Abstract
Cell-surface proteins are central for the interaction of cells with their surroundings and are also associated with numerous diseases. These molecules are exposed to mechanical forces, but the exact relation between force and the functions and pathologies associated with cell-surface proteins is unclear. An important cell-surface protein is CD4, the primary receptor of HIV-1. Here we show that mechanical force activates conformational and chemical changes on CD4 that may be important during viral attachment. We have used single-molecule force spectroscopy and analysis on HIV-1 infectivity to demonstrate that the mechanical extension of CD4 occurs in a time-dependent manner and correlates with HIV-1 infectivity. We show that Ibalizumab, a monoclonal antibody that blocks HIV-1, prevents the mechanical extension of CD4 domains 1 and 2. Furthermore, we demonstrate that thiol/disulfide exchange in CD4 requires force for exposure of cryptic disulfide bonds. This mechanical perspective provides unprecedented information that can change our understanding on how viruses interact with their hosts.
Keywords: atomic force spectroscopy, mechanochemistry, CD4 receptor, HIV-1, cell-surface proteins
Proteins located in the extracellular matrix perform important functions in cell signaling, motility, adhesion, cell–cell interaction, and antigen recognition.1 These proteins are generally exposed to mechanical forces that are believed to trigger chemical and structural alterations crucial for their function.2−4 In addition, many of these proteins are key players in diseases such as cancer metastasis, viral infections, and nervous disorders, but it is not known how mechanical force relates to medical conditions associated with these extracellular proteins. Understanding the effect of force on cell-surface proteins may have profound implications in physiology and medicine.5,6
A relevant cell-surface protein is the cluster of differentiation 4 (CD4), a four-domain protein that serves as a mechanical anchor of HIV-1. The virus initiates infection when the viral glycoprotein gp120 interacts with CD4 on the surface of T-cells.7,8 Upon gp120 binding to CD4 a cascade of conformational changes occurs in the gp120–CD4 complex that facilitates interaction with a secondary surface coreceptor (CCR5 or CXCR4), exposing the membrane-anchored gp41 subunit for final membrane fusion.9−11 Structural studies of the gp120–CD4 complex using X-ray, cryo-electron tomography or theoretical modeling have provided relevant information about the conformational changes;10,12−14 however, the key factors that trigger these large structural adjustments as well as their implications are still under debate. Similarly, the precise mechanisms by which some antibodies block HIV-1 infection,15,16 or the underlying mechanism by which oxidoreductases enzymes seem to regulate the redox state of disulfide bonds in CD4 and gp120 during infection are yet to be defined.17 We suggest that mechanical force can trigger structural and chemical alterations on CD4 and gp120 that may be relevant for the initial interaction of virus and host cell.
Here, we have used a combination of single-molecule force spectroscopy experiments and analysis on HIV-1 infectivity data to investigate the effect of force on domains 1 and 2 of human CD4. Our findings show that mechanical extension of CD4 domains may occur within a broad range of forces. We also show that mechanical forces may be relevant to better understand the mechanism by which antibodies block HIV-1, as well as the process of thiol/disulfide exchange on CD4 assisted by oxidoreductases enzymes.
Results
Nanomechanics of CD4D1 and CD4D2 Revealed by Single-Molecule Force Spectroscopy
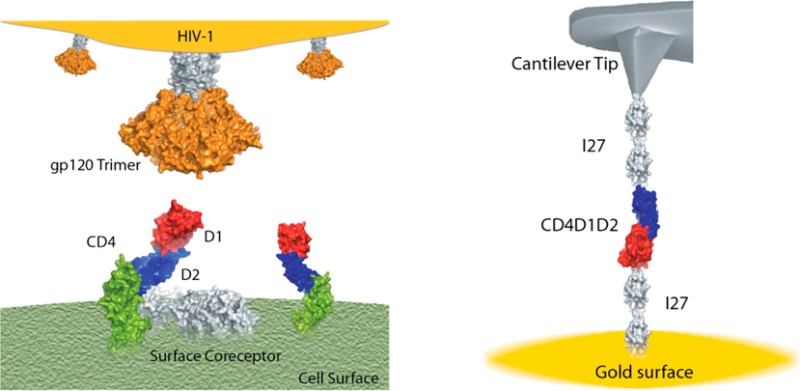
To investigate the effect of force on CD4 modules, we constructed a polyprotein composed of domains 1 and 2 of CD4 bracketed by dimeric handles made of the 27th domain of human cardiac titin used as molecular fingerprint, (I27)2-CD4D1D2-(I27)2 (Figure 1a). We used an atomic force microscope (AFM) to stretch the CD4 construct at a constant speed in the force extension mode. Mechanical unfolding of I27 modules provides a clear fingerprint distinguished by the unfolding force of ∼200 pN and contour length (ΔLc) of ∼28 nm for each module.18 This approach allows us direct identification of any other unfolding event that can be attributed to CD4 modules. A typical force–extension trace resulting from the unfolding of (I27)2-CD4D1D2-(I27)2 shows two peaks corresponding to CD4 domains prior to the unfolding of the four I27 modules (Figure 1b).
Figure 1.
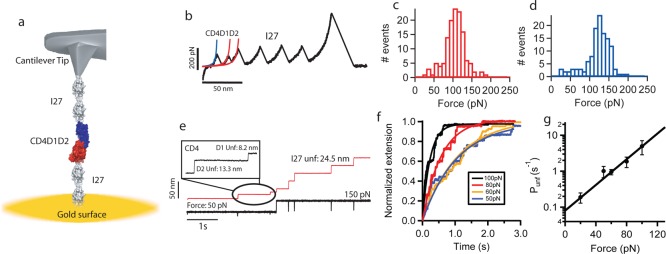
Nanomechanics of Human CD4. (a) Schematic representation of an AFM experiment of the polyprotein (I27)2-CD4D1D2-(I27)2. The I27 modules are used as molecular fingerprint. The polyprotein is attached to a gold-covered coverslide in one end and a cantilever tip in the other end. Notice the similarity between the artificial AFM experiments and the HIV-1 infection process (see the graphic in the Abstract section). (b) Force–extension curve of the polyprotein (I27)2-CD4D1D2-(I27)2. Two peaks corresponding to domains CD4D2 and CD4D1 precede the unfolding of the four I27 domains. The curves are fitting to the worm-like chain model from where we derive the contour length of CD4D1 (red) and CD4D2 (blue). (c) Histogram of unfolding force for CD4D1 (n = 132) and (d) CD4D2 (n = 125). (e) Force–clamp trace of the polyprotein (I27)2-CD4D1D2-(I27)2. The unfolding of CD4D1D2 is monitored at 50 pN, and the unfolding of I27 domains is monitored at 150 pN. (f) Exponential fitting to summed and averaged unfolding traces of CD4D1D2 at different forces. From this fitting we obtain the unfolding rate at a given force. We use a single-exponential fit to provide an approximated idea of the time scale of the CD4D1D2 extension. (g) Force-dependency of unfolding of CD4D1D2. An extrapolation to zero force predicts an unfolding rate of 0.08 s–1.
The contour lengths measured are 10 ± 1 nm and 16 ± 4 nm for CD4D1 and CD4D2, respectively (Supporting Information, Figure 1) which are in close agreement with the maximum theoretical values for unfolding up to their disulfide bonds (12 and 20 nm, respectively, considering 0.4 nm/residue). We have measured mean unfolding forces of 101 ± 30 pN for CD4D1 and 119 ± 32 pN for CD4D2 at a pulling speed of 400 nm/s (Figure 1c,d). The mechanical stability of proteins depends on the speed at which the proteins are stretched. We do not really know what the pulling speed could be in a biological context such as the interaction of an HIV-1 particle with CD4; therefore, we do not really know the force that CD4 experiences. For this reason, we performed experiments at a much lower pulling speed, 10 nm/s. At this speed we measured an unfolding force of 57 ± 21 pN for CD4D1 and 75 ± 23 pN for CD4D2 (Supporting Information, Figure 2).
We observe that the unfolding of CD4D2 normally occurs prior to the unfolding of CD4D1 even though the unfolding force of CD4D1 is lower (Supporting Information, Figure 3). This hierarchical behavior suggests a protective role of CD4D2 over D1. Both domains act in unity,19 sharing structural elements that confer mechanical rigidity.
To investigate the time scale at which the mechanical extension of CD4D1D2 occurs, we used the force–clamp technique, which allows the application of a well-controlled force to a single polyprotein over a period of time.20 We applied a double-pulse force protocol that allows the separation of the unfolding of CD4D1D2 from that of I27 domains. We first applied a force-pulse of 20–100 pN to trigger the extension of CD4D1 and CD4D2. We measured a step size of ∼13 nm for CD4D2 and ∼8 nm for CD4D1 (Supporting Information, Figure 4). A second pulse of 150 pN was applied for 4s to unfold I27 modules, ∼24.5 nm (Figure 1e), which is used as a molecular fingerprint.21 We have accumulated numerous unfolding traces of CD4D1D2 at different forces from 20 to100 pN, from where we can obtain the unfolding rate at a given force. As a first approximation we have used single-exponential fits to estimate the time scale for CD4D1D2 mechanical extension (Figure 1f and Supporting Information, Figure 5 for 20 pN). The force dependence of the rate of unfolding of CD4D1D2 is shown in Figure 1g. The extrapolation to low forces allows us to predict the unfolding rates (0.08 s–1 at F = 0 pN). Therefore, the mechanical extension of CD4D1D2 may occur even at very low forces. At these low forces this extension may proceed through intermediates. In fact, we have observed some traces (∼5%) in which CD4D2 unfolds in two steps (Supporting Information, Figure 6). We also carried out experiments in the force–ramp mode in which the force is changed linearly at a constant speed (33 pN/s) allowing the separation of the different unfolding events while controlling the force. We obtained the distribution of the initial unfolding force of CD4D1D2 which in this mode peaks at ∼80 pN (Supporting Information, Figure 7). Although we have not studied experimentally domains 3 and 4, they have some structural similarities with domains 1 and 2. In fact, computational analyses have shown that they unfold at similar forces as those of domains 1 and 2 (data not shown). Similarly to D1 and D2, D3 and D4 also act as a unity sharing a continuous β-strand, but are separated from domains D1 and D2 by a hinge-like variability (pdb code: 1wiq). Although some residues from D2 and D3 interact, the junction has been shown to be highly flexible,22 and therefore no mechanical rigidity is expected within that region.
Extension of CD4 Correlates with HIV-1 Infectivity
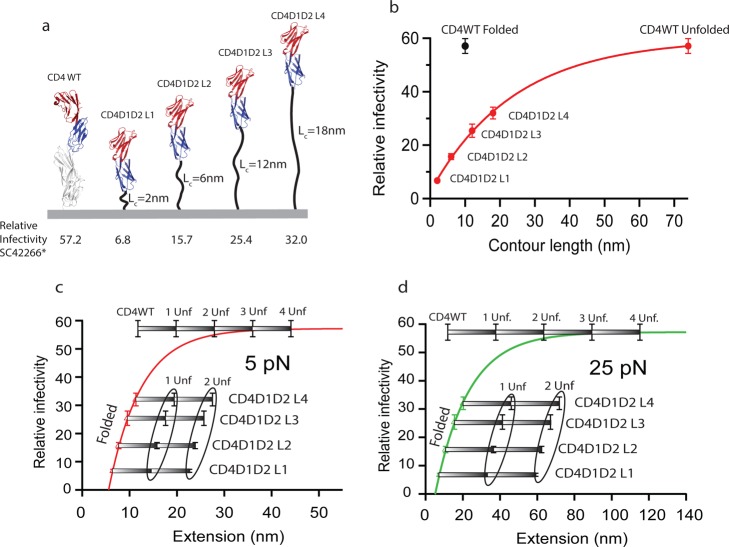
Independent experiments measuring HIV-1 infectivity of cells expressing variants of CD4 with extended linkers can offer information regarding CD4 extensibility. In recent work, Freeman et al.,23 expressed domains CD4D1D2 directly tethered to the transmembrane fragment of 293T cells throughout a flexible linker. Several constructs with linkers of 5, 15, 30, and 45 residues (repeats of the pentapeptide GGGGS) were designed and expressed in the surface of 293T cells. All variants supported HIV-1 infection but with different efficiency (Figure 2a). Using this data, we performed some calculations to correlate infectivity and construct extension. Considering a length of 0.4 nm/residue, the total length of the unstructured linkers is 2, 6, 12, and 18 nm. The total extension of the CD4 construct is the sum of the length of the linker plus the folded CD4D1D2 domains which is ∼6 nm, as determined from the CD4 X-ray structure (pdb: 1wip). The infectivity was plotted against the predicted length demonstrating a striking correlation with CD4D1D2-linker length; longer linkers support higher infectivity (Figure 2b). On the contrary, CD4WT infectivity does not follow the correlation of the CD4D1D2-linker variants (black dot in Figure 2b). However, if we consider, for instance, that two CD4WT domains are extended and two folded, the total extension is now longer. The correlation is now consistent and can be fitted to a single exponential (Figure 2b).
Figure 2.
Correlation between HIV-1 infectivity and CD4 extensibility. (a) Schematic representation of CD4D1D2 plus linkers tethered to cell membrane. The linkers are composed of repeats of the pentapetide GGGGS; one repeat for L1, three repeats for L2, six repeats for L3, and nine repeats for L4. The relative infectivity (relative light unit, RLU) in SC42266 isolates is indicated for each construct and CD4WT (data extracted from ref (23)). (b) Plot of relative infectivity versus contour length of CD4-linker constructs. The length of the constructs is estimated by adding the length of the folded domains to the length of linkers (number of residues times 0.4 nm/residue). The black dot corresponds to CD4WT in the folded state. The red dot for CD4 WT considers two domains unfolded and two folded. The red line is simple exponential fit intended to guide the eye. (c, d) Plots of infectivity versus extension calculated from the freely jointed chain model at 5 and 25 pN, respectively. Different scenarios regarding domain unfolding are considered and indicated in the gray bars, from all domains folded to all domains unfolded. The line is an exponential fitting to the empirical equation described in the text and in the Supporting Information.
Nevertheless, it has to be considered that the extension of a polypeptide under mechanical loads depends on the force applied and is better described by models of polymer elasticity such as the worm-like chain (WLC) or the freely jointed chain (FJC). In addition, the mechanical unfolding of different domains may occur independently which will impact the total extension. Since we do not know the force that might be acting on CD4, we applied the FJC model to predict the extension of the CD4D1D2-linker constructs at different forces: 5, 15, 25, and 40 pN (Supporting Information, Figure 8). We considered different unfolding possibilities, such as one or two unfolded domains or even three or four unfolded domains for CD4WT. By fitting an exponential, I(x) = I0 – A exp[x(F)/x0], we obtained an empirical correlation between the infectivity, I(x), and the force-dependent extensibility, x(F), in which CD4WT infectivity is considered an asymptote that represents maximum infectivity (Figure 2c,d). Here, I0 is the wild-type infectivity taken as 57.16 RLU;23A is a fitting parameter that defines the folded length at zero infectivity as x0ln(I0/A) ≈ 5.3 nm; and x0 is a length-scale coefficient of the infectivity at a given force. The force-dependent extensibility is described by the FJC phenomenological model, x(F) = xC[tanh–1(Fa/kBT) −kBT/Fa], where xC is the molecule’s combined contour length for the contributions of all the folded and unfolded domains, F is the applied force, a is the Kuhn length, kB is the Boltzmann constant, and T is the temperature (see Supporting Information, Table 1 for fitting parameters). The different scenarios for domain extension in CD4WT and CD4D1D2-linker variants demonstrate that at least 2 or 3 unfolded domains are necessary to follow the correlation at 5 pN and 25 pN (Figure 2c,d).
Mechanical Effect of HIV-1 Neutralizing Antibody Ibalizumab on CD4 Domains
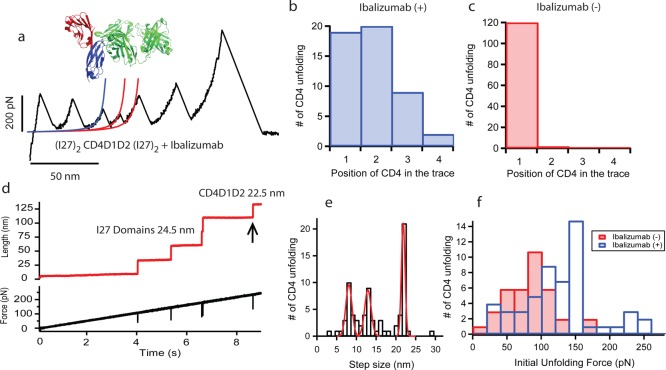
The calculations above suggest a correlation between the extensibility of CD4 and HIV-1 infectivity. We reasoned that an increase in the mechanical stability of CD4 should prevent extensibility, potentially blocking HIV-1 entry. The mechanical stability of a protein can be modified by introducing mutations in specific locations;18 however, introducing mutations is not reversible and the effect generally goes in the destabilizing direction. A strategy that has been proven successful in protein mechanical stabilization is antibody binding.16 This is important considering that the use of antibodies is a common strategy to fight HIV-1.15 We decided to test whether an anti-CD4 antibody able to block HIV-1 infectivity can actually affect the mechanics of CD4 modules. We tested the neutralizing antibody Ibalizumab, a humanized monoclonal antibody that specifically binds in the interface between CD4D1 and CD4D2 with very high affinity, (KD = 8.25 × 10–11 M) ref (24), without interfering gp120 binding. Ibalizumab has been shown to be effective against a wide range of HIV-1 isolates;23 however, the critical factors that make Ibalizumab a good inhibitor are not yet clear.24 The crystal structure of CD4 bound to Ibalizumab (pdb: 3O2D) shows critical residues that form the binding epitope in CD4.23
We performed experiments with (I27)2-CD4D1D2-(I27)2 in the force–extension mode in the presence of Ibalizumab (Figure 3a). The unfolding force of CD4D2 is slightly higher than in the absence of Ibalizumab, 138 ± 23 pN, which represents an increment of ∼20 pN. For CD4D1 the measured force was 100 ± 24 pN, similar to that observed with no Ibalizumab (Figure S7). Surprisingly, we observed that ∼60% of the traces showed the unfolding of the CD4 domains after the unfolding of one, two, or even three I27 peaks (Figure 3b), a phenomenon rarely observed in the absence of Ibalizumab (Figure 3c). This observation contradicts the logic of the force extension experiments in which one always expects the weaker domains to unfold first. From these experiments it is clear that Ibalizumab has a mechanical effect on CD4. However, it is difficult to assess the nature of this effect because the unfolding force of the CD4 domains is not much different than that without Ibalizumab. Surprisingly, CD4 domains are able to survive up to three consecutive I27 unfolding events which occur at forces of ∼200 pN. A possible scenario is that CD4D1D2 is very mechanically stable when Ibalizumab is bound, and only when the antibody leaves do they undergo mechanical extension at a force similar to their normal unfolding force.
Figure 3.
Mechanical effect of the HIV-1 entry inhibitor Ibalizumab in human CD4. (a) Force–extension trace of (I27)2-CD4D1D2-(I27)2 in the presence of Ibalizumab. The structure of the complex CD4D1D2-Ibalizumab is also shown. CD4D1D2 unfolding is detected in the third position after surviving the mechanical unfolding of two I27 modules. The lines represent fittings to the WLC model. (b and c) Histograms of CD4 domains position in the trace in the presence and absence of Ibalizumab, respectively. (d) Force–ramp trace of the (I27)2-CD4D1D2-(I27)2 polyprotein in the presence of Ibalizumab. The mechanical extension of CD4D1D2 is detected after all I27 domains at a force of ∼250 pN. (e) Histogram of step size for the unfolding of CD4D1D2 in force–ramp mode and in the presence of Ibalizumab (n = 76). The unfolding of CD4D1 and CD4D2 are measured at 8.6 ± 0.7 nm and 13.3 ± 1.1 nm, respectively. We monitored a dominant peak at around 22.2 ± 0.9 nm corresponding to the simultaneous unfolding of CD4D1 and CD4D2. Compare this histogram with the one with no Ibalizumab in Supporting Information, Figure 4. (f) Histogram of initial unfolding force of CD4D1D2 and in the absence (red, n = 37) and presence (blue, n = 54) of Ibalizumab. A displacement toward higher force is observed when Ibalizumab is present.
Interestingly, in the force–ramp mode the initial unfolding force of the tandem CD4D1D2 in the presence of Ibalizumab very often occurred at elevated forces (we collected traces with CD4 unfolding at ∼250 pN) and as a single step of ∼22 nm (Figure 3d,e). This is consistent with the simultaneous unfolding of domains both at high force and in sharp contrast with experiments in the absence of Ibalizumab, in which we observed one step for each CD4 domain (Supporting Information, Figure 7). A histogram of initial unfolding force demonstrates that the main peak of force shifts from ∼80 pN for CD4 with no Ibalizumab to ∼150 pN as a consequence of adding Ibalizumab (Figure 3f). We suggest that this force might be the release force of Ibalizumab indicating that CD4D1D2 only unfolds when Ibalizumab unbinds. The phenomenon of a release force in protein–protein interaction has been already observed in other molecules exposed to mechanical forces such as catenin and vinculin.25
We observed that in the presence of Ibalizumab, CD4D1D2 often unfolds alongside with an I27 domain (Supporting Information, Figure 10), indicating that Ibalizumab release force is similar or slightly lower than the unfolding force of I27, measured at 155 ± 22 pN. This may explain, in part, the effect of Ibalizumab in the force–extension traces. In this mode, the unfolding of I27 may be accompanied by Ibalizumab release, leaving CD4D1D2 free of antibodies for subsequent unfolding. However, in force–extension the force drops abruptly after I27 unfolding which allows CD4D1D2 to unfold at their normal forces. Nevertheless, we still observed a high number of CD4D1D2 unfolding peaks before I27 at forces below the suggested release force, which might correspond to Ibalizumab-free polyproteins. We also tested whether the release of Ibalizumab might be affected over time by applying a multiforce protocol using three forces, 20, 60, and 150 pN, with different application times (Supporting Information, Figure 11). We observed that CD4D1D2 only unfolds when the force applied is 150 pN. It does not do it at 15 nor 60 pN, irrespective of the time. From these experiments we infer that Ibalizumab makes CD4 stronger.
Our experiments do not address if the increment in CD4 mechanical stability is directly related to the inhibiting power of Ibalizumab, but we suspect that increasing mechanical rigidity in CD4 will have an effect in the cascade of conformational rearrangements necessary to bring the viral particle close to the surface.
Mechanochemical Cleavage of CD4 Disulfide Bonds by Thioredoxin Enzymes
Besides introducing external molecules, a natural way of increasing mechanical resistance is throughout disulfide bonds. Interestingly, thiol/disulfide exchange by oxidoreductase enzymes in both gp120 and CD4 seems to be important during HIV-1 infection contributing to the conformational changes in the gp120-CD4 complex required for membrane fusion and infection.17,26 Importantly, inhibition of oxidoreductase enzymes seems to prevent HIV-1 infection.27,28 In the case of CD4, thiol–disulfide exchange in CD4D2 seems to be carried out by thioredoxin enzymes (Trx).28 Surprisingly, the reduced form of CD4D2 is the one preferred for HIV-1 to infect cells. However, the disulfide bond in CD4D2 is buried and therefore inaccessible to Trx enzymes.29 We reasoned that force might trigger the mechanochemical cleavage of the buried disulfide bond.
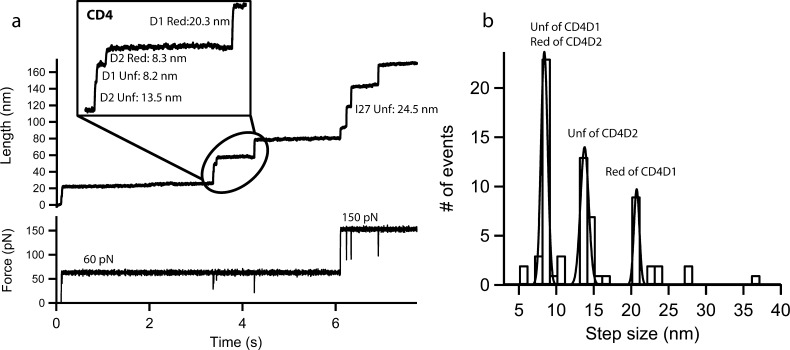
In a previous work, we used force spectroscopy to investigate the reduction of single disulfide bonds under force catalyzed by Trx enzymes.30,31 We demonstrated that force is required for exposure of cryptic disulfides and also for regulation of the chemical mechanisms utilized by Trx enzymes for disulfide bond reduction. Here, we have investigated the mechanoenzymatic cleavage of disulfide bonds in CD4D1D2. We used a two-pulse protocol to monitor the unfolding of CD4D1D2 and I27 modules. In the presence of 10 μM human Trx we observe extra steps following the unfolding of the CD4 domains. The release of the sequestered residues upon Trx reduction will lead to an extra extension of ∼20 and ∼8 nm for CD4D1 and CD4D2, respectively. We have observed such steps following the unfolding of CD4 domains (Figure 4a,b and Supporting Information, Figure 6). We have also observed a number of traces showing the reduction of disulfide bond in CD4D2 without the reduction of CD4D1 (Supporting Information, Figure 12), which might be due to different force dependence. Steps corresponding to the unfolding of reduced CD4D1 and CD4D2 at ∼29 and ∼22 nm, respectively, are barely observed, indicating that disulfide bonds remain oxidized and buried within the structure of the domains in the absence of mechanical exposure.
Figure 4.
Mechanochemical cleavage of human CD4D1D2 by Trx enzymes. (a) Mechanical unfolding of CD4D1 and CD4D2 (Unf) and subsequent disulfide bond reduction in both domains by 10 μM human Trx (Red). The reductions, ∼8 nm for CD4D1 and ∼20 nm for CD4D2, occur after unfolding, which demonstrates that mechanical exposition of disulfides is required. (b) Histogram of step size observed for the unfolding and reduction of CD4D1D2 by human Trx (n = 25). Three main peaks are observed. The first one at 8.0 ± 1.0 nm corresponds to both unfolding of CD4D1 and reduction of CD4D2 and it is higher because it collects these two events. The second peak at 13.4 ± 0.7 nm is the unfolding of CD4D2, and the third one at 20.2 ± 1.8 nm corresponds to the reduction of CD4D1.
Conclusion
Our results show that the mechanical extension of CD4 domains may occur within a broad range of forces. The interaction of a virus with the cell surface has been shown to be highly dynamic,32 but the forces involved in the interaction of the virus and its receptor are unknown. Mechanical forces may come from different sources such as Brownian motion,33 cellular uptake dynamics,32 viral surface movements, and cell–cell interaction.34 We show that even at low forces mechanical extension of CD4 domains may occur at longer time scales. This might have implications in the interaction of HIV-1 with T cells. Besides virus-to-cell transmission, HIV-1 can also be transmitted cell-to-cell throughout a virological synapse that seems to involve gp120, CD4, and others adhesion proteins.35−37 In this case, the interaction between cells could generate mechanical forces in the proteins of the adhesion junction that can reach 10–100 pN, or even higher.3,38 Therefore, the range of mechanical forces considering both virus-to-cell and cell-to-cell transmission may be enough to trigger mechanical extension of CD4 domains in a time-dependent manner. Mechanical extensibility of CD4 domains increases the flexibility which may facilitate the conformational rearrangements and interaction with surface coreceptors.8,39
Successful infection requires proper attachment starting with at least one single CD4 molecule. Partial or total mechanical extension of CD4 domains may help viruses to remain attached by acting as a shock absorber that permits receptor ligation to the virus. This mechanism has been already suggested for CD4.24,40 Similarly, the shock-absorber mechanism has been proposed for tenascin and Mel-CAM, modular proteins from the extracellular matrix that are important in cell–cell interaction. These proteins undergo mechanical extension to release tensile stress and prevent cell detachment.3,38 Mechanochemical cleavage of CD4 disulfide bonds would also facilitate structural adjustments and would help to increase extension.
The experiments with Ibalizumab suggest a possible connection between mechanical stability and HIV-1 entry. The interactions that the antibody establishes with CD4 generate a molecular complex with increased mechanical stability. To the best of our knowledge, this is the first experimental observation of protein mechanical alteration from a drug effective against viral infection. This opens the door for considering new strategies and treatments based on the manipulation of the mechanics of proteins, namely mechanopharmaceuticals. Molecules that increase the mechanical stability of CD4 and also gp120 will introduce steric constraints and rigidity that might be detrimental for virus attachment and conformational changes. This mechanical effect could also be achieved by manipulating the redox state of disulfide bonds in CD4 and gp120.
There are many viruses such as Epstein–Barr virus, rhinovirus, and poliovirus using proteins with similar features than those of CD4 also acting as mechanical anchors. The reason why viruses prefer these multimodular Ig proteins might be related to their recognition and adhesive properties,41 but we hypothesize that these proteins are crucial due to their unique mechanical properties. The role of mechanical forces has been experimentally tested in a small number of cell-surface and cytoskeletal proteins but never in the context of viral infection. We are aware that our results are an initial approximation and further experimentation will be necessary to extend these studies to the cellular level. However, we believe that this mechanical view offers new avenues for a better understanding of viral infections and also expands our knowledge about the effect of force on the regulation of biochemical and biological processes.
Methods
Protein Expression and Purification
The gene encoding the polyprotein (I27)2-CD4D1D2-(I27)2 was constructed as described elsewhere.18 The insert (I27)2-CD4D1D2-(I27)2 is constructed following a multistep cloning process using I27 and CD4D1D2 inserts with restriction sites BamHI, BglII, and KpnI. The final gene was cloned into pQE80L vector (Qiagen) and transformed in DH5α cells (Invitrogen). Protein expression was induced with 1 mM IPTG, and cells were incubated overnight in LB medium at 20 °C to avoid degradation. Cell pellets were disrupted using a French press and the His6-tagged proteins were loaded onto a His GraviTrap affinity column (GE Healthcare). The protein was further purified by size exclusion chromatography using a Superdex 200 HR 10/30 column from GE Healthcare. The protein was eluted in 10 mM HEPES at pH 7.2 containing 150 mM NaCl and 1 mM EDTA. The purified protein was verified by SDS-PAGE. The same protocol was used to express and purify human thioredoxin although expression was at 37 °C, and the cells used were E. coli BL21 (DE3) from Invitrogen.
AFM Experiments and Data Analysis
We used a custom-made atomic force microscope20 as well as a commercial version AFS-1 from Luigs & Newmann, GmbH. Cantilevers were from Bruker, model MLCT, with a typical spring constant of 15–20 pN/nm, measured using the equipartition theorem. The buffer used was 10 mM HEPES at pH 7.2 containing 150 mM NaCl and 1 mM EDTA. About 10 μL of the solution containing (I27)2-CD4D1D2-(I27)2 protein was deposited on a gold-covered coverslide. We allowed several minutes for protein adsorption. In the experiment using Ibalizumab, the sample was first incubated for 1 h at room temperature with an excess of antibody (proportion >1:5). In the experiments with human Trx, about 100 μL of solution containing Trx to a final concentration of 10 μM, 50 nM human Trx reductase, and 2 mM NADPH was added to the CD4 solution. Force–extension experiments were performed at 400 and 10 nm/s piezo movement speed. The data were analyzed using the worm-like chain model of polymer elasticity. In the force–clamp mode our AFM has a length resolution of 0.5 nm and the feedback response is 5 ms. We use a three-pulse protocol in the force–clamp experiments, first pressing the surface at 800 pN for 0.2 s, then retracting the tip at a stretching force of 20–100 pN for CD4 unfolding detection and finally increasing the stretching force to 150 pN for I27 unfolding probe. The time of the second and third pulses varies from 2 to 10 s. Data analysis was carried out using Igor software (Wavemetrics). Summed and averaged traces were fitted to single exponential functions. Figures were prepared using Igor software and Adobe Illustrator.
Acknowledgments
Research was supported by U.S. National Institutes of Health Grants HL061228 and HL066030 to J.M.F, and funding from the Spanish Ministry of Economy and Competitiveness BIO2013–46163-R to R.P.-J. We thank all Fernandez lab members for helpful discussions and critiques. R.P.-J. thanks Fundación Caja Madrid (Spain) and Ibercaja Obra Social (Spain) for financial support. R.P.-J. and A. A.-C. thank CIC nanoGUNE for financial support.
Supporting Information Available
Material and methods, supplementary figures, and supplementary table. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
# Department of Chemical-Engineering & Ilze Katz Institute for Nanoscience and Technology, Ben-Gurion University of the Negev, Beer-Sheva, Israel.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Deller M. C.; Jones E. Y. Cell Surface Receptors. Curr. Opin. Struct. Biol. 2000, 10, 213–219. [DOI] [PubMed] [Google Scholar]
- Bershadsky A. D.; Balaban N. Q.; Geiger B. Adhesion-Dependent Cell Mechanosensitivity. Annu. Rev. Cell Dev. Biol. 2003, 19, 677–695. [DOI] [PubMed] [Google Scholar]
- Oberhauser A. F.; Marszalek P. E.; Erickson H. P.; Fernandez J. M. The Molecular Elasticity of the Extracellular Matrix Protein Tenascin. Nature 1998, 393, 181–185. [DOI] [PubMed] [Google Scholar]
- Wang X.; Ha T. Defining Single Molecular Forces Required to Activate Integrin and Notch Signaling. Science 2013, 340, 991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V.; Sheetz M. In Nanotechnology; Vogel V., Ed.; Wiley-VCH: 2009; Vol. 5. [Google Scholar]
- Bao G. Protein Mechanics: A New Frontier in Biomechanics. Exp. Mech. 2009, 49, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R.; Kwong P. D.; Desjardins E.; Sweet R. W.; Robinson J.; Hendrickson W. A.; Sodroski J. G. The Antigenic Structure of the HIV gp120 Envelope Glycoprotein. Nature 1998, 393, 705–711. [DOI] [PubMed] [Google Scholar]
- Levy J. A.HIV and the Pathogenesis of Aids, 3rd ed.; ASM Press: Washington, DC, 2007. [Google Scholar]
- Wu L.; Gerard N. P.; Wyatt R.; Choe H.; Parolin C.; Ruffing N.; Borsetti A.; Cardoso A. A.; Desjardin E.; Newman W.; et al. CD4-Induced Interaction of Primary HIV-1 Gp120 Glycoproteins with the Chemokine Receptor Ccr-5. Nature 1996, 384, 179–183. [DOI] [PubMed] [Google Scholar]
- Zhou T.; Xu L.; Dey B.; Hessell A. J.; Van Ryk D.; Xiang S. H.; Yang X.; Zhang M. Y.; Zwick M. B.; Arthos J.; et al. Structural Definition of a Conserved Neutralization Epitope on HIV-1 gp120. Nature 2007, 445, 732–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachou A.; Sekaly R. P. Binding of Soluble Recombinant HIV Envelope Glycoprotein gp120 Induces Conformational Changes in the Cellular Membrane-Anchored CD4Molecule. Biochem. Biophys. Res. Commun. 1999, 265, 428–433. [DOI] [PubMed] [Google Scholar]
- Chen L.; Kwon Y. D.; Zhou T.; Wu X.; O’Dell S.; Cavacini L.; Hessell A. J.; Pancera M.; Tang M.; Xu L.; et al. Structural Basis of Immune Evasion at the Site of CD4 Attachment on HIV-1 gp120. Science 2009, 326, 1123–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Bartesaghi A.; Borgnia M. J.; Sapiro G.; Subramaniam S. Molecular Architecture of Native HIV-1 gp120 Trimers. Nature 2008, 455, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkut A.; Hendrickson W. A. Structural Plasticity and Conformational Transitions of HIV Envelope Glycoprotein gp120. PLoS One 2012, 7, e52170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F.; Mouquet H.; Dosenovic P.; Scheid J. F.; Scharf L.; Nussenzweig M. C. Antibodies in HIV-1 Vaccine Development and Therapy. Science 2013, 341, 1199–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R.; Franco D.; Kao C. Y.; Yu F.; Huang Y.; Ho D. D. Epitope Mapping of Ibalizumab, a Humanized Anti-CD4Monoclonal Antibody with Anti-HIV-1 Activity in Infected Patients. J. Virol 2010, 84, 6935–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J.; Isacsson O.; Soderlund J.; Balzarini J.; Johansson M.; Lundberg M. Human Glutaredoxin-1 Catalyzes the Reduction of HIV-1 gp120 and CD4 Disulfides and Its Inhibition Reduces HIV-1 Replication. Int. J. Biochem. Cell Biol. 2009, 41, 1269–1275. [DOI] [PubMed] [Google Scholar]
- Perez-Jimenez R.; Garcia-Manyes S.; Ainavarapu S. R.; Fernandez J. M. Mechanical Unfolding Pathways of the Enhanced Yellow Fluorescent Protein Revealed by Single Molecule Force Spectroscopy. J. Biol. Chem. 2006, 281, 40010–40014. [DOI] [PubMed] [Google Scholar]
- Ryu S. E.; Kwong P. D.; Truneh A.; Porter T. G.; Arthos J.; Rosenberg M.; Dai X. P.; Xuong N. H.; Axel R.; Sweet R. W.; et al. Crystal-Structure of an HIV-Binding Recombinant Fragment of Human CD4. Nature 1990, 348, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J. M.; Li H. Force-Clamp Spectroscopy Monitors the Folding Trajectory of a Single Protein. Science 2004, 303, 1674–1678. [DOI] [PubMed] [Google Scholar]
- del Rio A.; Perez-Jimenez R.; Liu R.; Roca-Cusachs P.; Fernandez J. M.; Sheetz M. P. Stretching Single Talin Rod Molecules Activates Vinculin Binding. Science 2009, 323, 638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Kwong P. D.; Hendrickson W. A. Dimeric Association and Segmental Variability in the Structure of Human CD4. Nature 1997, 387, 527–530. [DOI] [PubMed] [Google Scholar]
- Freeman M. M.; Seaman M. S.; Rits-Volloch S.; Hong X.; Kao C. Y.; Ho D. D.; Chen B. Crystal Structure of HIV-1 Primary Receptor CD4 in Complex with a Potent Antiviral Antibody. Structure 2010, 18, 1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch G. W.; Turville S.; Carter B.; Sloane A. J.; Chan A.; Muljadi N.; Li S.; Low L.; Armati P.; Raison R.; et al. Marked Differences in the Structures and Protein Associations of Lymphocyte and Monocyte CD4: Resolution of a Novel CD4 Isoform. Immunol Cell Biol. 2006, 84, 154–165. [DOI] [PubMed] [Google Scholar]
- Yao M.; Qiu W.; Liu R.; Efremov A. K.; Cong P.; Seddiki R.; Payre M.; Lim C. T.; Ladoux B.; Mege R. M.; et al. et al. Force-Dependent Conformational Switch of Alpha-Catenin Controls Vinculin Binding. Nat. Commun. 2014, 5, 4525. [DOI] [PubMed] [Google Scholar]
- Ryser H. J.; Fluckiger R. Progress in Targeting HIV-1 Entry. Drug Discovery Today 2005, 10, 1085–1094. [DOI] [PubMed] [Google Scholar]
- Gallina A.; Hanley T. M.; Mandel R.; Trahey M.; Broder C. C.; Viglianti G. A.; Ryser H. J. Inhibitors of Protein-Disulfide Isomerase Prevent Cleavage of Disulfide Bonds in Receptor-Bound Glycoprotein 120 and Prevent HIV-1 Entry. J. Biol. Chem. 2002, 277, 50579–50588. [DOI] [PubMed] [Google Scholar]
- Matthias L. J.; Yam P. T.; Jiang X. M.; Vandegraaff N.; Li P.; Poumbourios P.; Donoghue N.; Hogg P. J. Disulfide Exchange in Domain 2 of CD4 Is Required for Entry of HIV-1. Nat. Immunol 2002, 3, 727–732. [DOI] [PubMed] [Google Scholar]
- Matthias L. J.; Hogg P. J. Redox Control on the Cell Surface: Implications for HIV-1 Entry. Antioxid. Redox. Signaling 2003, 5, 133–138. [DOI] [PubMed] [Google Scholar]
- Ainavarapu S. R.; Brujic J.; Huang H. H.; Wiita A. P.; Lu H.; Li L.; Walther K. A.; Carrion-Vazquez M.; Li H.; Fernandez J. M. Contour Length and Refolding Rate of a Small Protein Controlled by Engineered Disulfide Bonds. Biophys. J. 2007, 92, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Jimenez R.; Li J.; Kosuri P.; Sanchez-Romero I.; Wiita A. P.; Rodriguez-Larrea D.; Chueca A.; Holmgren A.; Miranda-Vizuete A.; Becker K.; et al. Diversity of Chemical Mechanisms in Thioredoxin Catalysis Revealed by Single-Molecule Force Spectroscopy. Nat. Struct. Mol. Biol. 2009, 16, 890–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsher K.; Yang H. Multi-Resolution 3d Visualization of the Early Stages of Cellular Uptake of Peptide-Coated Nanoparticles. Nat. Nanotechnol. 2014, 9, 198–203. [DOI] [PubMed] [Google Scholar]
- English T. J.; Hammer D. A. Brownian Adhesive Dynamics (Brad) for Simulating the Receptor-Mediated Binding of Viruses. Biophys. J. 2004, 86, 3359–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt C. J.; Greber U. F. Virus Movements on the Plasma Membrane Support Infection and Transmission between Cells. PLoS Pathog. 2009, 5, e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes W.; Sherer N. M.; Jin J.; Zhong P. Virus Cell-to-Cell Transmission. J. Virol. 2010, 84, 8360–8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner W.; McNerney G. P.; Chen P.; Dale B. M.; Gordon R. E.; Chuang F. Y.; Li X. D.; Asmuth D. M.; Huser T.; Chen B. K. Quantitative 3d Video Microscopy of HIV Transfer across T Cell Virological Synapses. Science 2009, 323, 1743–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigdomenech I.; Massanella M.; Izquierdo-Useros N.; Ruiz-Hernandez R.; Curriu M.; Bofill M.; Martinez-Picado J.; Juan M.; Clotet B.; Blanco J. HIV Transfer between CD4 T Cells Does Not Require Lfa-1 Binding to Icam-1 and Is Governed by the Interaction of HIV Envelope Glycoprotein with CD4. Retrovirology 2008, 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl P.; Kwok C. H.; Manderson G.; Speicher D. W.; Discher D. E. Forced Unfolding Modulated by Disulfide Bonds in the Ig Domains of a Cell Adhesion Molecule. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 1565–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham P. R.; McKnight A. Cell Surface Receptors, Virus Entry and Tropism of Primate Lentiviruses. J. Gen. Virol. 2002, 83, 1809–1829. [DOI] [PubMed] [Google Scholar]
- Discher D. E.; Bhasin N.; Johnson C. P. Covalent Chemistry on Distended Proteins. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 7533–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody T. S.; Kirchner E.; Guglielmi K. M.; Stehle T. Immunoglobulin Superfamily Virus Receptors and the Evolution of Adaptive Immunity. PLoS Pathog. 2009, 5, e1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.