Abstract
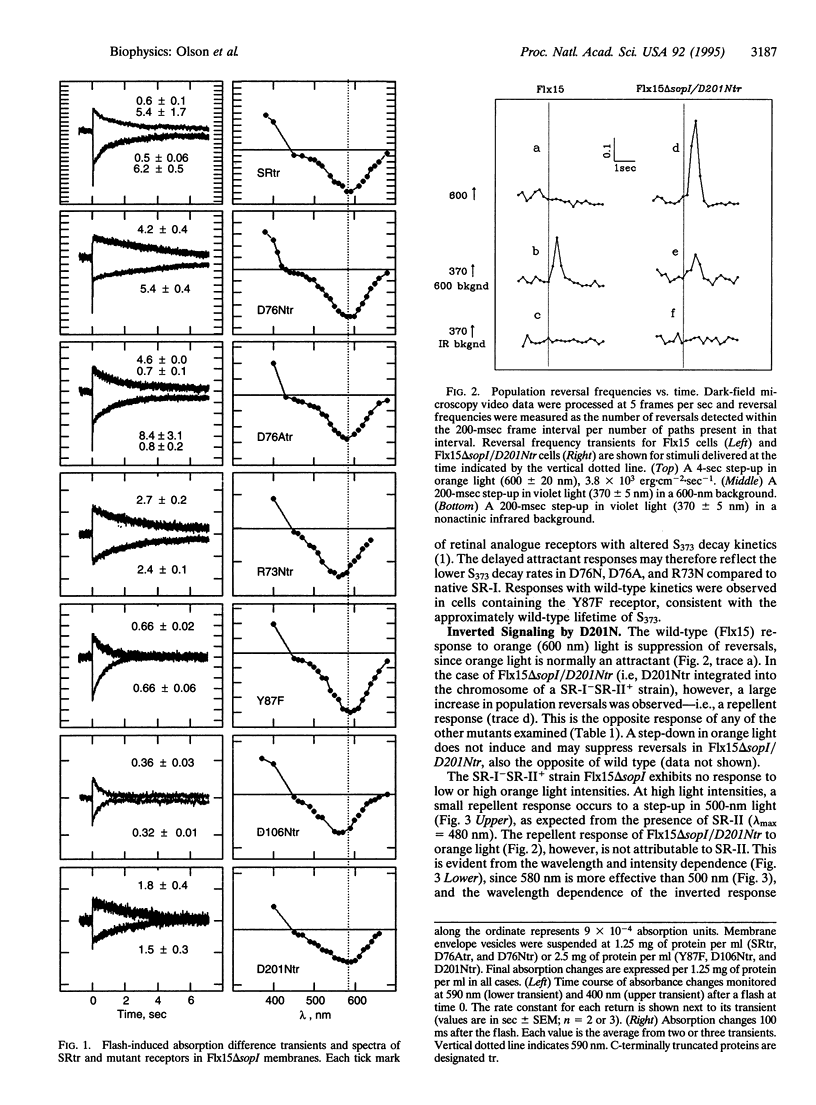
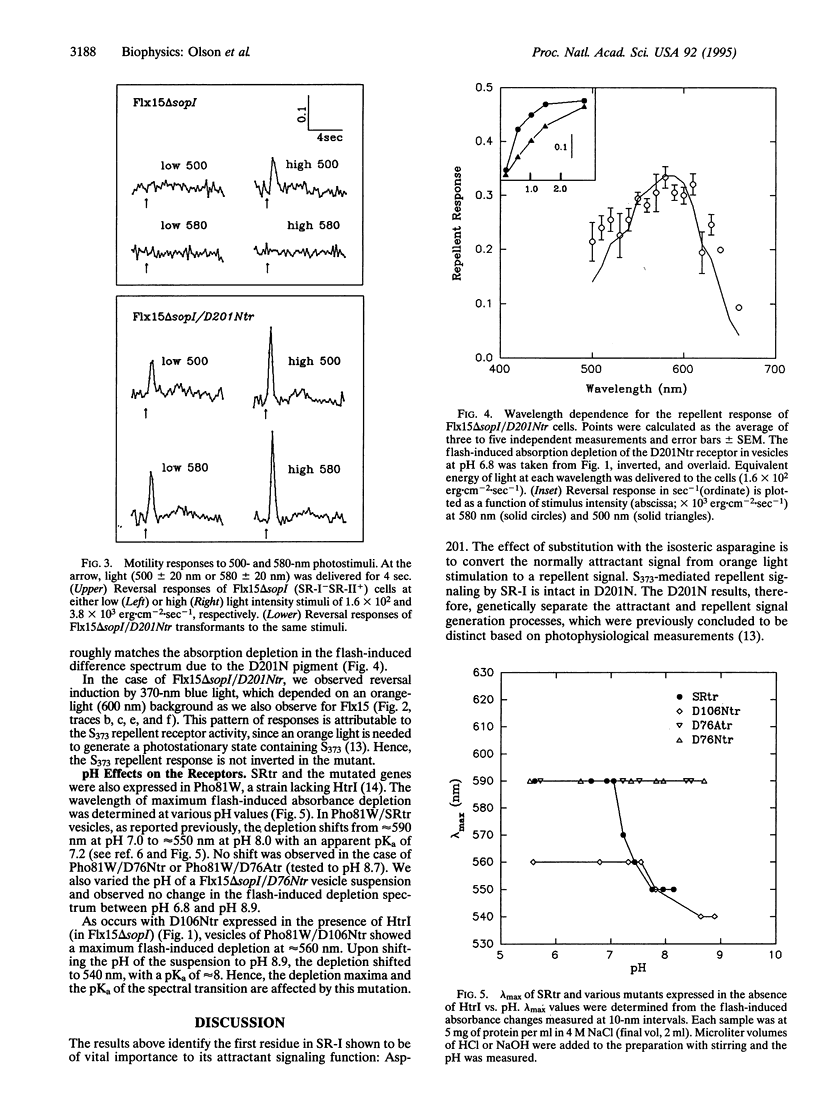
Residue replacements were made at five positions (Arg-73, Asp-76, Tyr-87, Asp-106, and Asp-201) in the Halobacterium salinarium phototaxis receptor sensory rhodopsin I (SR-I) by site-specific mutagenesis. The sites were chosen for their correspondence in position to residues of functional importance in the homologous light-driven proton pump bacteriorhodopsin found in the same organism. This work identifies a residue in SR-I shown to be of vital importance to its attractant signaling function: Asp-201. The effect of the substitution with the isosteric asparagine is to convert the normally attractant signal of orange light stimulation to a repellent signal. In contrast, similar neutral substitution of the four other ionizable residues near the photoactive site allows essentially normal attractant and repellent phototaxis signaling. Wild-type two-photon repellent signaling by the receptor is intact in the Asp-201 mutant, genetically separating the wild-type attractant and repellent signal generation processes. A possible explanation and implications of the inverted signaling are discussed. Results of neutral residue substitution for Asp-76 confirm our previous evidence that proton transfer reactions involving this residue are not important to phototaxis but that Asp-76 functions as the Schiff base proton acceptor in proton translocation by transducer-free SR-I.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bogomolni R. A., Stoeckenius W., Szundi I., Perozo E., Olson K. D., Spudich J. L. Removal of transducer HtrI allows electrogenic proton translocation by sensory rhodopsin I. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):10188–10192. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braiman M. S., Mogi T., Marti T., Stern L. J., Khorana H. G., Rothschild K. J. Vibrational spectroscopy of bacteriorhodopsin mutants: light-driven proton transport involves protonation changes of aspartic acid residues 85, 96, and 212. Biochemistry. 1988 Nov 15;27(23):8516–8520. doi: 10.1021/bi00423a002. [DOI] [PubMed] [Google Scholar]
- Dér A., Száraz S., Tóth-Boconádi R., Tokaji Z., Keszthelyi L., Stoeckenius W. Alternative translocation of protons and halide ions by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4751–4755. doi: 10.1073/pnas.88.11.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwert K., Hess B., Soppa J., Oesterhelt D. Role of aspartate-96 in proton translocation by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4943–4947. doi: 10.1073/pnas.86.13.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand E., Schimz A. Role of the response oscillator in inverse responses of Halobacterium halobium to weak light stimuli. J Bacteriol. 1987 Jan;169(1):254–259. doi: 10.1128/jb.169.1.254-259.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz M., Drachev L. A., Mogi T., Otto H., Kaulen A. D., Heyn M. P., Skulachev V. P., Khorana H. G. Replacement of aspartic acid-96 by asparagine in bacteriorhodopsin slows both the decay of the M intermediate and the associated proton movement. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2167–2171. doi: 10.1073/pnas.86.7.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krah M., Marwan W., Verméglio A., Oesterhelt D. Phototaxis of Halobacterium salinarium requires a signalling complex of sensory rhodopsin I and its methyl-accepting transducer HtrI. EMBO J. 1994 May 1;13(9):2150–2155. doi: 10.1002/j.1460-2075.1994.tb06491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. P., Khorana H. G. Mechanism of light-dependent proton translocation by bacteriorhodopsin. J Bacteriol. 1993 Mar;175(6):1555–1560. doi: 10.1128/jb.175.6.1555-1560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. P., Spudich E. N., Khorana H. G., Spudich J. L. Synthesis of a gene for sensory rhodopsin I and its functional expression in Halobacterium halobium. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3486–3490. doi: 10.1073/pnas.90.8.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi J. K. Proton translocation mechanism and energetics in the light-driven pump bacteriorhodopsin. Biochim Biophys Acta. 1993 Dec 7;1183(2):241–261. doi: 10.1016/0005-2728(93)90226-6. [DOI] [PubMed] [Google Scholar]
- McCain D. A., Amici L. A., Spudich J. L. Kinetically resolved states of the Halobacterium halobium flagellar motor switch and modulation of the switch by sensory rhodopsin I. J Bacteriol. 1987 Oct;169(10):4750–4758. doi: 10.1128/jb.169.10.4750-4758.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson K. D., Spudich J. L. Removal of the transducer protein from sensory rhodopsin I exposes sites of proton release and uptake during the receptor photocycle. Biophys J. 1993 Dec;65(6):2578–2585. doi: 10.1016/S0006-3495(93)81295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H., Marti T., Holz M., Mogi T., Lindau M., Khorana H. G., Heyn M. P. Aspartic acid-96 is the internal proton donor in the reprotonation of the Schiff base of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9228–9232. doi: 10.1073/pnas.86.23.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath P., Olson K. D., Spudich J. L., Rothschild K. J. The Schiff base counterion of bacteriorhodopsin is protonated in sensory rhodopsin I: spectroscopic and functional characterization of the mutated proteins D76N and D76A. Biochemistry. 1994 May 10;33(18):5600–5606. doi: 10.1021/bi00184a032. [DOI] [PubMed] [Google Scholar]
- Robinson P. R., Cohen G. B., Zhukovsky E. A., Oprian D. D. Constitutively active mutants of rhodopsin. Neuron. 1992 Oct;9(4):719–725. doi: 10.1016/0896-6273(92)90034-b. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. The photochemical reactions of sensory rhodopsin I are altered by its transducer. J Biol Chem. 1993 Aug 5;268(22):16095–16097. [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984 Dec 6;312(5994):509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Sensory rhodopsin I: receptor activation and signal relay. J Bioenerg Biomembr. 1992 Apr;24(2):193–200. doi: 10.1007/BF00762677. [DOI] [PubMed] [Google Scholar]
- Spudich J. L. Protein-protein interaction converts a proton pump into a sensory receptor. Cell. 1994 Dec 2;79(5):747–750. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- Subramaniam S., Marti T., Khorana H. G. Protonation state of Asp (Glu)-85 regulates the purple-to-blue transition in bacteriorhodopsin mutants Arg-82----Ala and Asp-85----Glu: the blue form is inactive in proton translocation. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1013–1017. doi: 10.1073/pnas.87.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittor J., Schweiger U., Oesterhelt D., Bamberg E. Inversion of proton translocation in bacteriorhodopsin mutants D85N, D85T, and D85,96N. Biophys J. 1994 Oct;67(4):1682–1690. doi: 10.1016/S0006-3495(94)80642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao V. J., Spudich E. N., Spudich J. L. Identification of distinct domains for signaling and receptor interaction of the sensory rhodopsin I transducer, HtrI. J Bacteriol. 1994 Nov;176(22):6931–6935. doi: 10.1128/jb.176.22.6931-6935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao V. J., Spudich J. L. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]



