Abstract
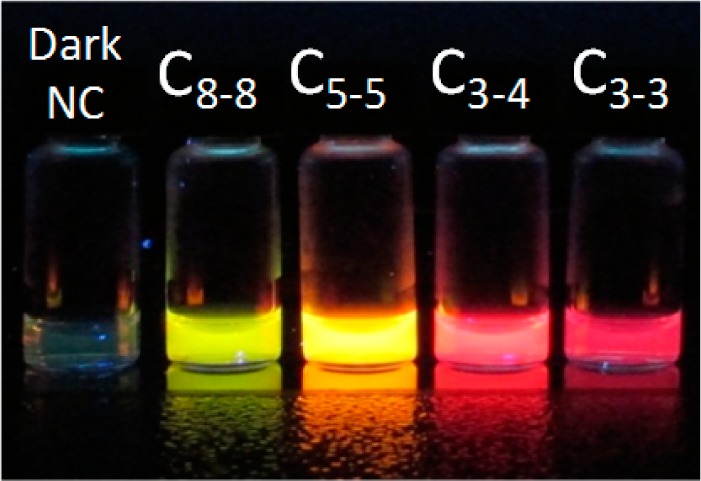
NanoCluster Beacons (NCBs), which use few-atom DNA-templated silver clusters as reporters, are a type of activatable molecular probes that are low-cost and easy to prepare. While NCBs provide a high fluorescence enhancement ratio upon activation, their activation colors are currently limited. Here we report a simple method to design NCBs with complementary emission colors, creating a set of multicolor probes for homogeneous, separation-free detection. By systematically altering the position and the number of cytosines in the cluster-nucleation sequence, we have tuned the activation colors of NCBs to green (C8–8, 460 nm/555 nm); yellow (C5–5, 525 nm/585 nm); red (C3–4, 580 nm/635 nm); and near-infrared (C3–3, 645 nm/695 nm). At the same NCB concentration, the activated yellow NCB (C5–5) was found to be 1.3 times brighter than the traditional red NCB (C3–4). Three of the four colors (green, yellow, and red) were relatively spectrally pure. We also found that subtle changes in the linker sequence (down to the single-nucleotide level) could significantly alter the emission spectrum pattern of an NCB. When the length of linker sequences was increased, the emission peaks were found to migrate in a periodic fashion, suggesting short-range interactions between silver clusters and nucleobases. Size exclusion chromatography results indicated that the activated NCBs are more compact than their native duplex forms. Our findings demonstrate the unique photophysical properties and environmental sensitivities of few-atom DNA-templated silver clusters, which are not seen before in common organic dyes or luminescent crystals.
Keywords: NanoCluster Beacons, DNA-templated silver nanoclusters, activatable probes, multicolor probes, multiplexed detection
Activatable fluorescent probes, which can be turned on in specific environments but otherwise remain dark, have great value for diverse applications in chemistry, biology, and medicine.1−3 Whereas no need to remove unbound probes and much improved target-to-background ratio in detection are the advantages of activatable probes, developing new activatable probes is difficult and producing a set of multicolor activatable probes is even more challenging. In the previous decades, researchers have employed different strategies to produce multicolor activatable probes for biosensing, such as intercalating dyes,4 molecular beacons,5 caged compounds,6 fluorescent proteins,7 and aptamer systems.8 While they have made substantial impact, current multicolor probes have many issues, including low detection specificity, low brightness, poor photostability, high manufacturing cost, low binding affinity, and low fluorescence enhancement upon activation. As a result, the community is constantly looking for new strategies to produce the next-generation of multicolor activatable probes to satisfy unmet challenges in biosensing.
Recently, Yeh et al. turned an emerging class of fluorescent nanomaterials, DNA-templated silver nanoclusters, into new activatable probes, termed NanoCluster Beacons (NCBs),9−11 using a proximal guanine-rich sequence as “the activator”. Owing to advantages such as (1) simple synthesis protocol (one-step process at room temperature), (2) low cost (no purification needed and intrinsic nucleobases used as activators to light up), (3) great enhancement ratio (activation mechanism not based on FRET), and (4) brighter and more photostable as compared to organic dyes,11−29 NCBs have been broadly applied to the detection of proteins,30,31 metabolites,32 and cancer cells,33 other than nucleic acids.9,10 The environmental sensitivity of NCBs has been further explored to create a new type of single-nucleotide polymorphism (SNP) probe, termed a chameleon NCB (cNCB),10 that can differentiate the 4 single-nucleotide variants into 3 groups by cNCB’s emission spectra, whereas all other SNP probes can only differentiate a match variant from 3 other mismatch variants (therefore only 2 groups).11 While Yeh et al. have shown that (1) reprograming the enhancer sequence (i.e., the proximal sequence that is used for fluorescence activation of dark silver clusters)9 and (2) altering the alignment of the enhancer sequence10 with respect to the cluster-nucleation sequence can both modify the activation colors of NCBs,11 a systematic approach to search for more activation colors has never been carried out. Here we focus our investigation on the cluster-nucleation sequence of NCB. By tuning the cluster-nucleation sequence in a variety of ways, we intend to observe how effectively the NCB activation color can be changed.
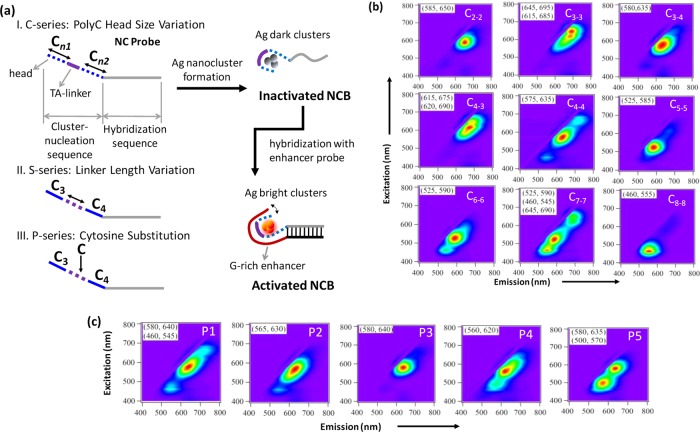
As depicted in Figure 1a, three series of experiments were designed: (1) the C-series, where the size of the “polycytosine heads” is changing in the cluster-nucleation sequence (see Supporting Information Figure S2); (2) the S-series, where the length of the “TA-linker” is changing (Supporting Information Figure S3); and (3) the P-series, where one of the linker nucleotides is replaced by a cytosine (Supporting Information Figure S4). Totally, 25 different cluster-nucleation sequences (Supporting Information Table S1) were synthesized and individually hybridized with the common enhancer probe. The activation fluorescence and enhancement ratio were then characterized for each combinatorial sample. From a separate set of experiments where we varied the composition of the linker (5 nt long and made only of T or A) and generated 32 (a permutation of 25) distinct cluster-nucleation sequences for investigation (see Supporting Information, section II), we found linker composition could have significant influence on the enhancement ratio (Supporting Information Figure S8) but little effect on the activation color (Supporting Information Figure S9). To find alternative ways of tuning the activation color, we then turned our investigation to the lengths of the polycytosine heads and the linker. The size of polycytosine heads, where the interactions with silver ions are thought to be the strongest,12 has previously been shown to affect the stability, dynamics, and emission spectra of the templated silver clusters.34 It is therefore very likely that polycytosine heads of various sizes can stabilize different silver cluster species, leading to different emission colors upon activation—this is the hypothesis behind the C-series experiment design.
Figure 1.
(a) Schematic showing various cluster-nucleation sequence design strategies in the 3 series of experiments (C-, S-, and P-series). (I) C-series, where the size of the “polycytosine heads” is changing in the cluster-nucleation sequence; (II) S-series, where the length of the “TA-linker” is changing; and (III) P-series, where one of the linker nucleotides is replaced by a cytosine. A NanoCluster Beacon (NCB) consists of a nanocluster (NC) probe (having a C-rich cluster-nucleation sequence at the 5′-end) and an enhancer probe (having a G-rich enhancer sequence at the 3′-end). When the enhancer probe is brought close to the NC probe through hybridization, the templated dark silver clusters are activated and become highly emissive through the interactions with the nearby enhancer sequence. We call this process “the guanine-proximity-induced activation of silver clusters” or, in short, “the activation of NCBs”. (b) Normalized 2D fluorescence contour plots of the NCBs in C-series. Here C3–4 represents a cluster-nucleation sequence design with a C3 and a C4 polycytosine heads (separated by the TA-linker). (c) 2D spectra of the P-series NCBs. P3 denotes a design with the third linker nucleotide being substituted with a cytosine. The 2D spectra of the S-series NCBs are shown in Supporting Information Figure S1. For each sample, the 2D fluorescence measurement started exactly at 1 h after the addition of the enhancer probe.
On the other hand, altering the length of the linker between the two polycytosine heads can lead to a change in the alignment state between the templated silver clusters and the guanine-rich enhancer sequence. Alignment state (i.e., the distance between silver clusters on the cluster-nucleation sequence and important interacting nucleobases on the enhancer sequence) manipulation has previously been employed as a strategy to generate multicolor cNCBs for SNP detection.10 Here we design a new set of experiments, the S-series (Supporting Information Figure S3), to test how effectively the activation color can be tuned by sliding one of the polycytosine heads with respect to the enhancer sequence.
Another way that can possibly perturb the activation emission spectra of NCBs is to substitute one of the weakly silver-interacting nucleotides (i.e., T or A) in the linker with a strongly silver-interacting nucleotide (i.e., C), as depicted in the P-series experiments (Supporting Information Figure S4). Several prior reports have already addressed the environmental sensitivity of silver clusters down to the single-nucleotide level.10,35−37 Here we would like to test this environmental sensitivity again with a different approach, aiming to find a simpler and more effective way to reprogram the activation colors of NCBs.
Results and Discussion
As our goal was to search for new NCBs with different emission colors, we eliminated the target strands9,10 from our experiments and focused our investigation only on the interplay between various cluster-nucleation sequences (carrying dark silver clusters) and the common enhancer sequence (G-rich) that were brought together through hybridization. The detailed sample preparation and fluorescence measurement procedures of NCBs are presented in the Materials and Methods section. We measured the 2D fluorescence spectrum and performed the emission pattern, brightness, and enhancement ratio analyses on each combinatorial sample.
Spectral Purities, Emission Patterns, and Tuning Ranges of Various NCB Designs
The normalized 2D fluorescence contour plots of the C-, P-, and S-series are presented in Figure 1, panels b and c, and Supporting Information Figure S1, respectively. Among the nine C-series NCBs, designs C2–2 (ex/em (nm), 585/650), C3–4 (580/635), C5–5 (525/585), and C8–8 (460/555) exhibited relatively pure 2D spectra (i.e., fluorescence intensity of the secondary (2°) peak is less than 25% of the major peak intesnity, Supporting Information Figure S2), indicating a dominant fluorescent species in these samples. The major spectral peaks of designs C2–2 (585/650), C5–5 (525/585), and C6–6 (525/590) had relatively symmetric profiles (eccentricity < 0.66, see Supporting Information Figure S2 and section III), with FWHM (estimated from 2D Gaussian fit, R2 > 0.994) ranging from 72 to 81 nm. On the other hand, designs C3–3, C4–3, C4–4, C6–6, and C7–7 clearly contained several less abundant species that could not be ignored. Asymmetric spectral profiles (eccentricity ≥ 0.66) of the major spectral peaks in designs C3–4, C4–3, C4–4, and C7–7 might be due to the coexistence of multiple fluorescent species with very close emission peaks. The C-series results demonstrated that the 2D emission patterns of NCBs are heavily influenced by the size of the polycytosine heads.
The 2D spectra of the S-series NCBs were quite similar (Supporting Information Figure S1) and the emission patterns were relatively pure (Supporting Information Figure S3) as compared to the C- and P-series. But their differences in spectral profiles were clearly noted, showing symmetric profiles for designs S1–S4 and S8 (eccentricity < 0.66, Supporting Information Figure S3) and asymmetric profiles for designs S0, S5–S7, S9, and S10 (eccentricity ≥ 0.66). On the other hand, the P-series NCBs exhibited distinct emission patterns (Figure 1c), in which only P3 was relatively spectrally pure (Supporting Information Figure S4). The S- and P-series results indicated that the length of the linker does not significantly alter the emission patterns of NCBs (S-series), but small modifications of the linker nucleotides can have a remarkable impact on the emission patterns of NCBs (P-series).
Among the three design series, the C-series showed the largest spectral tuning range, with NCB activation color changing from 555 nm (C8–8) to 695 nm (C3–3) (only the major spectral peaks were considered here). Such a wide tuning range (140 nm) is expected as polycytosine templates of various lengths (C8, C12, C24) have previously been shown to stabilize different silver cluster species, therefore giving different emission colors.34 Here we demonstrate that these various silver cluster species (templated on the polycytosine heads of different lengths) can also be activated through guanine proximity, leading to multiple NCB activation colors. Upon close examination of the resulting C-series spectra (Figure 1b and Supporting Information Figure S2), an interesting trend was noted: in general, the major spectral peaks tend to blue shift as the polycytosine heads become larger (C3–3, 645/695; C4–4, 575/635; C5–5, 525/585; C8–8, 460/555). This result coincides with the previous report34 which suggested that longer polycytosine templates tend to stabilize emitters with shorter emission wavelengths. Therefore, to design red NCBs, short polycytosine heads are desired in the cluster-nucleation sequence.
Unlike the C-series, the P-series showed a narrower spectral tuning range, with NCB activation color changing only from 570 to 640 nm. When the linker cytosine was placed close to either of the polycytosine heads, some green cluster species were activated and appeared in the spectra (most obvious in P5; also see Supporting Information Figure S5). Such green activated species were suppressed in the P3 design, where the linker cytosine was centered in the linker sequence (Figure 1c and Supporting Information Figure S4). We noticed that the emission pattern of P3 is similar to that of the C3–4 (also S5, the first reported NCB9), with the only difference being that the spectral profile of P3 is more symmetric (eccentricity = 0.58).
Although the alignment shifting method employed by the S-series had no significant effect on tuning the activation color (tuning range of only 40 nm, emission changing from 630 to 670 nm, Supporting Information Figures S1 and S3), we noticed the emission peaks (under 580 nm excitation) migrated in a periodic fashion as T or A was alternatively added to the 3′ side of the linker. As shown in Figure 2a, adding one thymine (T) to the 3′ side of the linker (such as S0–S1, S4–S6, and S8–S10) can red shift the emission peaks of NCBs by as much as 18 nm (from S9 to S10). On the contrary, adding one adenine (A) to the 3′ side of the linker (S3–S4 and S6–S8) tends to blue shift the emission peaks. Among the four nucleobases, thymine has the weakest binding affinity for silver ions (interaction strength: C > G > A > T).38 It is not clear at this moment how these subtly different weak cluster-nucleotide interactions affect the activation colors of NCBs (stabilize different cluster species or change the electronic structures of clusters). But we do know such an emission peak migration is very reliable. We saw a similar trend in the 32 linker variation experiments (Supporting Information Tables S4 and S5). By systematically replacing thymines with adenines in the linker (Figure 2b) and vice versa (Supporting Information Figure S10), highly reproducible periodic migration patterns of the emission peaks were also obtained. While the emission tuning range was limited to ∼20 nm here (e.g., from no. 1 NCB to no. 10 NCB), we emphasize that this tuning range was achieved under a fixed excitation wavelength of 580 nm. In addition, the reliable periodic migration patterns clearly demonstrated that the interactions between the clusters and the nucleotides are distance related and work at short ranges (<1 nm).
Figure 2.
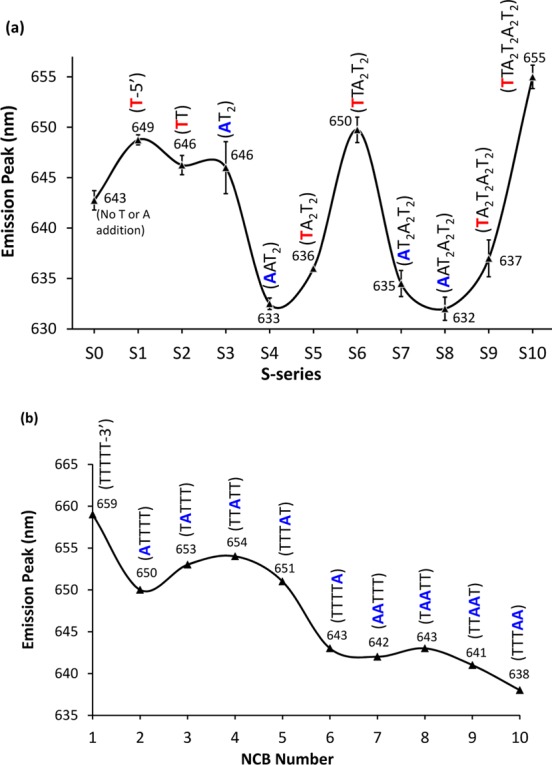
(a) Emission peaks of the S-series NCBs migrating in a periodic fashion. Here the excitation wavelength was fixed at 580 nm. Addition of a thymine (T, red) to the 3′ side of the linker (S0–S1, S4–S6, and S8–S10) generally tends to red shift the emission peaks of NCBs, whereas addition of an adenine (A, blue) shifts emission peak to shorter wavelengths (S3–S4 and S6–S8). (b) Emission peaks of selected NCBs in the 32 linker variation experiments also showing a periodic migration pattern (also see Supporting Information, section II, Table S4, and Figure S10). When the adenine sits right next to the polycytosine heads (nos. 2 and 6 NCBs), the adenine-cluster distance should be the shortest. As a result, adenine’s “cluster-emission-blue-shifting power” is the strongest. Interestingly, the blue-shifting power is greater when adenine sits next to the C4 head (nos. 6 and 10 NCBs) as compared to the C3 head (nos. 2 and 7 NCBs).
Color, Relative Brightness, and Enhancement Ratios of NCBs
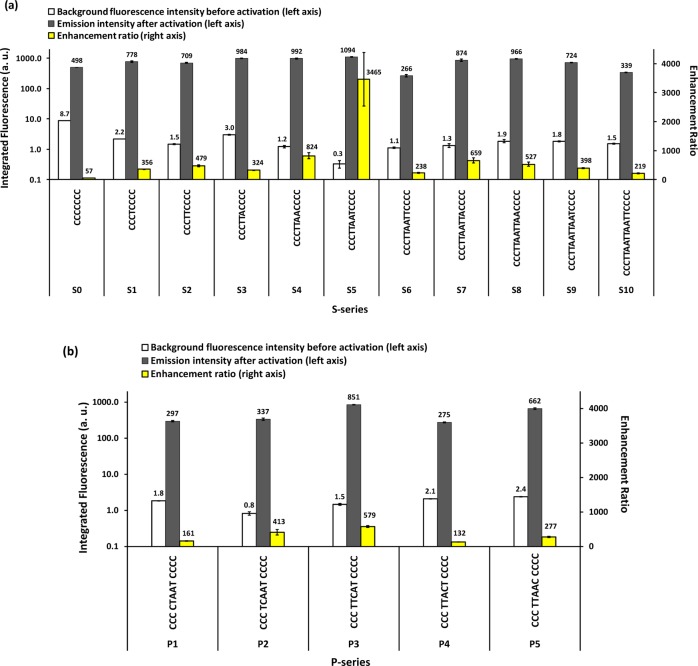
Here we used C3–4 (also S5) as the “gold standard” for color, brightness, and enhancement ratio comparison, as design C3–4 was the first NCB reported.9Supporting Information Figure S5 shows the color photos of C-, S-, and P-series NCBs under UV excitation.10,39 Clearly, the C-series designs tuned the activation colors of NCBs most effectively, creating multiple activation colors. The S- and P-series showed much less color variation, with red being the dominant activation color. S3 and P5 appeared orange under UV light due to the activation of yellow and green species in these samples. We found that the fluorescence intensity could vary significantly from one NCB to another in S- and P-series. For instance, S6 and P4 were much dimmer compared to other designs in the same series, presumably due to short shelf life21 of the activated cluster species. Figure 3 summarizes the fluorescence intensities of the S- and P-series NCBs before and after guanine-proximity-induced activation (also refer to Supporting Information Tables S2 and S3). These quantitative results agreed well with the photos presented in Supporting Information Figure S5.
Figure 3.
Enhancement ratios of the (a) S- and (b) P-series. The enhancement ratios were estimated based on the 1D scans using a fixed excitation wavelength of 580 nm and calculated as (Iactivated – Ibuffer)/(Iinactivated – Ibuffer). Fluorescence intensities before and after activation were integrated from 595 to 740 nm.
The performance of NCBs was evaluated by comparing their bulk enhancement ratios, which are defined as ER = (Iactivated – Ibuffer)/(Iinactivated – Ibuffer), where Iactivated, Ibuffer, and Iinactivated are the integrated fluorescence intensities of the activated NCB (i.e., the duplex), the sodium phosphate buffer, and the inactivated NCB (i.e., NC probe only), respectively. Since the major spectral peaks in both P- and S-series remained nearly unchanged, the brightness and enhancement ratio were compared at a fixed excitation wavelength of 580 nm. Although most of the S- and P-series NCBs reached comparably high fluorescence intensities after activation, only the gold standard S5 design gave an enhancement ratio greater than a 1000-fold (∼3500×). This was due to S5′s substantially lower background fluorescence before activation.
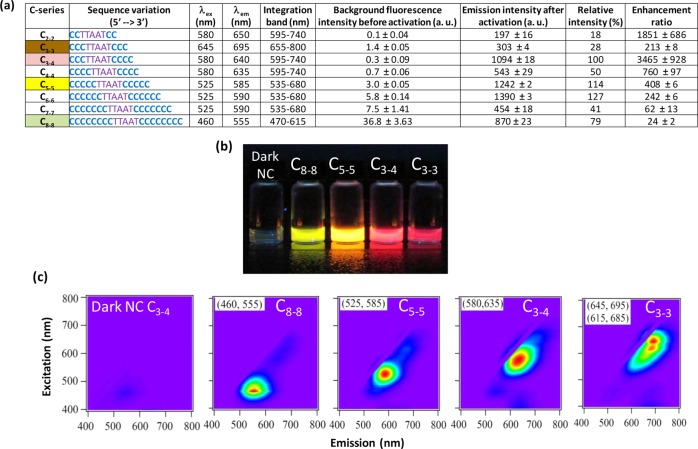
As the emission peaks varied greatly in the C-series, the brightness and enhancement ratios of NCBs were compared at the peak excitation wavelengths of individual designs and fluorescence intensities were integrated over different spectral bands (Figure 4a). Among the C-series designs, yellow C6–6 and C5–5 NCBs had the highest emission intensities once activated (C6–6 is ∼1.3× brighter than the traditional red C3–4 NCB). But due to their higher background fluorescence before activation, the enhancement ratios of C6–6 and C5–5 were only 242- and 408-fold, respectively, much lower than that of C3–4 (ER ∼3500×). When the size of polycytosine heads was reduced (from C8–8 to C3–4), the background fluorescence decreased rapidly, leading to a dramatic improvement in the enhancement ratios (Figure 4a and Supporting Information Figure S6). The background fluorescence reached a minimum at the gold standard design C3–4. While the background fluorescence of C2–2 was extremely low and that led to a high enhancement ratio (∼2000×), C2–2 had a much weaker emission intensity after activation (bulk intensity ∼20% of C3–4 and 15% of C6–6, Figure 4a). Since longer polycytosine templates are known to give a higher synthesis yield and better protect the templated clusters from the environment,34 it is not surprising to see that C6–6 had a much higher emission intensity after activation than C2–2. This trend did not continue as the size of the cluster-nucleation sequence grew beyond C6–6, however. With a fixed enhancer sequence, there is clearly a maximum activation intensity that NCB can achieve. One possible explanation is that the C7–7 and C8–8 designs were too large to support the right kind of clusters that could later be effectively activated by the enhancer. Another possibility is these large cluster-nucleation sequences did not match well with the enhancer sequence, moving the clusters far away from the guanines on the enhancer sequence and leading to reduced guanine-cluster interactions (see C7–7 and C8–8 sequence alignment in Supporting Information Figure S2).
Figure 4.
(a) Enhancement ratios of the C-series NCBs. Here λex and λem represent excitation and emission wavelengths of the major spectral peak for each NCB. The brightness and enhancement ratio of each NCB were estimated based on the 1D scans using the associated λex for excitation. Fluorescence intensities before and after activation were integrated over different spectral bands but with a fixed bandwidth of 145 nm. The integrated intensity of the gold standard, C3–4, after activation was set to 100% for comparison. (b) Color photo of four selected NCBs under UV (365 nm) excitation. The vial on the left was the inactivated C3–4 NCB (i.e., C3–4 NC probe with dark silver clusters, without the enhancer probe). (c) Normalized 2D fluorescence contour plots of the selected NCBs. Three of the four NCBs (green/C8–8, yellow/C5–5, and red/C3–4) were relatively spectrally pure.
Our systematic search in the cluster-nucleation sequences has resulted in four distinct NCB activation colors–green (C8–8, 460 nm/555 nm), yellow (C5–5, 525 nm/585 nm), red (C3–4, 580 nm/635 nm), and near-infrared (C3–3, 615 nm/685 nm, 645 nm/695 nm) (Figure 4b). Three of the four colors (green, yellow, and red) were relatively spectrally pure (Figure 4c). The ability to obtain multiple activation colors using the same enhancer probe (i.e., the activator) but different NC probes is unique to NCBs. This feature is not commonly shared by the existing fluorophores, opening opportunities for NCB’s use in multiplexed assays. For the 4 selected NCBs presented in Figure 4b, the relative intensities ranged from 28% to 114% and the enhancement ratios ranged from 24× to 3465×. We emphasize that NCBs with large emission intensity after activation (e.g., C5–5) and NCBs with high enhancement ratio (e.g., C3–4) suit different applications. Large emission intensity is ideal for use in complex biological systems where autofluorescence is high, whereas high enhancement ratio is suitable for single-molecule quantification where background fluorescence from impurities (e.g., nonfunctional NCBs) should be as low as possible.
Global Structures and Retention Times of NCBs
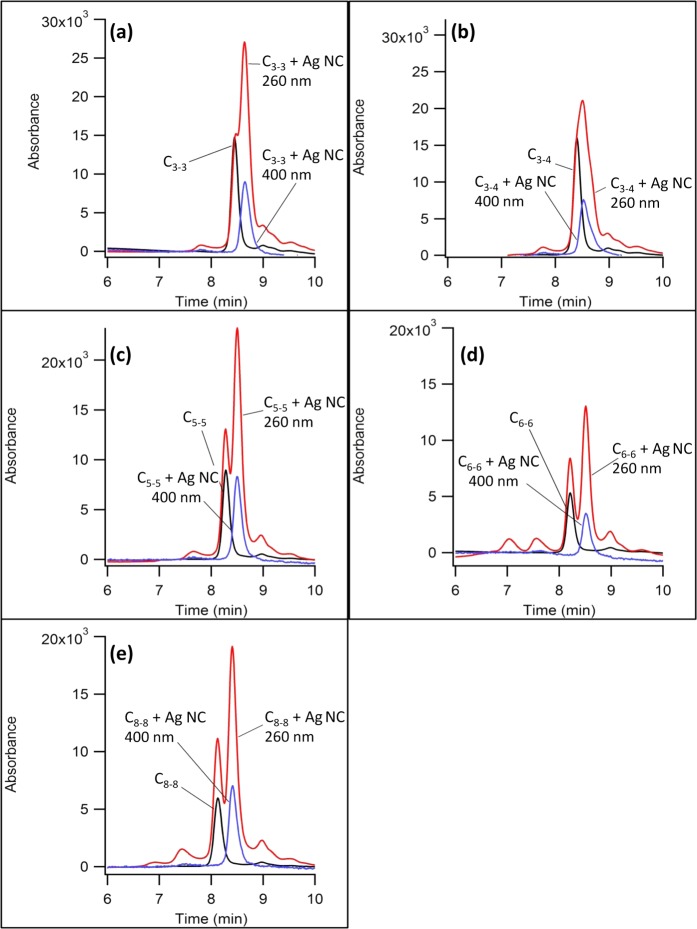
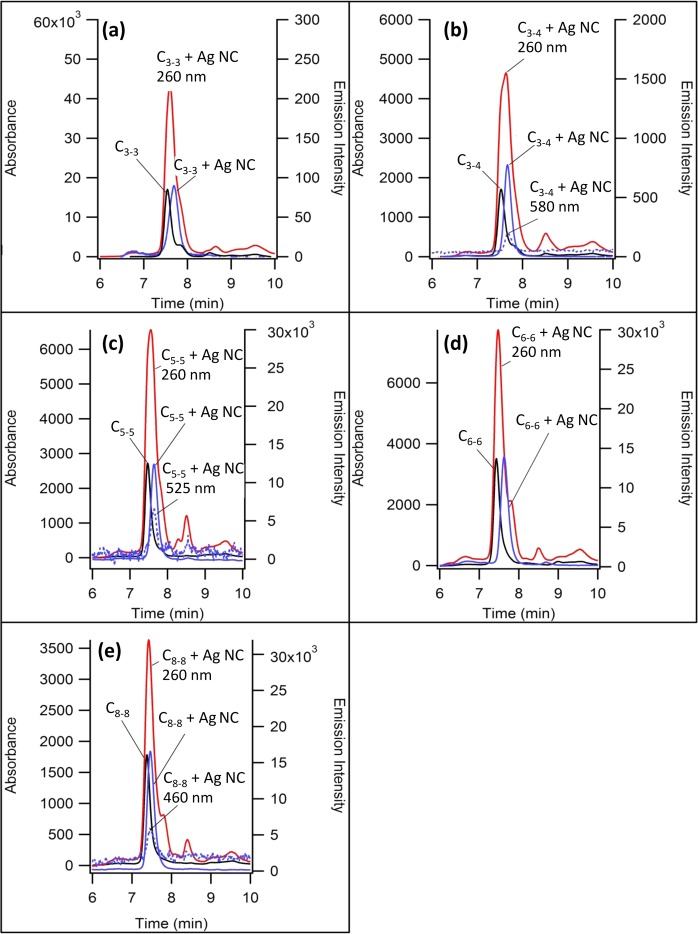
The global structures of the activated and inactivated NCBs were evaluated by size exclusion chromatography. This chromatographic technique distinguishes solutes based on their sizes because smaller solutes partition more effectively into the porous stationary phase and elute more slowly. We first evaluated the inactivated NCBs (i.e., NC probes only). Two oligonucleotide forms were resolved (Figure 5). The leading species exclusively absorbed in the ultraviolet region with λmax ∼260 nm and it eluted with the same time as the native NC probe (i.e., without silver clusters). Thus, the spectra and the retention times support an unlabeled, native NC probe in the sample. The lagging species absorbed in both the ultraviolet region with λmax ∼260 nm and the visible region with λmax ∼400 nm. These absorptions are associated with the DNA and silver clusters, respectively, and correlated elution supports formation of a silver cluster adduct with the DNA host (i.e., labeled NC probe).
Figure 5.
Size exclusion chromatograms for the NC probes without (black lines) and with (red and blue lines) dark silver clusters. The blue lines are cluster-specific because the absorption was collected at 400 nm. The red lines are DNA-specific because the absorption was collected at 260 nm. The NC probes are (a) C3–3, (b) C3–4, (c) C5–5, (d) C6–6, and (e) C8–8.
Although the lagging species was laden with a silver cluster, it eluted later and is thus more compact. The impact of the cluster adduct on DNA shape was assessed by measuring the hydrodynamic radii of the native and labeled NC probes. Five hydrodynamic radius standards were derived from the single-stranded homothymine oligonucleotides dT10, dT15, dT20, and dT30. These oligos adopted random coil conformations and had hydrodynamic radii that progressively increased with the oligonucleotide length.40 The logarithmic relationship between the retention time and the hydrodynamic radius allowed the sizes of the native and labeled NC probes to be determined.41 The native NC probe alone eluted earlier and had sizes that tracked their lengths. The labeled NC probes had sizes that were 6–18% smaller than sizes of the native counterparts. Prior studies have observed similar levels of condensation, and condensation suggests that the clusters contract the DNA strand by coordinating multiple and distant nucleobases.42,43
We now consider the activated NCBs. After hybridizing the NC probes with the common enhancer probe, both the cluster absorption spectra and the host DNA structure transformed (Figure 6). Interactions with the enhancer sequence yielded new clusters with distinct excitation peaks ranging from 460 to 645 nm. Hybridization with the enhancer probe reduced the elution time from ∼8.5 min for the inactivated NCBs (NC probes only) to ∼7.5 min for the activated NCBs (duplexes). Together, the concomitant spectral and structural changes suggested that the dark silver clusters were converted to new clusters with red-shifted excitation and enhanced emission. As with the NC probes, the size exclusion chromatograms exhibited two peaks but with lower resolution. The leading species eluted with the same time as the native duplex forms, which suggested that those were duplexes without a silver cluster. The lagging species exhibited correlated elution of the DNA absorption at ∼260 nm and cluster emission at their respective excitation and emission maxima. These species were smaller than the native duplex forms, which again suggested that the silver clusters condensed their DNA hosts. The extent of condensation was smaller as compared to that in the NC probes, and this distinction may arise because a large proportion of the nucleobase-cluster binding sites are blocked by hybridization.44
Figure 6.
Size exclusion chromatograms for the duplexes without (black lines) and with (red and blue lines) silver clusters. The solid blue lines are cluster-specific because the emission was collected with (a) ex/em (nm): 645/695 for C3–3, (b) 580/640 for C3–4, (c and d) 525/585 for both C5–5 and C6–6, and (e) 460/490 for C8–8. The red lines are DNA-specific because the absorption was collected at 260 nm. The NCBs are (a) C3–3, (b) C3–4, (c) C5–5, (d) C6–6, and (e) C8–8.
Cluster size and stoichiometry dictate the electronic structure of small silver clusters.45 One method to assess cluster:DNA stoichiometry is mass spectrometry. In earlier studies, the gas-phase clusters are unstable, and the clusters fragmented.12,46 Recent studies have diminished this fragmentation, and we are now pursuing such measurements.47,48 Alternatively, cluster:DNA complexes can be chromatographically isolated, and elemental analysis provides the silver:oligonucleotide stoichiometry.49,50 The resolution in Figure 5 demonstrates that the dark silver clusters (on NC probes) can be isolated, so we are also pursuing these complementary measurements. Our SEC studies showed that the activated NCBs are more compact than their native duplex forms. Earlier studies made a similar observation and furthermore determined that the cluster stoichiometry was conserved.50 These results suggested that the clusters switch binding sites. We are now pursuing analogous stoichiometry measurements with the present conjugates. Raman spectroscopy could be used to distinguish forms of the DNA bound clusters.51,52
The utility of multicolor NCBs developed here is defined by their use in biological environments. The present studies make significant progress toward this goal. Specifically, these NCBs are chemically stable in a biologically compatible phosphate buffer despite the favorability of Ag+ precipitation with phosphates. Additionally, they are thermally stable at 95 °C, which is used to promote hybridization. Prior studies have established that Ag+ further stabilizes cluster–DNA conjugates in serum solutions, so such strategies could be used with the present conjugates.44
Conclusion
We have explored various NC probe designs and found that the most effective way to tune the emission colors of NCBs is by changing the size of polycytosine heads in the cluster-nucleation sequences. NCBs with 3 new activation colors (green, yellow, near-IR), good enhancement ratios (24× to 408×) and reasonable spectral separation (Supporting Information Figure S7) are demonstrated in this report. Large emission intensity after activation often comes with the price of the reduced enhancement ratio (e.g., C5–5 and C6–6). The first reported NCB, C3–4, turns out to be a nearly optimized design in this study, having the highest enhancement ratio and strong fluorescent intensity after activation. Emission peak migration controlled by the spatial arrangement of the weakly interacting nucleotides (T or A) can be used to design new molecular rulers. Size exclusion chromatography studies show that the duplex conjugates are more compact than their native duplex forms. These results suggest that the clusters switch binding sites.
Materials and Methods
Materials
Sodium phosphate dibasic anhydrous (Na2HPO4; F.W. 141.96), sodium phosphate monobasic monohydrate (NaH2PO4·H2O; F.W. 137.99) and sodium borohydride (NaBH4) were purchased from Fisher Scientific, whereas silver nitrate (AgNO3) was acquired from Sigma-Aldrich. All oligonucleotides were purchased from Integrated DNA Technologies and were purified by desalting. Deionized (DI) water (18 MΩ·cm) was used for all solution preparations.
Preparation of Nanocluster Probes (NC Probes)
In a typical preparation,9,10 a 15 μM (final concentration) NC probe solution was prepared by adding 12.5 μL of 1.2 mM NC probe (Supporting Information Tables S1 and S4) to 940 μL of 20 mM sodium phosphate buffer (pH 6.7). The solution was vortexed for 2 s and centrifuged at 14 000 rpm for 30 s. Then, 45 μL of 4 mM silver nitrate solution was added, and the mixture was again vortexed and centrifuged. The solution was allowed to sit in the dark for 10 min at room temperature. For silver cluster formation, 7 μL of freshly prepared 13.2 μM NaBH4 solution was added, resulting in a pale yellow mixture which was then stored in the dark overnight. The resulting NC probe solution had the NC probe:Ag+:NaBH4 molar ratio of 1:12:6.
Activation of NanoCluster Beacons
An NCB consists of an NC probe (originally carrying dark silver clusters) and an enhancer probe (guanine-rich). When the enhancer probe is brought close to the NC probe through hybridization, dark silver clusters are activated and become highly emissive through the interactions with the nearby guanine-rich enhancer sequence. We call this process “the guanine-proximity-induced activation of silver clusters” or, in short, “the activation of NCBs”. To activate NCBs, 1.5 μL of 1.2 mM G-rich enhancer probe solution was added to a 120 μL aliquot of the previously prepared 15 μM NC probe solution. The mixture was vortexed, centrifuged, and immersed in a hot water bath (90–95 °C) for 1 min, followed by gradually cooling to room temperature for 1 h. The activated NCB had the NC probe:enhancer probe molar ratio of 1:1. The fluorescence measurements started exactly at 1 h after the addition of the enhancer probe.
Fluorescence Measurements
All fluorescence emission and excitation scans were performed on a FluoroMax-4 spectrofluorometer from Horiba Scientific unless otherwise stated. Each 120 μL NCB sample was placed in a 100 μL quartz cuvette (16.100F-Q-10/Z15, Starna Cells) for fluorometer measurements. For 1D emission scans, the excitation wavelength was set to 580 nm and the emission wavelength was scanned from 595 to 800 nm using 5 nm slit size, 1 nm increment step, 1000 detector gain, and 0.5 s integration time. A blank sample, 20 mM sodium phosphate buffer pH 6.7, was also measured.
For 2D measurements, both emission and excitation were scanned from 400 to 800 nm using 5 nm slit size, 5 nm increment step, and 0.1 s integration time. To avoid saturation, lower NCB concentration (10 μM) and detector gain (775) were used here. For each sample, the 2D scan was completed in 45 min.
Color photos of inactivated (NC probe only) and activated (the duplex) NCBs were acquired with a digital camera (Canon PowerShot SX 500 IS) on a Syngene gel imager (365 nm excitation).
Size Exclusion Chromatography Analysis
Size exclusion chromatography was conducted with a Shimadzu Prominence high performance liquid chromatography system using a 300 × 7.8 mm BioSep-SEC-S2000 column (Phenomenex), having 5 μm particles and a pore size of 145 Å. The mobile phase was buffered at pH = 6.5 with 10 mM citrate/citric acid that was supplemented with NaClO4 to minimize solute interactions with the stationary phase.53 To assess hydrodynamic radii, size standards were based on the thymine oligonucleotides dT10, dT15, dT20, and dT30.40,54
Acknowledgments
We thank J. Werner and J. Martinez for helpful discussion. This work was financially supported by the Robert A. Welch Foundation (F-1833) and University of Texas at Austin to H.-C.Y. and by National Institutes of Health (R15GM102818-01A1 to J.T.P.). J.T.P. is grateful for the support provided by the Henry Keith and Ellen Hard Townes Professorship.
Supporting Information Available
Detailed preparation of NanoCluster Beacons (NCBs), fluorescence measurement procedure, 2D fluorescence contour plots, enhancement ratios, color photos of NCBs in C-, S-, P- series and in 32 linker variation experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
⊥ J.M.O. and M.C.B. contributed equally.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Kobayashi H.; Ogawa M.; Alford R.; Choyke P. L.; Urano Y. New Strategies for Fluorescent Probe Design in Medical Diagnostic Imaging. Chem. Rev. 2010, 110, 2620–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpashchikov D. M. Binary Probes for Nucleic Acid Analysis. Chem. Rev. 2010, 110, 4709–4723. [DOI] [PubMed] [Google Scholar]
- Vendrell M.; Zhai D. T.; Er J. C.; Chang Y. T. Combinatorial Strategies in Fluorescent Probe Development. Chem. Rev. 2012, 112, 4391–4420. [DOI] [PubMed] [Google Scholar]
- Glazer A. N.; Rye H. S. Stable Dye-DNA Intercalation Complexes as Reagents for High-Sensitivity Fluorescence Detection. Nature 1992, 359, 859–861. [DOI] [PubMed] [Google Scholar]
- Tyagi S.; Bratu D. P.; Kramer F. R. Multicolor Molecular Beacons for Allele Discrimination. Nat. Biotechnol. 1998, 16, 49–53. [DOI] [PubMed] [Google Scholar]
- Adams S. R.; Tsien R. Y. Controlling Cell Chemistry with Caged Compounds. Annu. Rev. Physiol. 1993, 55, 755–784. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Campbell R. E.; Ting A. Y.; Tsien R. Y. Creating New Fluorescent Probes for Cell Biology. Nat. Rev. Mol. Cell Biol. 2002, 3, 906–918. [DOI] [PubMed] [Google Scholar]
- Paige J. S.; Wu K. Y.; Jaffrey S. R. RNA Mimics of Green Fluorescent Protein. Science 2011, 333, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh H.-C.; Sharma J.; Han J. J.; Martinez J. S.; Werner J. H. A DNA-Silver Nanocluster Probe that Fluoresces upon Hybridization. Nano Lett. 2010, 10, 3106–3110. [DOI] [PubMed] [Google Scholar]
- Yeh H.-C.; Sharma J.; Shih I. M.; Vu D. M.; Martinez J. S.; Werner J. H. A Fluorescence Light-Up Ag Nanocluster Probe that Discriminates Single-Nucleotide Variants by Emission Color. J. Am. Chem. Soc. 2012, 134, 11550–11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obliosca J. M.; Liu C.; Yeh H.-C. Fluorescent Silver Nanoclusters as DNA Probes. Nanoscale 2013, 5, 8443–8461. [DOI] [PubMed] [Google Scholar]
- Petty J. T.; Zheng J.; Hud N. V.; Dickson R. M. DNA-Templated Ag Nanocluster Formation. J. Am. Chem. Soc. 2004, 126, 5207–5212. [DOI] [PubMed] [Google Scholar]
- Vosch T.; Antoku Y.; Hsiang J. C.; Richards C. I.; Gonzalez J. I.; Dickson R. M. Strongly Emissive Individual DNA-Encapsulated Ag Nanoclusters as Single-Molecule Fluorophores. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 12616–12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie C. M.; Johnsen K. R.; Kiser J. R.; Antoku Y.; Dickson R. M.; Petty J. T. Ag Nanocluster Formation Using a Cytosine Oligonucleotide Template. J. Phys. Chem. C 2007, 111, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty J. T.; Fan C. Y.; Story S. P.; Sengupta B.; Iyer A. S.; Prudowsky Z.; Dickson R. M. DNA Encapsulation of 10 Silver Atoms Producing a Bright, Modulatable, Near-Infrared-Emitting Cluster. J. Phys. Chem. Lett. 2010, 1, 2524–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J.; Yeh H.-C.; Yoo H.; Werner J. H.; Martinez J. S. A Complementary Palette of Fluorescent Silver Nanoclusters. Chem. Commun. 2010, 46, 3280–3282. [DOI] [PubMed] [Google Scholar]
- Yeh H.-C.; Sharma J.; Han J. J.; Martinez J. S.; Werner J. H. A Beacon of Light—A New Molecular Probe for Homogeneous Detection of Nucleic Acid Targets. IEEE Nanotechnol. Mag. 2011, 5, 28–33. [Google Scholar]
- Sharma J.; Yeh H.-C.; Yoo H.; Werner J. H.; Martinez J. S. Silver Nanocluster Aptamers: In Situ Generation of Intrinsically Fluorescent Recognition Ligands for Protein Detection. Chem. Commun. 2011, 47, 2294–2296. [DOI] [PubMed] [Google Scholar]
- Neidig M. L.; Sharma J.; Yeh H.-C.; Martinez J. S.; Conradson S. D.; Shreve A. P. Ag K-Edge EXAFS Analysis of DNA-Templated Fluorescent Silver Nanoclusters: Insight into the Structural Origins of Emission Tuning by DNA Sequence Variations. J. Am. Chem. Soc. 2011, 133, 11837–11839. [DOI] [PubMed] [Google Scholar]
- Yang S. W.; Vosch T. Rapid Detection of MicroRNA by a Silver Nanocluster DNA Probe. Anal. Chem. 2011, 83, 6935–6939. [DOI] [PubMed] [Google Scholar]
- Sharma J.; Rocha R. C.; Phipps M. L.; Yeh H.-C.; Balatsky K. A.; Vu D. M.; Shreve A. P.; Werner J. H.; Martinez J. S. A DNA-Templated Fluorescent Silver Nanocluster with Enhanced Stability. Nanoscale 2012, 4, 4107–4110. [DOI] [PubMed] [Google Scholar]
- Yau S. H.; Abeyasinghe N.; Orr M.; Upton L.; Varnavski O.; Werner J. H.; Yeh H.-C.; Sharma J.; Shreve A. P.; Martinez J. S.; Goodson T. Bright Two-Photon Emission and Ultra-Fast Relaxation Dynamics in a DNA-Templated Nanocluster Investigated by Ultra-Fast Spectroscopy. Nanoscale 2012, 4, 4247–4254. [DOI] [PubMed] [Google Scholar]
- O’Neill P. R.; Young K.; Schiffels D.; Fygenson D. K. Few-Atom Fluorescent Silver Clusters Assemble at Programmed Sites on DNA Nanotubes. Nano Lett. 2012, 12, 5464–5469. [DOI] [PubMed] [Google Scholar]
- Shah P.; Rorvig-Lund A.; Ben Chaabane S.; Thulstrup P. W.; Kjaergaard H. G.; Fron E.; Hofkens J.; Yang S. W.; Vosch T. Design Aspects of Bright Red Emissive Silver Nanoclusters/DNA Probes for MicroRNA Detection. ACS Nano 2012, 6, 8803–8814. [DOI] [PubMed] [Google Scholar]
- Dong H. F.; Jin S.; Ju H. X.; Hao K. H.; Xu L. P.; Lu H. T.; Zhang X. J. Trace and Label-Free MicroRNA Detection Using Oligonucleotide Encapsulated Silver Nanoclusters as Probes. Anal. Chem. 2012, 84, 8670–8674. [DOI] [PubMed] [Google Scholar]
- Obliosca J. M.; Liu C.; Batson R. A.; Babin M. C.; Werner J. H.; Yeh H.-C. DNA/RNA Detection Using DNA-Templated Few-Atom Silver Nanoclusters. Biosensors 2013, 3, 185–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Q.; Wang F.; Niazov-Elkan A.; Guo W. W.; Willner I. Probing Biocatalytic Transformations with Luminescent DNA/Silver Nanoclusters. Nano Lett. 2013, 13, 309–314. [DOI] [PubMed] [Google Scholar]
- Zhang Y. D.; Cai Y. A.; Qi Z. L.; Lu L.; Qian Y. X. DNA-Templated Silver Nanoclusters for Fluorescence Turn-On Assay of Acetylcholinesterase Activity. Anal. Chem. 2013, 85, 8455–8461. [DOI] [PubMed] [Google Scholar]
- Shah P.; Cho S. K.; Thulstrup P. W.; Bhang Y. J.; Ahn J. C.; Choi S. W.; Rorvig-Lund A.; Yang S. W. Effect of Salts, Solvents and Buffer on miRNA Detection Using DNA Silver Nanocluster (DNA/AgNCs) Probes. Nanotechnology 2014, 25, 045101. [DOI] [PubMed] [Google Scholar]
- Li J. J.; Zhong X. Q.; Zhang H. Q.; Le X. C.; Zhu J. J. Binding-Induced Fluorescence Turn-On Assay Using Aptamer-Functionalized Silver Nanocluster DNA Probes. Anal. Chem. 2012, 84, 5170–5174. [DOI] [PubMed] [Google Scholar]
- Zhang L. B.; Zhu J. B.; Zhou Z. X.; Guo S. J.; Li J.; Dong S. J.; Wang E. K. A New Approach To Light Up DNA/Ag Nanocluster-Based Beacons for Bioanalysis. Chem. Sci. 2013, 4, 4004–4010. [Google Scholar]
- Zhang M.; Guo S. M.; Li Y. R.; Zuo P.; Ye B. C. A Label-Free Fluorescent Molecular Beacon Based on DNA-Templated Silver Nanoclusters for Detection of Adenosine and Adenosine Deaminase. Chem. Commun. 2012, 48, 5488–5490. [DOI] [PubMed] [Google Scholar]
- Yin J. J.; He X. X.; Wang K. M.; Xu F. Z.; Shangguan J. F.; He D. G.; Shi H. Label-Free and Turn-On Aptamer Strategy for Cancer Cells Detection Based on a DNA-Silver Nanocluster Fluorescence upon Recognition-Induced Hybridization. Anal. Chem. 2013, 85, 12011–12019. [DOI] [PubMed] [Google Scholar]
- Antoku Y. Ph.D. Dissertation, Georgia Institute of Technology, 2007. [Google Scholar]
- Ma K.; Cui Q. H.; Liu G. Y.; Wu F.; Xu S. J.; Shao Y. DNA Abasic Site-Directed Formation of Fluorescent Silver Nanoclusters for Selective Nucleobase Recognition. Nanotechnology 2011, 22, 1–6. [DOI] [PubMed] [Google Scholar]
- Ma K.; Shao Y.; Cui Q. H.; Wu F.; Xu S. J.; Liu G. Y. Base-Stacking-Determined Fluorescence Emission of DNA Abasic Site-Templated Silver Nanoclusters. Langmuir 2012, 28, 15313–15322. [DOI] [PubMed] [Google Scholar]
- Guo W. W.; Yuan J. P.; Dong Q. Z.; Wang E. K. Highly Sequence-Dependent Formation of Fluorescent Silver Nanoclusters in Hybridized DNA Duplexes for Single Nucleotide Mutation Identification. J. Am. Chem. Soc. 2010, 132, 932–934. [DOI] [PubMed] [Google Scholar]
- Shukla S.; Sastry M. Probing Differential Ag+-Nucleobase Interactions with Isothermal Titration Calorimetry (ITC): Towards Patterned DNA Metallization. Nanoscale 2009, 1, 122–127. [DOI] [PubMed] [Google Scholar]
- O’Neill P. R.; Gwinn E. G.; Fygenson D. K. UV Excitation of DNA Stabilized Ag Cluster Fluorescence via the DNA Bases. J. Phys. Chem. C 2011, 115, 24061–24066. [Google Scholar]
- Doose S.; Barsch H.; Sauer M. Polymer Properties of Polythymine as Revealed by Translational Diffusion. Biophys. J. 2007, 93, 1224–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akers G. K.In the Proteins; Neurath H., Hill R. L., Eds.; Academic Press: New York, 1975; p 547. [Google Scholar]
- Petty J. T.; Story S. P.; Juarez S.; Votto S. S.; Herbst A. G.; Degtyareva N. N.; Sengupta B. Optical Sensing by Transforming Chromophoric Silver Clusters in DNA Nanoreactors. Anal. Chem. 2012, 84, 356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty J. T.; Giri B.; Miller I. C.; Nicholson D. A.; Sergev O. O.; Banks T. M.; Story S. P. Silver Clusters as Both Chromophoric Reporters and DNA Ligands. Anal. Chem. 2013, 85, 2183–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty J. T.; Sengupta B.; Story S. P.; Degtyareva N. N. DNA Sensing by Amplifying the Number of Near-Infrared Emitting, Oligonucleotide-Encapsulated Silver Clusters. Anal. Chem. 2011, 83, 5957–5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacic-Koutecky V.; Veyret V.; Mitric R. Ab Initio Study of the Absorption Spectra of Agn (n = 5–8) Clusters. J. Chem. Phys. 2001, 115, 10450–10460. [Google Scholar]
- O’Neill P. R.; Velazquez L. R.; Dunn D. G.; Gwinn E. G.; Fygenson D. K. Hairpins with Poly-C Loops Stabilize Four Types of Fluorescent Agn:DNA. J. Phys. Chem. C 2009, 113, 4229–4233. [Google Scholar]
- Schultz D.; Gwinn E. G. Silver Atom and Strand Numbers in Fluorescent and Dark Ag:DNAs. Chem. Commun. 2012, 48, 5748–5750. [DOI] [PubMed] [Google Scholar]
- Guo J. S.; Kumar S.; Bolan M.; Desireddy A.; Bigioni T. P.; Griffith W. P. Mass Spectrometric Identification of Silver Nanoparticles: The Case of Ag32(SG)19. Anal. Chem. 2012, 84, 5304–5308. [DOI] [PubMed] [Google Scholar]
- Petty J. T.; Story S. P.; Hsiang J. C.; Dickson R. M. DNA-Templated Molecular Silver Fluorophores. J. Phys. Chem. Lett. 2013, 4, 1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty J. T.; Sergev O. O.; Nicholson D. A.; Goodwin P. M.; Giri B.; McMullan D. R. A Silver Cluster-DNA Equilibrium. Anal. Chem. 2013, 85, 9868–9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. F.; Fields-Zinna C. A.; Murray R. W. The Story of a Monodisperse Gold Nanoparticle: Au25L18. Acc. Chem. Res. 2010, 43, 1289–1296. [DOI] [PubMed] [Google Scholar]
- Tlahuice-Flores A.; Whetten R. L.; Jose-Yacaman M. Vibrational Normal Modes of Small Thiolate-Protected Gold Clusters. J. Phys. Chem. C 2013, 117, 12191–12198. [Google Scholar]
- Leroy J. L.; Gehring K.; Kettani A.; Gueron M. Acid Multimers of Oligodeoxycytidine Strands—Stoichiometry, Base-Pair Characterization, and Proton-Exchange Properties. Biochemistry 1993, 32, 6019–6031. [DOI] [PubMed] [Google Scholar]
- Sengupta B.; Springer K.; Buckman J. G.; Story S. P.; Abe O. H.; Hasan Z. W.; Prudowsky Z. D.; Rudisill S. E.; Degtyareva N. N.; Petty J. T. DNA Templates for Fluorescent Silver Clusters and I-Motif Folding. J. Phys. Chem. C 2009, 113, 19518–19524. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.