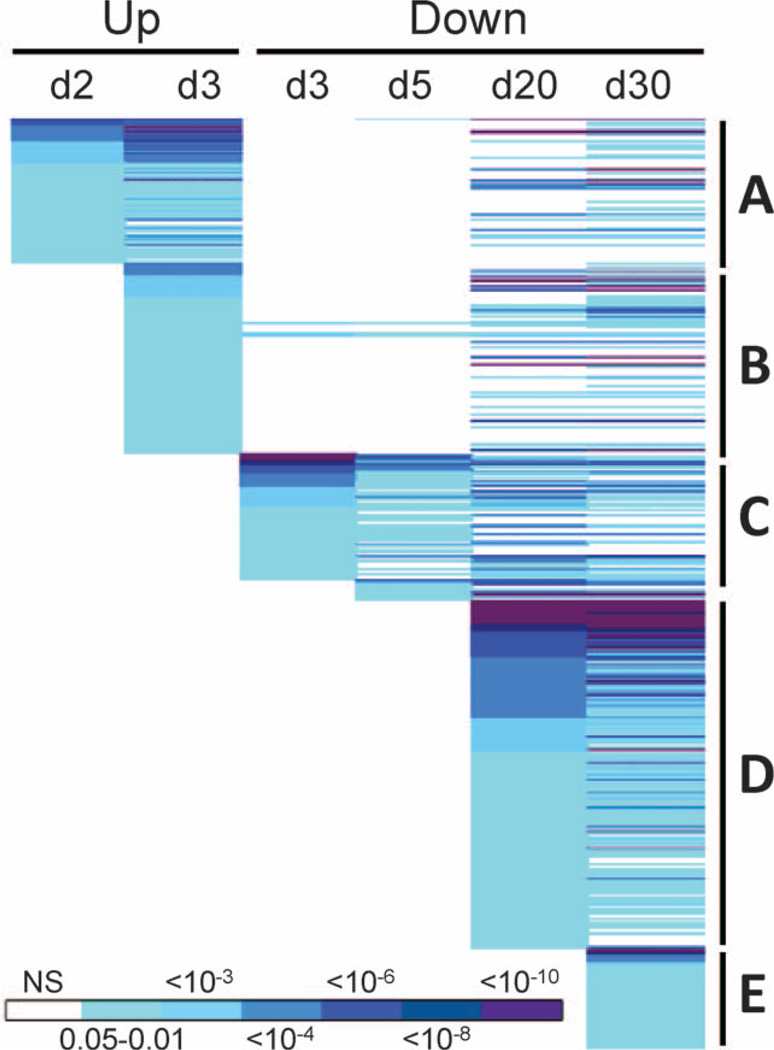
We next used the DAVID functional annotation tool (33, 34) to look for significantly overrepresented gene ontology (GO) categories among the genes significantly differentially expressed after 137CsCl administration. A large number of functional categories were found to be significantly enriched (Benjamini-corrected P < 0.05) among genes with differential expression at the individual times (Fig. 3; Supplementary Table S2; http://dx.doi.org/10.1667/RR13466.1.S2). Among genes expressed above control levels, significant GO categories were only found at days 2 and 3. In contrast, underexpressed genes yielded significant GO categories at all times except day 2. The biological functions implicated by the significant GO categories suggest a complex and evolving response to exposure to the internal emitter. Interestingly, some functions even appeared to be up-regulated at early times, and then suppressed at later times (Fig. 3).
FIG. 3.
Common GO terms significantly enriched among overexpressed genes (Up) from day 2 after 137Cs injection (A) included cytoskeleton, actin, plasma membrane, protein modification, nucleotide binding, blood clotting and adhesion functions. Beginning on day 3 (B) integrin signaling, cell adhesion, protein localization and transport, cell junctions, tubulin, differentiation and apoptosis were seen. Some of these same functions were later significantly enriched among underexpressed (Down) genes, including actin, cytoskeleton, apoptosis, protein localization, transport, and modification and nucleotide binding. Common GO terms significantly enriched among underexpressed genes (Down) from day 3 after 137Cs injection (C) included many immune functions, such as immune cell activation, proliferation, and differentiation, cytotoxicity and antigen processing. Beginning on day 20 (D) after 137Cs injection, functions related to mRNA processing, ribosome, spliceosome, mitochondria, catabolism, proteasome, apoptosis and additional immune functions were enriched. Functions associated with genes only underexpressed on day 30 after 137Cs injection (E) were similar to those seen at day 20 and included additional mitochondrial, ribosomal and apoptosis related functions. Benjamini-corrected P values for each function are color coded according to the scale bar at the bottom of the figure. The full annotation of all GO categories is available in Supplementary Table S2 (http://dx.doi.org/10.1667/RR13466.1.S2).
Network Analysis
We next ran an Ingenuity Pathway Analysis (IPA) core analysis using the differentially expressed genes from all times. From this we identified several IPA canonical pathways that were significantly overrepresented at the various times. These shared broad similarity with the enriched GO categories, and included actin cytoskeleton signaling and adherens junctions early on, with T- and B-cell pathways, and mitochondrial dysfunction emerging later on. Integrin signaling was the top canonical pathway at both days 2 and 3, dropped from significance at day 5, and then reemerged as a significant pathway at days 20 and 30, which was not seen in the GO analysis. Interestingly, the genes in this pathway were overexpressed at the early times, then underexpressed at the later times, following the pattern seen for many of the GO categories.
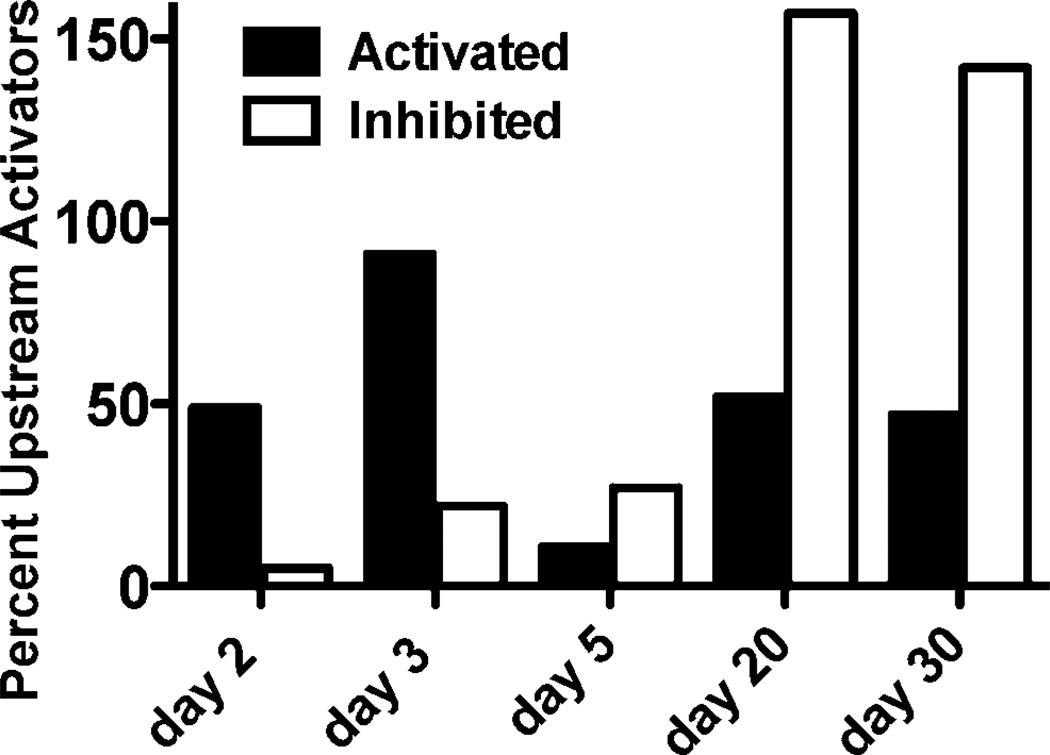
We also used the IPA transcription factor analysis to predict regulatory molecules that could be drivers of the time-dependent gene expression response to internal 137CsCl exposure. This analysis identified many potential upstream regulators of the gene expression responses observed at the different days after 137CsCl injection (Fig. 4). The largest number of activated regulatory factors was predicted at day 3, and the largest number of inhibited regulatory factors at day 20. Although IPA did not predict any single regulatory factor to be either activated or inhibited across the entire time course, it did predict activation of Trim24 and Mapk1, and inhibition of Irf7 and Parp9 at all times after day 2. IPA also predicted activation of the major radiation response transcription factor Tp53 at days 2 and 3 after 137CsCl injection, and inhibition of Tp53 at days 20 and 30 after exposure.
FIG. 4.
Number of potential upstream regulators predicted by the IPA analysis to be activated (filled bars) or inhibited (open bars) on each day of the study.
Quantitative Real-Time RT-PCR
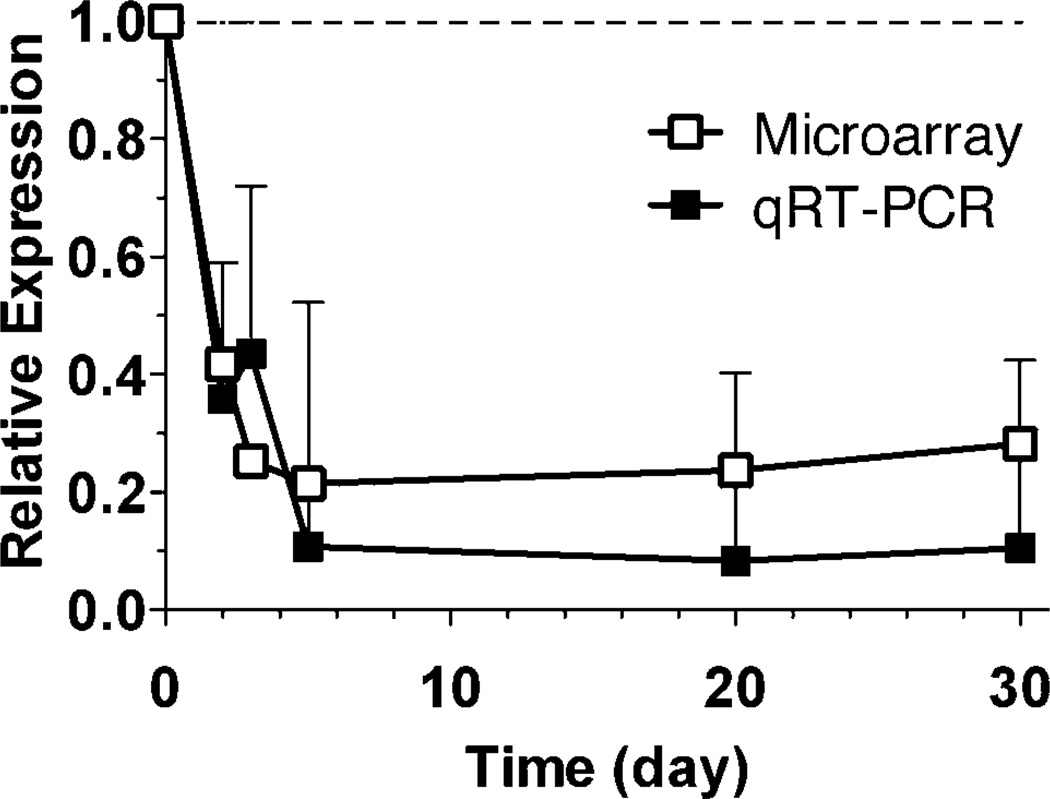
To confirm some of the prominent patterns of gene expression emerging from the microarray data, we selected several of the responding genes for analysis by quantitative real-time RT-PCR (qRT-PCR). Unc93b1 was selected as a representative of the small number of genes significantly underexpressed by microarray at all times assayed after 137Cs injection and showed significant underexpression by qRT-PCR (P < 0.05) at all times (Fig. 5).
FIG. 5.
Expression of Unc93b1 compared by microarray (open symbols) and quantitative real-time RT-PCR (filled symbols). The symbols are the average of six samples, error bars are standard error of the mean. All points were significantly below control values as measured by both microarray (P < 0.001) and qRT-PCR (P < 0.05). The dashed line represents the level in unexposed controls.
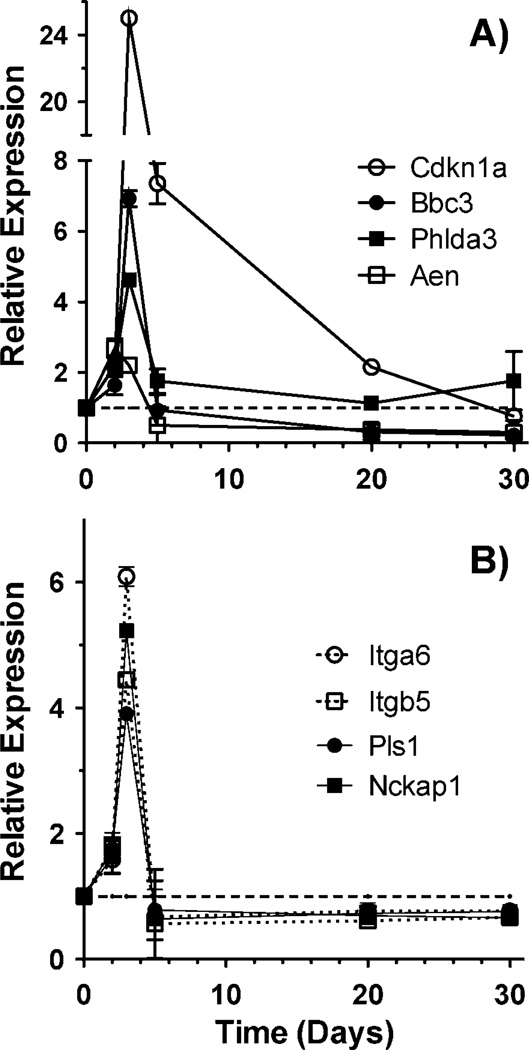
The Tp53 response genes Cdkn1a and Phlda3 also showed the same time-dependent pattern of relative expression evident from the microarray data, with expression rising over the first few days after injection to 25-fold and 4.6-fold by day 3, then dropping to near control levels by day 30 despite continued accumulation of total dose. Two other Tp53 response genes, Aen and Bbc3, after rising to peaks of 2.7-fold at day 2 and 6.9-fold at day 3, respectively, declined to around three- to fourfold below control expression levels, maintaining expression levels significantly below controls (P < 0.05) at days 20 and 30 post injection (Fig. 6A). This pattern was consistent with that of many Tp53 regulated genes on the microarray, and with the switch from Tp53 activation to inhibition predicted by the IPA transcription factor analysis.
FIG. 6.
Expression patterns of Tp53 regulated genes (panel A) and genes involved in integrin signaling or actin/cytoskeleton functions (panel B) measured by qRT-PCR. Points are the average of 6 samples, error bars are standard error of the mean. All points were significantly different from controls (P < 0.05) with the exception of Cdkn1a at day 30, Phlda3 at days 20 and 30, Bbc3 at days 2 and 5, and Aen, Itga6, Itgb5, Pls1 and Nckap1 at day 5. The dashed line represents the level in unexposed controls.
As our GO and network analyses indicated a similar switch from elevated expression to below control value expression levels for many functional categories, we also looked at genes representative of two of the broad categories showing this pattern, integrin signaling (Itga6 and Itgb5) and actin/cytoskeleton (Pls1 and Nckap1). These genes also showed the same pattern with qRT-PCR (Fig. 6B), a rapid significant increase in expression, followed by sustained expression levels significantly below those of controls (P < 0.05).
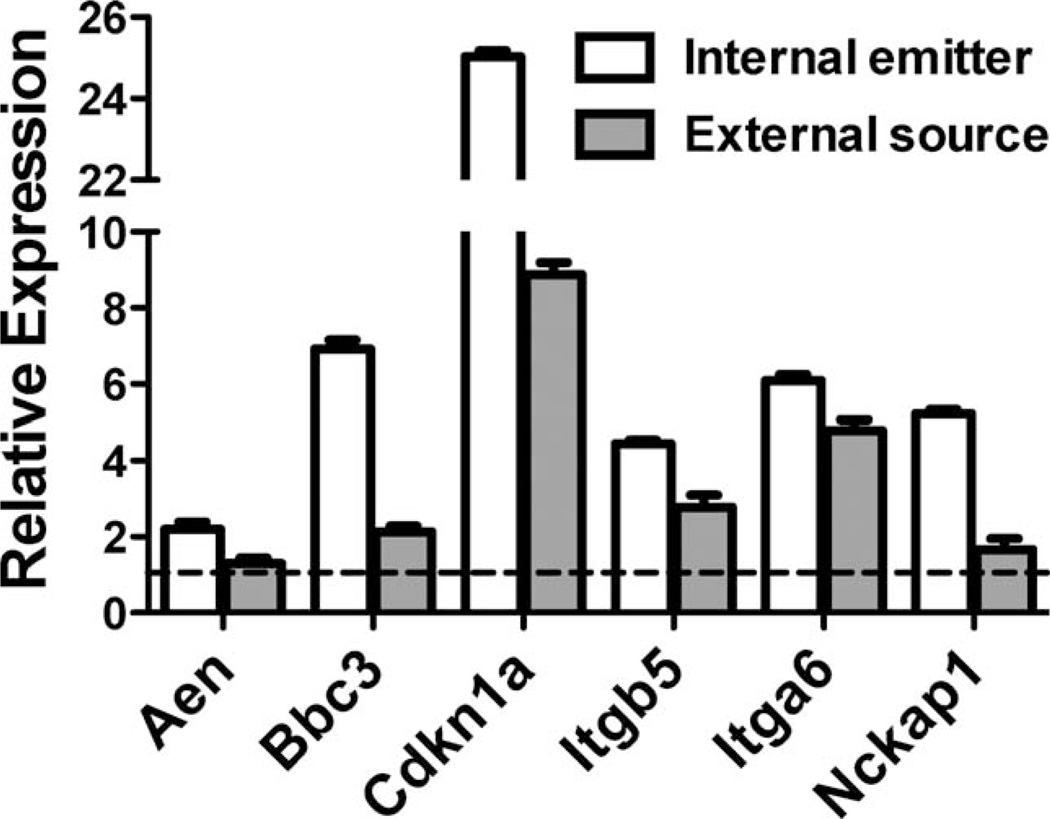
Finally, we measured expression of several of these genes in mice three days after a single acute external beam exposure to 2.8 Gy 137Cs gamma rays to provide an initial comparison of gene expression with the peak expression observed for these genes in our internal emitter study (Fig. 7). All genes measured were expressed above the levels in sham-irradiated mice, but the magnitude of overexpression was less in the mice receiving a single acute dose than in the mice receiving the dose as a chronic internal exposure, with Aen and Nckap1 not being significantly above control levels (P > 0.05).
FIG. 7.
Gene expression measured by qRT-PCR 3 days after administration of 137Cs as an internal emitter (open bars, data from Fig. 6) or 3 days after a single acute dose delivered externally (filled bars). Filled bars are the average of 5 samples, error bars are standard error of the mean. The total dose, 2.8 Gy, was the same in both cases. The dashed line represents the level in unexposed controls.
DISCUSSION
We have found that a single injection of 137CsCl resulted in altered expression of a large number of genes, and that perturbations in the pattern of gene expression continued throughout the first 30 days after exposure. Overall there was a striking difference between the response within the first 2–3 days and the response at 20–30 days post injection, with many biological processes and individual genes switching from overexpression relative to controls to underexpression. The day 5 time point showed fewer differentially expressed genes than other times, suggesting a general transition point at or around this time since the beginning of exposure.
The continual decrease in the rate at which the radiation exposure occurs may also affect the observed gene expression patterns. As the 137Cs is cleared from the body over time, dose accumulates at a lower rate. For instance, between days 3 and 5, the accrued whole-body dose increased by 1.4 Gy, representing an average rate of dose accumulation of approximately 0.49 mGy/min over the two-day interval. Similarly, between days 20 and 30, the accrued total body dose increased by 0.4 Gy, representing an average dose rate of 0.03 mGy/min during this ten-day interval. These are obviously crude estimates, as the actual dose rate is changing constantly throughout the experiment, but they give an idea of the range of dose rates involved in this experiment. Even aside from the complication of a changing dose rate, the effects of protracted irradiation on gene expression over time can be difficult to interpret and have not been studied in as much detail as the effects of single acute exposures.
Studies of gene expression after exposure to protracted external beam radiation have often focused on very low-dose rates and widely different assay times, making comparisons difficult. For instance, Gridley et al. exposed mice to a range of low doses of external beam 57Co at a dose rate of approximately 0.003 mGy/min (35), or to protons at approximately 0.02 mGy/min (36), and reported significant changes in the expression of several genes immediately after exposure. In another study, exposure to a range of low-dose rates at or below 0.013 mGy/min for more than 400 days resulted in small changes in a small number of genes (14). Another study exposed mice for 1–40 days to external gamma rays delivered at a constant 2.8 mGy/min (37) and reported a consistent overexpression of some p53 regulated genes. This may suggest the chronic activation of p53 above some dose rate threshold, in contrast to the reversal of p53 activity implied by our results as the dose rate declined.
Internal emitter studies of gene expression are even more limited. In a study using injection of 131I to produce a range of absorbed doses from 0.1–9.7 mGy (38), several hundred genes were found to be regulated in the lung and kidney of mice after 24 h. Although the doses were again much lower than those in the present study, the major biological process affected among responding genes was immune function, which was also prominent in our results. A study using injected 211At with absorbed doses from 0.05–32 Gy did span the dose range of our study, and at 24 h after exposure, detected effects on immune response, response to external stress, cell cycle regulation and proliferation (39), again consistent with our findings. Intriguingly, this study found many genes were regulated in the opposite direction by one of the intermediate doses (1.4 Gy) compared to all other doses tested, reminiscent of the reversal from overexpression to underexpression we found around day 5. This may suggest that the change from overexpression to underexpression observed in our study could be linked to a specific dose or dose rate. More large-scale experiments with multiple doses, dose rates and time points would be required to tease apart the factors that contribute to this response.
Despite significant differences in timing of exposure and expected response patterns, we have been able to confirm the response of several of the genes reported here to an acute external beam exposure (Fig. 7). Mice were exposed to an acute external dose of 137Cs gamma rays equivalent to the dose accrued by day 3 of the internal emitter study. Three days after the external gamma-ray exposure, we measured the expression of six genes that were overexpressed on day 3 of the internal emitter study, and found that all were expressed above baseline levels. Of the genes measured, Bbc3, Cdkn1a, Itgb5 and Itga6 were significantly over expressed (P < 0.05) three days after external beam exposure. In all cases, the fold change was less than that observed for the chronic exposure, possibly representing the return of gene expression toward baseline levels, as is commonly observed over time following acute exposures.
We also looked for reports in the literature of the five genes (Rnase6, March1, Bank1, Unc93b1 and Mmd) that were significantly differentially expressed at all times assayed. Although none were directly reported as radiation responsive, we used the Radiation Genes Database (40) to search published supplementary data. We found all of these genes changed by more than twofold in response to external beam X-ray or gamma-ray exposure in both human cancer cell lines (41, 42) and in mouse blood cells (43, 44). Together, this suggests that despite differences in the time course of response, many of the same genes likely respond to external acute and internal chronic exposures.
The effects of internal 137Cs on the mouse urinary metabolome have also recently been reported in a companion study to our gene expression study (45). The effect of 137Cs administration on metabolism was found to be most pronounced at earlier times, but changes were found to persist through day 30. Among the most prominent persistent changes were impaired mitochondrial function and fatty acid beta-oxidation. These metabolic changes are consistent with our gene expression findings that indicated significant decreases in biological functions including fatty acid metabolism, amino acid metabolism and the TCA cycle, as well as many mitochondrial functions at days 20 and 30.
Perhaps the most interesting pattern of response emerging from our gene expression study is the large number of biological processes that appear to be significantly activated at early times and then significantly inhibited at later times after internal deposition of 137Cs. One such pathway, integrin signaling, is known to play an important role in the response to radiation and other stresses, modifying cell–extracellular matrix (ECM) adhesion, cell survival and cell cycle arrest (46, 47). Integrins also play many important cell type-specific roles in lymphocytes and other immune cells, influencing processes such as cell migration, adhesion and development (48). Integrin signaling connects the ECM with the actin cytoskeleton to control some of these functions (49), and we found many GO categories related to actin and cytoskeletal functions overrepresented with the same pattern of early upregulation and later down-regulation, suggesting a major impact of this system in regulating both early and late responses. Interestingly, a study of clear-cell renal cell carcinomas from patients living in 137Cs contaminated areas near Chernobyl found decreased expression and altered distribution of ECM proteins compared to tumors from unexposed controls (50), suggesting that chronic exposure to 137Cs could result in broad remodeling of the ECM. The reported decreases in protein expression are consistent with the decreased gene expression at days 20 and 30 after 137Cs exposure in our study.
Network analysis-predicted inhibition of Tp53 transcriptional activity at days 20 and 30 represents yet another pathway with a reversal of its initial response. The Tp53 pathway is of special interest, as its stabilization and activation is known to be one of the major mediators of the early response to ionizing radiation. Less is known about its role in chronic or changing low-dose-rate exposures, however. The observed pattern of Tp53 response is consistent with some of the GO functions showing the same pattern, such as apoptosis and cell cycle. We also confirmed the pattern of early overexpression followed by late underexpression for the Tp53 regulated genes Aen and Bbc3, and the response of other genes, such as Cdkn1a and Phlda3, which returned to baseline levels without dropping significantly lower. The predicted Tp53 inhibition also coincided with a broad coordinate underexpression of transcripts for ribosomal proteins. Disruptions in rRNA synthesis or in the balance of ribosomal proteins have been shown to activate Tp53 and cell cycle arrest through interaction with the p53-mdm2 feedback pathway (51, 52). Conversely, depletion of Rpl6 was recently shown to lead to decreased levels of Tp53 through effects on protein stability (53), while expression of other ribosomal proteins was found to both stabilize Tp53 and alter the specific target genes transcribed (54). Ribosomal proteins have also recently been implicated in regulation of the response of Tp53 to DNA damage. Knockdown of Rps26 did not alter the stability of the Tp53 protein, but did impair its ability to activate its transcriptional target genes in response to the DNA damaging agent Doxorubicin (55). The significantly reduced expression of ribosomal protein transcripts observed at the times when Tp53’s transcriptional function was predicted to be impaired suggests the possibility that the ribosomal protein-mdm2-p53 regulatory pathway may play a role in the late response to chronic low dose-rate radiation damage in vivo. This will be an attractive area for future studies.
The effects of ongoing exposure at rapidly declining dose rates will likely complicate biodosimetry in instances where internal contamination occurs from exposure to radioactive fallout. The evolution of the gene expression response over time as revealed in this preliminary study suggests that gene expression could potentially provide information useful for the identification of internal emitter exposure. More detailed studies will obviously be required, but it may be possible to identify sets of genes that can serve as markers for different isotopes or levels of exposure. Much work is still needed to clarify both potential markers, and the mechanisms of biological response to these complex exposures.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr. Thomas Morgan of the Columbia University Radiation Safety Office for making special accommodations for receipt of the specimens. We would also like to acknowledge Drs. Michael Stabin and Luiz Bertelli for calculation of the murine dose coefficients, Dr. Norman Kleiman and Mr. Jeffrey D. Knotts for mouse irradiations and RNA isolation, and Ms. Mashkura Chowdhury for technical assistance with qRT-PCR. Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team. This work was supported by the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases grant no. U19 AI067773.
Footnotes
The online version of this article (DOI: 10.1667/RR13466.1) contains supplementary information that is available to all authorized users.
REFERENCES
- 1.Melo DR, Lipsztein JL, Oliveira CA, Lundgren DL, Muggenburg BA, Guilmette RA. A biokinetic model for 137Cs. Health Phys. 1997;73:320–332. doi: 10.1097/00004032-199708000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Williams JP, Huser AK, Brenner DJ. Overview of research from the centers for medical countermeasures against radiation: Introduction. Int J Radiat Biol. 2011;87:747. doi: 10.3109/09553002.2011.598263. [DOI] [PubMed] [Google Scholar]
- 3.Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, et al. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154:342–346. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 4.Amundson SA, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr Induction of gene expression as a monitor of exposure to ionizing radiation. Radiat Res. 2001;156:657–661. doi: 10.1667/0033-7587(2001)156[0657:iogeaa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 5.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 2008;71:1236–1244. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul S, Amundson SA. Gene expression signatures of radiation exposure in peripheral white blood cells of smokers and nonsmokers. Int J Radiat Biol. 2011;87:791–801. doi: 10.3109/09553002.2011.568574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul S, Smilenov LB, Amundson SA. Widespread decreased expression of immune function genes in human peripheral blood following radiation exposure. Radiat Res. 2013;180:575–583. doi: 10.1667/RR13343.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amundson SA, Grace MB, Mcleland CB, Epperly MW, Yeager A, Zhan Q, et al. Human in vivo radiation-induced biomarkers: Gene expression changes in radiotherapy patients. Cancer Res. 2004;64:6368–6371. doi: 10.1158/0008-5472.CAN-04-1883. [DOI] [PubMed] [Google Scholar]
- 9.Paul S, Barker CA, Turner HC, Mclane A, Wolden SL, Amundson SA. Prediction of in vivo radiation dose status in radiotherapy patients using ex vivo and in vivo gene expression signatures. Radiat Res. 2011;175:257–265. doi: 10.1667/RR2420.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Templin T, Paul S, Amundson SA, Young EF, Barker CA, Wolden SL, et al. Radiation-induced micro-RNA expression changes in peripheral blood cells of radiotherapy patients. Int J Radiat Oncol Biol Phys. 2011;80:549–557. doi: 10.1016/j.ijrobp.2010.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amundson SA, Lee RA, Koch-Paiz CA, Bittner ML, Meltzer P, Trent JM, et al. Differential responses of stress genes to low dose-rate gamma irradiation. Mol Cancer Res. 2003;1:445–452. [PubMed] [Google Scholar]
- 12.Sugihara T, Magae J, Wadhwa R, Kaul SC, Kawakami Y, Matsumoto T, et al. Dose and dose-rate effects of low-dose ionizing radiation on activation of Trp53 in immortalized murine cells. Radiat Res. 2004;162:296–307. doi: 10.1667/rr3223. [DOI] [PubMed] [Google Scholar]
- 13.Sokolov M, Panyutin IG, Neumann R. Genome-wide gene expression changes in normal human fibroblasts in response to low-let gamma-radiation and high-LET-like 125Iudr exposures. Radiat Prot Dosimetry. 2006;122:195–201. doi: 10.1093/rpd/ncl423. [DOI] [PubMed] [Google Scholar]
- 14.Uehara Y, Ito Y, Taki K, Nenoi M, Ichinohe K, Nakamura S, et al. Gene expression profiles in mouse liver after long-term low-dose-rate irradiation with gamma rays. Radiat Res. 2010;174:611–617. doi: 10.1667/RR2195.1. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhry MA, Omaruddin RA, Kreger B, De Toledo SM, Azzam EI. Micro RNA responses to chronic or acute exposures to low dose ionizing radiation. Mol Biol Rep. 2012 doi: 10.1007/s11033-012-1589-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, et al. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 2007;4:e106. doi: 10.1371/journal.pmed.0040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, et al. Radiation metabolomics. 1. identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meadows SK, Dressman HK, Daher P, Himburg H, Russell JL, Doan P, et al. Diagnosis of partial body radiation exposure in mice using peripheral blood gene expression profiles. PLoS One. 2010;5:e11535. doi: 10.1371/journal.pone.0011535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston CJ, Hernady E, Reed C, Thurston SW, Finkelstein JN, Williams JP. Early alterations in cytokine expression in adult compared to developing lung in mice after radiation exposure. Radiat Res. 2010;173:522–535. doi: 10.1667/RR1882.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Tsai Y, Nowak I, Wang N, Hyrien O, Wilkins R, et al. Validating high-throughput micronucleus analysis of peripheral reticulocytes for radiation biodosimetry: Benchmark against dicentric and CBMN assays in a mouse model. Health Phys. 2010;98:218–227. doi: 10.1097/HP.0b013e3181abaae5. [DOI] [PubMed] [Google Scholar]
- 21.Filiano AN, Fathallah-Shaykh HM, Fiveash J, Gage J, Cantor A, Kharbanda S, et al. Gene expression analysis in radiotherapy patients and C57BL/6 mice as a measure of exposure to ionizing radiation. Radiat Res. 2011;176:49–61. doi: 10.1667/RR2419.1. [DOI] [PubMed] [Google Scholar]
- 22.Tucker JD, Grever WE, Joiner MC, Konski AA, Thomas RA, Smolinski JM, et al. Gene expression-based detection of radiation exposure in mice after treatment with granulocyte colony-stimulating factor and lipopolysaccharide. Radiat Res. 2012;177:209–219. doi: 10.1667/rr2749.1. [DOI] [PubMed] [Google Scholar]
- 23.Jacob NK, Cooley JV, Yee TN, Jacob J, Alder H, Wickramasinghe P, et al. Identification of sensitive serum microRNA biomarkers for radiation biodosimetry. PLoS One. 2013;8:e57603. doi: 10.1371/journal.pone.0057603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, Marchetti F, Chen Z, Zaric S, Wilson RJ, Hall DA, et al. Nanosensor dosimetry of mouse blood proteins after exposure to ionizing radiation. Sci Rep. 2013;3:2234. doi: 10.1038/srep02234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richmond CR. Retention and excretion of radionuclides of the alkalai metals by five mammalian species. Health Phys. 1980;38:1111–1153. [Google Scholar]
- 26.Stara JF, Nelson NS, Della Rosa RJ, Bustad LK. Comparative metabolism of radionuclides in mammals: A review. Health Phys. 1971;20:113–137. doi: 10.1097/00004032-197102000-00001. [DOI] [PubMed] [Google Scholar]
- 27.Mailhot H, Peters RH, Cornett RJ. The biological half-time of radioactive Cs in poikilothermic and homeothermic animals. Health Phys. 1989;56:473–484. doi: 10.1097/00004032-198904000-00009. [DOI] [PubMed] [Google Scholar]
- 28.Stabin MG, Peterson TE, Holburn GE, Emmons MA. Voxel-based mouse and rat models for internal dose calculations. J Nucl Med. 2006;47:655–659. [PubMed] [Google Scholar]
- 29.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-array tools. Cancer Informatics. 2007;2:11–17. [PMC free article] [PubMed] [Google Scholar]
- 31.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 33.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gridley DS, Rizvi A, Luo-Owen X, Makinde AY, Pecaut MJ. Low dose, low dose rate photon radiation modifies leukocyte distribution and gene expression in Cd4(+) T cells. J Radiat Res (Tokyo) 2009;50:139–150. doi: 10.1269/jrr.08095. [DOI] [PubMed] [Google Scholar]
- 36.Gridley DS, Pecaut MJ, Rizvi A, Coutrakon GB, Luo-Owen X, Makinde AY, et al. Low-dose, low-dose-rate proton radiation modulates Cd4(+) T cell gene expression. Int J Radiat Biol. 2009;85:250–261. doi: 10.1080/09553000902748609. [DOI] [PubMed] [Google Scholar]
- 37.Sugihara T, Murano H, Nakamura M, Ichinohe K, Tanaka K. P53-mediated gene activation in mice at high doses of chronic low-dose-rate gamma radiation. Radiat Res. 2011;175:328–335. doi: 10.1667/RR2446.1. [DOI] [PubMed] [Google Scholar]
- 38.Schuler E, Parris TZ, Rudqvist N, Helou K, Forssell-Aronsson E. Effects of internal low-dose irradiation from 131I on gene expression in normal tissues in BALB/c mice. EJNMMI Res. 2011;1:29. doi: 10.1186/2191-219X-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudqvist N, Parris TZ, Schuler E, Helou K, Forssell-Aronsson E. Transcriptional response of BALB/c mouse thyroids following in vivo astatine-211 exposure reveals distinct gene expression profiles. EJNMMI Res. 2012;2:32. doi: 10.1186/2191-219X-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiani F, Iannone C, Negri R, Paoletti D, D’antonio M, De Meo PD, et al. Radiation genes: A database devoted to microarrays screenings revealing transcriptome alterations induced by ionizing radiation in mammalian cells. Database (Oxford) 2009;2009 doi: 10.1093/database/bap007. bap007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto Y, Iwakawa M, Furusawa Y, Ishikawa K, Aoki M, Imadome K, et al. Gene expression analysis in human malignant melanoma cell lines exposed to carbon beams. Int J Radiat Biol. 2008;84:299–314. doi: 10.1080/09553000801953334. [DOI] [PubMed] [Google Scholar]
- 42.Rashi-Elkeles S, Elkon R, Shavit S, Lerenthal Y, Linhart C, Kupershtein A, et al. Transcriptional modulation induced by ionizing radiation: P53 remains a central player. Mol Oncol. 2011;5:336–348. doi: 10.1016/j.molonc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voy BH, Scharff JA, Perkins AD, Saxton AM, Borate B, Chesler EJ, et al. Extracting gene networks for low-dose radiation using graph theoretical algorithms. PLoS Comput Biol. 2006;2:e89. doi: 10.1371/journal.pcbi.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meadows SK, Dressman HK, Muramoto GG, Himburg H, Salter A, Wei Z, et al. Gene expression signatures of radiation response are specific, durable and accurate in mice and humans. PLoS One. 2008;3:e1912. doi: 10.1371/journal.pone.0001912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goudarzi M, Weber W, Chung J, Mak TD, Doyle-Eisele M, Melo D, et al. Metabolomic analysis of internal exposure by 137Cs. Radiat Res. 2014;181(1):54–64. doi: 10.1667/RR13479.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cordes N, Meineke V. Integrin signalling and the cellular response to ionizing radiation. J Mol Histol. 2004;35:327–337. doi: 10.1023/b:hijo.0000032364.43566.3a. [DOI] [PubMed] [Google Scholar]
- 47.Jean C, Gravelle P, Fournie JJ, Laurent G. Influence of stress on extracellular matrix and integrin biology. Oncogene. 2011;30:2697–2706. doi: 10.1038/onc.2011.27. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Wang H. Integrin signalling and function in immune cells. Immunology. 2012;135:268–275. doi: 10.1111/j.1365-2567.2011.03549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross TD, Coon BG, Yun S, Baeyens N, Tanaka K, Ouyang M, et al. Integrins in mechanotransduction. Curr Opin Cell Biol. 2013;25:613–68. doi: 10.1016/j.ceb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romanenko A, Morell-Quadreny L, Ramos D, Nepomnyaschiy V, Vozianov A, Llombart-Bosch A. Extracellular matrix alterations in conventional renal cell carcinomas by tissue microarray profiling influenced by the persistent, long-term, low-dose ionizing radiation exposure in humans. Virchows Arch. 2006;448:584–590. doi: 10.1007/s00428-006-0160-2. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Lu H. Signaling to p53: Ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: Probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 53.Bai D, Zhang J, Xiao W, Zheng X. Regulation of the HDM2-p53 pathway by ribosomal protein l6 in response to ribosomal stress. Nucleic Acids Res. 2014;42:1799–1811. doi: 10.1093/nar/gkt971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daftuar L, Zhu Y, Jacq X, Prives C. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS One. 2013;8:e68667. doi: 10.1371/journal.pone.0068667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui D, Li L, Lou H, Sun H, Ngai SM, Shao G, et al. The ribosomal protein S26 regulates p53 activity in response to DNA damage. Oncogene. 2013;33:2225–2235. doi: 10.1038/onc.2013.170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.