Abstract
The dismal outcome of laryngeal squamous cell carcinoma (SCC) patients highlights the need for novel prognostic biomarkers. The involvement of microRNAs in cancer and their potential as biomarkers of diagnosis and prognosis are becoming increasingly appreciated. We sought to identify microRNAs that exhibit altered expression in laryngeal SCC and to determine whether microRNA (miRNA) expression is predictive of disease progression and/or patient survival. The expression of two miRNAs, miR-21 and miR-375, was evaluated using total RNA isolated from freshly-frozen primary tumors and non-cancerous laryngeal squamous epithelial tissues and quantitative real-time polymerase chain reaction (qRT-PCR) analysis. We further analyzed the association between the expression of miRNAs and the clinicopathological features. A marked difference in the microRNA expression pattern was observed between tumors and non-cancerous tissue. MiR-21 and miR-375 were expressed at higher and lower levels, respectively, in the laryngeal SCC samples, compared to the normal samples (p < 0.01 and p < 0.001, respectively). There was no correlation between characteristics such as age, sex, clinical stage, and alcohol use, and the expression level of mir-21. The relative expression of mir-375 in laryngeal SCC was shown to be associated with localization of the tumor in these patients (p = 0.037) and with alcohol use (p < 0.05). Patients with high miR-21 or low miR-375 expression in tumor tissues had poorer prognoses compared to patients with lower miR-21 or higher miR-375 expression. Furthermore, the miR-21/miR-375 expression ratio was highly sensitive (0.94) and specific (0.94) for disease prediction. These data suggest that the pattern of microRNA expression in primary laryngeal SCC tissues is reflective of the disease status and that miR-21 and miR-375 expression levels, in particular, may serve as potential biomarkers with applications in the clinical setting.
Keywords: Laryngeal squamous cell carcinoma, microRNA, biomarker, prognosis
Introduction
Head and neck squamous cell carcinoma (HNSCC) includes squamous cell carcinoma (SCC) of the oral cavity, oropharynx, hypopharynx, and larynx. It is the sixth most common type of cancer worldwide, representing about 6% of all cancer cases [1]. Laryngeal SCC, which originates from the laryngeal epithelium, has the second highest incidence of all head and neck squamous cell carcinomas. In recent years, the incidence of laryngeal cancer has been relatively stable, with about 160,000 new cases diagnosed per year [2], and a particularly high incidence in northeast China, accounting for 5.7-7.6% of all malignancies [3]. While the major risk factors for laryngeal SCC are tobacco and alcohol use, there is very little information about the precise molecular pathways underlying the development of laryngeal SCC. Despite significant advances in surgery and radiotherapy over the last few decades, no treatment has been shown to achieve a satisfactory therapeutic outcome and the mortality rate of laryngeal SCC is still high, with a 5-year survival rate of 64% [4]. Therefore, there is an urgent need to develop novel and clinically useful markers to distinguish patients with poor prognosis or at higher risk of an early recurrence.
Accumulating knowledge about the role of genes in tumorigenesis has indicated a key role for microRNAs (miRNAs) in different kinds of cancers. MiRNAs are small non-coding RNAs of 18 to 24 nucleotides, originally discovered and described in 1993 in the nematode Caenorhabditis elegans. found that a gene crucial for post-embryonic development of C. elegans, lin-4, did not code for a protein, but rather is transcribed into a 22-nucleotide RNA molecule. This small RNA molecule could repress the expression of the lin-14 mRNA by directly interacting with its 3’-untranslated region (UTR).While this was recognized as a new method of gene regulation, it was initially considered as an oddity peculiar to C. elegans. To date, more than 300 miRNAs have been discovered in humans, and computational analyses predict that up to 1,000 miRNAs exist in the human genome [5]. Since miRNAs can regulate more than one target, estimates indicate they may be able to regulate up to 30% of the protein-coding genes in the human genome [6], highlighting their importance as global regulators of gene expression. Recently, several studies reported the involvement of let-7 and miR-155 miRNAs in lung cancer diagnosis and prognosis [7], and high expression of miR-21 was associated with poor survival and therapeutic outcome in colon cancer [8]. Other expression profiling studies identified miRNA signatures in hepatocellular carcinoma [9], pancreatic cancer [10], breast cancer [11], papillary thyroid cancer [12], and chronic lymphocytic leukemia [13]. Importantly, the successful use of antagomirs (anti-miRNAs) to silence miRNAs in mice [14] and non-human primates [15] suggests the possible therapeutic use of miRNAs.
Recently, we reported that miR-21 and miR-375, the expression of which is frequently reported in SCC tissues [16-21], were aberrantly expressed in laryngeal SCC patients [22]. Based on our findings, we selected one oncogenic miRNA, miR-21, and one tumor suppressor miRNA, miR-375, as candidates for prognostic biomarkers for laryngeal SCC in a miRNA assay. In this study, we examined alterations in the expression of these two miRNAs in laryngeal specimens from patients with laryngeal SCC and analyzed the correlation between the expression of these miRNAs and the clinicopathological features of this malignancy.
Patients and methods
Patients and samples
Between September 2005 and July 2007, a total of 46 patients with laryngeal SCC were enrolled in this study. Surgical resection specimens were collected from consecutive patients who underwent partial or total laryngectomy at the Shanghai Jiaotong University of Medicine. All patients were recruited and enrolled in the study after providing written informed consent for use of their tissues for research purposes and all study protocols were approved by the Institutional Review Board of Shanghai Jiaotong University of Medicine. Fresh laryngeal SCC tumor specimens were obtained during the tumor survey. Meanwhile, paired normal laryngeal squamous epithelial tissues derived from laryngectomy specimens that served as normal controls were also assayed. Tissue samples were immediately frozen in liquid nitrogen after resection until further processing. None of patients had received preoperative chemotherapy or radiotherapy.
The resected laryngeal SCC specimens were fixed in buffered formalin and embedded in paraffin for pathological examination using standard methods. The pathological stage, grade, and nodal status of the tumors were defined according to the revised International UICC/TMN Staging System. The clinical and pathological data of the laryngeal SCC patients enrolled in the study are displayed in Table 1.
Table 1.
Clinicopathologic characteristics in laryngeal SCC patients (Total N=46)
| Characteristics | N (%) |
|---|---|
| Gender | |
| Male | 42 (91.3) |
| Female | 4 (8.7) |
| Age(y), Mean(SD) | 59.2 (7.84) |
| Smokinga | |
| Negative | 12 (27.9) |
| Positive | 31 (72.1) |
| Alcoholb | |
| Negative | 22 (53.7) |
| Positive | 19 (46.3) |
| T stage | |
| T0/1/2 | 21 (45.7) |
| T3/4 | 25 (54.3) |
| N stage | |
| Negative | 32 (69.6) |
| Positive | 14 (30.4) |
| Stage | |
| Low (I-II) | 31 (67.4) |
| High (III-IV) | 15 (32.6) |
| Localization | |
| Glottic | 33 (71.7) |
| Supraglottic | 11 (23.9) |
| Subglottic | 2 (4.4) |
Note: We examined mir-21 and mir-375 by quantitative reverse transcription polymerase chain reaction (qRT-PCR) in 46 consecutive laryngeal specimens from patients with laryngeal SCC (as described below) and compared the results according to the clinicopathological characteristics of the patients. Forty-two patients were male. The mean age was 59.2 years (±7.84 years). Thirty-three patients had glottic tumors, while 11 had supraglottic and 2 had subglottic tumors. The pathological tumor TNM stages were I-II in 31 patients and III-IV in 15 patients. There was nodal involvement in 14 patients.
Data missing in 3 samples.
Data missing in 5 samples.
RNA extraction from tissue samples
Total RNA isolation from tissue samples was performed using the TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. The concentrations and quality of the RNA were determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, DE, USA) and agarose gel electrophoresis.
Quantitative real-time PCR of miRNAs
The expression of mature miRNAs was assayed using TaqMan miRNA reverse transcription assays (Applied Biosystems) and appropriate primers (Applied Biosystems) following the manufacturer’s instructions. The primers for miR-21 and miR-375 were 5’-GCCCGCTAGCTTATCAGACTGATG-3’ and 5’-UUUGUUCGUUCG-GCUCGCGUGA-3’, respectively. In brief, 10 ng of total RNA was used as the template in a 15-μL RT reaction using probes specially designed for specific mature miRNAs. For assessing the expression of each miRNA, reactions were performed in triplicate using the 7900 RT-PCR system (Applied Biosystems) and small nuclear RNA (snRNA) U6 (Applied Biosystems) was used as the internal control for normalization of expression. The reaction conditions indicated in the manufacturer’s manual were used and the reaction mixtures were incubated at 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The cycle threshold values were calculated with the SDS 1.4 software package. The ΔΔCt method for relative quantization was used to determine miRNA expression levels. The Ct is the fractional cycle number at which the fluorescence of each sample passes the fixed threshold. The ΔCt value was calculated by subtracting the Ct of snRNA U6 from the Ct of the miRNA of interest. The ΔΔCt value was calculated by subtracting the ΔCt of the reference sample (paired non-tumorous tissue for surgical samples) from the ΔCt of each sample. The fold-change was determined as 2-ΔΔCt.
Statistical analysis
Values are presented as mean ± standard error of the mean (SEM). The difference between experimental and control groups was evaluated using the paired t-test. The relationship between clinicopathological factors in laryngeal SCC patients and the expression of miRNA was analyzed using Fisher’s exact or chi-square tests. The last date of follow-up was either the date of death or the last date the patient was contacted. Overall survival was estimated using the Kaplan-Meier method with comparisons between groups made using the log-rank test. A p-value less than 0.05 was considered statistically significant. Statistical analysis was performed using SPSS 10.0 software program.
Results
Differential expression of miRNA in laryngeal SCC patients
To identify more sensitive diagnostic biomarkers in laryngeal SCCs, we selected the most overexpressed microRNA (mir-21) and underexpressed microRNA (mir-375) for further analysis, based on our previous findings. The expression of miR-21 and miR-375 in laryngeal SCCs and paired, adjacent, histologically normal mucosal samples was determined by qRT-PCR analysis. Our data indicate that miR-21 expression is significantly up-regulated (p < 0.01) (Figure 1A) and miR-375 expression is down-regulated (p < 0.001) (Figure 1B) in laryngeal SCC tissue relative to the paired normal tissues. We also analyzed the ratio of the two miRNAs (miR-21/miR-375) as a combined biomarker. The data indicate that the miR-21/miR-375 ratio is significantly higher in laryngeal SCC patients than in controls (p = 0.005) (Figure 1C). The receiver operating characteristic (ROC) curves for miR-21, miR-375, and miR-21/miR-375 were plotted, and the area under the curve (AUC) values were 0.772, 0.957, and 0.991, respectively (Figure 2).
Figure 1.
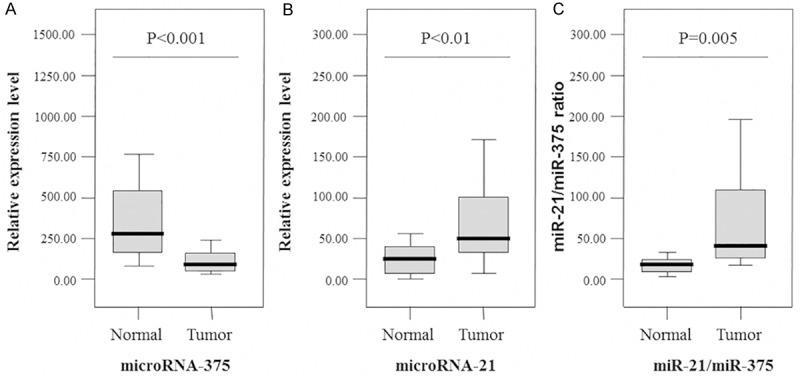
Validation of the differential expression of miR-21 and miR-375 by quantitative real-time PCR in tumor and normal samples (n = 46 for tumor samples and n = 46 for normal samples).The relative expression level of miR-21 tended to be higher in laryngeal SCC patients than in the controls (p < 0.01) (A), while the relative expression level of miR-375 was significantly lower in laryngeal SCC patients than in the controls (p < 0.001) (B). The combined biomarker based on the ratio of miR-21/miR-375 was significantly higher in laryngeal SCC patients than in controls (p = 0.005) (C). Boxes indicate the distribution of expression values from the 25th to 75th percentile for each microRNA. The lines inside the boxes indicate the median values and the upper and lower horizontal bars denote the 90th and 10th percentiles, respectively.
Figure 2.
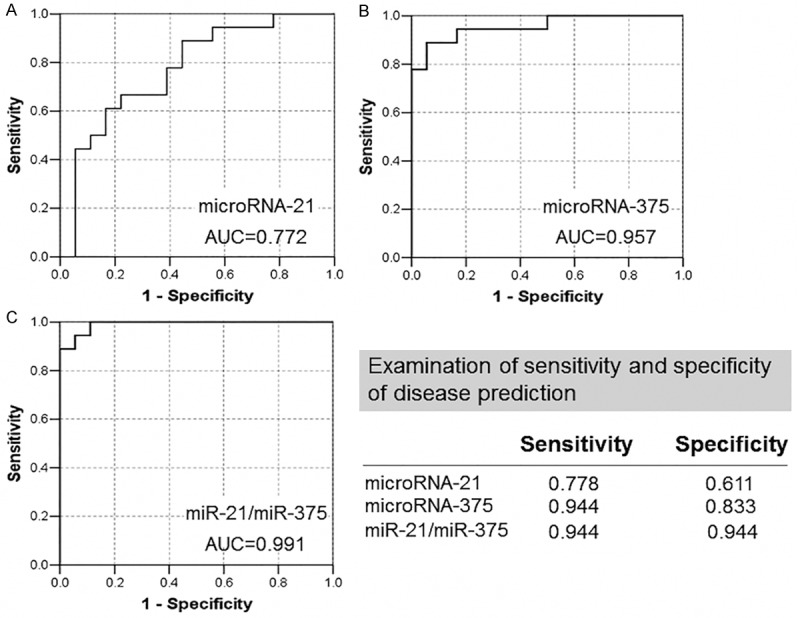
Receiver operating curve analysis of miR-21 and miR-375 expression levels and the ratio of miR-21/miR-375 for differentiation of laryngeal SCC tumors and normal tissues. AUC, Area under the curve.
Comparison of patients according to clinicopathological characteristics and the expression of miRNAs
Next, We determined whether there was any correlation between miR-21 and miR-375 levels and the clinicopathological features of the laryngeal SCC patients. The miRNA expression levels from the qRT-PCR analysis were dichotomized based on a within-cohort median cutoff. The major characteristics of the patients are shown in Table 2. When characteristics of the patients were compared, after stratification based on the expression levels of mir-21, no correlation was observed between miR-21 expression levels and the age, sex, differentiation, TNM stage, tobacco consumption, and alcohol use of the patient. Patients with T3-4 tumors exhibited a slight over-expression of mir-21, but the difference was not statistically significant (p > 0.05). The expression of mir-375 in patients with supraglottic tumors was significantly lower than that in those with glottic tumors (p = 0.037). Low expression of mir-375 was associated with the clinical feature of alcohol use (p < 0.05).
Table 2.
Correlation between the expression of miRNA-21 and miRNA-375 and clinicopathological factors in laryngeal SCC patients
| Relative expression of miR-21 | Relative expression of miR-375 | ||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| n | High | Low | P-value | High | Low | P-value | |
| Total | 46 | 24 (52.2%) | 22 (47.8%) | 23 (50.0%) | 23 (50.0%) | ||
| Gender | |||||||
| Male | 42 | 22 (52.4%) | 20 (47.6%) | 20 (47.6%) | 22 (52.4%) | ||
| Female | 4 | 2 (50.0%) | 2 (50.0%) | 1.000 | 3 (75.0%) | 1 (25.0%) | 0.608 |
| Age (y) | |||||||
| < 65 | 22 | 12(54.5%) | 10 (45.5%) | 12 (54.5%) | 10 (45.5%) | ||
| ≤ 65 | 24 | 12(50.0%) | 12 (50.0%) | 0.990 | 11 (45.8%) | 13 (54.2%) | 0.768 |
| T stage | |||||||
| T0/1/2 | 21 | 12 (57.1%) | 9 (42.9%) | 13 (61.9%) | 8 (38.1%) | ||
| T3/4 | 25 | 12 (48.0%) | 13 (52.0%) | 0.747 | 10 (40.0%) | 15 (60.0%) | 0.236 |
| N stage | |||||||
| Negative | 32 | 17 (53.1%) | 15 (46.9%) | 18 (56.3%) | 14 (43.8%) | ||
| Positive | 14 | 7 (50.0%) | 7 (50.0%) | 1.000 | 5 (35.7%) | 9 (64.3%) | 0.336 |
| Stage | |||||||
| Low (I-II) | 31 | 13 (41.9%) | 18 (58.1%) | 19 (61.3%) | 12 (38.7%) | ||
| High (III-IV) | 15 | 11 (73.3%) | 4 (26.7%) | 0.092 | 4 (26.7%) | 11 (73.3%) | 0.059 |
| Localization | |||||||
| Glottic | 33 | 18 (54.5%) | 15 (45.5%) | 20 (60.6%) | 13 (39.4%) | ||
| Supraglottic | 11 | 5 (45.5%) | 6 (54.5%) | 0.862 | 2 (18.2%) | 9 (81.8%) | 0.037 |
| Subglottic | 2 | 1 (50.0%) | 1 (50.0%) | 1 (50.0%) | 1 (50.0%) | ||
| Smokinga | |||||||
| Negative | 12 | 5 (41.7%) | 7 (58.3%) | 6 (50.0%) | 6 (50.0%) | ||
| Positive | 31 | 18 (58.1%) | 13 (41.9%) | 0.531 | 15 (48.4%) | 16 (51.6%) | 1.000 |
| Alcoholb | |||||||
| Negative | 22 | 11 (50.0%) | 11 (50.0%) | 15 (68.2%) | 7 (31.8%) | ||
| Positive | 19 | 10 (52.6%) | 9 (47.4%) | 1.000 | 6 (31.6%) | 13 (68.4%) | 0.043 |
Data missing in 3 samples;
Data missing in 5 samples; p < 0.05.
Note: Correlation between the expression of miRNA-21 and miRNA-375 and clinicopathological factors in laryngeal SCC patients. p values, as calculated using the χ2 or Fisher’s exact test, are shown, with p < 0.05 considered statistically significant.
The expression level of miRNAs is associated with survival in laryngeal SCC patients
The miRNA expression levels from the qRT-PCR analysis were dichotomized based on a within-cohort median cutoff. Kaplan-Meier analysis revealed a significant association between high expression of miR-21 in cancerous tissue and a worse prognosis (p = 0.045) (Figure 3A). In addition, low expression of miR-375 in cancerous tissue was associated with poor prognosis, but this correlation is not significant (p > 0.05) (Figure 3B). Univariate analysis showed that the life span of laryngeal SCC patients is correlated with tumor stage (p = 0.042) and expression level of miR-21 (p = 0.061). Multivariate analysis using the Cox regression model adjusted to clinicopathological variables showed that the expression level of miR-21 may serve as an independent prognostic factor for overall survival in laryngeal SCC patients (p = 0.001; 95% confidence interval [CI], 0.340-3.757) (Table 3).
Figure 3.
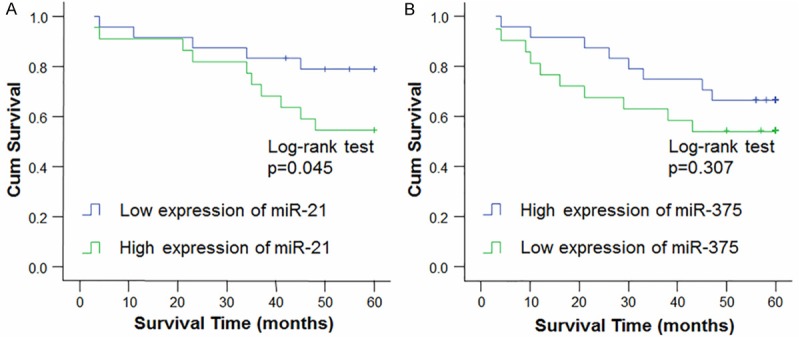
Comparison of cumulative five-year survival curves of patients with laryngeal SCC according to independent miRNA expression-related factors with the Kaplan-Meier method. The postoperative survival rate of patients with high expression levels of miR-21 (A) was poorer than those with lower expression of miR-21, while that of patients with high expression levels of miR-375 (B) group was better than those with lower expression of miR-375.
Table 3.
Univariate and multivariate analysis of prognostic factors for survival in laryngeal SCC patients
| Univariate analysis | multivariate analysis 95%CI | |||
|---|---|---|---|---|
|
| ||||
| p value | Lower | Upper | p value | |
| miR-375 expression level | ||||
| High versus Low | < 0.001 | 0.34 | 3.757 | 0.001 |
| Gender | ||||
| Male versus Female | 0.874 | 0.018 | 5.011 | 0.401 |
| Age (y) | ||||
| > 65 versus ≤ 65 | 0.596 | 0.728 | 0.993 | 0.041 |
| pTNM stage | ||||
| Low (I-II) versus | ||||
| High (III-IV) | 0.042 | 0.57 | 22.911 | 0.173 |
| Alcohol consumption | ||||
| Positive versus Negative | 0.715 | 0.233 | 31.331 | 0.427 |
| Tobacco use | ||||
| Positive versus Negative | 0.341 | 0.037 | 2.08 | 0.212 |
p < 0.05.
Discussion
Since the discovery of miRNAs, tremendous efforts have been devoted to determining their biological functions and their relevance to cancer diagnosis, prognosis, and therapy. Many studies have reported significant associations between miRNA expression profiles and important clinical features of tumors, as well as patient survival rates [23-28]. By elucidating the presence of miRNA expression signatures and their contribution to the development and progression of tumors in patients with laryngeal SCC, the present study has extended our knowledge of the role of miRNAs in the pathogenesis of this malignancy. In this study, we analyzed the expression of two miRNAs, miR-21 and miR-375, in laryngeal SCC tumors and found a significant correlation between the under-expressions of miR-375 with alcohol consumption, a major risk factor for laryngeal SCC. Our data also indicate that miR-21 over-expression might be an independent prognostic factor for overall survival.
Mir-21 is frequently overexpressed in a variety of malignancies, and this miRNA has been implicated in multiple malignancy-related processes, including cell proliferation, apoptosis, invasion, and metastasis [29-37]. Mir-21 has been previously identified as a putative oncogene in glioblastomas, and in pancreatic and breast cancers [38-40], whereas miR-21 was shown to be an anti-apoptotic factor in human gliomas [29]. Similar to the findings for mir-21 in other tumor types, mir-21 expression in the laryngeal SCC samples were high in both tumor and matched normal tissues, but the expression of mir-21 was twice as high in tumor samples as they were in the normal samples. Though the univariate log-rank test for survival was not significant, as it did not consider explanatory factors such as tumor localization and was limited in power by the sample size, the unadjusted Kaplan-Meier analysis showed a clear trend for worse survival in patients with high mir-21 expression. The multivariate analysis, which controlled for confounding factors, identified a significant association between high mir-21 expression in tumors and poor patient survival, and the same relationship that has been demonstrated in cancers of the breast and colon, as well as in non-small-cell lung cancer [8,30,41]. The strong association between the mir-21 levels and clinicopathologic characteristics indicates that it may serve as a new biomarker, indicative of a poor prognosis, in many cancers, including laryngeal SCCs.
A recent study [42] of human gastric cancers, published during the preparation of our manuscript, showed that the expression of miR-21 in cancer tissues was significantly higher than that in non-cancerous tissues, consistent with our data. Previous studies have demonstrated that the novel tumor suppressor protein programmed cell death 4 (PDCD4) is downregulated in several human solid cancers and is suppressed by miR-21 [43]. Phosphatase and tensin homolog (PTEN) was shown to be a direct target of mir-21 in cholangiocarcinoma cells [44]. Thus, the role of miR-21 is likely to be the inhibition of apoptosis.
Though miR-375 has mainly been studied in the context of diabetes, as it influences beta-cell mass and insulin levels [45], its expression has been shown to be decreased in a number of malignancies, including pancreatic adenocarcinomas and esophageal squamous cell and adenocarcinomas [46,47]. Additionally, the recent identification of a target for miR-375, phosphoinositide-dependent protein kinase-1, suggests a feasible role for miR-375 as a tumor suppressor since phosphoinositide-dependent protein kinase-1 is crucial for the activation of anti-apoptotic AKT protein [48]. In our study, lower expression of miR-375 was also found in laryngeal tumors, compared with normal tissues. We also found that miR-375 expression changed with alcohol consumption, with a significant trend for lower expression of miR-375 with increasing alcohol consumption. While it is known that alcohol is an independent risk factor for HNSCC, the mechanism for this association is poorly understood. Further studies are needed to address the potential role of miR-375 in alcohol use-mediated pathogenesis of HNSCC.
To improve the predictive potential of alterations in the expression of individual microRNAs, we also investigated the miR-21/miR-375 expression ratio as a diagnostic biomarker in laryngeal SCC. The ratio of miR-21/miR-375 showed a high discriminatory potential, with a sensitivity of 94% and specificity of 94%, in distinguishing tumor from normal tissue. These data suggest that the ratio of these microRNAs may hold significant clinical potential, but further validation is necessary in an independent series of laryngeal SCC tumors.
In conclusion, our results suggest that the expression level of miR-21 and miR-375 may have potential applications for more useful clinical stratification of laryngeal SCC patients and enable us to select prospective candidates for additional or alternative treatments in laryngeal SCC. The differential expression of miR-21 and miR-375 may have diagnostic implications for the management of laryngeal SCC patients. Our results also expand the known pathways and genes that promote the progression of this disease and represent an important new avenue of research in laryngeal SCC.
Acknowledgements
This study was supported by grants from Shanghai Gongli Hospital Youth Project (No. 2013GLQN04) and Medical Project of Pudong New Area Health Bureau of Shanghai (no. PWZxq2014-09). The authors thank the Immunology Institute of the Renji Hospital, School of Medicine, Shanghai JiaoTong University, especially Lijia Wang, MD, for their technical assistance.
Disclosure of conflict of interest
None to declare.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Ramroth H, Schoeps A, Rudolph E, Dyckhoff G, Plinkert P, Lippert B, Feist K, Delank KW, Scheuermann K, Baier G, Ott I, Chenouda S, Becher H, Dietz A. Factors predicting survival after diagnosis of laryngeal cancer. Oral Oncol. 2011;47:1154–1158. doi: 10.1016/j.oraloncology.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Ji W, Guan C, Pan Z. Analysis of curative effects on laryngeal carcinoma patients in the northeast region of China. Acta Otolaryngol. 2008;128:574–577. doi: 10.1080/00016480701596104. [DOI] [PubMed] [Google Scholar]
- 4.Boyle P, Ferlay J. Cancer incidence and mortality in Europe, 2004. Ann Oncol. 2005;16:481–488. doi: 10.1093/annonc/mdi098. [DOI] [PubMed] [Google Scholar]
- 5.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, Vallar L, Nashan D, Behrmann I, Kreis S. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70:4163–4173. doi: 10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- 7.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji J, Wang XW. New kids on the block: diagnostic and prognostic microRNAs in hepatocellular carcinoma. Cancer Biol Ther. 2009;8:1686–1693. doi: 10.4161/cbt.8.18.8898. [DOI] [PubMed] [Google Scholar]
- 10.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 12.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M, Maggio R, Peragine N, Santangelo S, Mauro FR, Landgraf P, Tuschl T, Weir DB, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Guarini A, Foa R, Macino G. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 14.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 15.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 16.Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, Shao K, Li N, Qiu B, Mitchelson K, Cheng J, He J. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 17.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–260. doi: 10.1016/j.jtcvs.2007.08.055. discussion 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123:2791–2797. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O’Sullivan B, Waldron J, Gullane P, Cummings B, Liu FF. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 21.Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, O’Brien C, Rose B. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- 22.Hu A, Jin X. [Expression of mir-21 and mir-375 in laryngeal squamous cell carcinoma] . Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2012;26:788–792. [PubMed] [Google Scholar]
- 23.Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, McClean MD, Marsit CJ, Kelsey KT. A let-7 microRNA-binding site polymorphism in the KRAS 3’ UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30:1003–1007. doi: 10.1093/carcin/bgp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV, Prystowsky MB, Belbin TJ, Schlecht NF. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 26.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S, Philip PA, Li Y. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res. 2013;3:465–477. [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Lu H, Li T, Yu L, Liu G, Peng X, Zhao J. Kruppel-like factor 8 promotes tumorigenic mammary stem cell induction by targeting miR-146a. Am J Cancer Res. 2013;3:356–373. [PMC free article] [PubMed] [Google Scholar]
- 29.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 30.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 31.Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M, Baba H. MicroRNA- 21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res. 2009;15:1915–1922. doi: 10.1158/1078-0432.CCR-08-2545. [DOI] [PubMed] [Google Scholar]
- 32.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T, Takahata S, Toma H, Nagai E, Tanaka M. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8:1067–1074. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 34.Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, Yamashita J, Saitoh H, Kameoka Y, Shimizu N, Ichinohasama R, Sawada K. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114:3265–3275. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Huang H, Sun L, Yang M, Pan C, Chen W, Wu D, Lin Z, Zeng C, Yao Y, Zhang P, Song E. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 36.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 37.Ren Q, Liang J, Wei J, Basturk O, Wang J, Daniels G, Gellert LL, Li Y, Shen Y, Osman I, Zhao J, Melamed J, Lee P. Epithelial and stromal expression of miRNAs during prostate cancer progression. Am J Transl Res. 2014;6:329–339. [PMC free article] [PubMed] [Google Scholar]
- 38.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 39.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 40.Fassan M, Baffa R, Palazzo JP, Lloyd J, Crosariol M, Liu CG, Volinia S, Alder H, Rugge M, Croce CM, Rosenberg A. MicroRNA expression profiling of male breast cancer. Breast Cancer Res. 2009;11:R58. doi: 10.1186/bcr2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motoyama K, Inoue H, Mimori K, Tanaka F, Kojima K, Uetake H, Sugihara K, Mori M. Clinicopathological and prognostic significance of PDCD4 and microRNA-21 in human gastric cancer. Int J Oncol. 2010;36:1089–1095. doi: 10.3892/ijo_00000590. [DOI] [PubMed] [Google Scholar]
- 43.Jin H, Kim TH, Hwang SK, Chang SH, Kim HW, Anderson HK, Lee HW, Lee KH, Colburn NH, Yang HS, Cho MH, Cho CS. Aerosol delivery of urocanic acid-modified chitosan/programmed cell death 4 complex regulated apoptosis, cell cycle, and angiogenesis in lungs of K-ras null mice. Mol Cancer Ther. 2006;5:1041–1049. doi: 10.1158/1535-7163.MCT-05-0433. [DOI] [PubMed] [Google Scholar]
- 44.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 45.Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, Altorki NK, Casson AG, Liu CG, Wang XW, Yanaihara N, Hagiwara N, Dannenberg AJ, Miyashita M, Croce CM, Harris CC. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 48.El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3’-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–2717. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]


