Abstract
Purpose
Extracellular proteins are easily accessible, which presents a sub-proteome of molecular targets that have high diagnostic and therapeutic potential. Efforts have been made to catalogue the cardiac extracellular matridome and analyze the topology of identified proteins for the design of therapeutic targets. Although many bioinformatics tools have been developed to predict protein topology, topology has been experimentally validated for only a very small portion of membrane proteins. The aim of this study was to use a glycoproteomics and mass spectrometry approach to identify glycoproteins in the extracellular matridome of the infarcted LV and provide experimental evidence for topological determination.
Experimental design
Glycoproteomics analysis was performed on eight biological replicates of day 7 post-MI samples from wild type mice using solid-phase extraction of glycopeptides, followed by mass spectrometric identification of N-linked glycosylation sites for topology assessment.
Results
We identified hundreds of glycoproteins and the identified N-glycosylation sites provide novel information on the correct topology for membrane proteins present in the infarct setting.
Conclusions and clinical relevance
Our data provides the foundation for future studies of the LV infarct extracellular matridome, which may facilitate the discovery of drug targets and biomarkers.
Keywords: Extracellular matridome, glycoprotein, membrane orientation, matrix metalloproteinase, proteomics, myocardial infarction, left ventricle
1 Introduction
The extracellular matridome consists of all proteins expressed outside of cells, including transmembrane proteins, cell surface proteins, extracellular matrix (ECM) proteins, and secreted proteins [1]. The composition includes receptors, growth factors, cytokines, chemokines, hormones, enzymes, and fibrillar components [2]. The cardiac extracellular proteome provides the left ventricle (LV) with mechanical support, coordinates the signal transduction capabilities, and regulates cell functions by modifying biological processes [3]. Myocardial infarction (MI) is associated with extensive extracellular protein turnover as old ECM is replaced by an infarct scar that is primarily composed of ECM. MI is a highly prevalent cardiovascular disease, with over 1.5 million new patients diagnosed each year in the U.S. [4]. LV remodeling following MI depends on the balance between ECM degradation and deposition, as too much degradation can lead to LV aneurysms or rupture and too much deposition can lead to a stiff LV that provides a substrate for the development of heart failure [5]. Matrix metalloproteinase-9 (MMP-9) is a member of the family of enzymes that break down ECM and has been shown to play a critical role in LV remodeling post-MI [6, 7].
Knowledge of protein structure provides crucial information for understanding protein function, including information on location and availability for post-translational modification that is necessary for selecting optimal antigen sites and identifying drug targets. A common feature for optimal antigens and drug targets is easy accessibility, making extracellular protein analysis highly relevant to drug development for LV remodeling post-MI [8]. Therefore, efforts have been made to determine the spatial orientation of target proteins. In addition to the traditional methods for topology evaluation, membrane protein side accessibility is becoming a tool to evaluate protein orientation. Examples of this concept include N-glycosylation sites, antibody epitopes, iodinatable sites, and proteolytic sites [8]. The reason N-glycosylation sites can be used for topology evaluation is based on the fact N-linked glycosylation occurs only in extracellular domains of membrane proteins [9].
The goal of this study was to examine the geographic location of proteins in the LV infarct region, focusing on the extracellular matridome. Glycoproteomic analysis was performed on eight biological replicates of day 7 post-MI samples from wild type mice using solid-phase extraction of glycopeptides, followed by mass spectrometric identification of N-linked glycosylation sites for topology assessment [10, 11]. The logic of this examination was based on the concept that extracellular proteins are frequently N-glycosylated [12–14]; therefore, preferential isolation of N-linked glycoproteins would greatly enrich for the extracellular matridome [1, 13, 15]. In the present study, we identified 1352 N-linked glycosylation sites, and 56% of them were from membrane proteins proteins. Since membrane N-glycans always face the extracellular space, the identification of these glycosites provides topological information and helps to determine the protein orientation.
2 Materials and methods
2.1 Mice
C57BL/6J male and female mice, 3–6 months of age, were used in this study (n=8, 4 male and 4 female). Mice were kept in a light-controlled environment with a 12:12 hour light-dark cycle and given free access to standard mice chow and water. All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center at San Antonio and the University of Mississippi Medical Center in accordance with the “Guide for the Care and Use of Laboratory Animals”. The mice underwent permanent coronary artery ligation surgery, to produce myocardial infarction, as described previously [16, 17]. At day 7 post-MI, the mice were sacrificed.
2.2 Tissue samples and protein extraction
The infarct region of the LV was collected at 7 days post-MI as described previously [17], and the LV tissue was homogenized first in phosphate buffered saline (PBS; 16 μL per mg LV wet weight) with 1x protease inhibitors (Roche, Basel, Switzerland) and centrifuged to remove the soluble fraction. The insoluble pellet was homogenized in Reagent 4 (16 μL per mg LV wet weight; Sigma, St. Louis, MO) with 1x protease inhibitors [18]. Because the insoluble fraction is enriched for ECM, we used that fraction for the glycoproteomic analysis.
2.3 Trypsin digestion
Total protein from the insoluble fraction (1 mg) was denatured and reduced by adding 8 M urea and 12 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) in 1 M ammonium bicarbonate and incubating at 37°C for 1 h. The samples were alkylated with 16 mM iodoacetamide at room temperature in the dark for 30 min. The samples were diluted 5-fold with water prior to the addition of trypsin (Promega, Madison, WI), which was added at a ratio of 1:50 (w/w, enzyme: protein). The samples were digested at 37°C overnight with gentle shaking. The samples were centrifuged to remove precipitate. Peptides were cleaned by Sep-Pak Vac C18 cartridge (Waters, Milford, MA). Peptide concentration was determined by BCA assay (Thermo Fisher Scientific, Rochford, IL).
2.4 N-Linked Glycopeptide Capture
N-linked glycopeptides were isolated from the tryptic peptides using the solid phase extraction of glycopeptides (SPEG) method previously reported [10, 11]. Briefly, 0.8 mg of peptides was oxidized by 10 mM sodium periodate (Bio-Rad, Hercules, CA) at room temperature for 1 h. Glycopeptides were covalently conjugated to a solid support via hydrazide chemistry (Bio-Rad, Hercules, CA), with incubation at room temperature overnight. The hydrazide beads were washed with 1.5 M NaCl, and water prior to release of formerly N-linked glycopeptides from solid support by PNGase F (New England Biolabs, Ipswich, MA) at 37°C overnight. The eluent was purified by Sep-Pak Vac C18 cartridge (Waters, Milford, MA) and re-suspended in 20 μL of 0.4% acetic acid.
2.5 Mass spectrometry analysis
The peptides were analyzed by LC-MS/MS using a Q Exactive (ThermoFisher, Waltham, MA) coupled with a 15 cm × 75 μm C18 column (5 μm particles with 100 angstrom pore size). A nano UPLC at 300 nL/min with a 90 min linear acetonitrile gradient (from 5–35% B over 90 min; A = 0.2% formic acid in water, B = 0.2% formic acid in 90% acetonitrile) was used. A top 20 data dependent MS/MS with exclusion for 25 s was set. The samples were run with HCD fragmentation at normalized collision energy of 30 and an isolation width of 2 m/z. A lock mass of the polysiloxane peak at 371.10123 was used to correct the mass in MS and MS/MS. Target values in MS were 1e6 ions at a resolution setting of 70,000 and in MS2 1e5 ions at a resolution setting of 17,500.
MS/MS spectra were searched with SEQUEST using Proteome Discoverer (version 1.3; Thermo Fisher) against the mouse IPI database (version 3.87) containing 59,534 sequences. For this database search, the precursor mass tolerance and fragment mass tolerance were set at 15 ppm and 0.05 Da, respectively. Trypsin was specified as the protease. The fixed modification was set as carbamidomethylation (C), and other modifications were set as flexible modification as follows: deamidation (N) and oxidation (M). Full-tryptic end and two missed cleavage sites were allowed. A decoy version of the IPI mouse database was used to estimate peptide and protein false discovery rate. The False Discovery Rate was set at 0.01 to eliminate low-probability protein identifications.
2.6 Protein classification
Signal peptides were predicted using SignalP 4.1 [19]. Transmembrane (TM) helices were predicted using the TMHMM program (version 2.0; CBS prediction servers), which predicts protein topology and the number of TM helices [20]. Information from SignalP and TMHMM were combined to classify proteins into one of the following 6 categories: (i) secreted proteins that contain predicted cleavable signal peptides and no predicted TM segments; (ii) single TM type I proteins that contain predicted single TM helix with an extracellular (or luminal) N-terminus; (iii) single TM type II proteins that contain predicted single TM helix with an extracellular (or luminal) C-terminus; (iv) multiple TM proteins that contain multiple TM helices; or (v) ambiguous proteins [21].
3 Results
3.1 Extracellular matridome of cardiac tissue
To profile the extracellular matridome of post-MI LV tissue, the infarct region was analyzed. Since extracellular proteins are more likely to be glycosylated compared to intracellular proteins [8], the glycopeptide capture method was used to enrich for extracellular proteins [10, 11]. Of the identified peptides, 95.6% were de-glycosylated and had N-linked glycosylation motif (NXS/T, where X is any amino acid except proline). We also determined the extent of spontaneous deamidation on Asn residues in the N-glycosylation motif by profiling the un-conjugated fraction of hydrazide beads with or without PNGase F treatment [22]. The rate of spontaneous deamidation was 1.5% in our sample sets. The deamidated peptides identified in this negative control were not included in the identification list (Supplementary Table S1).
A total of 1352 unique N-linked glycosylation sites were identified, with a 1% false discovery rate (FDR), and these sites represented 694 unique glycoproteins (Supplemental Table S1). A unique glycosylation site was defined as a peptide containing a unique glycosylation motif, regardless of the length of the peptide and other modification of amino acid other than on asparagine in the motif. As summarized in Figure 1A, 406 proteins (59%) were identified as having one unique glycosite, whereas 288 (41%) were identified as having two or more unique glycosites. The majority of single-glycosites (80%) were identified more than two times (either in multiple identifications of the same peptide sequence with varying charge states or in different peptides). For the single-glycosite peptides only identified once, the annotated MS/MS spectra are provided in Supplemental Table S2. Many ECM proteins were identified with multiple peptides, such as collagens, fibronectin, and laminins (Supplemental Table S2).
Figure 1.
(A) Pie chart showing the distribution of glycosites identified per protein. Of the proteins identified, 59% had 1 glycosite and 41% proteins had ≥2 glycosites. (B) Stacked column chart showing the prevalence of identified glycopeptides in each of the eight biological replicates. A total of 36% of the glycosites were identified in all eight samples. (C) Classification of identified glycopeptides according to SignalP and TNHMM. A total of 77% of the glycopeptides were classified as transmembrane (TM) or secreted proteins.
The prevalence of identified glycopeptides in eight biological replicates is illustrated in Figure 1B. A total of 907 glycosites (67%) were identified in at least 4 biological replicates, whereas 200 glycosites (15%) were only identified in one of the biological replicates. Notably, 486 glycosites (36%) were identified from all eight samples. This large overlap indicates that our data were highly consistent in biological replicates, implicating good coverage of the N-glycoproteome in the mouse LV infarct region.
3.2 Functional analysis and KEGG pathway analysis of identified glycoproteins
To obtain an overview of the molecular functions and associated pathways, the identified glycoproteins were analyzed using David bioinformatics tools (6.7) [23, 24]. With 1% FDR, many functions related to extracellular activities were significantly enriched (p < 0.01), including calcium ion binding, exopeptidase activity, endopeptidase inhibitor activity, peptidase activity, carboxypeptidase activity, ECM structural constituent, metalloexopeptidase activity, and metallocarboxypeptidase activity (Table 1 and Supplemental Table S3). KEGG pathway analysis using DAVID revealed similar results [25]. The N-glycoproteome enriched the pathways of ECM-receptor interaction, cell adhesion molecules, arrhythmogenic right ventricular cardiomyopathy, hypertrophic cardiomyopathy, and dilated cardiomyopathy (Table 1 and Supplemental Table S3). These results indicate that the N-glycoproteome of the LV infarct region has important myocardial functions, and the identified extracellular proteome contains a large number of cardiac specific glycoproteins.
Table 1.
Molecular functions and KEGG pathways enriched for the identified glycoproteins, according to Gene Ontology (GO) analysis. The molecular functions of the identified glycoproteins are ordered by p value, from lowest to highest value.
| Molecular functions enriched for the identified glycoproteins | ||||
|---|---|---|---|---|
| Term | P Value | FDR | Count | % |
| Calcium ion binding | 1.8E-35 | 2.6E-32 | 111 | 16.6 |
| Carbohydrate binding | 1.4E-28 | 2.0E-25 | 62 | 9.3 |
| Polysaccharide binding | 3.3E-24 | 4.9E-21 | 38 | 5.7 |
| Pattern binding | 3.3E-24 | 4.9E-21 | 38 | 5.7 |
| Glycosaminoglycan binding | 8.0E-23 | 1.2E-19 | 35 | 5.2 |
| Heparin binding | 1.8E-19 | 2.6E-16 | 28 | 4.2 |
| Exopeptidase activity | 1.6E-13 | 2.4E-10 | 21 | 3.1 |
| Endopeptidase inhibitor activity | 1.1E-11 | 1.5E-08 | 28 | 4.2 |
| Metallopeptidase activity | 1.3E-11 | 1.9E-08 | 30 | 4.5 |
| Peptidase inhibitor activity | 8.8E-11 | 1.3E-07 | 28 | 4.2 |
| Peptidase activity | 1.7E-10 | 2.5E-07 | 55 | 8.2 |
| Integrin binding | 2.5E-10 | 3.6E-07 | 12 | 1.8 |
| Peptidase activity | 2.8E-10 | 4.1E-07 | 56 | 8.4 |
| Carboxypeptidase activity | 1.2E-09 | 1.7E-06 | 13 | 1.9 |
| Extracellular matrix structural constituent | 2.6E-09 | 3.7E-06 | 12 | 1.8 |
| Serine-type endopeptidase inhibitor activity | 2.7E-09 | 3.9E-06 | 21 | 3.1 |
| Transmembrane receptor protein tyrosine kinase activity | 5.1E-09 | 7.4E-06 | 15 | 2.2 |
| Enzyme inhibitor activity | 7.4E-09 | 1.1E-05 | 30 | 4.5 |
| Growth factor binding | 2.1E-08 | 3.1E-05 | 16 | 2.4 |
| Extracellular matrix binding | 9.0E-08 | 1.3E-04 | 10 | 1.5 |
| ion binding | 9.2E-08 | 1.3E-04 | 191 | 28.6 |
| Cation binding | 9.9E-08 | 1.4E-04 | 189 | 28.3 |
| Metalloexopeptidase activity | 1.1E-07 | 1.6E-04 | 11 | 1.6 |
| Metallocarboxypeptidase activity | 4.4E-07 | 6.4E-04 | 9 | 1.3 |
| Scavenger receptor activity | 6.4E-07 | 9.2E-04 | 11 | 1.6 |
| Metal ion binding | 1.1E-06 | 1.5E-03 | 183 | 27.4 |
| Sugar binding | 1.4E-06 | 2.1E-03 | 22 | 3.3 |
| Protein complex binding | 2.8E-06 | 4.0E-03 | 14 | 2.1 |
| KEGG Pathway enriched for identified glycoproteins | ||||
| Term | P Value | FDR | Count | % |
| ECM-receptor interaction | 1.5E-35 | 1.8E-32 | 45 | 6.7 |
| Lysosome | 5.7E-20 | 6.6E-17 | 38 | 5.7 |
| Focal adhesion | 2.4E-17 | 2.8E-14 | 45 | 6.7 |
| Cell adhesion molecules | 1.1E-16 | 1.3E-13 | 39 | 5.8 |
| Complement and coagulation cascades | 1.2E-16 | 1.3E-13 | 28 | 4.2 |
| Arrhythmogenic right ventricular cardiomyopathy | 1.4E-14 | 1.7E-11 | 26 | 3.9 |
| Hypertrophic cardiomyopathy | 3.1E-14 | 3.6E-11 | 27 | 4.0 |
| Hematopoietic cell lineage | 2.3E-12 | 2.6E-09 | 25 | 3.7 |
| Dilated cardiomyopathy | 2.7E-12 | 3.1E-09 | 26 | 3.9 |
3.3 New information for protein topology provided by glycosite identification
Based on the information from SignalP and TMHMM, the proteins were classified into one of five categories: secreted, single transmembrane protein (TM) type I, single TM type II, multiple TM, or ambiguous [21]. Of 694 identified proteins, 388 (56%) were TM proteins including single TM type I, type II, and multiple TM (Figure 1C). The identification of N-linked glycosylation site provided topological information for the transmembrane proteins, since N-linked glycosylation only occurs in extracellular domains of plasma membrane proteins. Because the orientation of single TM protein determines the protein type, this information is important to corroborate the orientation prediction of single TM proteins. Our study identified 153 single TM type I proteins and 107 single TM type II proteins based on the prediction using TMHMM. The single TM type I proteins have extracellular (or luminal) N-terminus, while the single TM type II proteins have extracellular (or luminal) C-terminus. The identified N-glycosites matched with the TMHMM predicted single TM protein type for 252 of the 260 proteins (Supplementary Table S4). The remaining 8 single TM proteins had identified N-glycosites that TMHMM predicted to be cytoplasmic domains (Supplementary Table S4). For these 8 proteins, we checked the topology prediction in Swiss-Prot, and found that our results matched Swiss-Prot prediction (which was of TMHMM prediction) for 7 of the 8 proteins. The final protein was not recorded in Swiss-Prot.
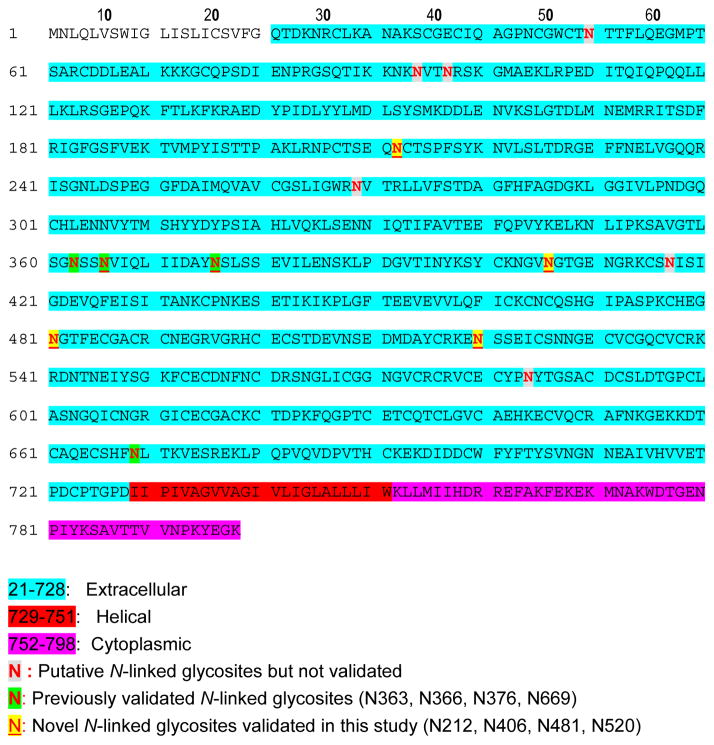
Many integrins were identified in the most enriched KEGG pathway, ECM-receptor interaction. They are the major adhesion receptor for many ECM proteins, and their primary role is to link ECM to the intracellular signaling network [26]. Therefore, it is very important to determine their topology in order to fully understand the signaling interactions. Fourteen integrins are predicted as single transmembrane proteins. Table 2 shows the predicted topology of these integrins, how many N-glycosites are predicted in the extracellular domain, and how many of the predicted N-glycosites identified in this study are in the extracellular domain. The identified glycosylation sites were all in the predicted extracellular domains, supporting the topology prediction. The β1 Integrin is further illustrated in Figure 2. The β1 Integrin has fourteen predicted N-linked glycosites, which are located in the potential extracellular domain (Figure 2). However, out of fourteen predicted N-linked glycosites in β1 integrin, only four have supporting experimental evidence according to Swiss-prot (highlighted in green). Our study identified four novel N-glycosites for β1 Integrin (namely, N212, N406, N481, N520, highlighted in yellow) in addition to one of the four N-glycosites recorded in Swiss-prot (Figure 2). These glycosites confirm the extracellular domain prediction of β1 integrin.
Table 2.
The topology of the integrins identified in the ECM-receptor interaction pathway. The numbers represent the amino acid position, with the bracketed numbers indicating the number of identified/predicted glycosites.
|
|
|||||
|---|---|---|---|---|---|
| Accession number | Protein description | cellular location | Confirmed topology (identified/predicted glycosites)
|
||
| Inside | Tmhelix | Outside | |||
| IPI00126077 | Integrin alpha-2 | Single TM type I | 1152–1178 | 1129–1151 | 1–1128 (1/9) |
| IPI00126090 | Integrin alpha-3 | Single TM type I | 606–642 | 583–605 | 1–582 (2/13) |
| IPI00121334 | Integrin alpha-4 | Single TM type I | 1008–1039 | 985–1007 | 1–984 (1/12) |
| IPI00115976 | Integrin alpha-5 | Single TM type II | 1026–1053 | 1003–1025 | 1–1002 (5/15) |
| IPI00227969 | Integrin alpha 6 | Single TM type I | 1038–1073 | 1015–1037 | 1–1014 (3/8) |
| IPI00345112 | Integrin alpha-8 | Single TM type I | 1034–1062 | 1011–1033 | 1–1010 (1/16) |
| IPI00417168 | Integrin alpha-11 | Single TM type I | 1165–1188 | 1142–1164 | 1–1141 (4/16) |
| IPI00315155 | Integrin alpha-IIb | Single TM type I | 1013–1033 | 990–1012 | 1–989 (1/5) |
| IPI00132286 | Integrin alpha-L | Single TM type I | 1109–1163 | 1086–1108 | 1–1085 (2/16) |
| IPI00120245 | Integrin alpha-V | Single TM type I | 1013–1044 | 990–1012 | 1–989 (5/13) |
| IPI00132474 | Integrin beta-1 | Single TM type I | 752–798 | 729–751 | 1–728 (5/14) |
| IPI00320605 | Integrin beta-2 | Single TM type I | 726–771 | 703–725 | 1–702 (2/6) |
| IPI00266264 | Integrin beta-3 | Single TM type I | 741–787 | 718–740 | 1–717 (1/6) |
| IPI00229516 | Integrin beta-5 | Single TM type I | 743–816 | 720–742 | 1–719 (1/7) |
Figure 2.
The glycopeptide identification supports the topology prediction for the β1 integrin, which is predicted to be a TM protein (with residues 21–728 as the extracellular domain, 729–751 as the TM helices, and 752–798 as the intracellular domain). There were four N-glycosites previously identified in other studies (highlighted in green). Five N-glycosites were identified in this study, and four of the five N-glycosites were not previously reported and highlighted in yellow.
4 Discussion
This study employed a glycoproteomic analysis followed by mass spectrometry and a topology bioinformatic assessment to profile the extracellular matridome of the LV infarct region. The most significant findings were: 1) 1352 unique N-linked glycosylation sites (representing 694 unique glycoprotein groups) were identified, with the majority (77%) being membrane proteins and secreted proteins; 2) the extracellular proteins identified included collagens, fibronectin, and laminins. These proteins were highly prevalent in the LV infarct; and 3) analysis of N-linked glycosites provides experimental evidence of membrane protein orientation. Out of 260 single TM proteins, the N-glycopeptides identified in the present study confirmed 259 proteins for the topology prediction using TMHMM or Swiss-Prot. TMHMM and Swiss-Prot had opposite topology predictions for seven proteins. Combined, these results provide the first glycoproteomic analysis of the extracellular matridom of the LV infarct region and validate the assumption that extracellular proteins are greatly enriched by the glycoprotein isolation approach used. These results also provide useful geographic information on membrane proteins, which will help in mechanistic and drug discovery studies, particularly for the proteins having controversial topology prediction.
Using a glycoproteomic approach to profile the extracellular matridome allows enrichment for extracellular proteins, targeting proteins with important signaling roles. In the functional analysis and pathway annotation of the identified proteins, functions related to extracellular proteins and cardiac signaling pathways were significantly enriched. For example, MMP-9 was identified as a central player, and roles for this metalloproteinase in the post-MI setting have been well documented [7, 27–29].
Using the N-linked glycosylation site analysis provided in vivo topologic evidence that has not been previously available. Zielinska et al demonstrated that glycosylation sites of membrane proteins always orient toward the extracellular space [30]. Gundry and co-workers found one glycoprotein transmembrane orientation was inconsistent with Swiss-Prot annotation using glycoproteomic analysis, providing complementary information to correct the protein orientation prediction [9]. Rossi et al studied the membrane-bound form of complement protein C9 using glycosylation mapping, anti-peptide antibody binding, and disulfide modification analyses [31]. By deleting two N-glycosites and introducing new N-glycosites, the authors determined the glycosylation required for human C9 activity and its membrane anchoring, which shows that the glycosite identification is a useful tool for protein orientation assignment. The current study identified 1352 unique N-linked glycosylation sites, providing new topologic information (Table 2 and Supplemental table S1). For β1 integrin, there were fourteen predicted N-linked glycosites, but only four of them (namely, N363, N366, N376, and N669) were identified previously according to Swiss-prot. In addition, these glycosites (N363, N366, and N376) are very close to each other, which is not good for supporting the large region (728 aa) of the extracelluar domain prediction. Our study identified four novel N-glycosites for β1 Integrin (namely N212, N406, N481, and N520) in addition to one of four N-glycosites (N669) recorded in Swiss-prot. These four novel glycosites were also recently identified by other groups [32, 33]. These five sites were distributed throughout the predicted extracellular domain, which provides supporting evidence for this section being the extracellular domain region of β1 integrin.
In the present data, 406 (59%) glycoproteins were identified by one unique glycosite, whereas 288 (41%) were identified by two or more unique glycosites. There are several possible reasons for a single identification. The first reason is that there really is only one potential N-glycosite in the protein. Out of the 406 proteins identified with one unique glycosite, 52 (12.8%) had only one single predicted glycosite, indicating that this reason accounted for a minority of the proteins. A second reason is that the peptide length of some glycopeptides may not be suitable for mass spectrometry analysis. Usually, a peptide length of 7–35 residues is most suitable for mass spectrometry sequencing technology [34]. A third reason is that some peptides may not be suitable for identification due to poor ionization and fragmentation. A fourth reason is the random sampling issue of mass spectrometry, which basically means that false negatives can occur. Our results indicate that all of the above reasons likely contribute to the results. To assess the quality of identification, the single identified glycosites were counted for the number of peptide-spectra matching. We found that the majority of single glycosites (80%) were identified more than two times (either in multiple identification of the same peptide sequence or in different glycopeptides), indicating the majority of the glycopeptide identification was reliable.
In conclusion, this study identified a large number of extracellular proteins in the LV infarct. The information of experimentally identified N-glycosites provides in vivo experimental evidence for topology prediction. Our data provides the foundation for future studies of the LV infarct extracellular matridome, which may facilitate the discovery of drug targets and biomarkers.
Supplementary Material
Clinical relevance.
The extracellular matridome plays a critical role in remodeling of the left ventricle (LV) following myocardial infarction (MI). The aim of this study was to use a glycoproteomics and mass spectrometry approach to identify glycoproteins in the extracellular matridome of the infarcted LV, to provide experimental evidence for topological determination. The information of protein topology is critical for selecting antigen sites and designing drug targets. The biological reproducibility was also investigated to assess the prevalence of the identified N-glycosylation sites. The identification of N-glycosylation sites supports the prediction of the membrane protein topology. Combined, this information may facilitate the discovery of drug targets and biomarkers for the post-MI patient.
Acknowledgments
We acknowledge support from AHA for POST14350034 and NIH/NHLBI T32 HL074464 to KYD-P, from NIH R00AT006704 to GVH, from Johns Hopkins Proteomics Center (N01-HV-00240) and Programs of Excellence in Glycosicences (PEG, P01HL107153) to HZ, from NIH/NHLBI HHSN 268201000036C (N01-HV-00244) for the San Antonio Cardiovascular Proteomics Center and R01 HL075360, HL051971, and GM104357 and from the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development Award 5I01BX000505 to MLL.
Abbreviations
- ECM
extracellular matrix
- LV
left ventricle
- TM
transmembrane
- PNGase F
Peptide-N-Glycosidase F
Footnotes
The authors have declared no conflict of interest.
References
- 1.Tian Y, Kelly-Spratt KS, Kemp CJ, Zhang H. Mapping tissue-specific expression of extracellular proteins using systematic glycoproteomic analysis of different mouse tissues. J Proteome Res. 2010;9:5837–5847. doi: 10.1021/pr1006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Didangelos A, Yin X, Mandal K, Baumert M, et al. Proteomics characterization of extracellular space components in the human aorta. Mol Cell Proteomics. 2010;9:2048–2062. doi: 10.1074/mcp.M110.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jourdan-Lesaux C, Zhang J, Lindsey ML. Extracellular matrix roles during cardiac repair. Life Sci. 87:391–400. doi: 10.1016/j.lfs.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frantz S, Bauersachs J, Ertl G. Post-infarct remodelling: contribution of wound healing and inflammation. Cardiovasc Res. 2009;81:474–481. doi: 10.1093/cvr/cvn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zamilpa R, Lindsey ML. Extracellular matrix turnover and signaling during cardiac remodeling following MI: causes and consequences. J Mol Cell Cardiol. 48:558–563. doi: 10.1016/j.yjmcc.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly D, Cockerill G, Ng LL, Thompson M, et al. Plasma matrix metalloproteinase-9 and left ventricular remodelling after acute myocardial infarction in man: a prospective cohort study. Eur Heart J. 2007;28:711–718. doi: 10.1093/eurheartj/ehm003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsey ML, Escobar GP, Dobrucki LW, Goshorn DK, et al. Matrix metalloproteinase-9 gene deletion facilitates angiogenesis after myocardial infarction. Am J Physiol Heart Circ Physiol. 2006;290:H232–239. doi: 10.1152/ajpheart.00457.2005. [DOI] [PubMed] [Google Scholar]
- 8.van Geest M, Lolkema JS. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol Mol Biol Rev. 2000;64:13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gundry RL, Raginski K, Tarasova Y, Tchernyshyov I, et al. The mouse C2C12 myoblast cell surface N-linked glycoproteome: identification, glycosite occupancy, and membrane orientation. Mol Cell Proteomics. 2009;8:2555–2569. doi: 10.1074/mcp.M900195-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 11.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roth J. Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem Rev. 2002;102:285–303. doi: 10.1021/cr000423j. [DOI] [PubMed] [Google Scholar]
- 13.Cordwell SJ, Thingholm TE. Technologies for plasma membrane proteomics. Proteomics. 2010;10:611–627. doi: 10.1002/pmic.200900521. [DOI] [PubMed] [Google Scholar]
- 14.McDonald CA, Yang JY, Marathe V, Yen TY, Macher BA. Combining results from lectin affinity chromatography and glycocapture approaches substantially improves the coverage of the glycoproteome. Mol Cell Proteomics. 2009;8:287–301. doi: 10.1074/mcp.M800272-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun B, Hood L. Protein-Centric N-Glycoproteomics Analysis of Membrane and Plasma Membrane Proteins. Journal of proteome research. 2014 doi: 10.1021/pr500187g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindsey ML, Escobar GP, Dobrucki LW, Goshorn DK, et al. Matrix metalloproteinase-9 gene deletion facilitates angiogenesis after myocardial infarction. Am J Physiol Heart Circ Physiol. 2006;290:H232–239. doi: 10.1152/ajpheart.00457.2005. [DOI] [PubMed] [Google Scholar]
- 17.Zamilpa R, Zhang J, Chiao YA, Bras LE, et al. Cardiac wound healing post-myocardial infarction: a novel method to target extracellular matrix remodeling in the left ventricle. Methods Mol Biol. 2013;1037:313–324. doi: 10.1007/978-1-62703-505-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Castro Bras LE, Ramirez TA, DeLeon-Pennell KY, Chiao YA, et al. Texas 3-step decellularization protocol: looking at the cardiac extracellular matrix. J Proteomics. 2013;86:43–52. doi: 10.1016/j.jprot.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 20.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 21.Chou KC, Elrod DW. Prediction of membrane protein types and subcellular locations. Proteins. 1999;34:137–153. [PubMed] [Google Scholar]
- 22.Palmisano G, Melo-Braga MN, Engholm-Keller K, Parker BL, Larsen MR. Chemical deamidation: a common pitfall in large-scale N-linked glycoproteomic mass spectrometry-based analyses. J Proteome Res. 2012;11:1949–1957. doi: 10.1021/pr2011268. [DOI] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pentassuglia L, Sawyer DB. ErbB/integrin signaling interactions in regulation of myocardial cell-cell and cell-matrix interactions. Biochim Biophys Acta. 2013;1833:909–916. doi: 10.1016/j.bbamcr.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halade GV, Jin YF, Lindsey ML. Matrix metalloproteinase (MMP)-9: a proximal biomarker for cardiac remodeling and a distal biomarker for inflammation. Pharmacol Ther. 2013;139:32–40. doi: 10.1016/j.pharmthera.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yabluchanskiy A, Ma Y, Iyer RP, Hall ME, Lindsey ML. Matrix metalloproteinase-9: Many shades of function in cardiovascular disease. Physiology (Bethesda) 2013;28:391–403. doi: 10.1152/physiol.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Castro Bras LE, Cates CA, Deleon-Pennell KY, Ma Y, et al. Citrate Synthase is a Novel In Vivo Matrix Metalloproteinase-9 Substrate that Regulates Mitochondrial Function in the Post-Myocardial Infarction Left Ventricle. Antioxid Redox Signal. 2014 doi: 10.1089/ars.2013.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Rossi V, Wang Y, Esser AF. Topology of the membrane-bound form of complement protein C9 probed by glycosylation mapping, anti-peptide antibody binding, and disulfide modification. Mol Immunol. 2010;47:1553–1560. doi: 10.1016/j.molimm.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaji H, Shikanai T, Sasaki-Sawa A, Wen H, et al. Large-scale identification of N-glycosylated proteins of mouse tissues and construction of a glycoprotein database, GlycoProtDB. Journal of proteome research. 2012;11:4553–4566. doi: 10.1021/pr300346c. [DOI] [PubMed] [Google Scholar]
- 33.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic acids research. 2011;39:D253–260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swaney DL, Wenger CD, Coon JJ. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. Journal of proteome research. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.