Abstract
Background
The functional polymorphism rs4938723 in the promoter region of pri-miR-34b/c is potentially associated with susceptibility to several cancers, including hepatocellular carcinoma, colorectal cancer, and breast cancer. Here we conducted a comprehensive meta-analysis to investigate the association between rs4938723 and cancer risk.
Material/Methods
Eligible studies extracted from the databases of PubMed, Web of Science, and Cochrane Library were evaluated. Statistical analysis was performed using Revman 5.2 and STATA 12.0 software.
Results
By characterizing the extracted data, a total of 11 studies reported in 10 publications including 6169 cases and 6337 controls were selected for further analysis. Our results revealed a significant association between the rs4938723 polymorphism and cancer risk in the codominant model (TC vs. TT: OR=1.10, 95% CI=1.02–1.19, P=0.009) but not in other genetic models. In the stratified analysis of different cancer types, a significant association was found in nasopharyngeal cancer, osteosarcoma, and renal cell cancer. Furthermore, stratified analysis of ethnicity indicated that a highly significant association was shown in the Asian population in a codominant model (TC vs. TT: OR=1.13, 95% CI=1.03–1.24, P=0.007) when compared with African-Americans and Caucasians.
Conclusions
Overall, the current study suggests that the miR-34b/c rs4938723 polymorphism may be associated with the risk of cancers, including nasopharyngeal cancer, osteosarcoma, and renal cell cancer, and to some extent this polymorphism is closely related to cancer susceptibility in Asians.
MeSH Keywords: Genes, Neoplasm, Meta-Analysis, MicroRNAs, Polymorphism, Genetic
Background
Cancers are still the leading cause of death worldwide and cancer burden continues to increase, receiving great public attention [1–3]. Investigators showed that genetic factors have significantly important functions in the development and progression of cancer.
MicroRNAs (miRNAs) are small, noncoding, single-stranded RNA molecules that participate in the transcriptional regulation of eukaryotic genes and lead to mRNA degradation or the translational repression of targeted genes [4–6]. MiRNAs were thought to be associated with many biological processes with critical roles in carcinogenesis, including cell differentiation, proliferation, and apoptosis. Furthermore, the variation of miRNAs through physiological processes may cause the incidence and development of tumors [7,8]. Recently, a potentially functional polymorphism rs4938723 has been discovered in the promoter region of miR-34b/c. The T to C shift of the rs4938723 polymorphism is thought to influence the GATA-X binding sites. It can bind to the GATA-X when the location is C; otherwise, it cannot bind to the GATA-X. In the past several years, many reported studies have focused on the association between miR34b/c rs4938723 polymorphism and cancer susceptibility in several populations and diverse types of cancer [9–18]. However, the results were inconclusive and controversial, and to the best of our knowledge no one has performed a meta-analysis to investigate the association of this polymorphism with cancer risk. Therefore, we conducted a meta-analysis of all eligible studies to further study the roles of miR-34b/c rs4938723 polymorphism in carcinogenesis.
Material and Methods
Search for study
A systematic search was conducted by 2 investigators independently. Studies were mainly searched in PubMed, Web of Science, and Cochrane Library databases from their inception to June 2014 with the following terms: ‘miR-34b/c’, ‘polymorphism’, and ‘cancer’. The search was limited to case-control studies in the English language. Reference lists from relevant articles were also examined to find additional publications. To avoid double-counting or other errors, 2 investigators compared their results discreetly and disagreements were resolved by consensus or by a third investigator.
Selection of study
All the included studies met the following criteria: 1) case-control study; 2) evaluation of the association between miR-34b/c rs4938723 polymorphism and cancer risks; 3) sufficient data for analysis, including genotype frequency in cases and controls; 4) genotype frequency in the control group was in Hardy-Weinberg equilibrium (HWE); and 5) the study was published in English. We excluded studies without eligible data for meta-analysis.
Data extraction
Two investigators who were blinded to each other abstracted the data in a traditional format and reached consensus on all items. The collected data included first author, publication year, country, ethnicity, cancer type, and available genotypes.
Statistical analysis
Statistical analysis was performed using Revman 5.2 and STATA 12.0 software. χ2 tests and I2 statistic were used to measure the study heterogeneity between trials. Both fixed- and random-effects models were used where appropriate [19,20]. I2 >50% was considered representative of significant statistical heterogeneity and the random-effects model was used; otherwise, the calculations were performed with the fixed-effects model. Odds ratio (OR) with 95% confidence interval (95% CI) was used to evaluate the association between polymorphism and cancer risk with the codominant model (TC vs. TT), codominant model (CC vs. TT), dominant model (TC+ CC vs. TT), recessive model (TC+TT vs. CC), and allele model (T vs. C). Subgroup analysis based on cancer type and ethnicity was also performed. Sensitivity analysis was used to identify sources of significant heterogeneity by removing individual studies and analyzing the effect on the overall results. Publication bias was further assessed by Begg’s test [21]. P value less than 0.05 was considered statistically significant in all statistics.
Results
Characteristics of the studies
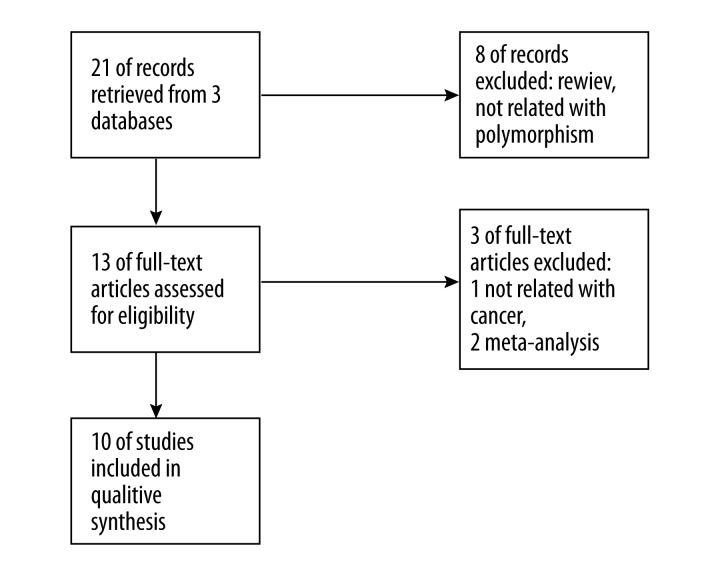
A study flow diagram is shown in Figure 1. Twenty-one studies of miR-34b/c polymorphism and cancers were found in a primary literature search in the PubMed, Web of Science, and Cochrane Library databases. After reviewing each publication, 11 articles were found to be inappropriate for the current meta-analysis because some of them were review articles, irrelevant to the current study or they contained duplicate data. Ten publications, including 11 studies with 6169 cases and 6337 controls (Table 1), were identified as being appropriate for inclusion in the current meta-analysis [9–18]. Of the 11 selected studies, 9 were matched for age and sex but 2 studies of breast cancer were not matched for sex. Furthermore, 9 studies were done in Asians and the other 2 studies of breast cancer were in Caucasians and African-Americans. The genotype distribution of control populations was in Hardy-Weinberg equilibrium in all 11 studies.
Figure 1.
Study flow diagram.
Table 1.
Characteristics of studies included in this meta-analysis.
| Author | Year | Country | Ethnicity | Type of cancer | No. (cases/controls) | Genotypes case (%) | Genotypes control (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | TT | TC | CC | ||||||
| Bensen et al. | 2013 | America | African American | Breast cancer | 742/658 | 362 (48.8) | 317 (42.7) | 63 (8.5) | 343 (52.1) | 257 (39.1) | 58 (8.8) |
| Bensen et al. | 2013 | America | Caucasian | Breast cancer | 1203/1088 | 496 (41.2) | 563 (46.8) | 144 (12.0) | 430 (39.5) | 503 (46.2) | 155 (14.2) |
| Gao et al. | 2013 | China | Asian | Colorectal cancer | 347/488 | 175 (50.4) | 144 (41.5) | 28 (8.1) | 216 (44.3) | 210 (43.0) | 62 (12.7) |
| Han et al. | 2013 | China | Asian | Hepatocellular carcinoma | 1013/999 | 451 (44.5) | 444 (43.8) | 118 (11.6) | 456 (45.6) | 424 (42.4) | 119 (11.9) |
| Li et al. | 2013 | China | Asian | Nasopharyngeal carcinoma | 217/360 | 82 (37.8) | 104 (47.9) | 31 (14.3) | 168 (46.7) | 155 (43.1) | 37 (10.3) |
| Oh et al. | 2014 | Korea | Asian | Colorectal cancer | 545/428 | 272 (49.9) | 233 (42.8) | 40 (7.3) | 216 (50.5) | 171 (40.0) | 41 (9.5) |
| Son et al. | 2014 | Korea | Asian | Hepatocellular carcinoma | 157/201 | 69 (43.9) | 75 (47.8) | 13 (8.3) | 110 (54.7) | 74 (36.8) | 17 (8.5) |
| Tian et al. | 2014 | China | Asian | Osteosarcoma | 133/133 | 41 (30.8) | 62 (46.6) | 30 (22.6) | 62 (46.6) | 53 (39.8) | 18 (13.5) |
| Xu et al. | 2011 | China | Asian | Hepatocellular carcinoma | 502/549 | 204 (40.64) | 236 (47.01) | 62 (12.35) | 266 (48.45) | 229 (41.71) | 54 (9.84) |
| Yin et al. | 2013 | China | Asian | Esophageal cancer | 600/673 | 277 (46.2) | 278 (46.3) | 45 (7.5) | 310 (46.1) | 290 (43.1) | 73 (10.8) |
| Zhang et al. | 2014 | China | Asian | Renal cell cancer | 710/760 | 302 (42.5) | 324 (45.6) | 84 (11.8) | 352 (46.3) | 344 (45.3) | 64 (8.4) |
Quantitative synthesis
We analyzed the association between miR-34b/c polymorphism and cancer risks within 5 genetic models, as mentioned in the Methods section. The main results of the meta-analysis are shown in Table 2. The data were extracted to estimate the association with the overall risk of all types of cancer. The pooled results revealed a significant association between rs4938723 genotype TC and increased cancer risk in the codominant model (TC vs. TT: OR=1.10, 95% CI=1.02–1.19, P=0.009). In contrast, no statistically significant association was found in the other 4 genetic models. We performed stratification analysis based on the cancer types and ethnicities. In the stratified analysis of cancer types, the significantly increased risk of nasopharyngeal cancer was found to be associated with the C allele carriers (TC + CC genotypes) in the dominant model (TC + CC vs. TT: OR=1.44, 95% CI=1.02–2.03, P=0.04) and allele comparison (C vs. T: OR=1.33, 95% CI=1.04–1.70, P=0.03). Except for the recessive model (CC vs. TC + TT), the other 4 compared models showed significant association with increased risk of osteosarcoma (TC vs. TT: OR=1.77, 95% CI=1.03–3.03, P=0.04; CC vs. TT: OR=2.52, 95% CI=1.25–5.10, P=0.01; TC+ CC vs. TT: OR=1.96, 95% CI=1.19–3.24, P=0.009; C vs. T: OR=1.68, 95% CI=1.19–2.39, P=0.004, respectively). We also found that the significantly increased risk of renal cell cancer was associated with C allele (C vs. T: OR=1.18, 95% CI=1.01–1.37, P=0.04) and homozygous genotype CC in the codominant (CC vs. TT: OR=1.53, 95% CI=1.07–2.19, P=0.02) and recessive (CC vs. TC+TT: OR=1.46, 95% CI=1.04–2.06, P=0.03) models. Stratified analysis of ethnicity showed a statistically increased cancer risk in Asians with heterozygous genotype TC in the codominant model (TC vs. TT: OR=1.13, 95% CI=1.03–1.24, P=0.007) but not in African-Americans or Caucasians.
Table 2.
Pooled ORs and 95% CIs of the overall and stratified meta-analysis.
| Variables | No. of studies | Codominant model: TC vs. TT | Codominant model: CC vs. TT | Dominant model: TC + CC vs. TT | Recessive model: CC vs. TC+TT | Allele: C vs. T | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | P value | I2 (%) | OR (95%CI) | P value | I2 (%) | OR (95%CI) | P value | I2 (%) | OR (95%CI) | P value | I2 (%) | OR (95%CI) | P value | I2 (%) | ||
| All cancers | 11 | 1.10 (1.02, 1.19) | 0.009 | 34 | 1.06 (0.85, 1.33) | 0.6 | 69 | 1.12 (1.00, 1.26) | 0.06 | 59 | 0.99 (0.82, 1.20) | 0.91 | 59 | 1.07 (0.97, 1.19) | 0.18 | 70 |
| Cancer type | ||||||||||||||||
| Digestive tract cancer | 6 | 1.10 (0.99, 1.22) | 0.07 | 42 | 0.90 (0.67, 1.20) | 0.46 | 61 | 1.07 (0.91, 1.26) | 0.39 | 60 | 0.87 (0.74, 1.03) | 0.1 | 47 | 1.01 (0.88, 1.16) | 0.88 | 67 |
| Breast cancer | 2 | 1.04 (0.91, 1.20) | 0.55 | 40 | 0.87 (0.70, 1.08) | 0.21 | 6 | 1.02 (0.84, 1.25) | 0.84 | 55 | 0.86 (0.70, 1.05) | 0.14 | 0 | 0.98 (0.84, 1.15) | 0.83 | 58 |
| Nasopharyngeal carcinoma | 1 | 1.37 (0.96, 1.98) | 0.09 | / | 1.72 (0.99, 2.96) | 0.05 | / | 1.44 (1.02, 2.03) | 0.04 | / | 1.45 (0.87, 2.42) | 0.15 | / | 1.33 (1.04, 1.70) | 0.03 | / |
| Osteosarcoma | 1 | 1.77 (1.03, 3.03) | 0.04 | / | 2.52 (1.25, 5.10) | 0.01 | / | 1.96 (1.19, 3.24) | 0.009 | / | 1.86 (0.98, 3.54) | 0.06 | / | 1.68 (1.19, 2.39) | 0.004 | / |
| Renal cell cancer | 1 | 1.10 (0.88, 1.36) | 0.4 | / | 1.53 (1.07, 2.19) | 0.02 | / | 1.17 (0.95, 1.43) | 0.15 | / | 1.46 (1.04, 2.06) | 0.03 | / | 1.18 (1.01, 1.37) | 0.04 | / |
| Ethnicity | ||||||||||||||||
| Asian | 9 | 1.13 (1.03, 1.24) | 0.007 | 37 | 1.12 (0.84, 1.49) | 0.44 | 72 | 1.16 (1.00, 1.33) | 0.05 | 61 | 1.03 (0.81, 1.31) | 0.83 | 64 | 1.10 (0.97, 1.25) | 0.14 | 72 |
| Caucasian | 1 | 0.97 (0.81, 1.16) | 0.74 | / | 0.81 (0.62, 1.05) | 0.1 | / | 0.93 (0.79, 1.10) | 0.41 | / | 0.82 (0.64, 1.04) | 0.11 | / | 0.92 (0.81, 1.04) | 0.16 | / |
| African American | 1 | 1.17 (0.94, 1.46) | 0.17 | / | 1.03 (0.70, 1.51) | 0.88 | / | 1.14 (0.93, 1.41) | 0.21 | / | 0.96 (0.66, 1.39) | 0.83 | / | 1.08 (0.91, 1.27) | 0.38 | / |
Sensitivity analysis
To assess the stability of the results, sensitivity analysis was performed by omitting 1 study at a time. The omission of any 1 study made no substantial change in the statistical results, which indicated that the results were statistically reliable.
Publication bias
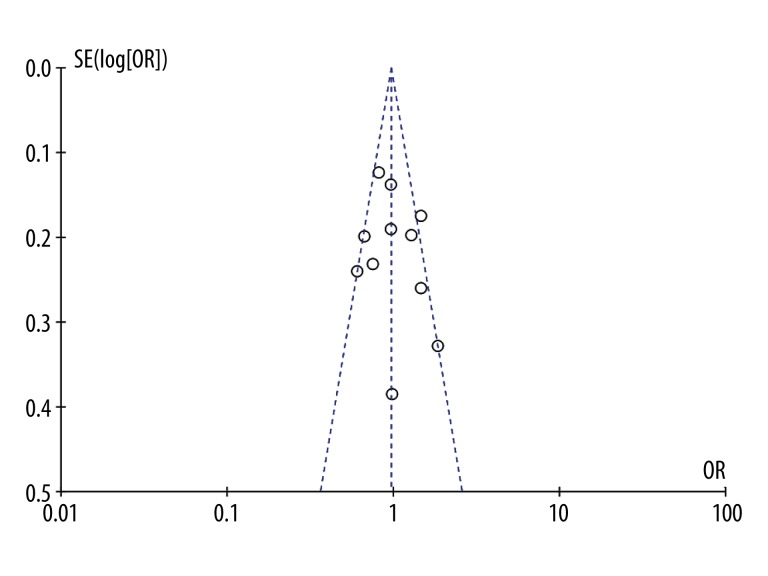
Funnel plots and Begg’s test were used to estimate the publication bias of the articles included in this study. There was no obvious evidence of asymmetry in the shape of funnel plots in the recessive model (CC vs. TC + TT) (Figure 2). In addition, the results of Begg’s test showed no evidence of publication bias (Z=0.16, P=0.876).
Figure 2.
Funnel plots for assessing publication bias in the meta-analysis.
Discussion
The reports from several laboratories demonstrated that miR-34 family members are the direct targets of TP53, and their upregulation induces cancer cell apoptosis and cell-cycle arrest [22–28]. miR-34 has been shown to be associated with carcinogenesis, prognosis, and survival of various cancers, including pancreatic cancer, gastric cancer, ovarian cancer, prostate cancer, and many other cancers [29–32]. Given that the polymorphism of rs4938723, which is capable of creating a predicted GATA-binding site, was found in the promoter region of miR-34b/c, this polymorphism may affect miR-34b/c expression via genetic and epigenetic mechanisms. Thus, several studies investigated the association between the polymorphism of rs4938723 and cancer risk. Three studies have reported that rs4938723 is associated with an increased risk of hepatocellular carcinoma [11,14,16], while Liang et al. presented a meta-analysis and disapproved this conclusion [33]. Furthermore, Gao et al. [10] and Oh et al. [13] have demonstrated that rs4938723 is associated with a reduced risk of colorectal cancer. Therefore, the results of association between rs4938723 and cancer risk are still controversial.
According to the current meta-analysis of 11 case-control studies of 6169 cases and 6337 controls, rs4938723 was found to be associated with increased cancer risk in a codominant model when compared TC with TT in the overall analysis. We further performed stratification analysis based on cancer type and ethnicity. The stratified analysis of cancer type revealed that a significantly elevated risk was associated with C allele and C allele carriers in nasopharyngeal carcinoma. Moreover, rs4938723 polymorphism was associated with osteosarcoma in 4 compared models (TC vs. TT; CC vs. TT; TC+CC vs. TT; and C vs. T). For renal cell cancer, the association analyses of codominant model (CC vs. TT), recessive model (CC vs. TC+TT), and allelic comparison (C vs. T) consistently supported that this polymorphism was statistically related to increased risks. A significant association was not observed in the digestive cancers, including hepatocellular carcinoma, colorectal cancer, and esophageal cancer, within 3164 cases and 3338 controls, although the original studies of these cancers supported a genetic association between rs4938723 and susceptibility to the respective cancer. In addition, the stratified analysis of ethnicity indicated that a statistically significant association was detected in Asians in the codominant model that compared TC with TT. These findings suggest that the polymorphism of miR34b/c rs4938723 may play an important role in carcinogenesis and its development.
However, limitations in our analysis should also be considered. First, as far as we know, the 10 publications, including 11 case-control studies, were relatively limited, especially for nasopharyngeal cancer, osteosarcoma, and renal cell cancer, which were only involved in a single study. Second, the ORs we got were unadjusted, and many other clinical factors such as age and sex in each study might lead to bias. Third, we restricted our included studies to English language. Fourth, our meta-analysis was limited by the quality of the original studies. Fifth, since 9 publications were done in Asians with only 1 study from a Western country, it is necessary to include investigations from other countries.
Conclusions
This meta-analysis investigated the relationship between rs4938723 and cancer risk. Although there are several limitations, our work indicates that the miR-34b/c rs4938723 polymorphism may be associated with risk of cancers in Asians.
Footnotes
Source of support: Self financing
Conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Zhao P, Dai M, Chen W, et al. Cancer trends in China. Jpn J Clin Oncol. 2010;40:281–85. doi: 10.1093/jjco/hyp187. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Dong J, Sui L, Wang Q, et al. MicroRNA-26a inhibits cell proliferation and invasion of cervical cancer cells by targeting protein tyrosine phosphatase type IVA 1. Mol Med Rep. 2014;10(3):1426–32. doi: 10.3892/mmr.2014.2335. [DOI] [PubMed] [Google Scholar]
- 5.Babashah S, Soleimani M. The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur J Cancer. 2011;47:1127–37. doi: 10.1016/j.ejca.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Niu H, Wang K, Zhang A, et al. miR-92a is a critical regulator of the apoptosis pathway in glioblastoma with inverse expression of BCL2L11. Oncol Rep. 2012;28:1771–77. doi: 10.3892/or.2012.1970. [DOI] [PubMed] [Google Scholar]
- 7.Ren Y, Han X, Yu K, et al. microRNA-200c downregulates XIAP expression to suppress proliferation and promote apoptosis of triple-negative breast cancer cells. Mol Med Rep. 2014;10:315–21. doi: 10.3892/mmr.2014.2222. [DOI] [PubMed] [Google Scholar]
- 8.Wang XH, Cai P, Wang MH, et al. microRNA25 promotes osteosarcoma cell proliferation by targeting the cellcycle inhibitor p27. Mol Med Rep. 2014;10:855–59. doi: 10.3892/mmr.2014.2260. [DOI] [PubMed] [Google Scholar]
- 9.Bensen JT, Tse CK, Nyante SJ, et al. Association of germline microRNA SNPs in pre-miRNA flanking region and breast cancer risk and survival: the Carolina Breast Cancer Study. Cancer Causes Control. 2013;24:1099–109. doi: 10.1007/s10552-013-0187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S, Qian J, Cao Q, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with renal cell cancer risk in a Chinese population. Mutagenesis. 2014;29:149–54. doi: 10.1093/mutage/geu001. [DOI] [PubMed] [Google Scholar]
- 11.Gao LB, Li LJ, Pan XM, et al. A genetic variant in the promoter region of miR-34b/c is associated with a reduced risk of colorectal cancer. Biol Chem. 2013;394:415–20. doi: 10.1515/hsz-2012-0297. [DOI] [PubMed] [Google Scholar]
- 12.Han Y, Pu R, Han X, et al. Associations of pri-miR-34b/c and pre-miR-196a2 polymorphisms and their multiplicative interactions with hepatitis B virus mutations with hepatocellular carcinoma risk. PLoS One. 2013;8:e58564. doi: 10.1371/journal.pone.0058564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Wu J, Sima X, et al. Interactions of miR-34b/c and TP-53 polymorphisms on the risk of nasopharyngeal carcinoma. Tumour Biol. 2013;34:1919–23. doi: 10.1007/s13277-013-0736-9. [DOI] [PubMed] [Google Scholar]
- 14.Oh J, Kim JW, Lee BE, et al. Polymorphisms of the pri-miR-34b/c promoter and TP53 codon 72 are associated with risk of colorectal cancer. Oncol Rep. 2014;31:995–1002. doi: 10.3892/or.2013.2926. [DOI] [PubMed] [Google Scholar]
- 15.Son MS, Jang MJ, Jeon YJ, et al. Promoter polymorphisms of pri-miR-34b/c are associated with hepatocellular carcinoma. Gene. 2013;524:156–60. doi: 10.1016/j.gene.2013.04.042. [DOI] [PubMed] [Google Scholar]
- 16.Tian Q, Jia J, Ling S, et al. A causal role for circulating miR-34b in osteosarcoma. Eur J Surg Oncol. 2014;40:67–72. doi: 10.1016/j.ejso.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Liu L, Liu J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer. 2011;128:412–17. doi: 10.1002/ijc.25342. [DOI] [PubMed] [Google Scholar]
- 18.Yin J, Wang X, Zheng L, et al. Hsa-miR-34b/c rs4938723 T>C and hsa-miR-423 rs6505162 C>A polymorphisms are associated with the risk of esophageal cancer in a Chinese population. PLoS One. 2013;8:e80570. doi: 10.1371/journal.pone.0080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–26. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 21.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101. [PubMed] [Google Scholar]
- 22.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–34. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corney DC, Flesken-Nikitin A, Godwin AK, et al. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–38. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 25.Wong MY, Yu Y, Walsh WR, et al. microRNA-34 family and treatment of cancers with mutant or wild-type p53 (Review) Int J Oncol. 2011;38:1189–95. doi: 10.3892/ijo.2011.970. [DOI] [PubMed] [Google Scholar]
- 26.Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka N, Toyooka S, Soh J, et al. Downregulation of microRNA-34 induces cell proliferation and invasion of human mesothelial cells. Oncol Rep. 2013;29:2169–74. doi: 10.3892/or.2013.2351. [DOI] [PubMed] [Google Scholar]
- 28.Tamura M, Uyama M, Sugiyama Y, et al. Canonical Wnt signaling activates miR-34 expression during osteoblastic differentiation. Mol Med Rep. 2013;8:1807–11. doi: 10.3892/mmr.2013.1713. [DOI] [PubMed] [Google Scholar]
- 29.Ji Q, Hao X, Zhang M, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji Q, Hao X, Meng Y, et al. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corney DC, Hwang CI, Matoso A, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–28. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng CY, Hwang CI, Corney DC, et al. miR-34 cooperates with p53 in suppression of prostate cancer by joint regulation of stem cell compartment. Cell Rep. 2014;6:1000–7. doi: 10.1016/j.celrep.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang TJ, Liu HJ, Zhao XQ, et al. Lack of association of MiR-34b/c polymorphism (rs4938723) with hepatocellular carcinoma: a meta-analysis. PLoS One. 2013;8:e68588. doi: 10.1371/journal.pone.0068588. [DOI] [PMC free article] [PubMed] [Google Scholar]