Abstract
We conducted a joint (pooled) analysis of three genome-wide association studies (GWAS) 1-3 of esophageal squamous cell carcinoma (ESCC) in ethnic Chinese (5,337 ESCC cases and 5,787 controls) with 9,654 ESCC cases and 10,058 controls for follow-up. In a logistic regression model adjusted for age, sex, study, and two eigenvectors, two new loci achieved genome-wide significance, marked by rs7447927 at 5q31.2 (per-allele odds ratio (OR) = 0.85, 95% CI 0.82-0.88; P=7.72x10−20) and rs1642764 at 17p13.1 (per-allele OR= 0.88, 95% CI 0.85-0.91; P=3.10x10−13). rs7447927 is a synonymous single nucleotide polymorphism (SNP) in TMEM173 and rs1642764 is an intronic SNP in ATP1B2, near TP53. Furthermore, a locus in the HLA class II region at 6p21.32 (rs35597309) achieved genome-wide significance in the two populations at highest risk for ESSC (OR=1.33, 95% CI 1.22-1.46; P=1.99x10−10). Our joint analysis identified new ESCC susceptibility loci overall as well as a new locus unique to the ESCC high risk Taihang Mountain region.
Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma are distinct diseases with different etiologies. ESCC remains the more common type of esophageal carcinoma in economically-developing countries as well as globally. Approximately half of the world's 500,000 new ESCC cases annually occur in China where the disease is a major public health problem. Three genome wide association studies (GWAS) have examined ESCC1-3 and two subsequent analyses used combinations of the three studies4, 5 to report as many as 12 loci associated with ESCC risk. Four additional loci have been reported based on an interaction with alcohol consumption, a known risk factor for ESCC4. Two of the GWAS1, 2 drew subjects primarily from the Taihang Mountain region of Henan and Shanxi provinces where ESCC occurs at very high rates. Total mortality due to ESCC and gastric cardia cancer can exceed 20% in the highest risk communities in this region6. The third GWAS3 drew subjects from a range of locations in China including a substantial collection from Beijing, where the population is comprised of people who originate from all provinces. The contribution of lifestyle risk factors for ESCC differs widely between locations in China, with alcohol consumption being a notable example. Heavy consumption of alcoholic beverages is a known cause of ESCC7, but historically was uncommon in the high incidence regions of China8. Therefore, alcoholic beverages played little role in the extraordinarily high rates, leaving the high incidence largely unexplained. To discover additional novel ESCC susceptibility alleles, we conducted a joint analysis of the three original GWAS and followed promising signals in an independent set of new cases and controls.
Supplementary Table 1 describes the subjects from the three underlying GWAS, which include additional subjects that were scanned after the first round publications plus the additional subjects used for replication of the top hits. In our joint analysis we investigated for the first time the individual GWAS data drawn from three studies in China, which consisted of 5,337 ESCC cases and 5,787 controls of Han Chinese ethnicity. The NCI GWAS used the Illumina 660W-Quad SNP microarray, the Henan GWAS used the Illumina 610-Quad SNP microarray, while the Beijing GWAS used the Affymetrix GeneChip Human Mapping 6.0 set. Supplementary Figure 1 shows eigenvector plots from a principal components analysis (PCA) of 25,676 uncorrelated genotyped SNPs (r2 < 0.01 in our combined control set). The results show distinct clusters corresponding to different source populations. However, only the first two eigenvectors were significant in the base cancer risk model and were thus used to adjust for population stratification in the final estimates for our joint analysis. To explore untyped common variants, we assessed variants based on the imputation of 40.5 million variants using version 3 of the 1000 Genomes data as the reference for each of the three data sets before combining them as described in the ONLINE METHODS and Supplementary Table 2. After filtering out SNPs with MAF < 1% or imputation information criteria (INFO) < 0.3, we advanced 7,556,215 SNPs to the association analysis. The inflation factor λ for the joint analysis is 1.01 for all SNPs, which indicates that population stratification should not be a concern. Supplementary Figure 2 provides a quantile-quantile plot for the joint case-control comparison with all SNPs and after exclusion of SNPs within 500 Kb of loci previously reported to be associated with ESCC. Supplementary Figures 3-5 provide individual quantile-quantile plots for each of the three underlying GWAS data sets. Fourteen promising SNPs based on the joint analysis (see ONLINE METHODS) were genotyped in an additional 9,654 ESCC cases and 10,058 controls divided between subjects from Henan Province and Beijing.
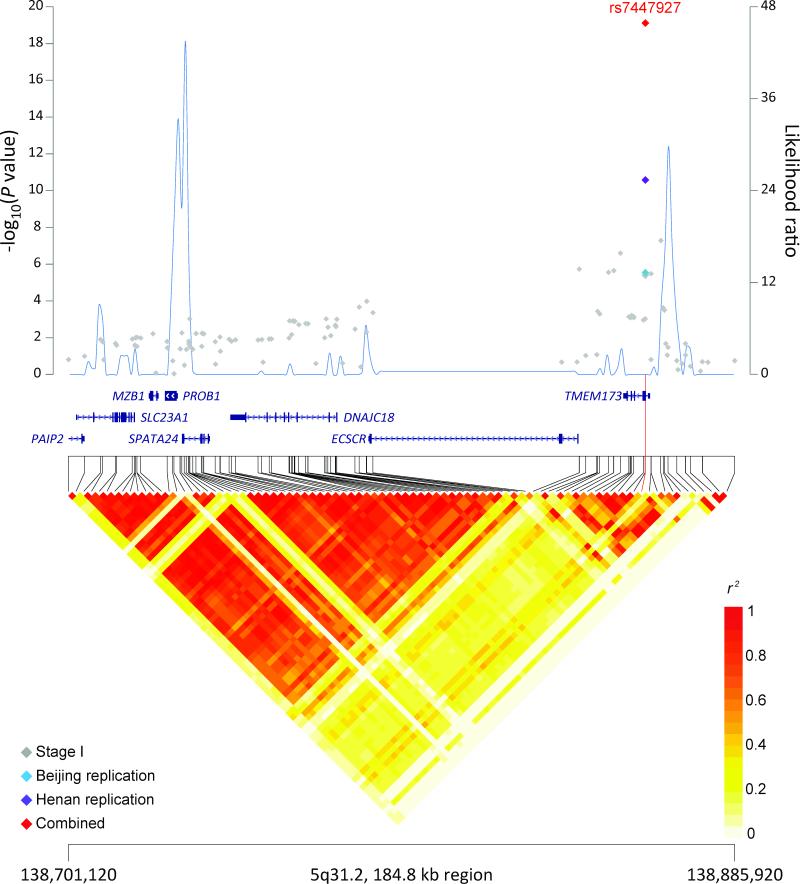
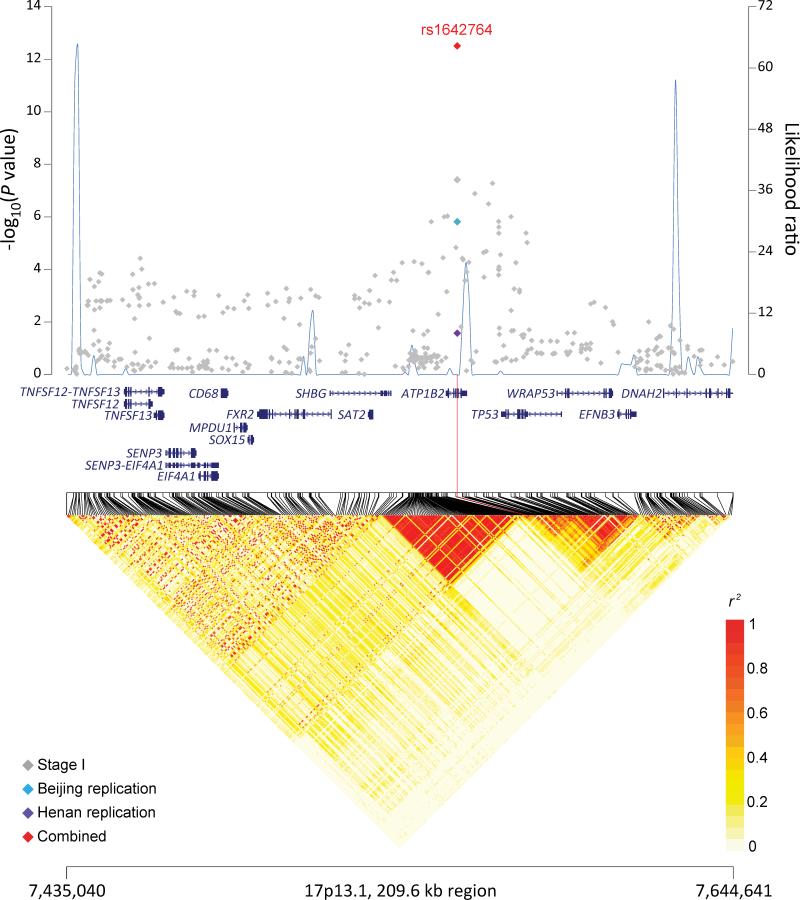
The joint analysis identified two new genome-wide significant loci at 5q31.2 (Figure 1a) and 17p13.1 (Figure 1b) associated with risk of ESCC in the pooled data of individual genotypes of all three GWAS and two replication studies (Table 1 and Supplementary Table 3). At 5q31.2, rs7447927, a synonymous SNP located in transmembrane protein 173 (TMEM173), had a combined per allele odds ratio (OR) and 95% confidence interval (95% CI) of 0.85 (0.82-0.88), P=7.72x10−20. At 17p13.1, rs1642764, an intronic SNP located in ATPase, Na+/K+ transporting, beta 2 polypeptide (ATP1B2), which is just telomeric to the tumor suppressor gene, TP53, had a combined per allele OR (95%CI) of 0.88 (0.85-0.91), P=3.10x10−13. No statistically significant heterogeneity was observed across the three pooled GWAS and two replication studies for either locus (Table 1). Furthermore, the associations were confirmed independently in the follow-up sets collected from both high- and low-risk populations.
Figure 1. Association results, recombination, and linkage disequilibrium plots for the 5q31.2 and 17p13.1 regions identified in stage 1 (joint analysis of three independent GWAS), for the two replications sets (Beijing and Henan), and the joint estimate from all data.
(a) 5q31.2:138,701,120–138,885,920 and (b) 17p13.1:7,435,040–7,644,641. Association results from a trend test in –log10 Pvalues (y axis, left; gray diamonds, stage I association result; sky blue diamonds, Beijing replication result; purple diamonds, Henan replication results; red diamonds, joint result) of the SNPs are shown according to their chromosomal positions (x axis). Linkage disequilibrium structure based on the 1000 Genomes CHB data (n=91) was visualized by snp.plotter software. The line graph shows likelihood ratio statistics (y axis, right) for recombination hotspot by SequenceLDhot software based on the background recombination rates inferred by PHASE v2.1 using the 1000 Genomes CHB data. Physical locations are based on NCBI human genome build 37. Gene annotation was based on the NCBI RefSeq genes from the UCSC Genome Browser.
Table 1.
Association between SNPs at three loci and risk for esophageal squamous cell carcinoma in a joint analysis of three independent genome-wide association studies of Han Chinese subjects.
| NCBI dbSNP 137 identifier (Reference Allele, Effect Allele) | Cytoband | Nearest Gene | Study | Controls | Cases | Effect Allele Frequency in Controls | Effect Allele Frequency in Cases | OR | (95% CI) | P-value | Pheterogeneity |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
All China
| |||||||||||
| rs7447927 (C,G) | 5q31.2 | TMEM173 | 3 scan analysis (Stage 1) | 5786 | 5336 | 0.460 | 0.451 | 0.87 | (0.82-0.92) | 4.07E-06 | |
| rs7447927 (C,G) | Henan Replication | 4802 | 4486 | 0.489 | 0.438 | 0.80 | (0.75-0.86) | 2.70E-11 | |||
| rs7447927 (C,G) | Beijing Replication | 5079 | 4797 | 0.431 | 0.397 | 0.87 | (0.82-0.92) | 2.92E-06 | |||
| rs7447927 (C,G) | Combined | 15667 | 14619 | 0.85 | (0.82-0.88) | 7.72E-20 | 1.31E-01 | ||||
| rs1642764(C,T) | 17p13.1 | ATP1B2,TP53,p53 | 3 scan analysis (Stage 1) | 5786 | 5336 | 0.499 | 0.456 | 0.84 | (0.79-0.89) | 3.91E-08 | |
| rs1642764(C,T) | Henan Replication | 4634 | 4694 | 0.484 | 0.470 | 0.93 | (0.87-0.99) | 2.67E-02 | |||
| rs1642764(C,T) | Beijing Replication | 5054 | 4777 | 0.520 | 0.483 | 0.86 | (0.81-0.92) | 1.55E-06 | |||
| rs1642764(C,T) | Combined | 15474 | 14807 | 0.88 | (0.85-0.91) | 3.10E-13 | 7.70E-02 | ||||
| rs35597309(G,A) | 6p21.32 | HLA class II genes | 3 scan analysis (Stage 1) | 5787 | 5336 | 0.072 | 0.094 | 1.32 | (1.18-1.47) | 6.17E-07 | |
| rs35597309(G,A) | Henan Replication | 4659 | 4597 | 0.067 | 0.085 | 1.23 | (1.09-1.38) | 1.00E-03 | |||
| rs35597309(G,A) | Beijing Replication | 5079 | 4786 | 0.075 | 0.080 | 1.05 | (0.94-1.17) | 3.59E-01 | |||
| rs35597309(G,A) | Combined | 15525 | 14719 | 1.19 | (1.12-1.27) | 1.18E-07 | 1.50E-02 | ||||
|
Taihang mountains only | |||||||||||
| rs35597309(G,A) | 6p21.32 | HLA class II genes | NCI Scan | 2707 | 2021 | 0.077 | 0.109 | 1.43 | (1.23-1.67) | 4.27E-06 | |
| rs35597309(G,A) | Henan Scan | 1082 | 1375 | 0.063 | 0.093 | 1.55 | (1.22-1.97) | 3.50E-04 | |||
| rs35597309(G,A) | Henan Replication | 4659 | 4597 | 0.067 | 0.085 | 1.23 | (1.09-1.38) | 1.00E-03 | |||
| rs35597309(G,A) | Combined | 8448 | 7993 | 1.33 | (1.22-1.46) | 1.99E-10 | 1.22E-01 | ||||
One of these new loci showed significant heterogeneity (p=0.015) among the three studies and the associations are also reported using only the two GWAS and one replication set that primarily used subjects from populations with high incidence in the Taihang Mountains of north central China.
On further analysis, we observed an additional susceptibility locus that showed geographic differences such that the significant association was observed only in the two GWAS1, 2 which included subjects from populations at the highest risk for ESCC. In joint analyses using all three GWAS, a SNP at 6p21.32 showed a nearly genome-wide significant association, however, statistically significant heterogeneity among studies (P=0.015) was evident when subjects from the Beijing GWAS and replication subjects were included (i.e., among the three pooled GWAS and two replication studies as shown in Table 1). The test for heterogeneity became non-significant when the Beijing GWAS and Beijing replication subjects were excluded (Table 1). Table 1 also shows that when the analyses were restricted to subjects from the high incidence regions, rs35597309, located in the HLA Class II gene region between HLA-DRB1 and HLA-DQA1, had a per allele OR (95% CI) of 1.33 (1.22-1.46), P=1.99 x 10-10 (Supplementary Figure 6). This heterogeneity between high- and low-risk regions was also evident when data from the three separate GWAS were examined (Supplementary Table 4). Further genotyping and possibly sequencing are necessary to map the susceptibility alleles across the HLA Class II region due to its complex structure defined by long-range haplotypes.
Finally, our joint analysis observed a promising association with rs61271866 (P=5.18 x 10−8), an intergenic SNP at 9p21.3 that includes the cyclin-dependent kinase inhibitor 2B (CDKN2B)-CDKN2A gene cluster (Supplementary Table 3). Variants in this region have been associated with risk of melanoma9, childhood acute lymphoblastic leukemia10, chronic lymphocytic leukemia11, and glioma12, as well as ESCC13 in a prior study using a subset of the samples examined here. This SNP showed heterogeneity among individual studies in the GWAS (Supplementary Table 4) and between the joint Stage 1 and replication phase results (Supplementary Table 3). The heterogeneity picture for the 9p21.3 locus is more varied as compared to the 6p21.32 HLA Class II locus, indicating that validation of this finding will require additional work.
For these four SNPs (rs7447927, rs1642764, rs35597309, and rs61271866), we tested for interactions by use of alcohol (Supplementary Table 5) or tobacco (Supplementary Table 6). Because of the substantial differences in the degree of alcohol and tobacco use by population, we did these tests separately for each of the three underlying studies. In total this constituted 24 tests and we found one that was nominally significant (rs35597309 and tobacco in the NCI GWAS, Supplementary Table 6), but the difference did not replicate in the two other studies and thus, is most likely due to chance. We further note that the number of GWAS subjects with these covariate data from Henan was limited.
Here we report a new finding of an association between rs7447927, a synonymous SNP in TMEM173, and ESCC risk. TMEM173 (also known as STING) facilitates innate immune reactions to viruses and bacteria through the production of type 1 interferon14. The only previous GWAS hit at this locus (rs13181561) was demonstrated to be associated with the modulation of interferon-α responses to the smallpox vaccine15 and is highly correlated (r2=0.956 in 1000 Genomes data for CHB population) with rs7447927. rs13181561 is also tagged as an expression quantitative trait locus in lymphoblastoid cells (Supplementary Table 7) that alters expression of genes in segment AC135457.2. This genomic region includes the sodium-dependent vitamin C transporter (SLC23A1), which is critical for vitamin C transport16. Interestingly, low vitamin C has been implicated in risk of ESCC17.
The new susceptibility locus at 17p13.1 is marked by rs1642764, an intronic SNP located within the ATP1B2 gene; this variant resides in an LD block that includes the 3’ region of TP53 (Figure 1b). Alteration of TP53 regulation or function could be a plausible explanation for the observed association between rs1642764 and risk of ESCC. It is noteworthy that no prior cancer GWAS has reported a conclusive association with a variant in or around TP53. Recently, a candidate gene study of genotyped and imputed SNPs across TP53 reported a strong association of a SNP with a low minor allele frequency, rs78378222, with glioma risk (P=6.86 x 10−24 with a MAF=0.013 in a population of European ancestry)18. We also observe that TP53 is frequently inactivated in ESCC19. Our target SNP in this region is in LD with several other SNPs, notably rs1050541 (r2=0.575), which alters a binding site for RAD21 (Supplementary Table 7). RAD21 is a key component of the cohesion complex, which binds to DNA and is essential for mitosis, homologous DNA repair, and enhancer activities and may be relevant to cancer20. Alternatively, this SNP falls between recombination hotspots that includes the SHBG gene, and variants bracketing this SNP are known to be associated with sex hormone binding globulin regulation and serum testosterone concentration21. Esophageal cancer is male predominant in low incidence populations, but this has typically been attributed to greater tobacco smoking and alcohol consumption by men compared to women. Recent studies have suggested that hormonal factors may play a role in the development of ESCC22, 23.
In published GWAS, HLA Class II genetic variants in close proximity (<20 kb) of rs35597309 on 6p21.32 have been associated with multiple cancers including nasopharyngeal carcinoma24, hepatocellular carcinoma25, lung cancer in never smokers26, and familial chronic lymphocytic leukemia27 as well as autoimmune diseases, including Crohn's disease28 and lupus 29. This SNP resides between HLA-DRB1 and HLADQA1, both MHC class 2 genes that function in antigen presentation. The HLA region is large, complex, and shows unusually long-range LD that makes the interpretation of GWAS hits in this region difficult. rs35597309 is in perfect LD (r2=1.0) with 34 other SNPs within 2 MB (Supplementary Table 7) including two missense mutations in HLADQA1 and a host of putative protein binding sites, enhancers, and regulatory motifs. But the LD with top hits in this region from previously reported studies of cancer was low; r2<0.1 for rs2860580 (nasopharyngeal carcinoma), rs9272105 (hepatocellular carcinoma), rs2395185 (lung cancer), and rs674313 (chronic lymphocytic leukemia) in 1000 Genomes data for CHB+JPT populations.
Our results provide evidence for an association between variants at 6p21.32 and risk of ESCC, although the association was restricted to the studies which examined subjects from the high incidence Taihang Mountain region. While it is plausible that our results could be due to a gene-environment interaction between HLA genotypes and an uncharacterized risk factor specific to the Taihang Mountains, it is also possible that chance could account for this finding. The result was, however, confirmed in the Henan replication set (P=0.001). We know that populations in this region suffer from very high rates of ESCC, which is likely multi-factorial and could involve immune challenges. It is also important to note that the Han Chinese population is genetically diverse and this difference in association may be due to true differences in the genomic structure of the HLA regions between subjects from the Taihang Mountains and other parts of China.
But we note that the allele frequency is similar between the Henan and Beijing replication sets (Table 1). Future work should use genomic methods specifically designed to investigate the HLA region to explore these results. We also note that a region at 6p22.1 that is linked to other HLA alleles has been associated with risk of Barrett's esophagus, the precursor lesion for esophageal adenocarcinoma30.
Supplementary Table 8 shows the individual GWAS and joint analysis results for the 12 main effect GWAS loci reported in five previous publications1-5. Joint analysis of the pooled data from the three GWAS did not strengthen all previously reported loci. We observed strong associations for SNPs in loci harboring PLCE1, CASP8, RUNX1, and CHEK2, but no signal for four loci. The Beijing GWAS showed geographic differences in associations for some SNPs3 and this variation may be attributed, in part, to differences in environmental exposures and habits, such as alcohol consumption. A previous analysis by Wu et al. 4 using partial data from two of the studies also showed substantial heterogeneity between GWAS. Of 18 hits identified in analyses of both main effects and alcohol interactions in the Beijing data, only four replicated in subjects from the Shanxi Upper Gastrointestinal Cancer Genetics Study1.
In conclusion, we present the joint analysis of the individual genotype data from three previously published GWAS in China and have established two new loci associated with risk of ESCC across all three studies, and two promising signals, the most notable, in the HLA class II region. The latter appears to be present only in subjects from the high incidence Taihang Mountain region of China. Environmental factors have previously been shown to be of varied relevance for ESCC in different Chinese populations and this may have led to differential GWAS findings. Here we find additional evidence for distinct results among these populations. Etiologic heterogeneity may play an important role in interpreting GWAS results and should be considered as GWAS are extended to understudied populations with distinct lifestyles. Lastly, further work is needed to fine-map the regions to identify the optimal alleles for laboratory studies that will provide an understanding of the basic biology underlying the ESCC susceptibility alleles and their interaction with environmental factors.
ONLINE METHODS
Subject selection and genotyping
This study pooled the individual genotype data of subjects from three independent GWAS of esophageal squamous cell carcinoma in Han Chinese populations, which were part of three earlier reports from the NCI1, Henan2, and Beijing study groups3. The numbers of subjects differs in some cases from those listed in the original publications because additional subjects were genotyped using the same platform subsequent to the original publications; these subjects were included in the joint analysis of the individual genotype data. For the NCI GWAS, subjects came from four prospective cohort studies and one large case-control study as reported in Abnet et al.1 and all subjects used in replication in the original paper were subsequently genotyped using the Illumina 660W-Quad microarray. For the Henan scan, subjects were collected from many hospitals in Henan Province and a smaller ‘genetically-matched’ subset2 (1,076 cases, 713 controls) was selected for this joint analysis. Furthermore, 299 cases and 370 controls were added who had subsequently been genotyped using the Illumina 610-Quad SNP microarray after the first publication. For the Beijing scan, subjects were collected from four different localities as previously reported in Wu et al.4 and all subjects were included in the current study. These subjects had been genotyped using the Affymetrix GeneChip Human Mapping 6.0 set. Subjects included in the replications from both Henan2 and Beijing3 were identified and recruited using the same approaches as for subjects included in the GWAS from each of these respective sites and as described in the initial publications from each of their GWAS. A description of the included subjects is given in Supplementary Table 1.
GWAS data
The details of the analytic pre-processing for the three GWAS were included in each of the primary papers. In addition to the quality control procedures performed in the previous primary publications for all three studies, SNPs with a call rate < 95% or Hardy-Weinberg proportion test P-value < 0.000001 or minor allele frequency < 1% were further removed prior to imputation for the current analysis (Supplementary Table 2). We also searched for potential duplicates or first degree relatives across all three GWAS with glu ibds module (http://code.google.com/p/glu-genetics/) using the set of 25,676 independent SNPs with pair-wise r2<0.01 estimated from the GWAS control population. A total of nine pairs of duplicates were found; all were between Henan2 and the NIT component of the NCI scan1, which enrolled subjects in the same high incidence area of Henan Province. As a result, we excluded nine individuals (one from each pair) from the Henan study for the joint analysis. Application of standardized QC procedures for subjects and for SNPs resulted in the exclusion of an additional small proportion of subjects such that the final numbers of subjects in the current analysis are slightly different from those reported previously.
Imputation analysis
Imputation was conducted separately for each scan using IMPUTE2 software version 2.2.2 (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html) and version 3 of the 1000 Genomes Project data as the reference set. First, the genomic coordinates were lifted over from NCBI human genome build 36 to build 37 using the UCSC lift over tool (http://hgdownload.cse.ucsc.edu/downloads.html). The few loci that failed to be lifted over were also excluded from the imputation. Second, the strand of the inference data was aligned with the 1000 Genomes data by simple allele state comparison or allele frequency matching for A/T and G/C SNPs. We implemented a 4-Mb sliding window to impute across the genome, resulting in 744 jobs running in parallel on the NIH BIOWULF cluster (http://biowulf.nih.gov/). A pre-phasing strategy with SHAPEIT software version 1 (http://www.shapeit.fr/) was adopted to improve the imputation performance. The phased haplotypes from SHAPEIT were fed directly into IMPUTE2. Imputed loci with INFO < 0.3 or MAF < 0.01 were excluded from further association analysis. To technically validate our imputation findings, we optimized TaqMan assays for rs7447927, rs1642764, and rs35597309. We genotyped a set of 892 samples from NCI and another set of 752 samples from Beijing. For the NCI set, the concordance rates between the imputed genotypes (using a posterior probability threshold of 0.95) and TaqMan genotypes were 99.3%, 96.5%, and 99.1%, respectively and for the Beijing set, the concordance rates between the imputed genotypes (using a posterior probability threshold of 0.95) and TaqMan genotypes were 96.7%, 90.4%, and 99.6%, respectively.
Association analysis
The imputed genotypes were merged using GTOOL software version 0.7.5 (http://www.well.ox.ac.uk/~cfreeman/software/gwas/gtool.html) and the association testing was performed using SNPTEST software version 2.2 (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html), with adjustment for age, sex, study, and the top two eigenvectors, which controls for population stratification. In the joint analysis baseline model (not including SNP effects) adjusting for age, sex, study, and all top ten eigenvectors (EVs), the top two eigenvectors were significantly associated with case status (P-value < 0.05), and were, therefore, included in the final joint analysis association models used to test SNP effects across all three studies. In a sensitivity analysis, we also conducted the association analyses in each of the three GWAS separately, followed by meta-analyses that used the fixed effect inverse variance method to combine the beta estimates and standard errors from each scan. In this second approach, we generated the set of eigenvectors based on each GWAS and identified significant eigenvectors to control for population stratification in each individual GWAS (EV1 for NCI; EV1, EV5 and EV7 for Beijing; no EV was needed for Henan). The meta-analysis (Supplementary Table 4) produced very similar results to our current joint analysis (Table 1), so we presented results for Stage 1 from the joint analysis as our primary analysis. The P for heterogeneity was calculated using Cochran's Q, which is distributed as a chi-square statistic with (n-1) degrees of freedom where n is the number of sets included in the meta-analysis. For exploring the gene and environment interaction with use of alcohol or tobacco, we performed stratified analyses for the four novel SNP associations and assessed the risk heterogeneity between drinkers and nondrinkers (Supplementary Table 5), and between smokers and nonsmokers (Supplementary Table 6). To evaluate population stratification, we examined QQ plots before and after eigenvector adjustment for the joint analysis (Supplementary Figure 7), for Beijing (Supplementary Figure 8), and for NCI (Supplementary Figure 9). No figure is shown for Henan as no adjustment was required. Further, we examined the association with risk for the four novel SNP associations we report here before and after eigenvector adjustment (Supplementary Table 9).
Recombination hotspot inference
Likelihood ratio statistics for recombination hotspots were estimated by SequenceLDhot software based on background recombination rates inferred by PHASE v2.1 using the 1000 Genomes CHB data.
Replication genotyping and analysis
After SNPs from previously reported ESCC risk loci were excluded, the top SNPs with p values less than 1.0 x 10−5 (n=14) from our Stage 1 analysis were selected for replication testing in both Beijing and Henan (Supplementary Table 3). However, when the imputation was updated with the addition of more covariate data on subjects, the new Stage 1 p-value for rs4252725 was only 1.0 x 10−4. At that point, primers for those top 14 SNPs from the initial analysis had already been designed and validated, so we proceeded to test all 14 of these SNPs. Therefore, we reported replication results for all the SNPs that we advanced to replication despite the updated analysis of our initial results and the appearance of a shift in our criterion. All SNPs genotyped in samples from the additional Beijing subjects used optimized TaqMan assays, whereas the Henan replication subjects were genotyped using Sequenom (11 SNPs) and TaqMan (three SNPs) assays. Three Sequenom assays failed genotyping and one (rs7822239) was repeated using TaqMan because it was nominally significant (P = 0.02) in the Beijing replication set. Samples with completion rates less than 80% in either replication were excluded from association analysis. Association analyses used log additive models with a trend effect and were adjusted for sex and age. Replication and Stage 1 results were combined using a fixed effect meta-analysis.
In silico bioinformatics analysis
Using 1000 Genomes CHB data, we identified all SNPs with r2>0.8, 0.8, 0.5 (because no SNPs passed the 0.8 threshold), respectively, for the lead SNP in each of the three novel regions we identified. We then used HaploReg31 and RegulomeDB32 to explore potential functional annotations within the ENCODE data in the genome surrounding our lead SNPs (Supplementary Table 7).
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by the National High-Tech Research and Development Program of China (2009AA022706 to D.L.), the National Basic Research Program of China (2011CB504303 to D.L. and W.T.), the National Natural Science Foundation of China (30721001 to D.L., Q.Z. and Z.L.).
We thank all the patients and their family members whose contributions made this work possible; medical students from Zhengzhou University, Xinxiang Medical University, Zhengzhou Medical School, and Henan University of Science and Technology for sample and data collections; Q.C. Kan (The First Affiliated Hospital of Zhengzhou University) and Y. Xing (Xinxiang Medical University) for organizing field work for sample collections and finding financial support for the study; the Key Laboratory of Dermatology (Anhui Medical University), Ministry of Education, China, Hefei, Anhui for genotyping; W. Huang (Health Department of Henan Province) for field work organization.
This work was supported by the Invitation Team of the Ministry of Education (2008IRTSTHN010), the National Nature Science Foundation of China (81071783), 863 High-Tech Key Projects (2012AA02A209, 2012AA02A503, 2012AA02A201), Innovation Scientists and Technicians Troop Construction Projects of Henan Province (3047), Key Disciplines Revitalization Plan of Zhengzhou University (20132016), and the Collaborative Innovation Center for Esophageal Cancer Research of Henan Province (20132016).
The Shanghai Men's Health Study (SMHS) was supported by the National Cancer Institute extramural research grant [R01 CA82729]. The Shanghai Women's Health Study (SWHS) was supported by the National Cancer Institute extramural research grant [R37 CA70837] and, partially for biological sample collection, National Cancer Institute Intramural Research Program contract NO2-CP-11010 with Vanderbilt University. The studies would not be possible without the continuing support and devotion from the study participants and staff of the SMHS and SWHS.
The Singapore Chinese Health Study (SCHS) was supported by the National Cancer Institute extramural research grants [R01 CA55069, R35 CA53890, R01 CA80205, and R01 CA144034]. We are indebted to the contributions of Drs Mimi C Yu and Hin-Peng Lee in the establishment of this cohort. The study would not be possible without the assistance with the identification of cancer cases through database linkage by the Ministry of Health in Singapore. We are indebted to the study subjects for their continuing participation and staff of the SCHS for their support.
The Shanxi Upper Gastrointestinal Cancer Genetics Project was supported by the National Cancer Institute Intramural Research Program contract NO2-SC-66211 with the Shanxi Cancer Hospital and Institute, Taiyuan, Shanxi, China.
The Nutrition Intervention Trials (NIT) were supported by National Cancer Institute Intramural Research Program contracts NO1-SC-91030 and HHSN261200477001C with the Cancer Institute & Hospital of the Chinese Academy of Medical Sciences, Beijing, China.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, the Division of Cancer Epidemiology and Genetics, and the Center for Cancer Research.
This project was funded in part with federal funds from the NIH, National Cancer Institute, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
AUTHOR CONTRIBUTIONS
C.C.A., S.J.C., N.D.F., A.M.G., N.H., D.L., X.S., P.R.T., L.D.W., Zhaoming Wang, and C. Wu organized and designed the study. L.B., S.J.C., A.H., D.L., X.S., P.R.T., L.D.W., Zhaoming Wang, C. Wu, J.Y., and M.Y. conducted and supervised the genotyping of samples. C.C.A., S.J.C., N.D.F., A.M.G., M.G., D.L., N.H., P.R.T., L.D.W., Zhaoming Wang, and K.Y. contributed to the design and execution of statistical analysis. C.C.A., S.J.C., N.D.F., X.S.F., A.M.G., J.H., N.H., D.L., X.S., P.R.T., W.T., L.D.W., Zhaoming Wang, C. Wu, and X.B.Z. wrote the first draft of the manuscript. C.Wu, Zhaoming Wang, X.S., X.S.F., C.C.A., J.H., N.H., X.B.Z., W.T., Q.Z., Z.Hu, Z.He, W.J., Y.Zhou, K.Y., X.O.S., J.M.Y, W.Zheng, X.K.Z., S.G.G, Z.Q.Y., F.Y.Z., Z.M.F, J.L.C., H.L.L, X.N.H., B.Li, X.C., S.M.D., L.L., M.P.L., T.D., Y.L.Q, Z.L., Y.Liu, D.Y., J.C., L.Wei, Y.T.G., W.P.K., Y.B.X, Z.Z.T, J.H.F, J.J.H, S.L.Z., P.Z., D.Y.Z., Y.Y., Y.H., C.L.L., K.Z., Y.Q., G.J., C.G., J.F., X.M., C.L., H.Y., C.Wang, W.A.W, M.G., M.Y., J.Y., E.T.G., A.L.L, W.Zhang, Xue-Min Li, L.D.S., B.G.M., Y.Li, S.T., XQ.P., J.L., A.H., K.J., C.G., L.B., J.F., H.S., Y.K., Y.Zeng, T.W., P.K., C.C.C., M.A.T., Z.C.H., Y.L.L., Y.L.H., Yu Liu, Li Wang, G.Y., L.S.C., X.L., T.M., H.M., L.S., Xin-Min Li, Xiu-Min Li, J.W.K., Y.F.Z., L.Q.Y., Zhou Wang, Yin Li, Q.Q., W.J.Y., G.Y.L., LQ.C., E.M.L., L.Y., W.B.Y., R.W., L.W.W., X.P.F., FH.Z., W.X.Z., Y.M.M., M.Z., G.L.X., J.L.L., M.H., J.L.R., B.Liu, S.W.R., Q.P.K., F.L., I.S., W.W., Y.R.Z., CW.F., J.W., Y.H.Y., H.Z.H., Q.D.B., B.C.L., A.Q.W., D.X., W.C.Y., Liang Wang, X.H.Z., S.Q.C., J.Y.H., X.J.Z., N.D.F., A.M.G., D.L., P.R.T., L.D.W. and S.J.C. contributed to the conduct of the epidemiological studies or contributed samples to the GWAS or follow-up genotyping. C.Wu, Zhaoming Wang, X.S., X.S.F., C.C.A., J.H., N.H., X.B.Z., W.T., Q.Z., Z.Hu, Z.He, W.J., Y.Zhou, K.Y., X.O.S., J.M.Y, W.Zheng, X.K.Z., S.G.G, Z.Q.Y., F.Y.Z., Z.M.F, J.L.C., H.L.L, X.N.H., B.Li, X.C., S.M.D., L.L., M.P.L., T.D., Y.L.Q, Z.L., Y.Liu, D.Y., J.C., L.Wei, Y.T.G., W.P.K., Y.B.X, Z.Z.T, J.H.F, J.J.H, S.L.Z., P.Z., D.Y.Z., Y.Y., Y.H., C.L.L., K.Z., Y.Q., G.J., C.G., J.F., X.M., C.L., H.Y., C.Wang, W.A.W, M.G., M.Y., J.Y., E.T.G., A.L.L, W.Zhang, Xue-Min Li, L.D.S., B.G.M., Y.Li, S.T., XQ.P., J.L., A.H., K.J., C.G., L.B., J.F., H.S., Y.K., Y.Zeng, T.W., P.K., C.C.C., M.A.T., Z.C.H., Y.L.L., Y.L.H., Yu Liu, Li Wang, G.Y., L.S.C., X.L., T.M., H.M., L.S., Xin-Min Li, Xiu-Min Li, J.W.K., Y.F.Z., L.Q.Y., Zhou Wang, Yin Li, Q.Q., W.J.Y., G.Y.L., LQ.C., E.M.L., L.Y., W.B.Y., R.W., L.W.W., X.P.F., FH.Z., W.X.Z., Y.M.M., M.Z., G.L.X., J.L.L., M.H., J.L.R., B.Liu, S.W.R., Q.P.K., F.L., I.S., W.W., Y.R.Z., CW.F., J.W., Y.H.Y., H.Z.H., Q.D.B., B.C.L., A.Q.W., D.X., W.C.Y., Liang Wang, X.H.Z., S.Q.C., J.Y.H., X.J.Z., N.D.F., A.M.G., D.L., P.R.T., L.D.W. and S.J.C. contributed to the writing of the manuscript.
We have no competing financial interests.
Reference List
- 1.Abnet CC, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat. Genet. 2010;42:764–767. doi: 10.1038/ng.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang LD, et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat. Genet. 2010;42:759–763. doi: 10.1038/ng.648. [DOI] [PubMed] [Google Scholar]
- 3.Wu C, et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat. Genet. 2011;43:679–684. doi: 10.1038/ng.849. [DOI] [PubMed] [Google Scholar]
- 4.Wu C, et al. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat. Genet. 2012;44:1090–1097. doi: 10.1038/ng.2411. [DOI] [PubMed] [Google Scholar]
- 5.Abnet CC, et al. Genotypic variants at 2q33 and risk of esophageal squamous cell carcinoma in China: a meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2012;21:2132–2141. doi: 10.1093/hmg/dds029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang CS. Research on esophageal cancer in China: a review. Cancer Res. 1980;40:2633–2644. [PubMed] [Google Scholar]
- 7.Kamangar F, Chow WH, Abnet CC, Dawsey SM. Environmental causes of esophageal cancer. Gastroenterol Clin North Am. 2009;38:27–57. doi: 10.1016/j.gtc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran GD, et al. Prospective study of risk factors for esophageal and gastric cancers in the Linxian general population trial cohort in China. Int. J Cancer. 2004;113:176–181. doi: 10.1002/ijc.20616. [DOI] [PubMed] [Google Scholar]
- 9.Bishop DT, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherborne AL, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat. Genet. 2010;42:492–494. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berndt SI, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat. Genet. 2013 doi: 10.1038/ng.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajaraman P, et al. Genome-wide association study of glioma and meta-analysis. Hum. Genet. 2012;131:1877–1888. doi: 10.1007/s00439-012-1212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu F, et al. Common genetic variants in the 9p21 region and their associations with multiple tumours. Br. J. Cancer.10. 2013 doi: 10.1038/bjc.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy RB, et al. Genome-wide analysis of polymorphisms associated with cytokine responses in smallpox vaccine recipients. Hum. Genet. 2012;131:1403–1421. doi: 10.1007/s00439-012-1174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eck P, et al. Genomic and functional analysis of the sodium-dependent vitamin C transporter SLC23A1-SVCT1. Genes Nutr. 2007;2:143–145. doi: 10.1007/s12263-007-0040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirvish SS. Role of N-nitroso compounds (NOC) and N-nitrosation in etiology of gastric, esophageal, nasopharyngeal and bladder cancer and contribution to cancer of known exposures to NOC. Cancer Lett. 1995;93:17–48. doi: 10.1016/0304-3835(95)03786-V. [DOI] [PubMed] [Google Scholar]
- 18.Enciso-Mora V, et al. Low penetrance susceptibility to glioma is caused by the TP53 variant rs78378222. Br. J. Cancer. 2013;108:2178–2185. doi: 10.1038/bjc.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu N, et al. Frequent inactivation of the TP53 gene in esophageal squamous cell carcinoma from a high-risk population in China. Clin. Cancer Res. 2001;7:883–891. [PubMed] [Google Scholar]
- 20.Remeseiro S, Losada A. Cohesin, a chromatin engagement ring. Curr. Opin. Cell Biol. 2013;25:63–71. doi: 10.1016/j.ceb.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Coviello AD, et al. A genome-wide association meta-analysis of circulating sex hormone-binding globulin reveals multiple Loci implicated in sex steroid hormone regulation. PLoS. Genet. 2012;8:e1002805. doi: 10.1371/journal.pgen.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freedman ND, et al. The association of menstrual and reproductive factors with upper gastrointestinal tract cancers in the NIH-AARP cohort. Cancer. 2010;116:1572–1581. doi: 10.1002/cncr.24880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang QM, Qi YJ, Jiang Q, Ma YF, Wang LD. Relevance of serum estradiol and estrogen receptor beta expression from a high-incidence area for esophageal squamous cell carcinoma in China. Med. Oncol. 2011;28:188–193. doi: 10.1007/s12032-010-9457-8. [DOI] [PubMed] [Google Scholar]
- 24.Bei JX, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat. Genet. 2010;42:599–603. doi: 10.1038/ng.601. [DOI] [PubMed] [Google Scholar]
- 25.Li S, et al. GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS. Genet. 2012;8:e1002791. doi: 10.1371/journal.pgen.1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan Q, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat. Genet. 2012;44:1330–1335. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slager SL, et al. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood. 2011;117:1911–1916. doi: 10.1182/blood-2010-09-308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okada Y, et al. HLA-Cw*1202-B*5201-DRB1*1502 haplotype increases risk for ulcerative colitis but reduces risk for Crohn's disease. Gastroenterology. 2011;141:864–871. doi: 10.1053/j.gastro.2011.05.048. [DOI] [PubMed] [Google Scholar]
- 29.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 30.Su Z, et al. Common variants at the MHC locus and at chromosome 16q24.1 predispose to Barrett's esophagus. Nat. Genet. 2012;44:1131–1136. doi: 10.1038/ng.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.