Diversification of anthocyanins in peach is attributed to glycosylation and methylation.
Abstract
Modification of anthocyanin plays an important role in increasing its stability in plants. Here, six anthocyanins were identified in peach (Prunus persica), and their structural diversity is attributed to glycosylation and methylation. Interestingly, peach is quite similar to the wild species Prunus ferganensis but differs from both Prunus davidiana and Prunus kansueasis in terms of anthocyanin composition in flowers. This indicates that peach is probably domesticated from P. ferganensis. Subsequently, genes responsible for both methylation and glycosylation of anthocyanins were identified, and their spatiotemporal expression results in different patterns of anthocyanin accumulation in flowers, leaves, and fruits. Two tandem-duplicated genes encoding flavonoid 3-O-glycosyltransferase (F3GT) in peach, PpUGT78A1 and PpUGT78A2, showed different activity toward anthocyanin, providing an example of divergent evolution of F3GT genes in plants. Two genes encoding anthocyanin O-methyltransferase (AOMT), PpAOMT1 and PpAOMT2, are expressed in leaves and flowers, but only PpAOMT2 is responsible for the O-methylation of anthocyanins at the 3′ position in peach. In addition, our study reveals a novel branch of UGT78 genes in plants that lack the highly conserved intron 2 of the UGT gene family, with a great variation of the amino acid residue at position 22 of the plant secondary product glycosyltransferase box. Our results not only provide insights into the mechanisms underlying anthocyanin glycosylation and methylation in peach but will also aid in future attempts to manipulate flavonoid biosynthesis in peach as well as in other plants.
Anthocyanins are important secondary metabolites and serve to protect plants against pathogenic attack and UV radiation and provide flowers and fruits with pigmentation to attract pollinators and seed dispersers (Winkel-Shirley, 2001). Anthocyanins are usually stored as glycosylated forms in vacuoles. The basic structure of anthocyanins is composed of an anthocyanidin aglycone and one or more sugar moieties linked to hydroxyl groups 3, 5, 7, 3′, and 5′, with the 3 position on the C-ring being dominant. The most common sugar moieties are Glc, Gal, Xyl, Ara, and Fru. With different types and/or numbers of sugar moieties attached to various positions, the structural diversity of anthocyanins increases significantly. Besides glycosylation, modifications such as methylation and acylation also contribute to the structural diversity of anthocyanins. To date, there have been more than 550 anthocyanins identified in nature (Kong et al., 2003).
The formation of glycosides is catalyzed by UDP-Glc:flavonoid glycosyltransferase (UFGT). Both monoglycosides and diglycosides are common in plants. For example, 3RT, 3GGT, and F3GGT1, responsible for further glycosylation of anthocyanidin 3-O-glycosides, have been reported in petunia (Petunia hybrida; Kroon et al., 1994), Japanese morning glory (Ipomoea nil; Morita et al., 2005), and kiwifruit (Actinidia chinensis; Montefiori et al., 2011), respectively. Glycosylation plays at least two roles in the accumulation of anthocyanins. One is to stabilize anthocyanins by attaching sugar moieties to the unstable anthocyanidin aglycones. Deficiency in UF3GT activity results in a significant reduction of anthocyanin accumulation in maize (Zea mays; Fedoroff et al., 1984) and Arabidopsis (Arabidopsis thaliana; Tohge et al., 2005), as anthocyanidins are highly unstable and easily susceptible to degradation. Another is to serve as a signal for transport of the anthocyanins to vacuoles (Ono et al., 2006). Anthocyanins are synthesized on the cytoplasmic face of the endoplasmic reticulum by metabolons, which are likely to function as a multienzyme complex (Winkel, 2004). Once synthesized, anthocyanins need to be transported to vacuoles by transporters such as multidrug and toxin extrusion transporters (Zhao et al., 2011). Vacuole localization is crucial for anthocyanins to function as pigments to attract pollinators and seed dispersers or as antioxidants to prevent photooxidation of the photosynthetic apparatus by lowering light intensity and screening out UV irradiation (Marrs et al., 1995). In short, glycosylation is essential for the accumulation of anthocyanins in plants.
UFGT belongs to family 1 glycosyltransferases, which are characterized by the presence of a conserved C-terminal domain termed the plant secondary product glycosyltransferase (PSPG) box (Yonekura-Sakakibara and Hanada, 2011). The PSPG motif is involved in binding to the UDP moiety of the sugar donor and thus plays an important role in anthocyanin biosynthesis (Caputi et al., 2012). To date, genes encoding UFGTs have been identified in a range of plant species, and they can be divided into two groups based on their specificity for sugar acceptors. The first group catalyze the glycosylation of anthocyanidins, including anthocyanidin 3-O-glycosyltransferase (F3GT; Fedoroff et al., 1984; Ford et al., 1998; Kubo et al., 2004; Tohge et al., 2005) and anthocyanidin 5,3-O-glucosyltransferase (Ogata et al., 2005). The second group are responsible for the further glycosylation of anthocyanins, including anthocyanidin 3-O-glucoside 6′′-O-rhamnosyltransferase (F3GGT; Kroon et al., 1994), anthocyanidin 3-O-glucoside 2′′-O-glucosyltransferase (F3GGT; Morita et al., 2005), anthocyanidin 3-O-glucoside 2′′-O-glucuronosyltransferase (Sawada et al., 2005), and anthocyanidin 3-O-glucoside 2′′-O-xylosyltransferase (F3GGT; Montefiori et al., 2011; Yonekura-Sakakibara et al., 2012). Among these UFGT genes, F3GTs can also catalyze the glycosylation of flavonols in some plant species, such as Arabidopsis (Tohge et al., 2005), petunia (Yamazaki et al., 2002), and grape (Vitis vinifera; Ford et al., 1998).
Methylation is known to affect both the stability and water solubility of anthocyanins; thus, it plays an important role in the accumulation of anthocyanins in plants (Sarni et al., 1995). Anthocyanin methylation is catalyzed by anthocyanin O-methyltransferase (AOMT), and AOMTs are responsible for the O-methylation of anthocyanins at either the 3′ or 5′ position (Wiering and deVlaming 1977). To date, two AOMT genes have been cloned and characterized in grapevine (Hugueney et al., 2009; Lücker et al., 2010; Fournier-Level et al., 2011).
Anthocyanin accumulation has a great impact on fruit quality, and many genes involved in the regulation of anthocyanin biosynthesis have been identified in fruit species such as grape (Kobayashi et al., 2004), apple (Malus domestica; Takos et al., 2006; Espley et al., 2007; Li et al., 2012), and pear (Pyrus communis; Feng et al., 2010). However, few studies have been reported on characterization of the UFGT and AOMT gene families in fruit trees. Functional characterization of UFGT and AOMT genes has so far only been reported in grape (Ford et al., 1998) and kiwifruit (Montefiori et al., 2011). Many questions related to the mechanism underlying the glycosylation and methylation of anthocyanins and the evolution of the UFGT and AOMT gene families in plants still remain unanswered.
Peach (Prunus persica), a member of the Rosaceae family, is the third most important of deciduous fruit trees worldwide. Peach is a diploid with a small genome size of approximately 230 Mb (Arús et al., 2012). To date, there are few reports on genes involved in the regulation of anthocyanin accumulation in peach. Recently, we sequenced peach transcriptomes using the Illumina RNA sequencing (RNA-Seq) strategy, and the results provide an opportunity to investigate genes involved in anthocyanin accumulation in peach. Here, we report on the mechanisms underlying the methylation and glycosylation of anthocyanins in peach. Our study is not only helpful for understanding the molecular mechanism of anthocyanin accumulation in fruit trees but also useful for future research to manipulate flavonoid biosynthesis in peach as well as in other plants.
RESULTS
The Anthocyanin Component in Red-Colored Tissues of Peach and Its Wild Relatives
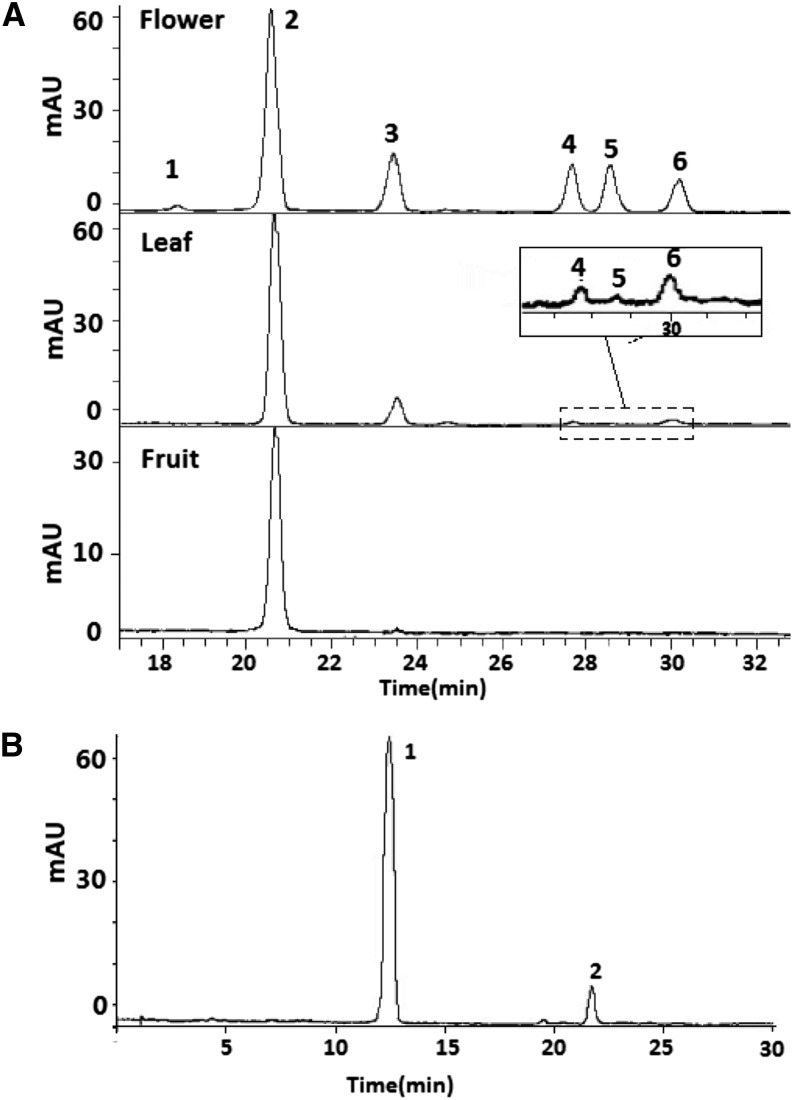
Peach cultivars Mantianhong (MT), Hongyetao (HY), and Wuhong9 (WH9) produce red-colored flowers, leaves, and fruits, respectively, and these red tissues were thus selected and subjected to an analysis of anthocyanin composition. As a result, a total of six peaks were identified in these three tissues (Fig. 1A). The flowers of cv MT contained all six peaks. The leaves of cv HY consisted of two major peaks, 2 and 3, and three traceable peaks, 4 to 6. The fruits of cv WH9 contained a major peak, 2, and a traceable peak, 3. Peaks 1 and 2 had the same mass spectrometry (MS) and tandem mass spectrometry (MS/MS) spectra, and peak 2 had the same retention time with the standard of cyanidin 3-glucoside (Cy-3-glu). Thus, peaks 1 and 2 were identified to be cyanidin 3-galactoside (Cy-3-gal) and Cy-3-glu, respectively (Table I). Based on the MS and MS/MS spectra, peaks 3 to 6 were deemed cyanidin 3-rutinoside (Cy-3-rut), peonidin 3-glucoside (Pn-3-glu), cyanidin 3-rhamnoside (Cy-3-rha), and peonidin 3-rutinoside (Pn-3-rut), respectively. Among these anthocyanins, Cy-3-rha was detected for the first time in Rosaceae, while Cy-3-gal, Pn-3-glu, and Pn-3-rut were identified for the first time in peach. To validate the anthocyanins identified in this study, anthocyanins in flowers of cv MT were further hydrolyzed to identify anthocyanidin composition using HPLC-electrospray ionization (ESI)-MS. Two peaks showing the same retention times with the standards of cyanidin and peonidin were detected (Fig. 1B). This is consistent with the results of HPLC-MS/MS analysis as mentioned above.
Figure 1.
HPLC scans of anthocyanin pigments in red-colored flower, leaf, and flesh from peach cv MT, cv HY, and cv WH9, respectively (A) and the component of anthocyanidins derived from red flower of cv MT (B). Individual anthocyanins corresponding to peaks 1 to 6 are listed in Table I. mAU, Milliabsorbance units.
Table I. Identification of individual anthocyanins in peach.
Peaks indicated with letters a and b are identified in Rosaceae and peach species, to our knowledge, for the first time, respectively. m/z, Mass-to-charge ratio.
To determine if there are other anthocyanins in peach, the identification of anthocyanin composition in flowers was further performed for six more cultivars and three wild species. As a result, only the six anthocyanins mentioned above were detected (Table II). Overall, all the cultivars showed a similarity in anthocyanin composition. The major component of anthocyanins in flowers of all the tested cultivars was Cy-3-glu, with an average content of 52.17%. The remaining components, including Cy-3-gal, Cy-3-rut, Pn-3-glu, Cy-3-rha, and Pn-3-rut, showed 1.5%, 12.43%, 12.97%, 10.93%, and 10.42% of average contents, respectively. Interestingly, Cy-3-glu also represents the major anthocyanin in flowers of Prunus ferganensis and accounts for approximately 61% of the total anthocyanins. In addition, four and two anthocyanins were detected in flowers of Prunus davidiana and Prunus kansueasis, respectively. The most abundant anthocyanin in flowers of P. davidiana is Pn-3-glu (40.92%), followed by Pn-3-rut (35.19%), Cy-3-glu (15.92%), and Cy-3-rut (7.96%). Two anthocyanins, Cy-3-rut and Cy-3-glu, account for 56.14% and 43.86% of the total anthocyanins in flowers of P. kansueasis, respectively. In summary, P. davidiana and P. kansueasis accumulate mainly two peonidin-based and two cyanidin-based anthocyanins in flowers, respectively, whereas the common major anthocyanin Cy-3-glu is synthesized in flowers of peach and P. ferganensis.
Table II. Comparison of anthocyanin components in flowers between peach and its wild relatives.
The relative content of each anthocyanin is calculated based on the value of the peach area. The major components are highlighted in boldface. ND, Not detectable.
| Species | Accession | Percentage |
|||||
|---|---|---|---|---|---|---|---|
| Cy-3-gal | Cy-3-glu | Cy-3-rut | Pn-3-glu | Cy-3-rha | Pn-3-rut | ||
| Peach | Juhuatao | ND | 50.00 | 14.00 | 17.00 | 8.00 | 11.00 |
| Fenhuashouxintao | ND | 48.54 | 9.22 | 19.42 | 6.31 | 16.50 | |
| Hongcuizhi | 2.13 | 53.19 | 9.57 | 13.30 | 13.30 | 8.51 | |
| Honghuashouxintao | 0.51 | 51.28 | 15.90 | 11.28 | 8.21 | 12.82 | |
| MT | 1.56 | 52.08 | 15.10 | 11.46 | 11.46 | 8.33 | |
| Hongbaihuatao | 1.51 | 50.25 | 10.05 | 13.57 | 13.07 | 11.56 | |
| HY | 1.80 | 59.88 | 13.17 | 4.79 | 16.17 | 4.19 | |
| P. ferganensis | Xinjiangtao | ND | 60.98 | 18.29 | 8.54 | 5.49 | 6.71 |
| P. davidiana | Shantao | ND | 15.92 | 7.96 | 40.92 | ND | 35.19 |
| P. kansueasis | Gansutao | ND | 43.86 | 56.14 | ND | ND | ND |
In summary, there are six anthocyanins in peach, and their structural diversity is attributed to glycosylation and/or methylation.
Identification of Genes Encoding UDP-Glycosyltransferase and O-Methyltransferase in the Peach Transcriptome
A total of 21.5 Gb of Illumina RNA-Seq reads, representing approximately 96 times the peach genome, were generated from different tissues, including red and green leaves, white- and red-fleshed fruits, and flowers (Wang et al., 2013). The RNA-Seq reads were assembled, and the consensus sequences were screened for the presence of the PSPG box, a characteristic of family 1 glycosyltransferases (UGTs). As a result, 79 genes encoding UGT were identified in the peach transcriptome (Supplemental Table S1). These peach UGT genes were named following the previously reported nomenclature system (Mackenzie et al., 1997), and they can be phylogenetically divided into eight orthologous groups according to the definition as described by Yonekura-Sakakibara and Hanada (2011). Of the eight orthologous groups, OG8 and OG23 consist of F3GT and F3GGT genes and are involved in the glycosylation of anthocyanins and anthocyanidins, respectively (Supplemental Fig. S1). Therefore, three F3GT genes, designated PpUGT78A1, PpUGT78A2, and PpUGT78B, and five F3GGT genes, designated PpUGT79A, PpUGT79B, PpUGT94A2, PpUGT94A3, and PpUGT94A5, were selected and subjected to expression profiling analysis.
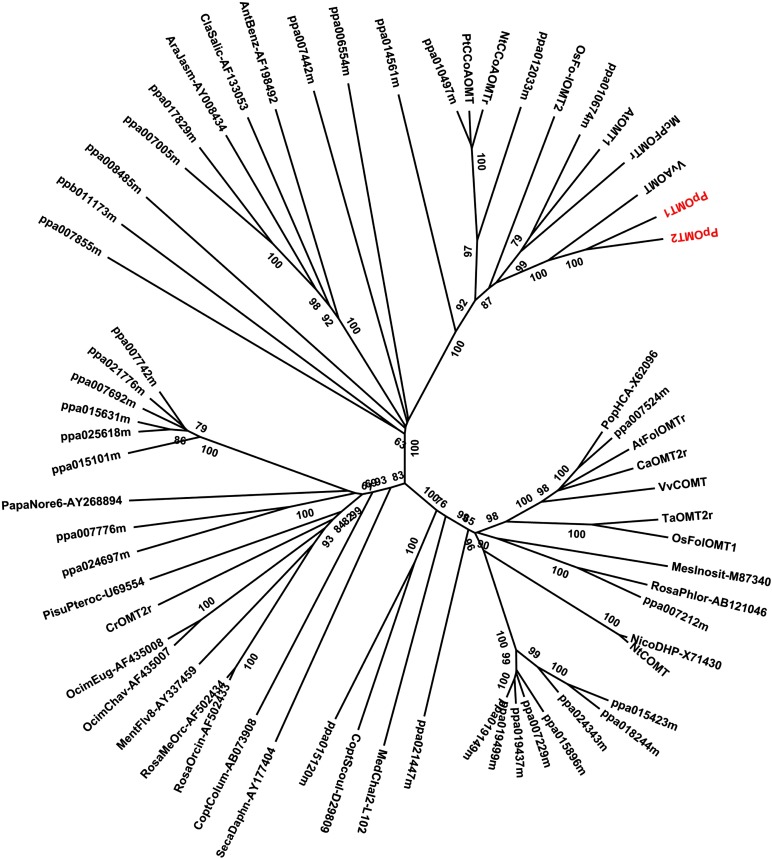
Similarly, 22 genes encoding O-methyltransferase (OMT) were identified in the peach transcriptome (Fig. 2). Phylogenetic analysis indicated that two OMT genes, designated PpAOMT1 and PpAOMT2, are closely related to VvAOMT, which is responsible for the methylation of anthocyanins in grapevine (Hugueney et al., 2009). In addition, an OMT gene (ppa010674m) also shows a close relationship with VvAOMT. However, RNA-Seq and reverse transcription-PCR analysis demonstrated that ppa010674m was highly expressed in fruits that accumulate only nonmethylated anthocyanins but was not detectable in flowers (Supplemental Fig. S2), and it was not included in further analysis.
Figure 2.
A phylogenetic tree of PpOMT genes in plants. Genes involved in anthocyanin methylation in peach are highlighted in red. The GenBank accession numbers are as follows: grapevine anthocyanin OMT (VvAOMT, FJ460168), grapevine caffeic acid OMT (VvCOMT, AF239740), tobacco caffeic acid OMT (NtCOMT, AF484252), Triticum aestivum OMT (TaOMT2r, DQ223971), Catharanthus roseus flavonoid OMT (CrOMT2r, AY127568), Chrysosplenium americanum OMT (CaOMT2r, U16793), Arabidopsis flavonol OMT (AtFolOMTr, U70424), rice flavonol OMT (OsFolOMT1, DQ288259), grapevine caffeoyl-CoA-OMT (VvCCoAOMTr, Z54233), tobacco caffeoyl-CoA-OMT (NtCCoAOMTr, U38612), Populus trichocarpa caffeoyl-CoA-OMT (PtCCoAOMT, AJ224896), rice flavonol OMT (OsFo-lOMT2, XM_483167), Mesembryanthemum crystallinum OMT (McPFOMTr, AY145521), and Arabidopsis OMT (AtOMT1, AY087244). [See online article for color version of this figure.]
Identification of Gene Families Encoding F3G, F3GGT, and AOMT in the Peach Genome
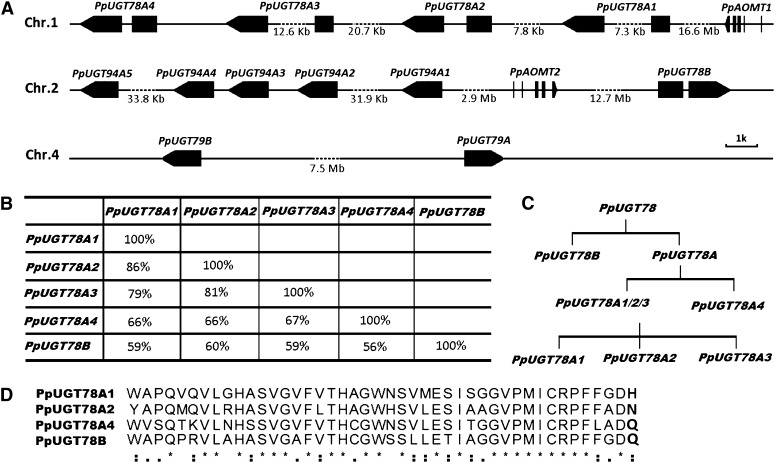
First, we screened the genome of peach cv Lovell (Verde et al., 2013), and two more UGT78 homologs designated PpUGT78A3 and PpUGT78A4 were identified. Of the five PpUGT78 genes, four (PpUGT78A1–PpUGT78A4) are clustered in a 56-kb region on chromosome 1, and one PpUGT78B is located on chromosome 2 (Fig. 3A). All the PpUGT78 genes except PpUGT78A3 have intact coding frames. PpUGT78A3 is a pseudogene due to a frameshift mutation induced by a 5-bp insertion in the coding region. The five PpUGT78 genes share 56% to 86% identities among their DNA coding sequences (CDSs; Fig. 3B). Of the four PpUGT78A genes, PpUGT78A1, PpUGT78A2, and PpUGT78A3 share 79% to 86% CDS identity, with approximately 66% CDS identity to PpUGT78A4. PpUGT78B shows the lowest CDS identity (56%–60%) with the four PpUGT78A genes. These results suggest that the PpUGT78A cluster is likely derived from two rounds of tandem gene duplication, while the ancestor of the cluster and PpUGT78B are probably derived following a genome duplication event during the speciation process of peach (Fig. 3C).
Figure 3.
Genes involved in glycosylation and methylation in peach. A, Structural features and chromosomal positions of PpUGT78, PpUGT79, PpUGT94, and PpAOMT genes. Black boxes represent exons. B, DNA sequence identity for the coding regions of the PpUGT78 genes. C, Duplication pattern of PpUGT78 genes in the peach genome. D, Alignment of the conserved PSPG motif sequences of the PpUGT78 genes. The amino acid residues at position 44 responsible for the sugar donor recognition are highlighted in boldface. Asterisks indicate that the residues in that column are identical; colons and periods indicate that conserved and semiconserved substitutions have been observed, respectively.
All four genes, PpUGT78A1, PpUGT78A2, PpUGT78A4, and PpUGT78B, contain a conserved 44-amino acid residue motif known as the PSPG box (Fig. 3D). The PpUGT78A1 and PpUGT78A2 proteins are similar, with equal length and 80% identity in amino acid sequences. The PpUGT78A1 and PpUGT78A2 proteins both show higher identities in amino acid sequences with PpUGT78A4 (53%) than with PpUGT78B (47%). The PpUGT78B and PpUGT78A4 proteins show a high degree of divergence and share the lowest identity (43%) in amino acid sequences.
Second, five copies of PpUGT94A genes are clustered in a 79-kb region on chromosome 2 (Fig. 3A). All five PpUGT94A genes show an average of 86% CDS identity, and PpUGT94A1 and PpUGT94A2 share the highest degree of CDS identity (97%).
Finally, both UGT79 and AOMT gene families contain two copies in the peach genome. PpUGT79A and PpUGT79B are both located on chromosome 4 and share 58% CDS identity. PpAOMT1 and PpAOMT2 are located on chromosomes 1 and 2, respectively, and share 79% CDS identity.
Expression Profiling of F3GT, F3GGT, and AOMT Genes in Peach
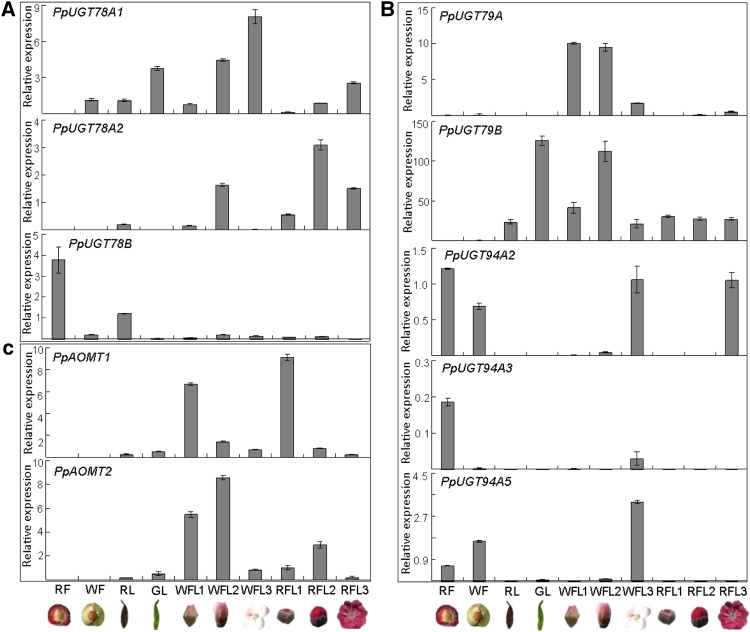
Gene expression levels were initially estimated using fragments per kilo bases per million reads values (Supplemental Fig. S2). Overall, F3GGT homologous genes showed very low levels of expression in all tested tissues, including red- and white-fleshed fruits, red flowers, and green and red leaves. In contrast, three F3GT homologous genes showed high levels of expression in red tissues. Two AOMT homologous genes were highly expressed in flowers at early stages of development but not expressed in fruit tissues. These results were subsequently validated using real-time PCR analysis (Fig. 4).
Figure 4.
Expression levels of UGT78A and UGT78B in different tissues of peach. The tissue samples are listed as follows: RF, fruits of red-fleshed cv WH9 at ripening stage; WF, fruits of white-fleshed cv BF at ripening stage; RL, young red leaves of cv HY; GL, young green leaves of cv BF; WFL1, flower buds of cv Shantao at pink stage; WFL2, flower buds of cv Shantao at balloon stage; WFL3, flowers of cv Shantao at full-bloom stage (open flowers); RFL1, flower buds of cv MT at pink stage; RFL2, flower buds of cv MT at balloon stage; RFL3, flowers of cv MT at full-bloom stage. [See online article for color version of this figure.]
PpUGT78A1 was expressed in fruits, leaves, and flowers. The expression levels of PpUGT78A1 increased during flower development and reached a maximum at full-bloom stage. Interestingly, PpUGT78A1 was expressed in both red and green leaves, and its level of expression in green leaves was higher than in red leaves. However, the transcripts of PpUGT78A1 were not detectable in red-fleshed fruits. PpUGT78A2 was predominantly expressed in flowers, and its level of expression in red flowers was significantly higher than in pale pink flowers. PpUGT78A2 transcripts were weakly accumulated in young red leaves but undetectable in green leaves and fruits. In contrast, PpUGT78B was mainly expressed in leaves and fruits, and its expression level in flowers was extremely low. PpUGT78B transcripts were significantly higher in red tissues than in nonred tissues. For example, the expression level of PpUGT78B in red-fleshed fruits of cv WH9 was 18.5-fold higher than that in white-fleshed fruits of cv Baifeng (BF). PpUGT78B transcripts were expressed at a 25.5-fold higher level in red leaves of cv HY than in green leaves of P. davidiana. In addition, PpUGT78A4 was not expressed in all analyzed tissues. In short, PpUGT78A1 was expressed in both green and red tissues, whereas PpUGT78A2 and PpUGT78B were predominantly expressed in red tissues.
PpUGT79A was exclusively expressed in flowers of P. davidiana. PpUGT79B transcripts were expressed in leaves and flowers, with high expression in nonred tissues, but were not detectable in fruits. For the PpUGT94A gene family, all three members were expressed in red-fleshed fruits, although both Cy-3-rut and Pn-3-rut were almost undetectable in red-fleshed fruits. Moreover, these PpUGT94A genes were highly expressed in flowers at full-bloom stage, but their expression levels were extremely low or undetectable in flower buds at pink and bloom stages. These results strongly suggest that the PpUGT94A gene family is unlikely to be related to the glycosylation of anthocyanins in peach.
The two PpAOMT genes were highly expressed in flowers but weakly or not expressed in leaves and fruits. The expression levels of PpAOMT1 in flowers had a maximum at pink stage and then decreased during flower development. The expression level of PpAOMT2 in flowers increased during flower development and reached a peak at bloom stage, then decreased at full-bloom stage. These results suggest that methylation of anthocyanidin would occur during flower development but prior to the full-bloom stage.
Functional Analysis of PpAOMT and PpUGT78 Genes
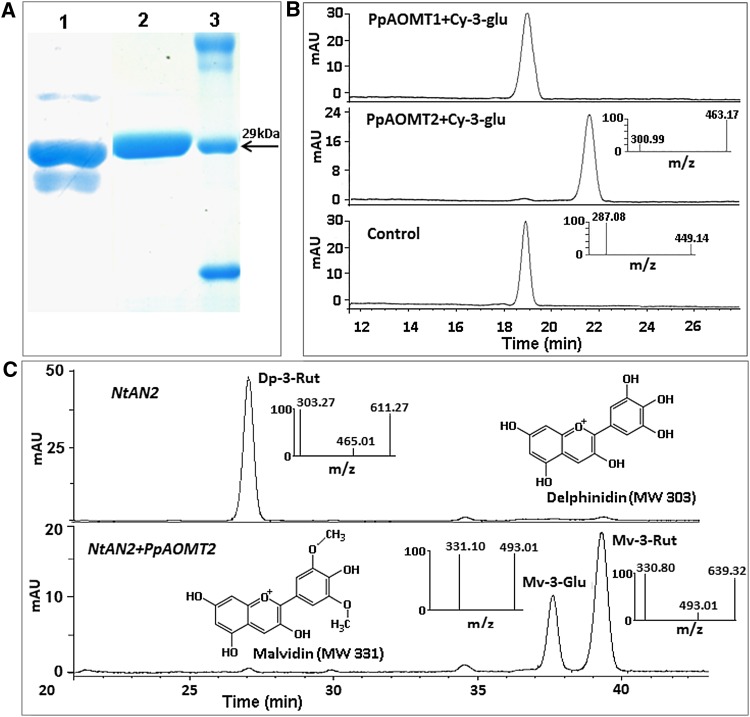
Enzyme activity in vitro was initially tested to determine the functionality of the PpAOMT genes. The recombinant proteins of both PpAOMT1 and PpAOMT2 were strongly induced in Escherichia coli, with an expected molecular size of approximately 29 kD (Fig. 5A). The recombinant protein was purified on an Ni2+-charged resin that binds the protein’s N-terminal His tag, and the activity was then assayed using Cy-3-glu as a substrate. No product was detected for the reaction of PpAOMT1 with Cy-3-glu (Fig. 5B). In contrast, the product of the reaction of PpAOMT2 with Cy-3-glu showed the same MS and MS/MS spectral properties as Pn-3-glu (Fig. 5B). This result shows that, in vitro, PpAOMT2 works well on Cy-3-glu, leading to the generation of the methylated anthocyanin Pn-3-glu.
Figure 5.
Functional analysis of the PpAOMT genes. A, Expression of PpAOMT1 and PpAOMT2 in E. coli. Lane 1, purified recombinant PpAOMT1 enzyme; lane 2, purified recombinant PpAOMT2 enzyme; lane 3, molecular mass marker proteins. B, HPLC-ESI-MS/MS analysis of the products catalyzed the PpAOMT enzymes. C, Transient expression of PpAOMT2 in N. benthamiana. mAU, Milliabsorbance units. [See online article for color version of this figure.]
The functionality of PpAOMT2 was further validated by transient expression in tobacco (Nicotiana tabacum). Two other discrete anthocyanins were detected in tobacco leaves coexpressing NtAN2 and PpAOMT2 genes (Fig. 5C). HPLC-MS/MS analysis indicated that two discrete anthocyanins were malvidin 3-glucoside and malvidin 3-rutinoside. This demonstrates that PpAOMT2 is functional in vivo.
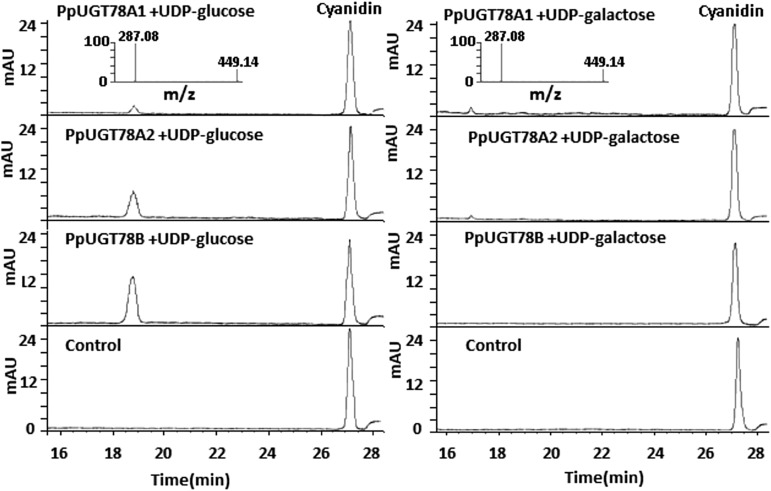
The functionality of PpUGT78 genes was also tested in vitro using cyanidin as acceptor and UDP-Glc or UDP-Gal as sugar donor (Fig. 6). PpUGT78A1 showed a very low activity toward cyanidin with either UDP-Glc or UDP-Gal as sugar donor. In contrast, both PpUGT78A2 and PpUGT78B had activity toward cyanidin, with a higher level of glucosyl transfer activity than galactosyl transfer activity.
Figure 6.
Functional assay of the PpUGT78 enzyme activity toward cyanidin with UDP-Glc or UDP-Gal as sugar donor. mAU, Milliabsorbance units.
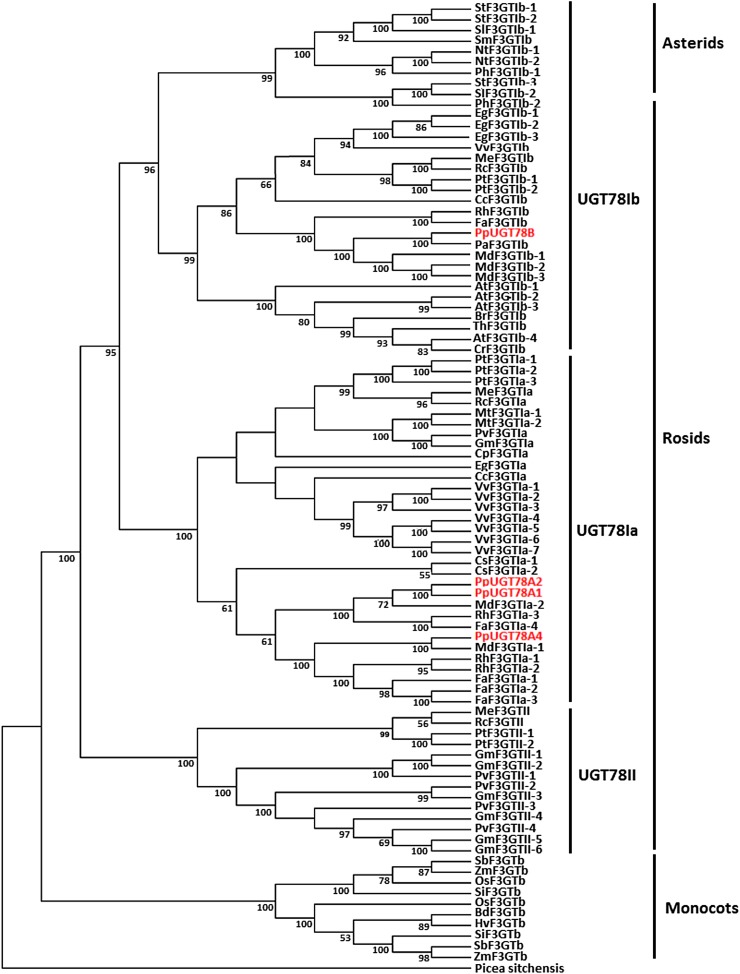
Phylogenetic Relationship between the Peach UGT78 Genes and Their Homologs in Other Plants
The UGT78 homologs were identified in sequenced genomes of plants, including monocots such as rice (Oryza sativa), maize, millet (Pennisitum americanum), and sorghum (Sorghum bicolor), and eudicots such as grapevine, poplar (Populus spp.), castor bean (Ricinus communis), cassava (Manihot esculenta), Medicago truncatula, soybean (Glycine max), common bean (Phaseolus vulgaris), apple, strawberry (Fragaria spp.), cucumber (Cucumis sativus), orange (Citrus spp.), papaya (Carica papaya), Eucalyptus globulus, Arabidopsis, Thellungiella halophila, Capsella rubella, Brassica rapa, tomato (Solanum lycopersicum), and potato (Solanum tuberosum). Phylogenetic analysis indicates that UGT78 genes in plants are grouped into monocot and eudicot clades (Fig. 7). All UGT78 genes identified in eudicots can be divided into two groups, denoted UGT78I and UGT78II, and the UGT78I group is composed of two subgroups, designated UGT78Ia and UGT78Ib. The three peach genes PpUGT78A1, PpUGT78A2, and PpUGT78A3 belong to the UGT78Ia subgroup, while the peach gene PpUGT78B belongs to the UGT78Ib group. The UGT78Ia subgroup consists of characterized UGT78 genes showing activity toward flavonols, including VvF3GTIa-1 and VvF3GTIa-2 in grapevine (Ono et al., 2010), MtF3GTIa-1 in M. truncatula (Modolo et al., 2007), and RhF3GTIa-3 in rose (Rosa spp.; Fukuchi-Mizutani et al., 2011). However, PtF3GTIa-2, which does not accept any flavonoids as a substrate, is also grouped into the UGT78Ia subgroup (Veljanovski and Constabel, 2013). The UGT78Ib group contains characterized UGT78 genes showing activity toward anthocyanins and flavonols, including VvF3GT1b in grapevine (Ford et al., 1998), AtF3GT1b-4 in Arabidopsis (Kubo et al., 2007), RhF3T1b in rose (Fukuchi-Mizutani et al., 2011), and FaF3GT1b in strawberry (Griesser et al., 2008), but two Arabidopsis genes, AtF3GTIb-1 and AtF3GTIb-3, within the same subgroup accept only flavonols (Jones et al., 2003; Yonekura-Sakakibara et al., 2008).
Figure 7.
Phylogenetic tree derived from amino acid sequences of UGT78 genes in both monocots and eudicots. The GenBank accession numbers of previously reported F3GT genes are as follows: petunia PhF3GT1b-2 (Q9SBQ8) and PhF3GT1b-1 (BAA89008); potato StF3GT1b-1 (AAX63403) and StF3GT1b-2 (AEN83502); tobacco NtF3GT1b-1 (BAM37965) and NtF3GT1b-2 (BAM37964); Solanum melongena SmF3GT1b (Q43641); Rosa × hybrida RhF3GT1b (BAF80946), RhF3GT1a-1 (BAF80947), RhF3GT1a-2 (BAK09602), and RhF3GT1a-3 (BAE72453); and Hordeum vulgare HvF3GTa (P14726). The genome sequences of grapevine, poplar, castor bean, cassava, M. truncatula, soybean, common bean, cucumber, papaya, E. globulus, Arabidopsis, T. halophila, C. rubella, B. rapa, tomato, potato, rice, maize, sorghum, citrus, apple, and strawberry were searched for UGT78 homologs through the Phytozome version 5.0 database (http://www.phytozome.net). The peach UGT78 genes are highlighted in light gray. [See online article for color version of this figure.]
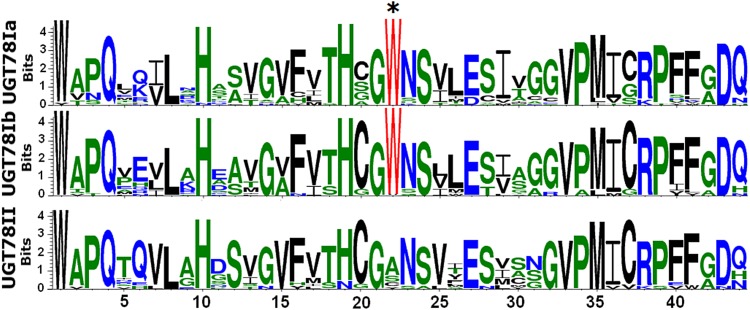
Moreover, all the UGT78II genes except GmF3GT2-2 lack introns, while both UGT78Ia and UGT78Ib genes have one intron. The GmF3GT2-2 gene contains a single intron. The UGT78I genes are present in all analyzed eudicot plants, while the UGT78II genes are identified only in five eudicot plant species: poplar, castor bean, cassava, common bean, and soybean (Table III). To investigate whether specific amino acid residues within the PSPG motif could serve as sequence logos to differentiate one UGT78 gene group from another in eudicots, logos for the conserved 44-amino acid residue motif of each UGT78 gene group were analyzed using the online WebLogo program (http://weblogo.berkeley.edu). Interestingly, the amino acid residue at position 22 is very highly conserved in both UGT78Ia and UGT78Ib subgroups, whereas it is subjected to variation to Ala (A), Ser (S), or Cys (C) in the UGT78II gene group (Fig. 8). Thus, it seems that the residue Trp (W) at position 22 is a sequence logo for the UGT78I gene group.
Table III. Copy numbers of intact UGT78 homologs identified in the sequenced genomes of eudicots.
| Eudicots | Species | UGT78Ia | UGT78Ib | UGT78II | ||
|---|---|---|---|---|---|---|
| Rosids | Vitales | Vitaceae | Grapevine | 7 | 1 | 0 |
| Eurosids I | Salicaceae | Poplar | 3 | 2 | 2 | |
| Euphorbiaceae | Castor bean | 1 | 1 | 1 | ||
| Cassava | 1 | 1 | 1 | |||
| Fabaceae | Soybean | 1 | 0 | 3 | ||
| Common bean | 1 | 0 | 4 | |||
| M. truncatula | 2 | 0 | 0 | |||
| Cucurbitaceae | Cucumber | 2 | 0 | 0 | ||
| Rosaceae | Apple | 2 | 3 | 0 | ||
| Peach | 3 | 1 | 0 | |||
| Strawberry | 4 | 1 | 0 | |||
| Eurosids II | Rutaceae | Orange | 1 | 1 | 0 | |
| Myrtaceae | E. globulus | 1 | 3 | 0 | ||
| Caricaceae | Papaya | 1 | 0 | 0 | ||
| Brassicaceae | Arabidopsis | 0 | 4 | 0 | ||
| C. rubella | 0 | 1 | 0 | |||
| B. rapa | 0 | 1 | 0 | |||
| T. halophila | 0 | 1 | 0 | |||
| Asterids | Solanaceae | Tomato | 0 | 2 | 0 | |
| Potato | 0 | 3 | 0 | |||
Figure 8.
WebLogo of the plant-specific PSPG motif constructed from the UGT78 genes encoding UDP-Glc:flavonoid 3-O-glycosyltransferase in eudicots. Letter size is proportional to the degree of amino acid conservation. The star indicates a sequence logo that differentiates the UGT78II genes from both UGT78Ia and UGT78Ib genes. [See online article for color version of this figure.]
DISCUSSION
The Complexity of Glycosylation and Methylation Leads to the Diversification of Anthocyanins in Peach
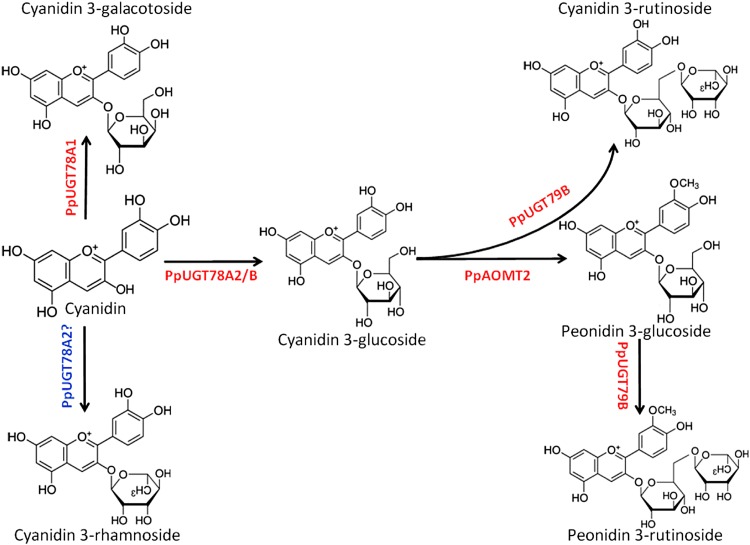
F3GTs catalyze the glycosylation of anthocyanidins. In this study, three F3GT genes, PpUGT78A1, PpUGT78A2, and PpUGT78B, are identified in the peach transcriptome. Of these genes, PpUGT78B is expressed in red-fleshed fruits, where only one anthocyanin Cy-3-glu accumulates predominantly. PpUGT78B contains the residue Gln (Q) at position 44 of the PSPG box, suggesting that it has glucosyl transfer activity (Kubo et al., 2004). This deduced functionality also has been validated in vitro. Thus, there is no doubt that PpUGT78B is involved in the synthesis of Cy-3-glu. PpUGT78A1 is closely related to the rose RhF3GTIa-1, which can use both flavonols and anthocyanins as substrates, but with extremely low activity toward anthocyanins (Fukuchi-Mizutani et al., 2011). We also found that PpUGT78A1 has very low activity toward cyanidin. Moreover, PpUGT78A1 contains the residue His (H) at position 44 of the PSPG box, which is required for recognition of the sugar donor UDP-Gal (Kubo et al., 2004). Therefore, the low activity of PpUGT78A1 for anthocyanidins is likely responsible for the traceable amount of Cy-3-gal in peach flowers.
In flowers, both PpUGT78A1 and PpUGT78A2 are expressed, but the transcripts of PpUGT78B are almost undetectable. As mentioned above, PpUGT78A1 has very low activity toward cyanidin, while PpUGT78A2 has high activity toward cyanidin with UDP-Glc as sugar. Therefore, PpUGT78A2 is responsible for the accumulation of Cy-3-glu in flowers. However, more studies are still needed to address the mechanism of functional divergence between the two clustered UGT78 genes PpUGT78A1 and PpUGT78A2. In addition, PpUGT78A2 contains the residue Asn (N) at position 44 of the PSPG box, whose activity toward the recognition of the sugar donor has not been reported. It is unclear if PpUGT78A2 also can recognize the UDP-Rha donor, leading to the accumulation of Cy-3-rha in flowers of peach.
PpUGT79A and PpUGT79B are identified in the peach transcriptome, and they are homologs of the kiwifruit and petunia F3GGTs responsible for the glycosylation of anthocyanidin 3-O-glycosides (Morita et al., 2005; Montefiori et al., 2011). PpUGT79B is not expressed in fruits, but its transcripts are abundant in leaves and flowers. Both flowers and leaves accumulate Cy-3-rut and Pn-3-rut, but they are almost undetectable in fruits. Therefore, there is no doubt that PpUGT79B is involved in the accumulation of Cy-3-rut and Pn-3-rut in peach. Moreover, transcripts of PpUGT79A are also found in the flowers of P. davidana, but its expression is not in accordance with the accumulation of Cy-3-rut and Pn-3-rut in red-colored tissues (Supplemental Fig. S3A). In contrast, the expression of PpUGT79B is well in accordance with the accumulation of Cy-3-rut and Pn-3-rut. Taken together, these results suggest that PpUGT79B is involved in the biosynthesis of Cy-3-rut and Pn-3-rut in peach. It is worth noting that functional redundancy is often observed for secondary metabolite genes in plants (Hanada et al., 2011). We cannot exclude the possibility that PpUGT79A also can catalyze the glycosylation of anthocyanins in P. davidana.
Two genes in the peach genome, PpAOMT1 and PpAOMT2, are closely related to VvAOMT, which has been shown to participate in the methylation of anthocyanins in grapevine (Hugueney et al., 2009). A significant correlation is observed between the expression level of PpAOMT2 and the accumulation of peonidin-based anthocyanins in red-colored tissues, but not for PpAOMT1 (Supplemental Fig. S3B). Moreover, PpAOMT2 has been shown to be functional in vitro and in vivo. These results strongly suggest that PpAOMT2 is responsible for anthocyanin methylation in peach. Besides PpAOMT2, PpAOMT1 also is expressed in flowers and leaves. However, the functionality of PpAOMT1 remains unclear, as it shows no activity toward cyanidin in vitro.
Taking all the results above, we propose a model underlying the structural diversification of anthocyanins in peach (Fig. 9).
Figure 9.
A proposed model of the glycosylation and methylation of anthocyanins in peach. [See online article for color version of this figure.]
Differential Expression Pattern of Genes Involved in Anthocyanin Modification in Plants
Tissue-specific expression profiles of regulatory genes involved in anthocyanin biosynthesis have been reported in previous studies. For example, four R2R3 MYB transcription factors (TFs), Anthocyanin2 (AN2), AN4, PURPLE HAZE (PHZ), and DEEP PURPLE (DPL), have been isolated in petunia. AN2 and AN4 are responsible for the coloration of floral tissues, including petal limb, flower tube, and anther (Quattrocchio et al., 1999), while PHZ and DPL mainly control anthocyanin accumulation in vegetative tissues such as leaves and stems (Albert et al., 2011). Similarly, two basic helix-loop-helix genes, B-I and B-Peru, confer anthocyanin pigmentation on vegetative tissues and seeds in maize, respectively (Selinger and Chandler, 1999). However, there are few reports of tissue-specific expression profiles of structural genes involved in anthocyanin modification.
Here, we characterize the expression profiles of gene families encoding F3GT, F3GGT, and AOMT in peach. Like the R2R3 MYB TFs in petunia and maize, two F3GT genes, PpUGT78A2 and PpUGT78B, also show tissue-specific expression. PpUGT78A2 is predominantly expressed in flowers, while PpUGT78B is mainly expressed in leaves and fruits. Similarly, two F3GGT genes, PpUGT79A and PpUGT79B, are not expressed in fruits but are expressed in leaves and/or flowers. Two AOMT genes, PpAOMT1 and PpAOMT2, are mainly expressed in flowers, but with no or low expression in fruits and leaves. Spatiotemporal control of the expression of genes involved in the methylation and glycosylation of anthocyanins results in the differences of anthocyanin composition in flower, leaf, and fruit tissues of peach. Moreover, UFGT is a key gene in anthocyanin biosynthesis, and its expression is induced by MYB TFs (Kobayashi et al., 2002). To date, seven R2R3 MYB TFs have been identified in the peach genome (Rahim et al., 2014; Uematsu et al., 2014). It is worth studying whether the expression of PpUGT78 genes is induced by specific MYB TFs, leading to the spatial and temporal patterns of anthocyanin synthesis in peach.
The UGT78II Genes Represent a Novel Branch of the UGT78 Gene Family in Plants
In this study, we have comprehensively investigated the evolutionary development of UGT78 genes in 20 sequenced plant genomes. In eudicots, UGT78 genes can be divided into two groups, UGT78I and UGT78II. It is well known that the Arabidopsis UGT intron 2 is highly conserved among the UGT78 gene family (Li et al., 2001; Paquette et al., 2003). Interestingly, a single intron equivalent to the Arabidopsis UGT intron 2 has been detected in the UGT78I group but is nearly absent in the UGT78II group. Only one gene, GmF3GT2-2, in the UGT78II group contains intron 2. Moreover, the PSPG motif of the UGT78I group contains a highly conserved amino acid residue, Trp (W), at position 22, while residue 22 has great variation in the UGT78II group. In monocots, the UGT78 genes, like the eudicot UGT78I genes, also contain intron 2 and the conserved amino acid residue Trp at position 22. This suggests that the UGT78I genes in eudicots and the UGT78 genes in monocots are derived from a common ancestor. However, the UGT78II genes can be well distinguished from other UGT78 genes in plants based on the combination of intron 2 and the amino acid residue at position 22. Thus, the UGT78II genes represent a novel branch of the UGT78 gene family in plants.
As mentioned above, four of the five characterized UGT78Ia genes show activity toward flavonols (Modolo et al., 2007; Ono et al., 2010; Fukuchi-Mizutani et al., 2011), while four of the six characterized UGT78Ib genes accept both anthocyanins and flavonols (Ford et al., 1998; Kubo et al., 2007; Griesser et al., 2008; Fukuchi-Mizutani et al., 2011). To date, two UGT78II genes, GmF3GT2-6 in soybean and PtF3GTII-2 in poplar, have been characterized to function as a 3-O-glycosyltransferase in anthocyanin and flavonol biosynthesis (Kovinich et al., 2010; Veljanovski and Constabel, 2013). This indicates that the UGT78II genes are more closely related to the UGT78Ib genes than to the UGT78Ia genes. Thus, it is reasonable to speculate that the common ancestor of UGT78I and UGT78II groups is likely to have activity toward anthocyanins and flavonols. The UGT78II group retains the ancestral function, while the UGT78I group may have undergone functional divergence, dividing genes into the two subgroups UGT78Ia and UGT78Ib. In addition, the UGT78II genes are only found in five of 10 plant species in eurosids I and are absent in the genomes of 10 plant species in eurosids II, vitales, and asterids. It is unclear if the UGT78II genes tend to be lost in eudicots.
The Difference of Anthocyanin Composition between Peach and Its Wild Relatives and Its Implications for Inferring Peach Domestication History
In this study, six anthocyanins are identified in flowers of peach and its wild relatives. Interestingly, peach and P. ferganensis show a similar anthocyanin composition in flowers. However, peach is different from both P. davidiana and P. kansueasis in terms of anthocyanin composition in flowers. These results clearly demonstrate that peach is closely related to P. ferganensis, native to Xinjiang Province of northwestern China and the Ferghana and Zeravshan valleys in central Asia (Scorza and Okie, 1990). Peach was first domesticated in the region of northwestern China between the Tarim basin and the north slopes of the Kunlun Shan Mountains (Faust and Timon, 1995). Thus, it seems likely that peach is domesticated from P. ferganensis. This finding is supported by the latest study of peach genome sequencing, which indicates that P. ferganensis represents an intermediate genome in peach domestication (Verde et al., 2013). In addition, all seven of the tested peach cultivars share a similar pattern of anthocyanin composition in flowers, which is consistent with the fact that peach cultivars have a very narrow genetic base (Scorza et al., 1985).
The peach blossom has always been a sacred symbol in Chinese culture. Peach and P. ferganensis produce red or purple flowers, while both P. davidiana and P. kansueasis have pink or pale pink flowers. In other words, the flowers of peach and P. ferganensis are more attractive than those of P. davidiana and P. kansueasis. If peach is indeed domesticated from P. ferganensis, then the question arises of the purpose of the original domestication of peach. Both P. davidiana and P. kansueasis bear edible fruits, and P. kansueasis is also native to Xinjiang Province of northwestern China. The original cultivation of peach might not have been very important as a source of fruits but could be important as a source of valuable ornamental plants due to the beauty of flowers. Moreover, our study shows that the flowers of peach and P. ferganensis are richer in anthocyanin components than those of P. davidiana and P. kansueasis. Thus, the complexity of anthocyanin composition is likely related to the diversity of flower color in peach and its wild relatives.
Peach flowers are a common ingredient used in traditional Chinese medicine. Here, we show that peach flowers accumulate a variety of anthocyanins, whereas the fruits and leaves predominantly accumulate a single anthocyanin, Cy-3-glu. Several studies have demonstrated that both peonidin-based and cyanidin-based anthocyanins can inhibit cancer cell growth (Chen et al., 2005; Ho et al., 2010). Therefore, peach flowers are a more suitable ingredient to use in medicine than leaves and fruits in terms of the anthocyanin benefits.
CONCLUSION
Methylation and glycosylation contribute to the diversification of anthocyanins in peach. Peach and P. ferganensis share a similar anthocyanin composition in flowers, suggesting that they are closely related to each other. Four UGT genes, PpUGT78A1, PpUGT78A2, PpUGT78B, and PpUGT79B, are likely responsible for the glycosylation of anthocyanins, while one gene, PpAOMT2, is involved in the methylation of anthocyanins in peach. The spatiotemporal control of expression of these anthocyanin modification genes is responsible for the different patterns of anthocyanin accumulation in flower, leaf, and fruit tissues. In addition, our study reveals the divergent evolution of both UGT78 and AOMT genes in peach.
MATERIALS AND METHODS
Plant Materials
Peach (Prunus persica) accessions used in this study consisted of seven ornamental cultivars, Juhuatao, Fenhuashouxintao, Hongcuizhi, Honghuashouxintao, MT, Hongbaihuatao, and HY; two other cultivars, red-fleshed WH9 and white-fleshed BF; and three wild relatives, Prunus davidiana, Prunus ferganensis, and Prunus kansueasis. All the peach accessions are maintained at the Wuhan Botanical Garden of the Chinese Academy of Sciences in Hubei Province. Leaves were collected during juvenile stage in spring. Fruits were harvested at mature stage. Flowers were collected at pink, balloon, and blossom stages. All samples were immediately frozen in liquid nitrogen and then stored at −75°C until use.
Extraction and HPLC-ESI-MS/MS Analysis of Anthocyanins
The anthocyanins were extracted according to a previously reported method (Huang et al., 2009). Briefly, an approximately 0.5-g sample was ground in liquid nitrogen and then added to 25 mL of extraction solution (80:20 [v/v] methanol:water mixture containing 1.18 mm HCl). The mixture was incubated on a steam bath in the dark at 30°C with gentle shaking for 16 h, sonicated for 15 min, and centrifuged at 10,000g for 10 min. An aliquot of 10 mL of supernatant was collected and evaporated under vacuum at 30°C using a rotary evaporator. The residual was resuspended in acidified water (1.18 mm HCl) and added to a CNWBOND LC-C18 SPE column that was preconditioned with 5 mL of methanol followed by 5 mL of acidified water. The SPE column was then washed three times with acidified water.
Anthocyanins were eluted with 1 mL of methanol, filtered through 0.22-μm Millipore membranes, and analyzed using an HPLC-ESI-MS/MS system (ThermoFisher Scientific). The analytical column was Zorbax Extend C18, 4.6 × 250 mm, with a particle size of 5 μm (Agilent Technologies). The analytical column was sequentially eluted using mobile phase A (formic acid:water, 5:95, v/v) and mobile phase B (methanol) with a flow rate of 0.8 mL min−1. The linear gradient of phase B was as follows: 0 to 2 min, 5%; 2 to 7 min, 5% to 15%; 7 to 20 min, 15% to 20%; 20 to 25 min, 20% to 27%; 25 to 32 min, 27%; 32 to 41 min, 27% to 35%; and 41.01 to 43 min, 5%. A UV-visible light detector wavelength was set at 520 nm. Mass spectra were acquired in positive ion mode. Ion was scanned from 200 to 1,000 mass-to-charge ratio at a speed of 1,000 atomic mass units s−1. The flow rate of nebulizing gas was 1.5 L min−1, and the drying gas (N2) pressure was 0.1 MPa. The components of anthocyanins were determined according to the retention time of standards and Mr and subsequently quantified according to cyanidin equivalents.
Hydrolyzation of Anthocyanins
Anthocyanins were hydrolyzed into anthocyanidins under acidic and high-temperature conditions according to a previously reported procedure (Pinho et al., 2011). Approximately 1 g of ground sample was mixed with 10 mL of 6 m HCl and 40 mL of methanol. The mixture was flushed with nitrogen and incubated in a water bath at 100°C for 2 h. After acid hydrolysis, the mixture was cooled, mixed with 50 mL of methanol, sonicated for 15 min, and centrifuged at 10,000g for 10 min. The supernatant was purified using the SPE column as mentioned above. The components of anthocyanidins were analyzed using the HPLC-ESI-MS system. The mobile phase was composed of 3:57:40 (v/v/v) formic acid:water:methanol solution. The column flow rate was 0.8 mL min−1. The sample running time was 35 min. Anthocyanidins were recalled according to MS data, retention time, and Mr of internal standards.
Expression of the Peach AOMT and UGT78 Genes in Escherichia coli
The full complementary DNA (cDNA) sequences of PpAOMT1 and PpAOMT2 and were amplified using two pairs of primers (the XbaI and Xhol sites at the 5′ end of the forward and reverse primers, respectively, are indicated in bold), 5′-GGGAATTCCATATGATGGCAGAAGAAGGCAAACA-3′/5′-AGCCGCTCGAGCCACATTATTTGAAGACAACAGC-3′ and 5′-GGGAATTCCATATGTTGGCTCTTCTGTTCAACATGG-3′/5′-AGCCGCTCGAGCTAGTATCCTAATAGAGACGCCTGC-3′, respectively, and cloned individually into pET-28b(+) vector (Novagen). Clone authenticity was confirmed by sequencing. The constructed vector was then transferred into E. coli BL21 (DE3) cells (Transgen). The transformed cells were cultured at 37°C in Luria-Bertani medium containing 25 μg mL−1 kanamycin to an optical density at 600 nm of 0.4 to 0.6. The culture was adjusted to 18°C, and recombinant AOMT expression was induced by the addition of isopropylthio-β-galactoside to a final concentration of 5 mm. The culture was incubated for 20 h at 18°C (250 rpm) prior to harvesting the cells. Then, recombinant AOMT was purified using His•Bind Resin (Novagen). The activity of purified recombinant AOMT (5 μg) was measured in a final volume of 200 μL containing 200 μm S-adenosyl-l-methionine, 200 μm anthocyanin substrate, 0.1 m HEPES, pH 7.5, 10 mm MgCl2, and 14 mm 2-mercaptoethanol. The reaction was incubated for 30 min at 35°C and stopped with 400 μL of methanol containing 200 mm HCl. All reactions were filtered through 0.22-μm Millipore membranes and analyzed by HPLC-ESI-MS/MS. Assays were performed in triplicate, and experiments were repeated three times. Likewise, the full coding sequences of three genes, PpUGT78A1, PpUGT78A2, and PpUGT78B, were cloned into pET-28b(+) vector using three pairs of primers, 5′-GGGGATCCATGGTAGGAAACCCCAAACCC-3′/5′-GCCTCGAGCGTTTTGGACCTAGATCTAAGAAGC-3′, 5′-GGGAATTCATGGCAGGAAACCCCAAACA-3′/5′-AGCTCGAGCCTTTTTCACTGCCCATTCA-3′, and 5′-CCGAATTCATGGCACCACAACCGATT-3′/5′-GCCTCGAGGCGAGATAAATAGTAGAAGCAAAGC-3′, respectively. The activity of recombinant UGT78 was measured in a final volume of 200 μL containing 5 mm UDP-Glc or UDP-Gal, 250 mm substrate, 0.1 m HEPES, pH 7, 14 mm 2-mercaptoethanol, and raw protein extract with approximately 1 μg of recombinant protein. Enzyme assays were incubated for 30 min at 30°C and stopped by the addition of 250 μL of 0.6 m HCl.
Transient Expression in Tobacco
For Agrobacterium tumefaciens-mediated transient expression, PpAOMT2 from peach and NtAN2 from tobacco (Nicotiana tabacum) were amplified using two pairs of primers, 5′-GGGAATTCCATATGATGGCAGAAGAAGGCAAACA-3′/5′-AGCCGCTCGAGCCACATTATTTGAAGACAACAGC-3′ and 5′-CCGCTCGAGATGAATATTTGTACTAATAAG-3′/5′-GCTCTAGATCAACTGAGAAGTGGCATTTC-3′, respectively, and inserted individually into the modified binary vector pSAK277. All constructs were introduced into A. tumefaciens strain GV3101 by electroporation. Nicotiana benthamiana leaves were infiltrated with A. tumefaciens cultures (optical density at 600 nm of 0.1–0.3). Discs were punched from N. benthamiana leaves 1 week after A. tumefaciens infiltration and analyzed for anthocyanin content.
Gene Expression Analysis Using Real-Time PCR
Total RNA was extracted using the ZP401 kit (Beijing Zoman Biotechnology) according to the manufacturer’s instructions and was treated with DNase I (Takara) to remove any contamination of genomic DNAs. Approximately 3 μg of total RNA per sample was then used for cDNA synthesis. A SYBR Green-based real-time PCR assay was carried out in a total volume of 20 µL of reaction mixture containing 10 µL of 2× SYBR Green I Master Mix (Takara), 0.2 µm of each primer, and 100 ng of template cDNA. An actin gene encoding glyceraldehyde-3-phosphate dehydrogenase, PpGAPDH, in peach was used as a constitutive control.
Amplifications were performed using the StepOnePlus Real-Time PCR System (Applied Biosystems). The amplification program consisted of one cycle of 95°C for 3 min followed by 40 cycles of 95°C for 30 s and 60°C for 34 s. The fluorescent product was detected at the second step of each cycle. Melting curve analysis was performed at the end of 40 cycles to ensure the proper amplification of target fragments. Fluorescence readings were consecutively collected during the melting process from 60°C to 90°C at the heating rate of 0.5°C s−1. All analyses were repeated three times using biological replicates. Primer sequences used for real-time PCR are listed in Supplemental Table S2.
Sequence Alignment and Phylogenetic Analysis
The amino acid sequences of genes encoding UGT and AOMT from various plants were used for phylogenetic analysis. Sequences were aligned using ClustalX and adjusted manually as necessary. The resulting data were analyzed using equally weighted neighbor joining. Neighbor-joining trees were sought using the heuristic search strategies of MEGA version 5. Bootstrap values (Felsenstein, 1985) were calculated from 1,000 replicate analyses, and only those values compatible with the 50% majority-rule consensus tree were recorded. A UGT gene of Picea sitchensis (GenBank accession no. ABK24936) was used as an outgroup.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree derived from amino acid sequences of genes encoding UGT in the peach transcriptome.
Supplemental Figure S2. Estimation of gene expression levels of F3GT, F3GGT, and AOMT homologs using fragments per kilobases per million reads values in peach.
Supplemental Figure S3. Correlation between the expression of the anthocyanin modification genes and anthocyanin accumulation in different tissues of peach.
Supplemental Table S1. Genes encoding UDP identified in the peach transcriptome.
Supplemental Table S2. Primers used for real-time PCR analysis of genes encoding UGT78, UGT79, UGT94, and OMT in peach.
Supplementary Material
Glossary
- PSPG
plant secondary product glycosyltransferase
- RNA-Seq
RNA sequencing
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- ESI
electrospray ionization
- Cy-3-glu
cyanidin 3-glucoside
- Cy-3-gal
cyanidin 3-galactoside
- Cy-3-rut
cyanidin 3-rutinoside
- Pn-3-glu
peonidin 3-glucoside
- Cy-3-rha
cyanidin 3-rhamnoside
- Pn-3-rut
peonidin 3-rutinoside
- CDS
coding sequence
- MT
Mantianhong
- HY
Hongyetao
- BF
Baifeng
- TFs
transcription factors
- cDNA
complementary DNA
Footnotes
This work was supported by the National Program on Key Basic Research Projects of China (973 Program; grant no. 2011CB100600) and the National 863 Program of China (grant no. 2011AA10020600–02).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. (2011) Members of an R2R3-MYB transcription factor family in petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65: 771–784 [DOI] [PubMed] [Google Scholar]
- Arús P, Verde I, Sosinski B, Zhebentyayeva T, Abbott AG. (2012) The peach genome. Tree Genet Genomes 8: 1–17 [Google Scholar]
- Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S. (2012) A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J 69: 1030–1042 [DOI] [PubMed] [Google Scholar]
- Chen PN, Chu SC, Chiou HL, Chiang CL, Yang SF, Hsieh YS. (2005) Cyanidin 3-glucoside and peonidin 3-glucoside inhibit tumor cell growth and induce apoptosis in vitro and suppress tumor growth in vivo. Nutr Cancer 53: 232–243 [DOI] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust M, Timon B (1995) Origin and dissemination of peach. Hort Rev 17: 331–379 [Google Scholar]
- Fedoroff NV, Furtek DB, Nelson OE. (1984) Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element Activator (Ac). Proc Natl Acad Sci USA 81: 3825–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phytogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Feng S, Wang Y, Yang S, Xu Y, Chen X. (2010) Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232: 245–255 [DOI] [PubMed] [Google Scholar]
- Ford CM, Boss PK, Hoj PB. (1998) Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J Biol Chem 273: 9224–9233 [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, Hugueney P, Verriès C, This P, Ageorges A. (2011) Genetic mechanisms underlying the methylation level of anthocyanins in grape (Vitis vinifera L.). BMC Plant Biol 11: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M, Akagi M, Ishiguro K, Katsumoto Y, Fukui Y, Togami J, Nakamura N, Tanaka Y. (2011) Biochemical and molecular characterization of anthocyanidin/flavonol 3-glucosylation pathways in Rosa × hybrid. Plant Biotechnol 28: 239–244 [Google Scholar]
- Griesser M, Hoffmann T, Bellido ML, Rosati C, Fink B, Kurtzer R, Aharoni A, Muñoz-Blanco J, Schwab W. (2008) Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol 146: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Sawada Y, Kuromori T, Klausnitzer R, Saito K, Toyoda T, Shinozaki K, Li WH, Hirai MY. (2011) Functional compensation of primary and secondary metabolites by duplicate genes in Arabidopsis thaliana. Mol Biol Evol 28: 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho ML, Chen PN, Chu SC, Kuo DY, Kuo WH, Chen JY, Hsieh YS. (2010) Peonidin 3-glucoside inhibits lung cancer metastasis by downregulation of proteinases activities and MAPK pathway. Nutr Cancer 62: 505–516 [DOI] [PubMed] [Google Scholar]
- Huang Z, Wang B, Williams P, Pace RD. (2009) Identification of anthocyanins in muscadine grapes with HPLC-ESI-MS. LWT-Food Sci Technol 42: 819–824 [Google Scholar]
- Hugueney P, Provenzano S, Verriès C, Ferrandino A, Meudec E, Batelli G, Merdinoglu D, Cheynier V, Schubert A, Ageorges A. (2009) A novel cation-dependent O-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol 150: 2057–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima J, Schäffner AR, Saito K. (2003) UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278: 43910–43918 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H. (2004) Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Ishimaru M, Hiraoka K, Honda C. (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215: 924–933 [DOI] [PubMed] [Google Scholar]
- Kong JM, Chia LS, Goh NK, Chia TF, Brouillard R. (2003) Analysis and biological activities of anthocyanins. Phytochemistry 64: 923–933 [DOI] [PubMed] [Google Scholar]
- Kovinich N, Saleem A, Arnason JT, Miki B. (2010) Functional characterization of a UDP-glucose:flavonoid 3-O-glucosyltransferase from the seed coat of black soybean (Glycine max (L.) Merr.). Phytochemistry 71: 1253–1263 [DOI] [PubMed] [Google Scholar]
- Kroon J, Souer E, de Graaff A, Xue Y, Mol J, Koes R. (1994) Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: characterization of insertion sequences in two mutant alleles. Plant J 5: 69–80 [DOI] [PubMed] [Google Scholar]
- Kubo A, Arai Y, Nagashima S, Yoshikawa T. (2004) Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch Biochem Biophys 429: 198–203 [DOI] [PubMed] [Google Scholar]
- Kubo H, Nawa N, Lupsea SA. (2007) Anthocyaninless1 gene of Arabidopsis thaliana encodes a UDP-glucose:flavonoid-3-O-glucosyltransferase. J Plant Res 120: 445–449 [DOI] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim EK, Bowles DJ. (2001) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, Shu HR, Hao YJ. (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160: 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücker J, Martens S, Lund ST. (2010) Characterization of a Vitis vinifera cv. Cabernet Sauvignon 3′,5′-O-methyltransferase showing strong preference for anthocyanins and glycosylated flavonols. Phytochemistry 71: 1474–1484 [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Bélanger A, Fournel-Gigleux S, Green M, Hum DW, Iyanagi T, et al. (1997) The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7: 255–269 [DOI] [PubMed] [Google Scholar]
- Marrs KA, Alfenito MR, Lloyd AM, Walbot V. (1995) A glutathione S-transferase involved in vacuolar transfer encoded by the maise gene Bronze-2. Nature 375: 397–400 [DOI] [PubMed] [Google Scholar]
- Modolo LV, Blount JW, Achnine L, Naoumkina MA, Wang X, Dixon RA. (2007) A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol Biol 64: 499–518 [DOI] [PubMed] [Google Scholar]
- Montefiori M, Espley RV, Stevenson D, Cooney J, Datson PM, Saiz A, Atkinson RG, Hellens RP, Allan AC. (2011) Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J 65: 106–118 [DOI] [PubMed] [Google Scholar]
- Morita Y, Hoshino A, Kikuchi Y, Okuhara H, Ono E, Tanaka Y, Fukui Y, Saito N, Nitasaka E, Noguchi H, et al. (2005) Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2″-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J 42: 353–363 [DOI] [PubMed] [Google Scholar]
- Ogata J, Kanno Y, Itoh Y, Tsugawa H, Suzuki M. (2005) Plant biochemistry: anthocyanin biosynthesis in roses. Nature 435: 757–758 [DOI] [PubMed] [Google Scholar]
- Ono E, Fukuchi-Mizutani M, Nakamura N, Fukui Y, Yonekura-Sakakibara K, Yamaguchi M, Nakayama T, Tanaka T, Kusumi T, Tanaka Y. (2006) Yellow flowers generated by expression of the aurone biosynthetic pathway. Proc Natl Acad Sci USA 103: 11075–11080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E, Homma Y, Horikawa M, Kunikane-Doi S, Imai H, Takahashi S, Kawai Y, Ishiguro M, Fukui Y, Nakayama T. (2010) Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell 22: 2856–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette S, Møller BL, Bak S. (2003) On the origin of family 1 plant glycosyltransferases. Phytochemistry 62: 399–413 [DOI] [PubMed] [Google Scholar]
- Pinho C, Melo A, Mansilha C, Ferreira IM. (2011) Optimization of conditions for anthocyanin hydrolysis from red wine using response surface methodology (RSM). J Agric Food Chem 59: 50–55 [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim MA, Busatto N, Trainotti L. (May 15, 2014) Regulation of anthocyanin biosynthesis in peach fruits. Planta 10.1007/s00425-014-2078-2 [DOI] [PubMed] [Google Scholar]
- Sarni P, Fulcrand H, Souillol V, Souquet JM, Cheynier V. (1995) Mechanisms of anthocyanin degradation in grape must-like model solutions. J Sci Food Agric 69: 385–391 [Google Scholar]
- Sawada S, Suzuki H, Ichimaida F, Yamaguchi MA, Iwashita T, Fukui Y, Hemmi H, Nishino T, Nakayama T. (2005) UDP-glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers: enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J Biol Chem 280: 899–906 [DOI] [PubMed] [Google Scholar]
- Scorza R, Mehlenbacher SA, Lightner GW. (1985) Inbreeding and coancestry of freestone peach cultivars of the eastern United States and implications for peach germplasm improvement. J Am Soc Hortic Sci 110: 547–552 [Google Scholar]
- Scorza R, Okie WR. (1990) Peaches (Prunus): genetic resources of temperate fruit and nut crops. Acta Hortic 290: 177–231 [Google Scholar]
- Selinger DA, Chandler VL. (1999) Major recent and independent changes in levels and patterns of expression have occurred at the b gene, a regulatory locus in maize. Proc Natl Acad Sci USA 96: 15007–15012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR. (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142: 1216–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Uematsu C, Katayama H, Makino I, Inagaki A, Arakawa O, Martin C. (2014) Peace, a MYB-like transcription factor, regulates petal pigmentation in flowering peach ‘Genpei’ bearing variegated and fully pigmented flowers. J Exp Bot 65: 1081–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljanovski V, Constabel CP. (2013) Molecular cloning and biochemical characterization of two UDP-glycosyltransferases from poplar. Phytochemistry 91: 148–157 [DOI] [PubMed] [Google Scholar]
- Verde I, Abbott AG, Scalabrin S, Jung S, Shu S, Marroni F, Zhebentyayeva T, Dettori MT, Grimwood J, Cattonaro F, et al. (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45: 487–494 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao S, Gu C, Zhou Y, Zhou H, Ma J, Cheng J, Han Y. (2013) Deep RNA-Seq uncovers the peach transcriptome landscape. Plant Mol Biol 83: 365–377 [DOI] [PubMed] [Google Scholar]
- Wiering H, deVlaming P. (1977) Glycosylation and methylation patterns of anthocyanins in Petunia hybrida. II. The genes Mr1 and Mf2. Pflanzenzucht 78: 113–123 [Google Scholar]
- Winkel BSJ. (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55: 85–107 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Yamagishi E, Gong Z, Fukuchi-Mizutani M, Fukui Y, Tanaka Y, Kusumi T, Yamaguchi M, Saito K. (2002) Two flavonoid glucosyltransferases from Petunia hybrida: molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol Biol 48: 401–411 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromori T, Ito T, Shinozaki K, et al. (2012) Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J 69: 154–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Hanada K. (2011) An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J 66: 182–193 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huhman D, Shadle G, He XZ, Sumner LW, Tang Y, Dixon RA. (2011) MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23: 1536–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.