Abstract

A variety of bacterial pathogens use nanoscale protein fibers called type IV pili to mediate cell adhesion, a primary step leading to infection. Currently, how these nanofibers respond to mechanical stimuli and how this response is used to control adhesion is poorly understood. Here, we use atomic force microscopy techniques to quantify the forces guiding the adhesion of Pseudomonas aeruginosa type IV pili to surfaces. Using chemical force microscopy and single-cell force spectroscopy, we show that pili strongly bind to hydrophobic surfaces in a time-dependent manner, while they weakly bind to hydrophilic surfaces. Individual nanofibers are capable of withstanding forces up to 250 pN, thereby explaining how they can resist mechanical stress. Pulling on individual pili yields constant force plateaus, presumably reflecting conformational changes, as well as nanospring properties that may help bacteria to withstand physiological shear forces. Analysis of mutant strains demonstrates that these mechanical responses originate solely from type IV pili, while flagella and the cell surface localized and proposed pili-associated adhesin PilY1 play no direct role. We also demonstrate that bacterial–host interactions involve constant force plateaus, the extension of bacterial pili, and the formation of membrane tethers from host cells. We postulate that the unique mechanical responses of type IV pili unravelled here enable the bacteria to firmly attach to biotic and abiotic surfaces and thus maintain attachment when subjected to high shear forces under physiological conditions, helping to explain why pili play a critical role in colonization of the host.
Keywords: adhesion, AFM, chemical force microscopy, mechanics, pathogens, Pseudomonas aeruginosa, single-cell force spectroscopy, type IV pili
The adhesion of pathogens to tissues and implanted devices, and the subsequent formation of biofilms on these surfaces, represents the primary step of many microbial infections.1−4 Because biofilm-associated microbes are resistant to many antimicrobial agents, there is an urgent need to better understand the molecular basis of biofilm formation and to develop new therapeutic approaches, such as antiadhesion molecules. An example of a biofilm-forming species is Pseudomonas aeruginosa, an opportunistic pathogen that causes a variety of infections, including severe respiratory infections of cystic fibrosis patients.5 Several surface-associated adhesion molecules, including flagella, pili, outer membrane proteins, lipopolysaccharides, and exopolysaccharides, mediate attachment of P. aeruginosa to epithelial cells and to abiotic surfaces such as contact lenses and catheters, thus contributing to the virulence of this pathogen.5
Pili are nanoscale protein filaments that decorate the surface of many bacteria.6−10 While pili from Gram-positive bacteria are covalent polymers in which the attachment of the subunits (pilins) to each other is made through sortase-catalyzed covalent bonding, Gram-negative pili are formed by noncovalent interactions between pilins. Pili fulfill numerous important functions that determine the survival, spread, and success of bacteria in nature. Notably, pili can display bacterial cell adhesion proteins (adhesins) that bind to other bacteria, host tissue, and abiotic substrates, thereby leading to host colonization and biofilm formation.6,7 For example, the PilY1 protein of P. aeruginosa has been proposed to be a pilus-associated adhesin required for colonization of epithelial cells.11,12
One of the most extensively studied examples of Gram-negative pili is the type IV pilus, known to play key roles in the pathogenicity of species like Neisseria gonorrheae and P. aeruginosa.6 These pili are strong, flexible rod-like filaments of 5–8 nm in diameter and 1–2 μm in length. Pilin subunits are assembled through interactions between their conserved N-terminal α-helices, forming a hydrophobic core in the filament that is believed to provide extreme mechanical strength. In P. aeruginosa, type IV pili are believed to be a major virulence-associated adhesin. Recent models suggest that the pilin subunits from P. aeruginosa are organized along either a right-handed one-start helix with a 41 Å pitch and four subunits per turn or a left-handed three-start helix with also four subunits per turn. Because of its extension from the cell surface, the pilus is believed to be responsible for the initial contact between the bacterium and the epithelial cell surface. Type IV pili bind to the glycolipids asialo-GM1 and asialo-GM2 on epithelial cell surfaces.6,13 The pilin receptor-binding site is only exposed at the tip of the pilus filaments.6 Pili represent potential vaccine targets because specific antibodies could potentially block the attachment of P. aeruginosa to the host cell receptors.6 To date, we know little about the adhesion and nanomechanics of type IV pili.
Single-molecule techniques have provided new insights into the nanobiophysics of pili from various species.14−21 Specifically, type IV pili from N. gonorrheae have been shown to exert retractile forces involved in twitching motility and host cell adhesion, presumably through filament disassembly into the inner membrane.20 Cooperative retraction of bundled pili can generate forces in the nanonewton range that could be critical for bacterial surface interactions.21 Here, we use atomic force microscopy (AFM) to quantify the nanoscale forces guiding the adhesion of P. aeruginosa type IV pili to biotic and abiotic surfaces. Chemical force microscopy (CFM)22 and single-cell force spectroscopy (SCFS)23,24 experiments show that type IV pili display constant force responses that are engaged in the adhesion of the bacteria to hydrophobic substrates and to host epithelial cells.
Results and Discussion
Chemical Force Microscopy Unravels the Mechanical Response of Type IV Pili
Previous work has shown that P. aeruginosa binds to various abiotic surfaces such as contact lenses and catheters.25−31 Yet, whether the direct adhesion and nanomechanics of type IV pili could play a role in such nonspecific interactions is unclear. To address this issue, we used CFM with tips functionalized with hydrophobic groups to quantify the mechanical properties of individual pili (Figure 1).
Figure 1.
Quantifying the nanomechanics of type IV pili on living bacteria using chemical force microscopy. (a,b) AFM deflection images of wild-type P. aeruginosa bacteria recorded in buffer (a) and in air (b), showing that surface appendages can only be visualized in air. Adhesion force histogram (c) and rupture length histogram (d) obtained by recording force curves in M63 medium between a hydrophobic tip and the polar region of a wild-type P. aeruginosa cell (n = 1024 curves). Adhesion values in (c) correspond to the largest adhesion forces observed in each curve, while rupture lengths in (d) correspond to the last rupture events. All curves were obtained using a contact time of 100 ms, a maximum applied force of 250 pN, and approach and retraction speeds of 1.0 μm s–1. Similar data were obtained using three different tips and three cells from different cultures. (e,f) Stretching individual pili yields constant force plateaus: (e) typical plateau curves composed of a region at zero force followed by a progressive, nonlinear increase in the force (red arrows) to reach the constant force regime, and (f) histogram of the average plateau forces (n = 90 curves from three experiments). As illustrated in the right panel, force plateau signatures are believed to result from force-induced conformational changes within the pili. (g,h) Stretched pili also show single linear force peaks, indicating that they behave as nanosprings: typical linear force peak signatures (g) and histogram of maximum adhesion forces (n = 61 curves from three experiments) (h) as well as quantification of spring-like properties, estimation of pilus spring constant kp (h; inset). Superimposition of 10 curves shows that spring-like properties are highly reproducible.
Before performing the CFM analyses, we checked for the presence of pili on P. aeruginosa by imaging wild-type (WT) bacteria with a silicon nitride tip (Figure 1a,b). Pili were never observed when the cells were imaged in a liquid medium, as they were likely too flexible (Figure 1a). However, bacteria imaged in air clearly showed pili on the cell poles. Pili showed an average length of 1.1 ± 0.3 μm and diameter of 4.2 ± 1.1 nm. These dimensions are consistent with previous electron microscopy and AFM analyses showing that P. aeruginosa type IV pili have a diameter of 4–6 nm.15 Note, however, that we cannot exclude that pili might have become thinner upon drying.
Multiple force–distance curves were recorded in liquid medium between the poles of WT P. aeruginosa bacteria and hydrophobic tips (Figure 1c–h). As can be seen in the adhesion force and rupture length histograms of Figure 1c,d, a substantial fraction of the force curves (15% from n = 1024 curves) showed adhesion forces of 50–250 pN magnitude and 50–2000 nm rupture length. The shape of the curves did not substantially change from one cell to another. We attribute the measured adhesive signatures to the hydrophobic binding of cell surface pili as (i) these features were never observed on pili-less mutant cells (see Figure 2 and text below for details), and (ii) the extended rupture lengths (up to 2000 nm) are consistent with the average pili length. Consistent with our AFM images, type IV pili in P. aeruginosa have been reported to be at one end of the cell and relatively low in number (∼5),15 as opposed to bacterial species with peritrichous pili, which can have tens to hundreds of such pili. It is therefore very likely that the force signatures are associated with individual pili. Hence, our CFM data demonstrate that (i) P. aeruginosa type IV pili mediate bacterial attachment to hydrophobic surfaces, and (ii) they are mechanically strong as they can sustain forces up to 250 pN. This latter finding agrees well with earlier single-molecule work on type IV pili from N. gonorrheae, showing that pili can resist forces in the range of 100 pN.20,32
Figure 2.
Control experiments demonstrate that constant force plateaus and nanospring behaviors originate from type IV pili. Representative forces curves obtained in M63 medium between hydrophobic tips and the polar region of different mutant strains of P. aeruginosa: (a) ΔflgK strain (7% plateau forces and 12% single adhesions; n = 1024 curves), (b) ΔpilA flgK::Tn5 strain (0% adhesion), (c) WT/pPilY1 strain (6% plateau forces and 7% single adhesions), and (d) ΔpilY1 flgK::Tn5 strain (0% adhesion). All curves were obtained using a contact time of 100 ms, a maximum applied force of 250 pN, and approach and retraction speeds of 1.0 μm s–1. Similar data were obtained using three different tips and three cells from different cultures.
Notably, a portion (4% of the curves) of the force signatures obtained for stretched pili displayed constant force plateaus with a mean magnitude of 154 ± 43 pN (mean ± SD from n = 90 plateaus from several independent experiments) (Figure 1e,f). Most plateaus were preceded by a region of zero force followed by a progressive, nonlinear increase in the force (Figure 1e, red arrows) to reach the constant force region. Several observations suggest that this unique mechanical response is due to force-induced conformational changes within individual pili. First, earlier AFM studies on P-pili and type I pili from Gram-negative bacteria14,16,33 have shown that they readily elongate under force to yield constant force plateaus, a behavior resulting from the uncoiling (unfolding) of their helical quaternary structure. However, as type IV pili do not show such helical conformation, it is unclear whether they are capable of unfolding at constant force. Second, using a combination of optical and magnetic tweezers and AFM, Biais et al.(32) showed that when type IV pili from N. gonorrheae are subjected to a force of ∼100 pN, they transition into a unique conformation, much longer and narrower than the original structure. They also showed that the force-induced conformation reveals hidden epitopes previously buried in the pili, providing a means for the bacteria to maintain attachment to the host while withstanding intermittent forces. Accordingly, our data are consistent with a mechanism in which P. aeruginosa type IV pili respond to force by transitioning into an extended quaternary structure that may expose hidden residues capable of promoting adhesion. Alternatively, it is possible that this change in pilus structure could act as a mechanical signal that the microbe has engaged and is firmly attached to a surface.
Another mechanical signature of special interest (11% of the curves) was the existence of single linear force peaks of 92 ± 41 pN (n = 61 curves from independent experiments) (Figure 1g,h), suggesting that the pili behave as nanosprings. We further quantified the mechanical behavior of individual pili by estimating their spring constant (Figure 1h). Using the slope (s) of the linear portion of the raw deflection versus piezo displacement curves and the following equation, kp = (kc × s)/(1 – s), in which kp and kc are the pili and AFM cantilever spring constants, we found that the pilus spring constant was kp = 2.0 ± 0.5 pN nm–1. The high reproducibility of these values suggests that they reflect an intrinsic mechanical extension of the pilus. Our results are reminiscent of the behavior of type IV pili from N. gonorrheae, which also showed linear force peaks suggested to originate from the stacking energy of the α-helices in the center of the fiber.32 Pili from the Gram-positive bacterium Lactobacillus rhamnosus GG (LGG) also showed nanospring properties with spring constants in the range of 4–15 pN nm–1,18 thus not too different from the values of P. aeruginosa pili. However, the nanosprings of LGG pili are able to sustain larger forces compared to those of P. aeruginosa, consistent with the notion that, unlike Gram-negative pili, Gram-positive pili are formed by covalent polymerization and are stabilized by internal isopeptide bonds. We suggest that the spring behavior of type IV pili may be of biological importance as it may help P. aeruginosa to withstand physiological shear forces while being attached to its host.
Plateau and Spring Signatures Are Associated with Type IV Pili
Do the observed force signatures (force plateaus, spring behaviors) represent the direct mechanical response of type IV pili, or could they also originate from other cell surface components? To answer this question, several control experiments were performed using different bacterial mutants. To rule out the possible contribution of flagella, we first analyzed a mutant strain impaired in flagellar production (ΔflgK; Figure 2a). Similar levels of force plateaus (7% from n = 1024 curves) and spring signatures (12%) were observed, indicating that flagella do not influence the measured adhesion forces. By contrast, a double mutant also lacking the structural pilin PilA did not show any adhesion (0% adhesion events from n = 1024 curves) (Figure 2b), revealing that plateau and spring signatures originate from type IV pili.
Next, we asked whether the cell surface protein PilY1 could play a role in the measured nanoscale adhesion forces. PilY1 has been demonstrated to be required for type IV pilus biogenesis and for a robust pili-associated attachment of P. aeruginosa.12 PilY1 is present at the cell surface and has also been proposed to be a pilus-associated adhesin.11,12 A strain overexpressing PilY1 (WT/pPilY1, Figure 2c) showed the same adhesion profiles as the WT (6% force plateaus, 7% spring behaviors; n = 1024), thus suggesting that PilY1 per se does not directly contribute to adhesion. Additionally, a strain impaired in PilY1 production (ΔpilY1 flgK::Tn5, Figure 2d) led to a complete loss of adhesion, an effect that we attribute to the lack of pili in view of the role of PilY1 in pilus assembly.34 All together, these experiments demonstrate that plateau and spring signatures originate from type IV pili, with flagella and PilY1 playing no direct role in these events.
Single-Cell Analysis Shows That Type IV Pili Mediate Adhesion to Hydrophobic Surfaces
We next used SCFS to quantify the forces engaged in the adhesion of single bacteria to abiotic surfaces (Figure 3). Single cells were picked up with colloidal probe cantilevers coated with polydopamine, which does not alter their viability.24 Force–distance curves were measured between these individual cells and methyl- (Figure 3a, inset) or hydroxyl-terminated substrates. These hydrophobic (water contact angle of 110°) and hydrophilic (30°) surfaces reflect both natural surfaces and surfaces associated with infection. For example, catheters and plastic contact lenses are hydrophobic, while metal implants are hydrophilic.
Figure 3.
Single-cell force spectroscopy of the interaction between P. aeruginosa and hydrophobic substrates. (a, Inset) To quantify single-cell adhesion forces, living P. aeruginosa bacteria (blue) were attached on polydopamine-coated colloidal probes (pink). (a,c) Adhesion force histograms, (b,d) rupture length histograms, and representative retraction force profiles obtained by recording multiple force–distance curves between three different WT cells (red, green, and blue) and hydrophobic substrates at 100 ms contact time (a,b; n = 495, 957, and 442 curves for each cell) and 1.1 s contact time (c,d; n = 396, 177, and 863 curves). The inset in (b) shows an enlarged view of the force plateau signatures. The curves were obtained using a maximum applied force of 250 pN and approach and retraction speeds of 1.0 μm s–1.
Figure 3a,b shows the adhesion force and rupture length histograms and representative force curves obtained at short contact time (100 ms) between three different WT cells and hydrophobic substrates. The general features of the curves did not substantially change when recording consecutive force curves on different spots of the substrate or when using cells from independent cultures. Also, only small variations of adhesion frequency, mean adhesion force, and mean rupture length were observed from one cell to another. All of the curves showed well-defined, single or multiple force peaks of 250–750 pN magnitude with 50–250 nm rupture lengths (Figure 3b, bottom curve). In addition, some curves (4%; n = 950) showed constant force plateaus of 100–250 pN magnitude (Figure 3b, top curves and inset) similar to those seen by CFM, presumably reflecting the mechanical response of pili.
As cell adhesion often strengthens with increased time of interaction,23,35,36 we also measured single-cell adhesion forces using a contact time of 1.1 s. For the interaction between WT bacteria and hydrophobic substrates, Figure 3c,d shows that increasing the contact time generally increased the mean adhesion force, up to 3000 pN, while the rupture lengths were not really affected and the number of force plateaus did not significantly increased (6%, n = 396). We note that there was substantial variation of adhesion forces from one cell to another, suggesting some heterogeneity in the cell population at these longer times. Further increase of the interaction time had little effect on adhesion.
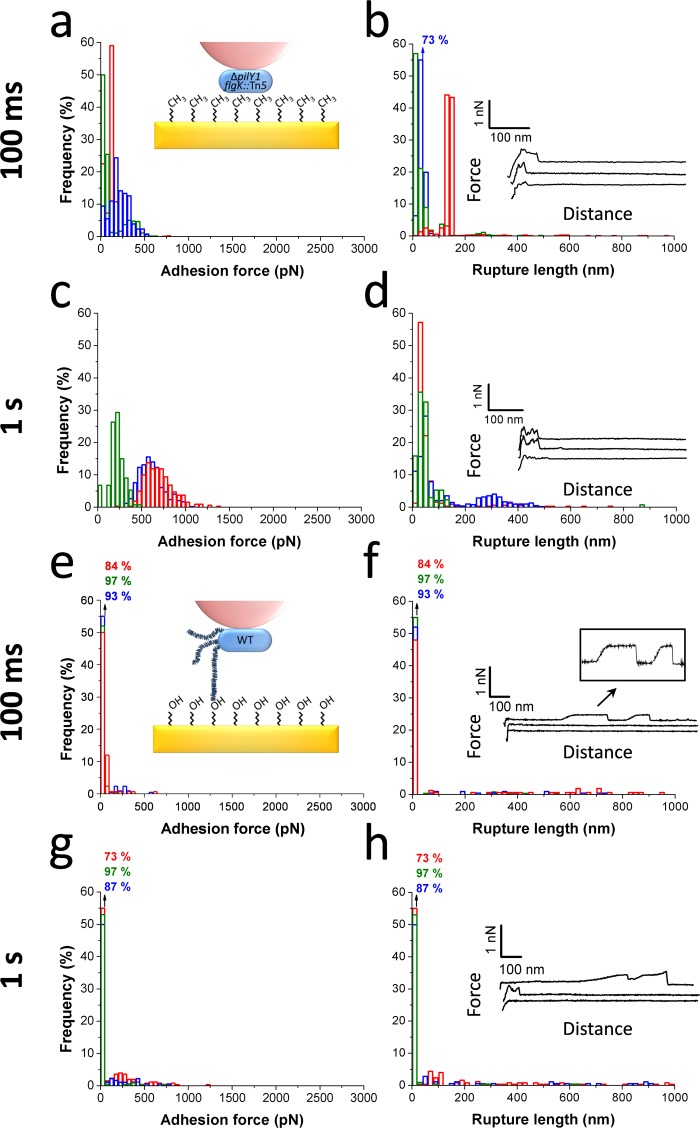
To gain further insight into the nature of the short-range (adhesion peaks) and long-range (force plateaus) adhesion forces, two additional experiments were conducted (Figure 4). Although there were some variations from one cell to another, some interesting observations emerged. Measuring the forces between pili-less bacteria (ΔpilY1 flgK::Tn5 mutant) and hydrophobic substrates had a modest effect on the magnitude of the short-range adhesion forces (50–500 pN vs 250–750 pN) but completely abolished the long-range force plateaus (Figure 4b), thus indicating that pili play a major role in long-range interactions. Other cell surface constituents such as lipopolysaccharides and outer membrane proteins may play a role in mediating short-range attachment to hydrophobic substrates.5 Consistent with the findings for the WT strain, the interaction between pili-less bacteria (ΔpilY1 flgK::Tn5) and hydrophobic substrates (Figure 4c,d) strengthened with time; however, compared to WT bacteria, the effect was less pronounced as the largest adhesion forces were ∼1000 pN (vs ∼3000 pN).
Figure 4.
Role of pili and substrate chemistry in controlling P. aeruginosa single-cell adhesion. (a,c) Adhesion force histograms, (b,d) rupture length histograms, and representative retraction force profiles obtained by recording multiple force–distance curves between cells from the pili-less mutant (ΔpilY1 flgK::Tn5) and hydrophobic substrata at 100 ms (a,b; n = 906, 539, and 502 curves for each cell) and 1.1 s (c,d; n = 132, 592, 320 curves) contact time. (e,g) Adhesion force histograms, (f,h) rupture length histograms, and representative retraction force profiles obtained by recording multiple force–distance curves between WT cells and hydrophilic substrata at 100 ms (e,f; n = 159, 250, 217 curves for each cell) and 1.1 s (g,h; n = 211, 179, and 251 curves for each cell) contact time.
Interestingly, force curves obtained between WT bacteria and hydrophilic substrates showed poor short-range adhesion (7–16% vs 94–100% on hydrophobic substrates), while force plateaus were still observed (Figure 4f). Contact time had little effect on hydrophilic substrates (Figure 4g,h), implying that time-dependent adhesive interactions are hydrophobic in nature.
In light of these single-cell data, we propose a mechanism for the adhesion of P. aeruginosa to hydrophobic surfaces involving two types of interactions: strong, short-range cohesive interactions originating from tight, time-dependent hydrophobic interactions between cell surface constituents (e.g., membrane proteins) and the substrate, and weaker, long-range interactions involving extension and force-induced conformational changes of type IV pili.
Role of Type IV Pili in the Attachment to Pneumocytes
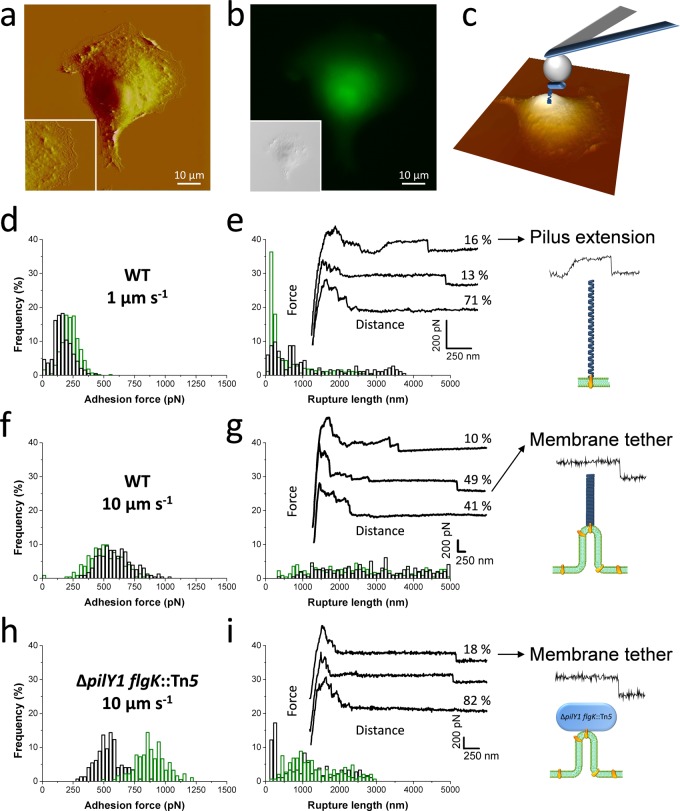
As a key function of P. aeruginosa type IV pili is to promote bacterial attachment to host cells, we also measured the interaction forces between single bacteria and the surface of living pneumocytes (Figure 5). Figure 5a,b shows correlative AFM and fluorescence images of an A549 pneumocyte in buffer, respectively. The AFM image reveals the central round nucleus surrounded by the flattened cytoplasm and membrane and the underlying cytoskeleton structures.
Figure 5.
Single-cell force spectroscopy of the P. aeruginosa–host interaction. (a) AFM deflection image a living A549 pneumocyte cell in buffer (inset: higher magnification of the smooth area). (b) Correlative fluorescence image (inset: corresponding DIC image) of the cell labeled with the fluorescent dye CFSE (green). (c) SCFS was used to quantify the adhesion forces between single P. aeruginosa bacteria (blue) and pneumocytes. For each pneumocyte tested, force curves were recorded on at least three different spots. (d,f) Adhesion force histograms, (e,g) rupture length histograms, and representative retraction force profiles obtained by recording multiple force–distance curves at 1.1 s contact time between WT cells and A549 pneumocytes using a pulling speed of 1.0 μm s–1 (d,e; n = 192 and 434 curves for each cell) and 10.0 μm s–1 (f,g; n = 231 and 223 curves). (h,i) Force data obtained for ΔpilY1 flgK::Tn5 mutant bacteria at 10.0 μm s–1 pulling speed (n = 180 and 146 curves). As illustrated in the cartoons, long-range constant force plateau interactions of two types are observed with WT bacteria: “rough” plateaus reflecting the extension of bacterial pili (e, right panel) and “smooth” plateaus associated with the extraction of host membrane tethers (g, right panel). With ΔpilY1 flgK::Tn5 bacteria, however, only smooth plateaus are observed (i, right panel).
Force–distance curves were obtained between single WT bacteria and flat regions of the pneumocytes (Figure 5c) while varying the pulling speed as this parameter is known to be important for animal cell adhesion.37 At low pulling speed (1.0 μm s–1; Figure 5d,e), many curves showed adhesion force peaks of 50–500 pN magnitude with sequential rupture events and rupture distances of 250–500 nm. Similar behaviors were observed from one bacterium to another. This profile, different from the “hydrophobic profile” (Figure 3), indicates that multiple bonds were formed and did not all rupture simultaneously during detachment.
Following the short-range adhesion peak, constant force plateaus were frequently observed (29% from a total of 434 curves), with rupture lengths up to 4000 nm. Importantly, force plateaus with two different shapes were observed, that is, “rough plateaus” that showed substantial force fluctuations along the plateaus and were preceded by a region of zero force (16%; Figure 5e, top curve) and “smooth plateaus” that showed less fluctuations and directly originated from the short-range adhesion peaks without being preceded by a region of zero force (13%; Figure 5e, middle curve). We suggest that rough plateaus are due to the force-induced extension of type IV pili, just as observed for hydrophobic substrates, while smooth plateaus would reflect the extraction of membrane tethers from the host cells. This latter interpretation is supported by previous investigations showing that the characteristics of our smooth plateaus are very similar to those reported for membrane tethers on different cell lines,19,37,38 including A549 pneumocytes.39,40 Specifically, the mean tether force (i.e., force step before rupture) was 62 ± 11 pN (n = 20 curves), which is close to values reported for A549 tethers at a similar pulling speed.39,40 The lifetime of the bacterial–host bond is given by the tether length divided by the tip velocity. Therefore, the formation of membrane tethers between the pathogens and host cells may substantially increase the lifetime of the interaction, thus favoring host colonization.
As shown in Figure 5f,g, increasing the pulling speed (10.0 μm s–1) led to an increase of the short-range adhesion forces (250–1000 pN vs 50–500 pN). In addition, force plateaus were more frequently observed (59%; n = 231), as reflected by the substantial increase in rupture lengths (up to 5000 nm). Close inspection of these plateaus revealed that many of them were smooth, thus presumably reflecting the formation of membrane tethers. This is consistent with earlier work showing that tethers are preferably formed at high pulling velocity.39,40
Finally, force curves were obtained for pili-less bacteria (ΔpilY1 flgK::Tn5) to further substantiate the origin of the force plateaus (Figure 5h,i). Mutant bacteria showed adhesion frequency and maximum adhesion forces that were similar to WT bacteria. However, rough plateaus were lacking while smooth plateaus were still observed, thus demonstrating that the two signatures are associated with host cell membranes and bacterial pili, respectively.
Taken together, our findings favor a model in which the adhesion of P. aeruginosa to pneumocytes involves a complex interplay of molecular interactions: (i) short-range cohesive interactions, most likely hydrophobic in nature, and originating from the tight contact between the bacterial surface and the host membrane; (ii) long-range constant force interactions originating from the binding and force-induced extension of type IV pili; and (iii) long-range constant force interactions due to the formation of host tethers. We postulate that both pili extension and tether formation provide a means for P. aeruginosa to increase the lifetime of the bacterial–host interaction and to withstand shear stress conditions. As shown for N. gonorrheae pili,32 extended pili may expose hidden epitopes; these newly exposed regions may include adhesive residues capable of binding to host cells.
Conclusions
An important challenge in biology is to understand how bacterial pili respond to mechanical force and how this response is used to achieve cellular functions. Our nanoscale experiments demonstrate that, upon interaction with a hydrophobic surface, P. aeruginosa type IV pili exhibit unique mechanical properties, that is, constant force plateaus most likely resulting from force-induced conformational changes, and linear force peaks reflecting nanospring behaviors. These features enable individual pili to sustain forces up to 250 pN, thus helping the bacteria to withstand shear forces in the physiological environment. Adhesion of P. aeruginosa to host cells involves a complex mix of interactions: short-range cohesive interactions between the bacterial outer membrane and the host membrane, and long-range constant force interactions reflecting the extension of bacterial pili and the extraction of host tethers. These findings emphasize the key role that type IV pili mechanics play in controlling bacterial attachment to biotic and abiotic surfaces. Our work may have important implications in nanomedicine, for the development of vaccines and antiadhesion molecules capable of blocking the pilus activity.
Methods
Bacterial Strains and Cell Cultures
The following P. aeruginosa strains were used in this study: P. aeruginosa PA14 wild-type carrying plasmid pMQ80 (WT, SMC3018); the ΔflgK deletion strain carrying vector pMQ80 (ΔflgK, SMC6103); the WT strain carrying a plasmid with the pMQ80 backbone expressing a C-terminally His-tagged PilY1 (WT/pPilY1; SMC3733); the double mutant flgK::Tn5 del-pilA (ΔpilA flgK::Tn5; SMC6589); and the double mutant flgK::Tn5 del-pilY1 (ΔpilY1 flgK::Tn5; SMC6076).
Strains SMC3018 and SMC3733 were constructed as part of a previous study,34 while strains SMC6103, SMC6589, and SMC6076 were constructed for this study. In-frame deletion mutants were constructed via allelic exchange as described by Shanks et al.(41) The ΔpilA flgK::Tn5 mutant (SMC6589) was generated by conjugating Escherichia coli S17 transformed with a pMQ30-pilA knockout construct (SMC3772)34 with P. aeruginosaflgK::Tn5 (SMC 257).42 Integrants were isolated on gentamicin (30 μg/mL) and nalidixic acid (20 μg/mL), followed by sucrose counter-selection. Resolved integrants were confirmed by PCR and sequencing.
Strains were cultivated overnight in lysogeny broth (LB, Sigma) at 37 °C with shaking at 200 rpm. Gentamycin (20 μg mL–1, Sigma) was added for culturing strains WT/pMQ80, WT/pPilY1, and ΔflgK/pMQ80. For biofilm induction,35 overnight cultures were diluted 1:100 in M63 complete medium containing 3 g L–1 KH2PO4 (Fisher), 7 g L–1 K2HPO4 (Fisher), 2 g of (NH4)2SO4 (Sigma), supplemented with 1 mM MgSO4 (Sigma), 0.2% glucose (Sigma), 0.5% Casamino acids + 0.2% arabinose, and incubated 10 h at 37 °C and 200 rpm.
Human A-549 type II pneumocytes were obtained from the American Type Culture Collection (ATCC, Manassas, VA). They were cultivated in Ham’s F-12K medium (Sigma) supplemented with 10% (v/v) heat-inactivated bovine serum (Sigma) containing 50 U mL–1 of penicillin-G and 50 μg mL–1 of streptomycin. Twenty-four hours before the AFM experiment, the cells were transferred into glass-bottomed Petri dishes (Willco Wells, The Netherlands). One hour prior to bacterial deposition in the Petri dish, medium was replaced by Ham’s F-12K without antibiotics. For live staining imaging, 10 μM of carboxyfluorescein succinimidyl ester (CFSE green, excitation 492 nm/emission 517 nm, Molecular Probes) was added 30 min before microscopic analysis following the supplier recommendation. Correlative microscopy images were obtained on live cells incubated in prewarmed PBS.
Preparation of Hydrophobic Tips and of Hydrophobic/Hydrophilic Substrates
To prepare hydrophobic and hydrophilic substrates, glass coverslips were coated by electron beam thermal evaporation with a 5 nm thick Cr layer followed by a 30 nm thick Au layer. Gold surfaces were immersed overnight in solutions of 1 mM 1-dodecanethiol (Sigma) or 1 mM of 11-mercapto-1-undecanol, respectively. Samples were then rinsed with ethanol and dried under N2. To obtain hydrophobic tips, gold-coated cantilevers (OMLC-TR4, Olympus Ltd., Tokyo, Japan; nominal spring constant ∼0.02 N/m) were immersed for 12 h in a 1 mM 1-dodecanethiol for 12 h, rinsed with ethanol, and dried with N2.
Atomic Force Microscopy Imaging
For imaging in air, AFM contact mode images were obtained at room temperature using a Nanoscope VIII Multimode AFM (Bruker Corporation, Santa Barbara, CA) with oxide-sharpened microfabricated Si3N4 cantilevers with a nominal spring constant of 0.01 N m–1 (MSCT, Bruker Corporation). Force data were analyzed using the Nanoscope software (version 8.15, Bruker) and Matlab software (version R2013b). One hundred microliters of biofilm-induced cells was put in contact with freshly cleaved mica supports mounted on steel pucks. The samples were incubated for 2 h at 37 °C, gently rinsed in three successive baths of ultrapure water (Elga, purelab water), and allowed to dry at 30 °C overnight. For imaging in liquid, bacteria were immobilized by mechanical trapping into a polycarbonate porous membrane (Millipore) with a pore size similar to the cell size. After the cell suspension was filtered, the filter was gently rinsed in M63, carefully cut (1 cm × 1 cm), attached to a steel sample puck using a small piece of double-faced adhesive tape, the mounted sample transferred into the AFM liquid cell while avoiding dewetting, and imaged under minimum applied force using MSCT cantilevers.
Chemical Force Microscopy
Force measurements using hydrophobic tips were performed on a Nanoscope VIII Multimode AFM (Bruker Corporation, Santa Barbara, CA) at room temperature (20 °C) in M63 complete medium. Bacteria were immobilized by mechanical trapping (see above). A single cell was first localized using a silicon nitride tip, after which the tip was exchanged with a hydrophobic tip. Adhesion and rupture length histograms were obtained by recording 32 × 32 force–distance curves on areas of 500 × 500 nm on the bacterial pole surface. All force curves were recorded with a maximum applied force of 250 pN using a constant approach and retraction speed of 1.0 μm s–1 and a contact time of 0.1 s.
Single-Cell Force Spectroscopy
For single bacterial cell force spectroscopy, cell probes were prepared using a recently developed protocol that combines colloidal probe cantilevers and bioinspired polydopamine wet adhesives.24,43 Briefly, silica microspheres (6.1 μm diameter, Bangs Laboratories) were attached on triangular-shaped tipless cantilevers (NP-O10, Microlevers, Bruker Corporation) using UV-curable glue (NOA 63, Norland Edmund Optics). The cantilevers were then immersed for 1 h in a 10 mM Tris buffer solution (pH 8.5) containing 4 mg mL–1 dopamine hydrochloride (99%, Sigma) and dried with N2 flow. Single bacteria were then attached onto polydopamine-coated colloidal probes using a Bioscope Catalyst AFM (Bruker Corporation). To this end, overnight bacterial cultures were diluted to 1:100 in the corresponding medium, and 50 μL of the diluted suspension was deposited in a glass Petri dish containing hydrophobic and hydrophilic substrates or added to the Petri dish containing pneumocytes. Four milliliters of complete M63 medium and of Ham’s F-12K medium without antibiotics was added for experiments on abiotic surfaces and on pneumocytes, respectively. A colloidal probe was brought into contact with an isolated bacterium for 3 min, and the obtained cell probe was then transferred over a solid substrate or a pneumocyte for further force measurements.
SCFS measurements were performed at room temperature (20 °C) using a Bioscope Catalyst AFM (Bruker AXS Corporation). The nominal spring constant of the colloidal probe cantilever was determined by the thermal noise method. For experiments on solid substrates, multiple force–distance curves were recorded on various spots of hydrophobic and hydrophilic substrates using a maximum applied force of 250 pN, a contact time of 100 ms or 1.1 s as mentioned, and constant approach and retraction speeds of 1.0 μm s–1. For pneumocyte experiments, multiple force–distance curves were recorded on various spots of the cell membrane using a maximum applied force of 500 pN, a contact time of 1.1 s, and retraction speeds of 1.0 or 10.0 μm s–1 as mentioned. The maximum downward force exerted on the cantilever is referred to as the adhesion force (measured relative to the baseline) and used to build the adhesion force histogram, whereas the last rupture peak is used to generate the rupture length histogram.
Acknowledgments
Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted Research Action). G.A.O. is supported by the NIH through Grant 2 R37 AI83256-06 and the Human Frontiers in Science Program. A.E.B. is supported by a NSF Graduate Research Fellowship. Y.F.D. is a Research Director at the FNRS. We thank Katy Forest for helpful discussions.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Costerton J. W.; Stewart P. S.; Greenberg E. P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [DOI] [PubMed] [Google Scholar]
- Douglas L. J. Candida Biofilms and Their Role in Infection. Trends Microbiol. 2003, 11, 30–36. [DOI] [PubMed] [Google Scholar]
- Finkel J. S.; Mitchell A. P. Genetic Control of Candida albicans Biofilm Development. Nat. Rev. Microbiol. 2011, 9, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R.; Greenberg E. P. Microbial Sciences: The Superficial Life of Microbes. Nature 2006, 441, 300–302. [DOI] [PubMed] [Google Scholar]
- Davey M. E.; O’Toole G. A. Microbial Biofilms: From Ecology to Molecular Genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L.; Pique M. E.; Tainer J. A. Type IV Pilus Structure and Bacterial Pathogenicity. Nat. Rev. Microbiol. 2004, 2, 363–378. [DOI] [PubMed] [Google Scholar]
- Telford J. L.; Barocchi M. A.; Margarit I.; Rappuoli R.; Grandi G. Pili in Gram-Positive Pathogens. Nat. Rev. Microbiol. 2006, 4, 509–519. [DOI] [PubMed] [Google Scholar]
- Kline K. A.; Dodson K. W.; Caparon M. G.; Hultgren S. J. A Tale of Two Pili: Assembly and Function of Pili in Bacteria. Trends Microbiol. 2010, 18, 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. J.; Baker E. N. Structure and Assembly of Gram-Positive Bacterial Pili: Unique Covalent Polymers. Curr. Opin. Struct. Biol. 2012, 22, 200–207. [DOI] [PubMed] [Google Scholar]
- Mandlik A.; Swierczynski A.; Das A.; Ton-That H. Pili in Gram-Positive Bacteria: Assembly, Involvement in Colonization and Biofilm Development. Trends Microbiol. 2008, 16, 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm R. A.; Hallinan J. P.; Watson A. A.; Mattick J. S. Fimbrial Biogenesis Genes of Pseudomonas aeruginosa: pilW and pilX Increase the Similarity of Type 4 Fimbriae to the GSP Protein-Secretion Systems and pilY1 Encodes a Gonococcal PilC Homologue. Mol. Microbiol. 1996, 22, 161–173. [DOI] [PubMed] [Google Scholar]
- Heiniger R. W.; Winther-Larsen H. C.; Pickles R. J.; Koomey M.; Wolfgang M. C. Infection of Human Mucosal Tissue by Pseudomonas aeruginosa Requires Sequential and Mutually Dependent Virulence Factors and a Novel Pilus-Associated Adhesin. Cell. Microbiol. 2010, 12, 1158–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivan H. C.; Roberts D. D.; Ginsburg V. Many Pulmonary Pathogenic Bacteria Bind Specifically to the Carbohydrate Sequence Galnac Beta 1-4Gal Found in Some Glycolipids. Proc. Natl. Acad. Sci. U.S.A. 1988, 85, 6157–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E.; Garcia T.; Hultgren S.; Oberhauser A. F. The Mechanical Properties of E. coli Type 1 Pili Measured by Atomic Force Microscopy Techniques. Biophys. J. 2006, 91, 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhami A.; Jericho M. H.; Boyd J. M.; Beveridge T. J. Nanoscale Characterization and Determination of Adhesion Forces of Pseudomonas aeruginosa Pili by Using Atomic Force Microscopy. J. Bacteriol. 2006, 188, 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugmaier R. A.; Schedin S.; Kühner F.; Benoit M. Dynamic Restacking of Escherichia coli P-pili. Eur. Biophys. J. 2008, 37, 111–120. [DOI] [PubMed] [Google Scholar]
- Castelain M.; Sjöström A. E.; Fällman E.; Uhlin B. E.; Andersson M. Unfolding and Refolding Properties of S Pili on Extraintestinal Pathogenic Escherichia coli. Eur. Biophys. J. 2010, 39, 1105–1115. [DOI] [PubMed] [Google Scholar]
- Tripathi P.; Beaussart A.; Alsteens D.; Dupres V.; Claes I.; Von Ossowski I.; De Vos W. M.; Palva A.; Lebeer S.; Vanderleyden J.; Dufrêne Y. F. Adhesion and Nanomechanics of Pili from the Probiotic Lactobacillus rhamnosus GG. ACS Nano 2013, 7, 3685–3697. [DOI] [PubMed] [Google Scholar]
- Sullan R. M. A.; Beaussart A.; Tripathi P.; Derclaye S.; El-Kirat-Chatel S.; Li J.; Schneider Y. J.; Vanderleyden J.; Lebeer S.; Dufrêne Y. F. Single-Cell Force Spectroscopy of Pili-Mediated Adhesion. Nanoscale 2014, 6, 1134–1143. [DOI] [PubMed] [Google Scholar]
- Merz A. J.; So M.; Sheetz M. P. Pilus Retraction Powers Bacterial Twitching Motility. Nature 2000, 407, 98–102. [DOI] [PubMed] [Google Scholar]
- Biais N.; Ladoux B.; Higashi D.; So M.; Sheetz M. Cooperative Retraction of Bundled Type IV Pili Enables Naonewton Force Generation. PLoS Biol. 2008, 6, 907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dague E.; Alsteens D.; Latgé J. P.; Verbelen C.; Raze D.; Baulard A. R.; Dufrêne Y. F. Chemical Force Microscopy of Single Live Cells. Nano Lett. 2007, 7, 3026–3030. [DOI] [PubMed] [Google Scholar]
- Helenius J.; Heisenberg C. P.; Gaub H. E.; Müller D. J. Single-Cell Force Spectroscopy. J. Cell Sci. 2008, 121, 1785–1791. [DOI] [PubMed] [Google Scholar]
- Beaussart A.; El-Kirat-Chatel S.; Herman P.; Alsteens D.; Mahillon J.; Hols P.; Dufrêne Y. F. Single-Cell Force Spectroscopy of Probiotic Bacteria. Biophys. J. 2013, 104, 1886–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher E. L.; Weissman B. A.; Efron N.; Fleiszig S. M. J.; Curcio A. J.; Brennan N. A. The Role of Pili in the Attachment of Pseudomonas aeruginosa to Unworn Hydrogel Contact Lenses. Curr. Eye Res. 1993, 12, 1067–1071. [DOI] [PubMed] [Google Scholar]
- Miller M. J.; Ahearn D. G. Adherence of Pseudomonas aeruginosa to Hydrophilic Contact Lenses and Other Substrata. J. Clin. Microbiol. 1987, 25, 1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel J. C.; Ruseska I.; Wright J. B.; Costerton J. W. Tobramycin Resistance of Pseudomonas aeruginosa Cells Growing as a Biofilm on Urinary Catheter Material. Antimicrob. Agents Chemother. 1985, 27, 619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel J. C.; Downey J. A.; Costerton J. W. Ultrastructural Study of Microbiologic Colonization of Urinary Catheters. Urology 1989, 34, 284–290. [DOI] [PubMed] [Google Scholar]
- Lyczak J. B.; Cannon C. L.; Pier G. B. Establishment of Pseudomonas aeruginosa Infection: Lessons from a Versatile Opportunist. Microbes Infect. 2000, 2, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Ahearn D. G.; Borazjani R. N.; Simmons R. B.; Gabriel M. M. Primary Adhesion of Pseudomonas aeruginosa to Inanimate Surfaces Including Biomaterials. Methods Enzymol. 1999, 310, 551–557. [DOI] [PubMed] [Google Scholar]
- Tran V. B.; Fleiszig S. M. J.; Evans D. J.; Radke C. J. Dynamics of Flagellum- and Pilus-Mediated Association of Pseudomonas aeruginosa with Contact Lens Surfaces. Appl. Environ. Microbiol. 2011, 77, 3644–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biais N.; Higashi D. L.; Brujić J.; So M.; Sheetz M. P. Force-Dependent Polymorphism in Type IV Pili Reveals Hidden Epitopes. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 11358–11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero M.; Yakovenko O.; Sokurenko E. V.; Thomas W. E.; Vogel V. Uncoiling Mechanics of Escherichia coli Type I Fimbriae Are Optimized for Catch Bonds. PLoS Biol. 2006, 4, 1509–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma S. L.; Ballok A. E.; Merritt J. H.; Hammond J. H.; Lu W.; Rabinowitz J. D.; O’Toole G. A. Cyclic-di-GMP-Mediated Repression of Swarming Motility by Pseudomonas aeruginosa: The pilY1 Gene and Its Impact on Surface-Associated Behaviors. J. Bacteriol. 2010, 192, 2950–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kirat-Chatel S.; Beaussart A.; Boyd C. D.; O’Toole G. A.; Dufrêne Y. F. Single-Cell and Single-Molecule Analysis Deciphers the Localization, Adhesion, and Mechanics of the Biofilm Adhesin LapA. ACS Chem. Biol. 2013, 9, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P.; El-Kirat-Chatel S.; Beaussart A.; Geoghegan J. A.; Foster T. J.; Dufrêne Y. F. The Binding Force of the Staphylococcal Adhesin SdrG Is Remarkably Strong. Mol. Microbiol. 2014, 93, 356–368. [DOI] [PubMed] [Google Scholar]
- Krieg M.; Helenius J.; Heisenberg C. P.; Müller D. J. A Bond for a Lifetime: Employing Membrane Nanotubes from Living Cells To Determine Receptor-Ligand Kinetics. Angew. Chem., Int. Ed. 2008, 47, 9775–9777. [DOI] [PubMed] [Google Scholar]
- Sun M.; Graham J. S.; Hegedüs B.; Marga F.; Zhang Y.; Forgacs G.; Grandbois M. Multiple Membrane Tethers Probed by Atomic Force Microscopy. Biophys. J. 2005, 89, 4320–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupres V.; Verbelen C.; Raze D.; Lafont F.; Dufrêne Y. F. Force Spectroscopy of the Interaction between Mycobacterial Adhesins and Heparan Sulphate Proteoglycan Receptors. ChemPhysChem 2009, 10, 1672–1675. [DOI] [PubMed] [Google Scholar]
- El-Kirat-Chatel S.; Mil-Homens D.; Beaussart A.; Fialho A. M.; Dufrêne Y. F. Single-Molecule Atomic Force Microscopy Unravels the Binding Mechanism of a Burkholderia cenocepacia Trimeric Autotransporter Adhesin. Mol. Microbiol. 2013, 89, 649–659. [DOI] [PubMed] [Google Scholar]
- Shanks R. M. Q.; Caiazza N. C.; Hinsa S. M.; Toutain C. M.; O’Toole G. A. Saccharomyces cerevisiae-Based Molecular Tool Kit for Manipulation of Genes from Gram-Negative Bacteria. Appl. Environ. Microbiol. 2006, 72, 5027–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G. A.; Kolter R. Flagellar and Twitching Motility are Necessary for Pseudomonas aeruginosa Biofilm Development. Mol. Microbiol. 1998, 30, 295–304. [DOI] [PubMed] [Google Scholar]
- Beaussart A.; El-Kirat-Chatel S.; Sullan R. M. A.; Alsteens D.; Herman P.; Derclaye S.; Dufrêne Y. F. Quantifying the Forces Guiding Microbial Cell Adhesion Using Single-Cell Force Spectroscopy. Nat. Protoc. 2014, 9, 1049–1055. [DOI] [PubMed] [Google Scholar]