A quantitative proteomic approach enables unambiguous protein localization to the plant Golgi apparatus and rank ordering of the Golgi resident proteins by abundance, providing a unique view on the organization of this organelle.
Abstract
The proteomic composition of the Arabidopsis (Arabidopsis thaliana) Golgi apparatus is currently reasonably well documented; however, little is known about the relative abundances between different proteins within this compartment. Accurate quantitative information of Golgi resident proteins is of great importance: it facilitates a better understanding of the biochemical processes that take place within this organelle, especially those of different polysaccharide synthesis pathways. Golgi resident proteins are challenging to quantify because the abundance of this organelle is relatively low within the cell. In this study, an organelle fractionation approach targeting the Golgi apparatus was combined with a label-free quantitative mass spectrometry (data-independent acquisition method using ion mobility separation known as LC-IMS-MSE [or HDMSE]) to simultaneously localize proteins to the Golgi apparatus and assess their relative quantity. In total, 102 Golgi-localized proteins were quantified. These data show that organelle fractionation in conjunction with label-free quantitative mass spectrometry is a powerful and relatively simple tool to access protein organelle localization and their relative abundances. The findings presented open a unique view on the organization of the plant Golgi apparatus, leading toward unique hypotheses centered on the biochemical processes of this organelle.
The plant Golgi apparatus plays an important role in protein and lipid glycosylation and sorting as well as biosynthesis of large amounts of extracellular polysaccharides. It contains a large and diverse set of glycosyltransferases and other enzymes that are required for the synthesis and modification of these polysaccharides (Parsons et al., 2012b; Oikawa et al., 2013). The protein composition of this organelle has been the focus of a number of studies; however, these studies largely report a catalog of Golgi-localized proteins, and to date, there are no comprehensive data on the relative abundance of the different protein constituents of the Golgi apparatus (Dunkley et al., 2004, 2006; Sadowski et al., 2008; Nikolovski et al., 2012; Groen et al., 2014). The quantification of the plant Golgi proteome has been considered challenging, because this organelle is proportionally of low abundance in the cell; therefore, its constituent proteins are rarely identified in conventional proteomics experiments. Investigation of such low-abundance proteins generally requires sample fractionation on the organelle, protein, or peptide level (Stasyk and Huber, 2004; Haynes and Roberts, 2007; Di Palma et al., 2012).
Here, an organelle fractionation approach in conjunction with label-free quantitative proteomic analysis was used to assess the localization and relative abundance of proteins within the plant Golgi apparatus. Label-free quantification is an increasingly popular alternative to isotopic tagging quantitative methods; it does not require labeling reagents and can be applied to an unlimited number of samples (Neilson et al., 2011; Evans et al., 2012). This is particularly appealing within plant proteomics, because the most conventional labeling strategy, Stable Isotope Labeling by Amino Acids in Cell Culture, is not easily suited for quantitative plant proteomic studies. The average labeling efficiency achieved using exogenous amino acid supply to Arabidopsis (Arabidopsis thaliana) cell cultures was found to be only 70% to 80% (Gruhler et al., 2005). Quantitative strategies with 15N metabolic labeling have been described for plant proteome analysis; however, care should be taken to ensure complete 15N incorporation, because even small amounts of 14N in the labeled sample can have significant detrimental effects on the number of peptide identifications (Nelson et al., 2007; Guo and Li, 2011; Arsova et al., 2012).
In all label-free methods, samples under comparison are analyzed during separate mass spectrometry (MS) experiments (Neilson et al., 2011). The information from identified peptides is then used for relative and/or absolute quantification. The simplest label-free method involves taking the number of spectra acquired and assigned to peptides from the same protein as a measure of abundance (Ishihama et al., 2005). In an alternative approach, ion current recorded for a peptide ion is used as a measure of its abundance. The assumption is made that ion intensity is proportional to peptide amount in the sample analyzed, which holds true for nanoflow and microflow liquid chromatography (LC) systems (Levin et al., 2011; Christianson et al., 2013). Comparing peptide ion current between samples is, thus, widely used for relative quantification (Silva et al., 2005). To allow such comparison, a peptide must be identified across all samples under investigation, which is often challenging in LC-MS experiments given the highly complex nature of proteomics samples that contain tens of thousands of different peptides (Michalski et al., 2011). Hence, most relative ion intensity-based label-free approaches usually involve a step of identification transfer (Pasa-Tolíc et al., 2004). This involves matching ions from different acquisitions (in one of which, the ion has not been identified and is assigned the sequence from its matching pair in the other acquisition).
Additionally, label-free proteomics can be used for absolute quantification (i.e. to estimate abundance of different proteins relative to each other within a given sample). Several different approaches have been suggested on how to convert peptide intensities to protein amounts (for comparison, see Wilhelm et al., 2014). One of the first such methods was Top-3 described by Silva et al. (2006b), who made a notable and unexpected observation, stating that the average MS signal response for the three most abundant peptides per 1 mol of protein is constant within a coefficient of variation of less than 10% (Silva et al., 2006b).
In all these approaches, the peptide ion current is typically computed as the area under the curve of the chromatographic elution profile that is reconstituted from separate MS1 survey scans in which intact precursors are recorded. Determining a chromatographic profile accurately requires that the MS1 scans are performed at optimal frequency (Lange et al., 2008) and for optimal duration to record the MS1 signal at a high signal-to-noise ratio. In typical data-dependent acquisitions, however, the mass spectrometer oscillates between MS1 survey scans recording the mass/charge (m/z) for precursor peptide ions and then, a series of MS2 scans fragmenting one peptide ion precursor at a time, producing fragmentation spectra necessary for identification (Sadygov et al., 2004). As a result, the duration and frequency of MS2 scans determine the identification rate in data-dependent acquisition experiments but compromise time spent in MS1 required for accurate area under the curve quantification. Several groups have suggested data-independent acquisition, in which individual peptide ions are not selected for fragmentation but rather, groups of peptides of similar m/z are fragmented together. The exact number of cofragmented precursors depends on the speed and sensitivity of instrument configuration (for review, see Law and Lim, 2013). The simplest approach involves alternating between low-energy and high-energy scans of equal duration; low-energy scans record precursor peptide ions, whereas in high-energy scans, all precursors entering the mass spectrometer are cofragmented, and their fragments are recorded simultaneously. The method was called MSE for Waters qTOF Mass Spectrometers (Geromanos et al., 2009) or all-ion fragmentation for Thermo Orbitrap Mass Spectrometers (Geiger et al., 2010). The analysis required downstream of this type of data acquisition is challenging given that the information of fragment origin (i.e. from what precursor peptide ion fragment was generated) is lost completely and that the high number of coeluting peptides is expected to create highly overlapping fragment spectra on fragmentation. To address this problem, Hoaglund-Hyzer and Clemmer (2001) have suggested fractionating peptides by ion mobility separation before fragmentation and MS and assigning fragments to precursors based on similarity of both chromatographic and mobility profiles (Hoaglund-Hyzer and Clemmer, 2001). The method was termed parallel fragmentation, and since that time, it has been commercialized by Waters as IMS-MSE or HDMSE (Shliaha et al., 2013).
To date, the application of label-free quantitative proteomics to plant biology has been very limited. Recently, Helm et al. (2014) applied the LC-IMS-MSE with Top-3 quantification to quantify the Arabidopsis chloroplast stroma proteome, allowing quantitative modeling of chloroplast metabolism. Two other works used the LC-MSE method to assess the quantitative changes of cytosolic ribosomal proteins in response to Suc feeding and the extracellular proteome in response to salicylic acid (Cheng et al., 2009; Hummel et al., 2012).
A number of proteomics approaches have been described to assess protein localization on a large scale (for review, see Gatto et al., 2010). Purification approaches attempt to isolate organelles to high levels of purity and subsequently identify and quantify proteins using LC-MS; however, such attempts yield limiting success and high false discovery rates (Andersen et al., 2002; Parsons et al., 2012a). A known limitation of this technique is the inability to completely isolate an organelle of interest, which combined with high proteome dynamic range, can result in some more abundant contaminants being identified and quantified at higher amounts than the target organelle residents. Moreover, even if a target organelle could be isolated to a certain degree of purity, it would still be impossible to deconvolute organelle residents from transient proteins that traffic through the target organelle. This becomes especially challenging for the organelles of the secretory pathway. To address these challenges, several groups applied fractionation of all organelles by gradient centrifugation and subsequent protein quantification by LC-MS. This produces distributions across the gradient for all quantified proteins, which are then used to assign organelle localization based on the specific distributions of organelle marker proteins. This effectively solves the problem of organelle contamination and protein trafficking, because a protein is expected to have a distribution characteristic of its organelle of residence, even if it is identified in all fractions, including those enriched in other organelles. Current variations of this method differ mostly by the LC-MS strategy used for quantification; for example, spectral counting was applied for protein-correlating profiles (Andersen et al., 2003), isobaric mass tagging (Nikolovski et al., 2012) and isotope-coded affinity tagging (Dunkley et al., 2004) were applied for localization of organelle proteins by isotope tagging (LOPIT), and Stable Isotope Labeling by Amino Acids in Cell Culture was applied for nucleolus/nucleus/cytosolic fractionation (Boisvert and Lamond, 2010).
Here, a label-free LC-IMS-MSE method was used for the analysis of density ultracentrifugation fractions enriched for the Golgi apparatus. First, we use relative label-free quantification involving identification transfer using the previously published synapter algorithm (Bond et al., 2013) to assess distributions of Golgi-localized proteins across the density gradient. These distributions are significantly different from those of residents of other organelles, which results in unambiguous protein assignment to the Golgi apparatus by multivariate data analysis. Second, the Top-3 absolute quantification method as implemented in Protein Lynx Global Server (PLGS) was used to rank order the Golgi-localized proteins by abundance in the fraction most enriched for Golgi apparatus. In conclusion, we present the analysis of protein distribution and abundances of the Golgi apparatus-enriched portion of the ultracentrifugation density gradient, allowing for simultaneous protein quantification and localization and leading to the assessment of relative abundances of 102 Golgi-localized proteins.
RESULTS
The strategy for quantification of Golgi apparatus proteins is described in Figure 1. Density gradient centrifugation allowed partial separation of the Golgi apparatus from other abundant organelles. The organelle fractions were subjected to data-independent acquisition LC-IMS-MSE. Individual protein abundance distributions across the gradient were used to determine the Golgi apparatus sedimentation profile. The fraction most enriched for the Golgi apparatus was then used for the analysis of protein abundances by the Top-3 method (Supplemental Table S1).
Figure 1.
Outline of the experimental design and the data processing workflow. Organelle separation was performed by density gradient ultracentrifugation. Two biological replicates were acquired. The Golgi apparatus was resolved in the upper part of the gradient, and the corresponding 10 fractions were used for LC-IMS-MSE data acquisition. The data from all acquisitions were used to reconstruct the protein abundance distribution along the density gradient. The profile data were normalized and used for classification of Golgi proteins versus non-Golgi proteins. The Top-3 quantities from the most enriched fraction for the Golgi apparatus were used to rank order the Golgi-localized proteins.
Label-Free Data-Independent Acquisition Allows Accurate Assignment of Golgi-Localized Proteins
Quantitative determination of gradient fractionation profiles of organelle proteins can be used to assign subcellular localization (Dunkley et al., 2004). If the measurement of protein abundance in each fraction is linear and quantitative, proteins in different organelles can be distinguished based on their different sedimentation profiles. To assess whether LC-IMS-MSE quantification could be used for localization of Golgi apparatus proteins, label-free quantitative proteomic analysis was performed of organelle fractionations targeted at Golgi enrichment in two biological replicates of Arabidopsis callus. The replication allowed estimation of the quality of the analysis. Stringent filtering criteria were applied for protein localization analysis: a protein needed to be quantified in eight of 10 analyzed fractions in both gradients with at least two fully tryptic, unmodified proteotypic peptides. Using these criteria, sedimentation profiles for 1,385 proteins were constructed (Supplemental Table S2).
To investigate the sedimentation profiles of Golgi-localized proteins in the gradients, a protein marker set was generated composed of proteins for which subcellular localization has been shown by prior cell biological (nonproteomic) studies and high homology to well-characterized proteins (Supplemental Table S3). The marker proteins were divided into two groups: Golgi (26 for Golgi proteins) and non-Golgi (226 proteins localized elsewhere). A principal component analysis plot of sedimentation profiles of both experiments combined, shown in Supplemental Figure S1A, indicates that these two groups of proteins can be distinguished based on their fractionation profiles. The sedimentation profiles for 26 Golgi marker proteins in both replicate experiments are shown in Figure 2B. These distributions are consistent with the distribution of the Galactosyltransferase-Like6 (Gtl6)/At2g22900 Golgi marker protein as measured by western blotting (Fig. 2A).
Figure 2.
Golgi apparatus protein abundance distribution profiles along the density gradients. A, Western-blot analysis of the Golgi marker protein Gtl6-myc. B, Relative protein abundance distribution of Golgi marker proteins along the upper 10 fractions of the density gradient as measured by LC-IMS-MSE. C, Relative protein abundance distribution of the Golgi-assigned proteins along the upper 10 fractions of the density gradient as measured by LC-IMS-MSE.
The marker protein distribution profiles were used to classify unlabeled proteins to either Golgi or non-Golgi classes using a support vector machine (SVM) classifier as implemented in the pRoloc software (Gatto and Lilley, 2012; Gatto et al., 2014). The optimal SVM parameters (σ = 0.01, cost = 0.0625), obtained as described above, yield a macro-F1 score of 0.94 for the marker protein set. Of 1,133 unlabeled proteins that were subjected to the SVM classifier, only proteins with an SVM score greater than or equal to 0.75 were considered reliably classified. At this threshold, 35 proteins were classified as Golgi residents (Supplemental Fig. S1B and Supplemental Table S4). The sedimentation profiles for the SVM-classified Golgi proteins in both replicate experiments are consistent with the sedimentation profiles of Golgi marker proteins (Fig. 2C). Of these 35 proteins classified as Golgi by LC-IMS-MSE, 28 proteins have been localized by LOPIT experiments reported previously (Nikolovski et al., 2012). Almost all of these proteins (25 of 28) were localized by LOPIT to the Golgi apparatus, showing high consistency between the two methods. Of three inconsistent assignments, one protein assigned in LOPIT to the endoplasmic reticulum (vacuolar sorting receptor1 [VSR1] /At3g52850) seems to be primarily localized to post-Golgi compartments based on recent literature, and two proteins are plastidic and thus, may be contaminants in this LC-IMS-MSE Golgi-assigned list (Ahmed et al., 2000); five proteins from this list of 35 SVM-assigned proteins are unique Golgi-assigned proteins not previously reported by a high-confidence assignment method (Supplemental Table S4).
Assessment of the Top-3 Quantification Accuracy
The Top-3 method of quantification is a method of converting peptide intensity to protein amount. This quantification method is based on a premise that the sum intensity of the three most intense peptides from any protein has a linear correlation to the given protein concentration, regardless of the protein sequence or size (Silva et al., 2006b). The method was initially applied on qTOF instruments and is a standard form of analysis of MSE acquisitions. This quantification method was later implemented on other platforms (Grossmann et al., 2010). The method has since been applied to assess protein abundances in a variety of systems, including Escherichia coli (Silva et al., 2006a), budding yeast (Saccharomyces cerevisiae; Carroll et al., 2011), human cell lines (Distler et al., 2014), and plants (Blackburn et al., 2010; Helm et al., 2014). To assess its accuracy in our experiments, we analyzed the commercial protein standards Universal Protein Standard1 (UPS1; 48 proteins at equimolar abundances) and UPS2 (48 proteins at six different concentrations). Supplemental Figure S2 shows protein quantification for the two standards. In general, the equimolar proteins in UPS1 tend to produce similar Top-3 values approximately within a factor of 3, and the method enables the inference of the correct abundance group to which the protein belongs in the UPS2 standard (Supplemental Fig. S2).
Quantification of Proteins
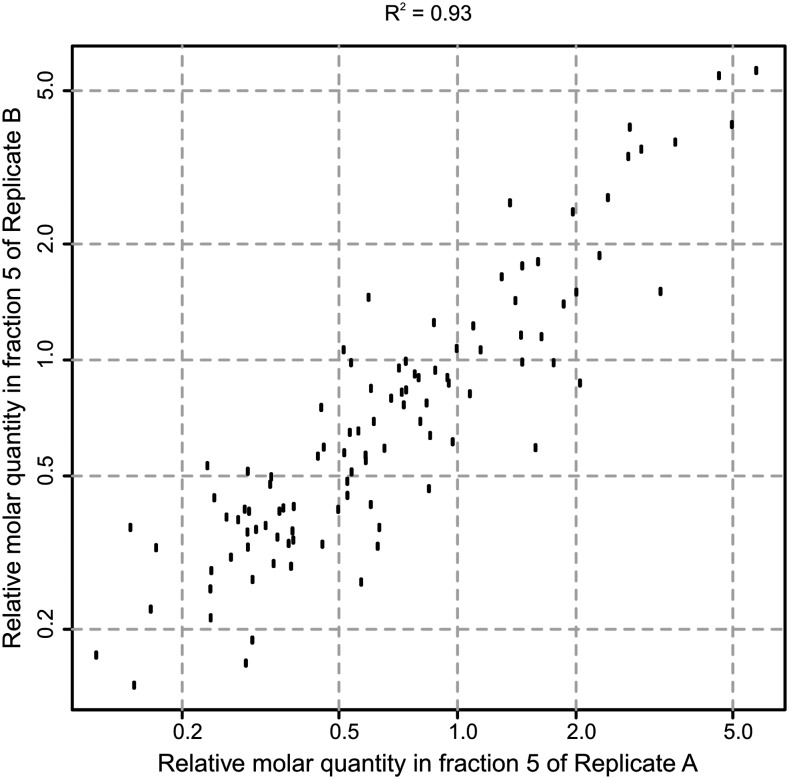
Given that accuracy and precision of quantification in proteomics experiments using the Top-3 method are known to correlate positively with protein abundance, for Golgi protein quantification, LC-IMS-MSE data were used from the most Golgi-enriched fraction. The data on organelle sedimentation presented above identified fraction number five as most enriched in Golgi proteins (Fig. 2). From this fraction, using the Top-3 method, the relative molar quantities of 102 Golgi proteins were determined of 1,266 quantified proteins from the most Golgi-enriched fraction (Fig. 3; Supplemental Table S5). These 102 Golgi proteins consisted of 37 proteins from the known Golgi proteins marker set, 33 additional proteins localized to the Golgi by SVM in this study, and 32 proteins localized to the Golgi in previous LOPIT experiments (Nikolovski et al., 2012). The Top-3 LC-IMS-MSE quantification analysis was carried out independently on two biological replicates of membranes and showed good reproducibility: the R2 was 0.93, and the median coefficient of variance of quantification was 13% (Fig. 4).
Figure 3.
Rank order distribution of 102 Golgi-localized proteins based on the abundance from Top-3 quantification of the fraction most enriched for the Golgi apparatus. The average value of the two biological replicates is plotted. The ranking of protein abundance forms an S-shaped curve on the logarithmic scale. Protein names referred to in “Discussion” are labeled. The nomenclature of the putative GTs is as in Nikolovski et al. (2012). COBRA is a family of extracellular glycosyl-phosphatidyl inositol-anchored proteins. The data are available in Supplemental Data S5.
Figure 4.
Correlation of Golgi protein abundance (measured by Top-3) between two biological replicates of the fraction most enriched for the Golgi apparatus.
The 102 Golgi proteins constitute approximately 7.7% mol of 1,266 quantified proteins in the most Golgi-enriched gradient fraction (Supplemental Table S6). The Golgi marker protein overexpressed in the cell cultures, Gtl6/At2g22900, was the most abundant protein at 5.7% mol of the quantified Golgi proteome. Notably, when the unfractionated whole-callus cell lysate was analyzed by LC-IMS-MSE, only four Golgi proteins (of 960 quantified proteins; Supplemental Table S7) were identified and quantified, and one of which was the Gtl6 marker protein. This shows that the Golgi is a very low-abundance organelle and also indicates that the organelle fractionation strategy provides significant enrichment of the Golgi apparatus.
DISCUSSION
We have developed and applied a label-free proteomic method for analysis of the relative abundance of over 100 Arabidopsis Golgi proteins. The method was also able to classify proteins as Golgi localized based on their subcellular fractionation profile. The assignment of Golgi localization is based on the label-free measurement of protein abundance distributions in the density gradient fractions. It is important to note that protein localizations inferred in this study are very consistent with those reported previously by LOPIT (Nikolovski et al., 2012). The fact that two very different MS techniques were performed on two different platforms not only validates the methods quantification accuracy but also, creates a high-confidence Golgi proteins dataset.
The Quantified Golgi Proteome
Quantification of Golgi proteins is especially challenging, because they are of low abundance. Indeed, only 0.5% of proteins identified during the LC-IMS-MSE analysis of whole-callus cell lysate were Golgi proteins (four Golgi resident proteins of 960 proteins in total; Supplemental Table S7). Even in the fraction most enriched for the Golgi, the quantified Golgi apparatus proteome contributed to only 7.7% of the total protein quantity (Supplemental Table S7). The relative abundance of 102 Golgi proteins ranged over two orders of magnitude (Supplemental Table S5). They are likely the most abundant Golgi proteins and represent just 10% to 20% of all Golgi proteins based on our unpublished LOPIT data and corresponding homologs deduced from sequence analysis. It can be inferred that the dynamic range of the Golgi proteome is higher than two orders of magnitude. This wide range may reflect the different protein quantities required for protein trafficking, metabolite transport, and flux through various high-abundance and low-abundance polysaccharide synthesis pathways.
Despite using a data-independent approach, we could not quantify all Golgi residents that are expected to be expressed in Arabidopsis callus. Quantifying the entire Golgi proteome will require significantly more sensitive and higher peak capacity LC-MS instrumentation or development of a targeted proteomics strategy. Although LC-MS instruments have shown truly remarkable improvements in sensitivity over the recent years, it is nonetheless challenging to quantify a proteome comprehensively because of its large dynamic range of greater than six orders of magnitude (Huh et al., 2003; Hebert et al., 2014). Several recent studies suggest that targeted proteomics will, in the near future, allow deeper coverage than any untargeted method (Gillet et al., 2012). Targeted proteomics also allow experimental designs that use isotopically labeled internal standard peptides, providing accurate quantification information. For example, a comparison of the accuracy of Top-3 label-free quantification and absolute quantification by a targeted method using isotopically labeled peptides for quantification of the yeast glycolytic pathway showed lower limits of detection in experiments using a targeted method (Carroll et al., 2011). Despite the fact that both targeted and untargeted methods having their own biases, they showed general agreement, but the studies suggest that using isotopic standards and targeted workflows should provide more accurate quantitative information. To comprehensively quantify the full Golgi apparatus enzyme complement with the highest possible accuracy, future studies will benefit from targeted proteomics incorporating isotopically labeled peptides as internal standards.
The Endomembrane Protein70 Family Members Are the Most Abundant Golgi Proteins
The Endomembrane Protein70 (EMP70) family members contribute to almost one-quarter of the quantified Golgi proteome and represent one-half of the top 10 most abundant proteins (Fig. 3). The EMP70 family is structurally highly conserved, being present in many eukaryotes, with limited functional characterization. The family contains nine potential transmembrane domains; hence, the characteristic protein domain for this family is termed nonaspanin or Transmembrane9. Previously, several EMP70 homologs have been detected by proteomic methods in the Golgi (Nikolovski et al., 2012), but their very high abundance was unexpected. Hegelund et al. (2010) investigated the physiological function of two Arabidopsis EMP70 proteins by heterologous expression in yeast. A yeast mutant line lacking all endogenous EMP70 homologs showed altered metal homeostasis with a reduction in the cellular copper content. Heterologous expression in yeast of AtTMN7 (Transmembrane Nine7, At3g13772) affected copper homeostasis similar to the overexpression of the yeast TMN1. It was concluded that EMP70 homologs from Arabidopsis have the ability to affect the intracellular copper balance (Hegelund et al., 2010). However, the observed high abundance of putative metal transporters in the Golgi is surprising. This observation may suggest a Golgi structural or membrane-trafficking function rather than an enzymatic role.
Quantities of the Cell Wall Synthesis Machineries in the Golgi Apparatus
The relative abundance of enzymes in different polysaccharide synthesis pathways gives an insight into the main activities of this organelle. The Arabidopsis liquid-grown callus is a tissue that synthesizes a typical primary cell wall. Its cell wall is composed of cellulose, abundant pectic components, such as homogalacturonan and rhamnogalacturonan I, the hemicellulose xyloglucan, and the cell wall extensin proteins (Goubet et al., 2002; Handford et al., 2003; Manfield et al., 2004; Barton et al., 2006; P. Dupree, unpublished data). Additionally, there are minor amounts of type II arabinogalactans, glucuronoarabinoxylan, rhamnogalacturonan II, and glucomannan. As expected, we identified and quantified several enzymes involved in the synthesis of the main primary cell wall components.
Cellulose Synthase A1 (CesA1), CesA3, and COBRA-Like7 proteins involved in primary cell wall cellulose synthesis were quantified. Each of these proteins was found with similar abundances within the experimental error of this technique (ranks 65, 75, and 73, respectively), which is consistent with the theory that the CesA proteins form a stoichiometric complex (Taylor et al., 2006). Three distinct CesA isoforms form the primary wall cellulose synthesis complex. A third CesA subunit was probably not quantified, because CesA6, CesA5, and CesA2 are expected to be present as the third subunit at lower abundances (Desprez et al., 2007).
Primary cell wall pectin is abundant and methylesterified in Arabidopsis callus (Barton et al., 2006). Consistently, many members of the Domain of Unknown Function248 putative pectin methyltransferase family were detected and quantified (14 of a total of 35 members) as well as cotton (Gossypium hirsutum) Golgi-related2 (At3g49720) belonging to another putative homogalacturonan methyltransferase family (Mouille et al., 2007; Held et al., 2011; Miao et al., 2011). In sum, they constitute 10% of the quantified Golgi proteome. However, no galacturonosyltransferases involved in homogalacturonan biosynthesis (belonging to the glycosyltransferase8 (GT8) family) were quantified. This may indicate that the galacturonosyltransferase activity is provided by less abundant but highly active machinery.
The extensin glycosylation pathway of the Golgi apparatus has been well described (Velasquez et al., 2011). We found that most of the enzymes involved in this type of O-glycosylation are abundant in the Golgi apparatus. Three prolyl 4-hydroxylase members (ranks 46, 47, and 89) as well as Hyp arabinosyltransferase1 (Ogawa-Ohnishi et al., 2013; rank 95) were quantified. Two arabinosyltransferases that extend this glycan (members of the GT77 family Reduced Residual Arabinose3 [rank 30] and Xyloglucanase113 [rank 42]) were also found. However, no enzymes known to be involved in arabinogalactan protein glycosylation were detected, suggesting that this pathway is of lower abundance.
Xyloglucan is the main hemicellulose of primary cell walls. We quantified two proteins, Xyloglucan XylosylTransferase1 (XXT1) and XXT2, that are involved in xyloglucan synthesis, and XXT2 showed slightly higher abundance (rank 66) than XXT1 (rank 101; Cavalier et al., 2008). However, no Cellulose Synthase-Like C (CslC) proteins that synthesize the xyloglucan backbone were quantified. Because glucomannan is a minor hemicellulose of the primary cell wall, it is perhaps surprising that proteins implicated in glucomannan synthesis were quantified by this method. CslA2 (rank 91) is a GT2 member that is required for glucomannan backbone synthesis (Liepman et al., 2005; Goubet et al., 2009). Mannan Synthesis-Related1 (MSR1; rank 58) and MSR2 (rank 19) are putative glycosyltransferases of the GT65R family involved in glucomannan production, but their exact biochemical role is unclear (Wang et al., 2012). The high MSR abundance may suggest additional function for these proteins in the Golgi apart from glucomannan synthesis.
High Abundance of the Nucleotide Sugar Conversion Enzymes
The nucleotide sugar interconversion enzymes, especially the members of UDP-Xyl synthase (UXS) and UDP-GlcA epimerase (GAE) protein families, contributed to up to 18% of the total quantified Golgi protein mass. Among the quantified Golgi proteins were UXS2 (rank 10), UXS4 (rank 15), UXS1 (rank 24), GAE1 (rank 51), and GAE6 (rank 102). One uncharacterized putative epimerase was also quantified (RmlC-R/At3G56820; rank 76). Additionally, a single apyrase, Apyrase1 (rank 98), involved in nucleoside diphosphate recycling in the Golgi lumen was also observed.
Only a single nucleotide sugar transporter was quantified in this study: At1G06890 (rank 55) may be involved in the transport of the nucleotide sugars with the highest metabolic flux, such as UDP-Gal and UDP-Glc. It is likely that most of the transporters required for polysaccharide synthesis were below the quantification threshold in this work, implying low abundance for these transporters.
Abundance of the Trans-Golgi Network Proteins
The Trans-Golgi Network was not separated from the Golgi apparatus, and we were able to assess the relative abundance of some well-established Trans-Golgi Network proteins. We observed high abundance of Vacuolar Sorting Receptor3 (VSR3; rank 9) and VSR4 (rank 12) as well as ECHIDNA, RAN1 (copper-transporting ATPase), Vacuolar Proton ATPase-a1, Yip1 integral membrane domain-containing protein, and Syntaxin43. VSR3 and VSR4 are two of the main isoforms that participate in vacuolar sorting in Arabidopsis (Zouhar et al., 2010).
CONCLUSION
To date, there have been no data that describe the distribution of abundances of proteins within the plant Golgi apparatus. Here, we give the first report, to our knowledge, of rank ordering of the proteins that are of high abundance within this organelle. Our results provide unique insight into the molecular organization of this organelle, illustrate the complexity of the biochemical processes present, and enable unique hypotheses centered on the biochemical and cell biology aspects of this organelle.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) Columbia-0 ecotype liquid-grown callus line was used for the preparation of the Golgi-enriched membranes and quantitative proteomic analysis. The transgenic line 35S::GTL6-myc was used, expressing a Golgi apparatus marker protein (Gtl6/At2g29900) fused with a c-Myc tag. The conditions of growth were as described in Prime et al., 2000.
Separation of Membrane Organelles, Protein Extraction, and In-Solution Digestion
Organelle separation was achieved by iodixanol density ultracentrifugation of the homogenized Arabidopsis liquid-grown callus line as described previously (Sadowski et al., 2006). In brief, the plant callus material was harvested 4 d after the culture medium was refreshed, and all subsequent steps were performed at 4°C. Approximately 60 g of callus tissue was homogenized in an equal volume of homogenization buffer (250 mm Suc, 25 mm HEPES, pH 7.5, 1 mm EDTA, 1 mm dithiothreitol, and Complete Protease Inhibitor Cocktail Tablets; Roche). The homogenate was centrifuged for 30 min at 2,200g two times to remove unbroken cells, cell wall components, and nuclei. The supernatant was centrifuged for 2 h at 4°C in an SW28 rotor at 100,000g onto a 6-mL cushion containing 18% (v/v) iodixanol in homogenization buffer. Concentrated membranes from the interface were collected with a syringe, adjusted to 12% (v/v) iodixanol, and spun for 4 h at 4°C in a VTi65.1 rotor in OptiSeal 11.2-mL tubes at 350,000g to separate organelles on a self-forming iodixanol density gradient; 0.5-mL fractions were collected from the upper part of the gradient, with fractions 1 to 10 enriched for the Golgi apparatus that was used in this study. Two independent biological replicates (named replicates A and B) were performed from separate callus cultures. The protein samples were precipitated in 10% (w/v) TCA, and the pellets were solubilized in 50 mm ammonium bicarbonate and 1% (w/v) RapiGest SF (Waters). After protein extraction from the density gradient fractions, protein quantities of each fraction were determined by the BCA assay (Thermo Scientific), and the sample volumes used in subsequent analysis were adjusted accordingly. Immunoblot analyses were used to study the sedimentation profile of the Golgi apparatus along the density gradient by tracking the abundance of the Gtl6-myc protein with a rabbit polyclonal anti-c-Myc antibody A-14 (Santa Cruz Biotech). For each sample, the proteins were reduced with 5 mm dithiothreitol for 1 h at 37°C, alkylated with 15 mm iodoacetamide for 1 h at room temperature, and digested with trypsin (porcine sequencing grade modified trypsin; Promega) by applying a 1:20 (w/w) trypsin:protein ratio and incubating at 37°C overnight. Concentrated HCl was added to the digestion solution until pH 2 was reached to hydrolyze the acid-labile detergent RapiGest SF. The solutions were centrifuged (20,000g for 15 min) to pellet-insoluble material before loading of the samples on the LC. The UPS1 and UPS2 protein standards (Waters), consisting of 48 human proteins in equimolar (for UPS1) and a dynamic range of six concentrations ranging from 500 attomol to 50 pmol (for UPS2), were used to assess the accuracy of the Top-3 quantification. The protein sample was diluted in 0.1% (w/v) RapiGest SF and digested with trypsin as described above.
Nano-LC-MS Analysis
Approximately 250 ng of peptides from 10 fractions of the upper part of the density gradient (Golgi-enriched part of the gradient) from both biological replicates was separated by the NanoACQUITY UPLC System (Waters). Loading was kept low to avoid detector the saturation issues described previously. The LC aqueous mobile phase contained 0.1% (v/v) formic acid in water, and the organic mobile phase contained 0.1% (v/v) formic acid in 100% (v/v) acetonitrile. The samples were injected on a fused silica (Symmetry C18 5 μm, 180 μm × 20 mm) trapping column (Waters) and trapped for 5 min at a 5 μL min−1 flow rate of aqueous mobile phase. The separation was performed on a T3 1.8 μm, 75-μm × 250-mm column (Waters) at 300 nL min−1 flow rates using a 90-min linear gradient elution (from 3%–35% [v/v] organic mobile phase). The column was washed with 80% (v/v) organic mobile phase for 5 min and reequilibrated with 3% (v/v) organic mobile phase for 15 min. The column temperature was maintained at 40°C.
Peptides fractionated on the nanoACQUITY were then analyzed in line with a SYNAPT G2 Hybrid IMS-MS System (Waters). The data reported were acquired in IMS-MSE mode with low-energy and high-energy scans of 900-ms duration each. During high-energy scans, the collision energy was linearly ramped in the Transfer region of TriWave from 21 to 44 V. Emitters were manufactured by etching a fused silica line with hydrofluoric acid as described by Kelly et al. (2006). To allow postacquisition lock mass correction, [Glu-1]-fibrinopeptide B (500 fmol μL−1) was infused through the lockspray ion source at a flow rate of 500 nL min−1 as an external reference compound (lock mass compound) through an auxiliary pump and acquired one time every 30 s for a 1-s period. The mass correction was applied to the spectra postacquisition. The full list of instrument settings is available in Supplemental Table S1.
Data Processing and Database Searching
The raw data were processed with the PLGS 2.5.2 Apex3D and Pep3D algorithms (64 bit; Waters) to generate precursor mass lists and associated product ion mass lists for subsequent protein identification and quantification. The thresholds for low-energy ions, high-energy ions, and low-energy exact mass retention time pairs were set to 100, 15, and 750 counts, respectively. The [Glu-1]-fibrinopeptide was specified as a lock mass compound with 785.8426 m/z for z = 2 and a 0.25-D tolerance window. A detailed description of the PLGS pipeline is in Li et al. (2009).
For Golgi proteins ranking, the database search was performed with the settings suggested by the manufacturer against the latest version of the nonredundant Arabidopsis genome annotation database (The Arabidopsis Information Resource 10; obtained from www.arabidopsis.org) containing 27,241 protein sequence entries, in which entries with entirely identical sequences were merged into single FASTA entries by an in-house R script. For Golgi-localized protein quantification, the most abundant Golgi fraction of both biological replicates gradients (fraction number 5) was searched with 4% false discovery rate (as suggested by the manufacturer), carbamidomethyl Cys as the fixed modification, and oxidation of Met as the variable modification. The database search was performed with the following settings: a peptide could be identified by a minimum of one fragment ion, and the protein identification required at least three fragment ions and at least one peptide per protein for identification. Only proteins with Green autocurate thresholds were used in the subsequent analysis. The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) through the PRIDE partner repository (Vizcaíno et al., 2014) with the dataset identifier PXD001056.
To obtain protein distributions across the gradient, the LOPIMS pipeline implemented in pRoloc (Gatto et al., 2014) and based on functionality from synapter (Shliaha et al., 2013) and MSnbase (Gatto and Lilley, 2012) was developed (a detailed description is in the documentation of lopims function in the pRoloc package; Gatto and Lilley, 2012; Shliaha et al., 2013). In brief, raw data analysis was performed in PLGS to get a list of exact mass retention times (unidentified peptide features) and identified peptides from both Arabidopsis forward and scrambled databases (The Arabidopsis Information Resource 10). Identified peptides from fractions 1, 3, and 5 from the first biological replicate gradient and fractions 4 and 9 from second biological replicate gradient were used to construct a list of m/z and retention times for 12,931 fully tryptic unmodified proteotypic peptides. These peptide identifications were then transferred to all the acquisitions based on similarity of m/z and retention time. This resulted in 8,156 and 8,805 peptides being quantified in the first and second gradient replicates, respectively. These peptides were present in at least 5 of 10 gradient fractions in both gradients. The peptide data were converted to protein intensities as follows. For each protein, a fraction with the highest number of peptides quantified was nominated as a reference fraction. The protein abundance in that reference fraction was taken as one. Then, other fractions were quantified against the reference fraction by finding the ratio of the combined intensity for peptides shared between the interrogated and reference fractions. When the quantities in all fractions for a given protein were computed, the values were renormalized to give a sum = 1 across all 10 fractions.
An SVM classifier as implemented in the pRoloc R package was used to infer protein localization (Gatto et al., 2014). Briefly, the labeled data were separated into stratified test (20%) and training (80%) partitions. The latter was submitted to another round of stratified 5-fold cross validation to optimize the SVM parameters (regularization parameter C and kernel radial basis function width σ). Model accuracy is evaluated using the macro-F1 score, F1 = 2 × (precision × recall)/(precision + recall), which was calculated as the harmonic mean of the precision, precision = true positives [tp]/(tp + false positives [fp]), a measure of exactness (i.e. returned output is a relevant result), and recall, recall = tp/(tp + false negatives [fn]), a measure of completeness (i.e. indicating how much was missed from the output). The best pair of parameters (i.e. the one yielding the highest macro-F1 score) was subsequently used in training a classifier on all training profiles before assessment on all test/validation profiles. This procedure was repeated 100 times to estimate generalization performance values and identify suitable model parameters, which were then applied to train a model on all marker profiles and classify unlabeled data.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Golgi protein localization by SVM.
Supplemental Figure S2. UPS1 and UPS2 standards.
Supplemental Table S1. Synapt G2 instrument settings.
Supplemental Table S2. Protein profiles.
Supplemental Table S3. Protein marker list.
Supplemental Table S4. Golgi protein localization by SVM.
Supplemental Table S5. Rank ordering of Golgi proteins.
Supplemental Table S6. Composition of the peak Golgi fraction.
Supplemental Table S7. Composition of the whole plant lysate.
Note Added in Proof
As this paper was in the proof stage, a publication by Rennie et al. (2014) about PGSIP6 identified this protein as an Inositol Phosphhorylceramide Glucuronosyltransferase1 (Rennie EA, Ebert B, Miles GP, Cahoon RE, Christiansen KM, Stonebloom S, Khatab H, Twell D, Petzold CJ, Adams PD, et al [2014] Identification of a sphingolipid α-glucuronosyltransferase that is essential for pollen function in Arabidopsis. Plant Cell.
Supplementary Material
Acknowledgments
We thank Zhinong Zhang and Xiaolan Yu for maintaining the Arabidopsis callus lines.
Glossary
- LC
liquid chromatography
- LOPIT
localization of organelle proteins by isotope tagging
- m/z
mass/charge
- MS
mass spectrometry
- PLGS
Protein Lynx Global Server
- SVM
support vector machine
Footnotes
This work was supported by the Darwin Trust of Edinburgh (to P.V.S.), by the European Union 7th Framework Program (PRIMEXS Project grant no. 262067 to L.G. and K.S.L.), and by the Biotechnology and Biological Sciences Research Council (grant nos. BB/G016240/1 Biotechnology and Biological Sciences Research Council Sustainable Energy Centre Cell Wall Sugars Programme to P.D. and K.S.L. and BBS/B/10684 to P.D. and K.S.L.).
The online version of this article contains Web-only data.
References
- Ahmed SU, Rojo E, Kovaleva V, Venkataraman S, Dombrowski JE, Matsuoka K, Raikhel NV. (2000) The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J Cell Biol 149: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. (2002) Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [DOI] [PubMed] [Google Scholar]
- Arsova B, Kierszniowska S, Schulze WX. (2012) The use of heavy nitrogen in quantitative proteomics experiments in plants. Trends Plant Sci 17: 102–112 [DOI] [PubMed] [Google Scholar]
- Barton CJ, Tailford LE, Welchman H, Zhang Z, Gilbert HJ, Dupree P, Goubet F. (2006) Enzymatic fingerprinting of Arabidopsis pectic polysaccharides using polysaccharide analysis by carbohydrate gel electrophoresis (PACE). Planta 224: 163–174 [DOI] [PubMed] [Google Scholar]
- Blackburn K, Cheng FY, Williamson JD, Goshe MB. (2010) Data-independent liquid chromatography/mass spectrometry (LC/MS(E)) detection and quantification of the secreted Apium graveolens pathogen defense protein mannitol dehydrogenase. Rapid Commun Mass Spectrom 24: 1009–1016 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Lamond AI. (2010) p53-Dependent subcellular proteome localization following DNA damage. Proteomics 10: 4087–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond NJ, Shliaha PV, Lilley KS, Gatto L. (2013) Improving qualitative and quantitative performance for MS(E)-based label-free proteomics. J Proteome Res 12: 2340–2353 [DOI] [PubMed] [Google Scholar]
- Carroll KM, Simpson DM, Eyers CE, Knight CG, Brownridge P, Dunn WB, Winder CL, Lanthaler K, Pir P, Malys N, et al. (2011) Absolute quantification of the glycolytic pathway in yeast: deployment of a complete QconCAT approach. Mol Cell Proteomics 10: 007633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng FY, Blackburn K, Lin YM, Goshe MB, Williamson JD. (2009) Absolute protein quantification by LC/MS(E) for global analysis of salicylic acid-induced plant protein secretion responses. J Proteome Res 8: 82–93 [DOI] [PubMed] [Google Scholar]
- Christianson CC, Johnson CJ, Needham SR. (2013) The advantages of microflow LC-MS/MS compared with conventional HPLC-MS/MS for the analysis of methotrexate from human plasma. Bioanalysis 5: 1387–1396 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Palma S, Hennrich ML, Heck AJ, Mohammed S. (2012) Recent advances in peptide separation by multidimensional liquid chromatography for proteome analysis. J Proteomics 75: 3791–3813 [DOI] [PubMed] [Google Scholar]
- Distler U, Kuharev J, Navarro P, Levin Y, Schild H, Tenzer S. (2014) Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat Methods 11: 167–170 [DOI] [PubMed] [Google Scholar]
- Dunkley TPJ, Watson R, Griffin JL, Dupree P, Lilley KS. (2004) Localization of organelle proteins by isotope tagging (LOPIT). Mol Cell Proteomics 3: 1128–1134 [DOI] [PubMed] [Google Scholar]
- Dunkley TPJ, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C, Noirel J, Ow SY, Salim M, Pereira-Medrano AG, Couto N, Pandhal J, Smith D, Pham TK, Karunakaran E, et al. (2012) An insight into iTRAQ: where do we stand now? Anal Bioanal Chem 404: 1011–1027 [DOI] [PubMed] [Google Scholar]
- Gatto L, Breckels LM, Wieczorek S, Burger T, Lilley KS. (2014) Mass-spectrometry-based spatial proteomics data analysis using pRoloc and pRolocdata. Bioinformatics 30: 1322–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto L, Lilley KS. (2012) MSnbase-an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics 28: 288–289 [DOI] [PubMed] [Google Scholar]
- Gatto L, Vizcaíno JA, Hermjakob H, Huber W, Lilley KS. (2010) Organelle proteomics experimental designs and analysis. Proteomics 10: 3957–3969 [DOI] [PubMed] [Google Scholar]
- Geiger T, Cox J, Mann M. (2010) Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol Cell Proteomics 9: 2252–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geromanos SJ, Vissers JP, Silva JC, Dorschel CA, Li GZ, Gorenstein MV, Bateman RH, Langridge JI. (2009) The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9: 1683–1695 [DOI] [PubMed] [Google Scholar]
- Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 11: 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F, Barton CJ, Mortimer JC, Yu X, Zhang Z, Miles GP, Richens J, Liepman AH, Seffen K, Dupree P. (2009) Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J 60: 527–538 [DOI] [PubMed] [Google Scholar]
- Goubet F, Jackson P, Deery MJ, Dupree P. (2002) Polysaccharide analysis using carbohydrate gel electrophoresis: a method to study plant cell wall polysaccharides and polysaccharide hydrolases. Anal Biochem 300: 53–68 [DOI] [PubMed] [Google Scholar]
- Groen AJ, Sancho-Andrés G, Breckels LM, Gatto L, Aniento F, Lilley KS. (2014) Identification of trans-golgi network proteins in Arabidopsis thaliana root tissue. J Proteome Res 13: 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann J, Roschitzki B, Panse C, Fortes C, Barkow-Oesterreicher S, Rutishauser D, Schlapbach R. (2010) Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J Proteomics 73: 1740–1746 [DOI] [PubMed] [Google Scholar]
- Gruhler A, Schulze WX, Matthiesen R, Mann M, Jensen ON. (2005) Stable isotope labeling of Arabidopsis thaliana cells and quantitative proteomics by mass spectrometry. Mol Cell Proteomics 4: 1697–1709 [DOI] [PubMed] [Google Scholar]
- Guo G, Li N. (2011) Relative and accurate measurement of protein abundance using 15N stable isotope labeling in Arabidopsis (SILIA). Phytochemistry 72: 1028–1039 [DOI] [PubMed] [Google Scholar]
- Handford MG, Baldwin TC, Goubet F, Prime TA, Miles J, Yu X, Dupree P. (2003) Localisation and characterisation of cell wall mannan polysaccharides in Arabidopsis thaliana. Planta 218: 27–36 [DOI] [PubMed] [Google Scholar]
- Haynes PA, Roberts TH. (2007) Subcellular shotgun proteomics in plants: looking beyond the usual suspects. Proteomics 7: 2963–2975 [DOI] [PubMed] [Google Scholar]
- Hebert AS, Richards AL, Bailey DJ, Ulbrich A, Coughlin EE, Westphall MS, Coon JJ. (2014) The one hour yeast proteome. Mol Cell Proteomics 13: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegelund JN, Jahn TP, Baekgaard L, Palmgren MG, Schjoerring JK. (2010) Transmembrane nine proteins in yeast and Arabidopsis affect cellular metal contents without changing vacuolar morphology. Physiol Plant 140: 355–367 [DOI] [PubMed] [Google Scholar]
- Held MA, Be E, Zemelis S, Withers S, Wilkerson C, Brandizzi F. (2011) CGR3: a Golgi-localized protein influencing homogalacturonan methylesterification. Mol Plant 4: 832–844 [DOI] [PubMed] [Google Scholar]
- Helm S, Dobritzsch D, Rödiger A, Agne B, Baginsky S. (2014) Protein identification and quantification by data-independent acquisition and multi-parallel collision-induced dissociation mass spectrometry (MS(E)) in the chloroplast stroma proteome. J Proteomics 98: 79–89 [DOI] [PubMed] [Google Scholar]
- Hoaglund-Hyzer CS, Clemmer DE. (2001) Ion trap/ion mobility/quadrupole/time-of-flight mass spectrometry for peptide mixture analysis. Anal Chem 73: 177–184 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Hummel M, Cordewener JH, de Groot JC, Smeekens S, America AH, Hanson J. (2012) Dynamic protein composition of Arabidopsis thaliana cytosolic ribosomes in response to sucrose feeding as revealed by label free MSE proteomics. Proteomics 12: 1024–1038 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Kelly RT, Page JS, Luo Q, Moore RJ, Orton DJ, Tang K, Smith RD. (2006) Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal Chem 78: 7796–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 4: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law KP, Lim YP. (2013) Recent advances in mass spectrometry: data independent analysis and hyper reaction monitoring. Expert Rev Proteomics 10: 551–566 [DOI] [PubMed] [Google Scholar]
- Levin Y, Hradetzky E, Bahn S. (2011) Quantification of proteins using data-independent analysis (MSE) in simple and complex samples: a systematic evaluation. Proteomics 11: 3273–3287 [DOI] [PubMed] [Google Scholar]
- Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. (2009) Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9: 1696–1719 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Orfila C, McCartney L, Harholt J, Bernal AJ, Scheller HV, Gilmartin PM, Mikkelsen JD, Paul Knox J, Willats WG. (2004) Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. Plant J 40: 260–275 [DOI] [PubMed] [Google Scholar]
- Miao Y, Li HY, Shen J, Wang J, Jiang L. (2011) QUASIMODO 3 (QUA3) is a putative homogalacturonan methyltransferase regulating cell wall biosynthesis in Arabidopsis suspension-cultured cells. J Exp Bot 62: 5063–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski A, Cox J, Mann M. (2011) More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res 10: 1785–1793 [DOI] [PubMed] [Google Scholar]
- Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hématy K, McCartney L, Truong HN, Gaudon V, Thibault JF, et al. (2007) Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J 50: 605–614 [DOI] [PubMed] [Google Scholar]
- Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC, Haynes PA. (2011) Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 11: 535–553 [DOI] [PubMed] [Google Scholar]
- Nelson CJ, Huttlin EL, Hegeman AD, Harms AC, Sussman MR. (2007) Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics 7: 1279–1292 [DOI] [PubMed] [Google Scholar]
- Nikolovski N, Rubtsov D, Segura MP, Miles GP, Stevens TJ, Dunkley TP, Munro S, Lilley KS, Dupree P. (2012) Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol 160: 1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y. (2013) Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat Chem Biol 9: 726–730 [DOI] [PubMed] [Google Scholar]
- Oikawa A, Lund CH, Sakuragi Y, Scheller HV. (2013) Golgi-localized enzyme complexes for plant cell wall biosynthesis. Trends Plant Sci 18: 49–58 [DOI] [PubMed] [Google Scholar]
- Parsons HT, Christiansen K, Knierim B, Carroll A, Ito J, Batth TS, Smith-Moritz AM, Morrison S, McInerney P, Hadi MZ, et al. (2012a) Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol 159: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons HT, Drakakaki G, Heazlewood JL. (2012b) Proteomic dissection of the Arabidopsis Golgi and trans-Golgi network. Front Plant Sci 3: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasa-Tolić L, Masselon C, Barry RC, Shen Y, Smith RD. (2004) Proteomic analyses using an accurate mass and time tag strategy. Biotechniques 37: 621–636 [DOI] [PubMed] [Google Scholar]
- Prime TA, Sherrier DJ, Mahon P, Packman LC, Dupree P. (2000) A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21: 3488–3499 [DOI] [PubMed] [Google Scholar]
- Sadowski PG, Dunkley TPJ, Shadforth IP, Dupree P, Bessant C, Griffin JL, Lilley KS. (2006) Quantitative proteomic approach to study subcellular localization of membrane proteins. Nat Protoc 1: 1778–1789 [DOI] [PubMed] [Google Scholar]
- Sadowski PG, Groen AJ, Dupree P, Lilley KS. (2008) Sub-cellular localization of membrane proteins. Proteomics 8: 3991–4011 [DOI] [PubMed] [Google Scholar]
- Sadygov RG, Cociorva D, Yates JR., III (2004) Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book. Nat Methods 1: 195–202 [DOI] [PubMed] [Google Scholar]
- Shliaha PV, Bond NJ, Gatto L, Lilley KS. (2013) Effects of traveling wave ion mobility separation on data independent acquisition in proteomics studies. J Proteome Res 12: 2323–2339 [DOI] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, et al. (2005) Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem 77: 2187–2200 [DOI] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ. (2006a) Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol Cell Proteomics 5: 589–607 [DOI] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. (2006b) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 5: 144–156 [DOI] [PubMed] [Google Scholar]
- Stasyk T, Huber LA. (2004) Zooming in: fractionation strategies in proteomics. Proteomics 4: 3704–3716 [DOI] [PubMed] [Google Scholar]
- Taylor LE, II, Henrissat B, Coutinho PM, Ekborg NA, Hutcheson SW, Weiner RM. (2006) Complete cellulase system in the marine bacterium Saccharophagus degradans strain 2-40T. J Bacteriol 188: 3849–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Pol-Fachin L, Egelund J, Gille S, Harholt J, Ciancia M, et al. (2011) O-glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mortimer JC, Davis J, Dupree P, Keegstra K. (2012) Identification of an additional protein involved in mannan biosynthesis. Plant J 73: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509: 582–587 [DOI] [PubMed] [Google Scholar]
- Zouhar J, Muñoz A, Rojo E. (2010) Functional specialization within the vacuolar sorting receptor family: VSR1, VSR3 and VSR4 sort vacuolar storage cargo in seeds and vegetative tissues. Plant J 64: 577–588 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.