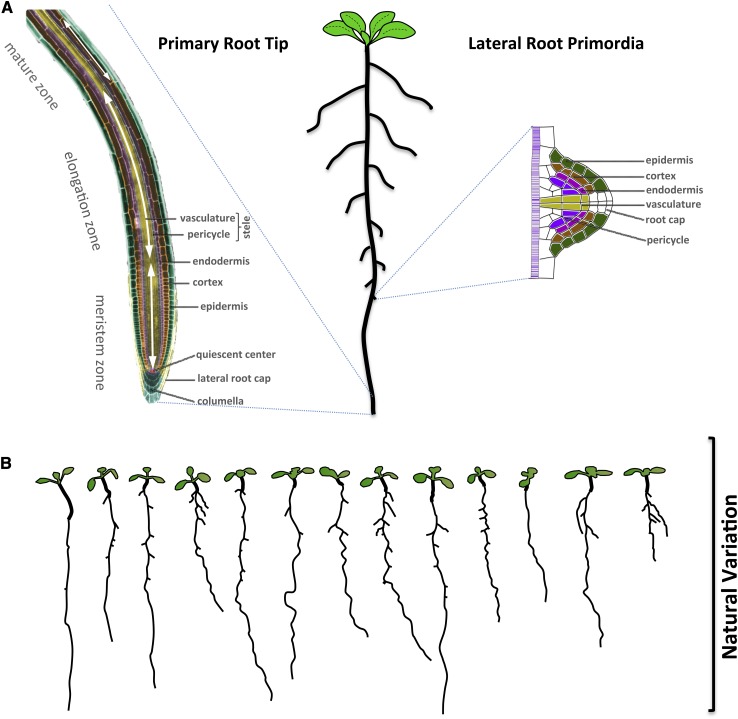
Natural variation of root growth informs on processes that govern root development, responses to nutrient availability, and ion uptake and homeostasis.
Abstract
The root system has a crucial role for plant growth and productivity. Due to the challenges of heterogeneous soil environments, diverse environmental signals are integrated into root developmental decisions. While root growth and growth responses are genetically determined, there is substantial natural variation for these traits. Studying the genetic basis of the natural variation of root growth traits can not only shed light on their evolution and ecological relevance but also can be used to map the genes and their alleles responsible for the regulation of these traits. Analysis of root phenotypes has revealed growth strategies and root growth responses to a variety of environmental stimuli, as well as the extent of natural variation of a variety of root traits including ion content, cellular properties, and root system architectures. Linkage and association mapping approaches have uncovered causal genes underlying the variation of these traits.
Since their advent more than 400 million years ago, vascular plants have drastically transformed the land surface of our planet and facilitated the dense colonization of its land masses (Algeo and Scheckler, 1998; Gibling and Davies, 2012). Key to this was the evolution of root systems that enable plants to forage their environment for nutrients and water and anchor themselves tightly in the soil substrate. Soils are very heterogeneous environments, and because of the constant need to optimize root distribution in the soil according to sometimes conflicting parameters, root growth and development are some of the most plastic traits in plants. This plasticity is guided by environmental information that is integrated into decisions regarding how fast and in which direction to grow and where and when to place new lateral roots (LRs; Malamy and Ryan, 2001; Malamy, 2005). The distribution and function of roots are of crucial importance for plants. In fact, they are considered the most limiting factors for plant growth in almost all natural ecosystems (Den Herder et al., 2010). Not surprisingly, the plant root system plays a major role in yield and overall plant productivity (Lynch, 1995; Den Herder et al., 2010).
The extent of plasticity is determined by genetic components (Pigliucci, 2005). For instance, one ecotype of a plant species may be able to increase root growth rate on a certain stimulus, whereas another ecotype lacks this characteristic (Gifford et al., 2013). The genetic components that govern traits in different ecotypes represent the outcome of adaptation arising from the selection of those traits that allow better adapted populations to reproduce more successfully (higher fitness) than less well-adapted populations (Trontin et al., 2011; Savolainen, 2013). Although local adaptation is common in plants and animals, its genetic basis is still poorly understood (Savolainen et al., 2013). Traits that drive local adaptation are often quantitative traits shaped by multiple genes. Therefore, phenotypic differences are often caused by allelic variation at several loci, each of them making small contributions to the trait (Weigel and Nordborg, 2005; Rockman, 2012). Studying the genetic basis of the natural variation of traits cannot only shed light on the evolution of these traits and their ecological relevance but also, can be used to map the genes responsible for the regulation of these traits.
Most efforts to study intraspecies genetic variation to find trait-governing genes or identify useful traits have been conducted in crop species and the model plant Arabidopsis (Arabidopsis thaliana). Whereas in crop species, traits that are used have been subjected to human-directed selection during domestication, often with the aim of increasing productivity, in Arabidopsis, it is mostly natural selection that is examined. Arabidopsis is widely distributed around the world, inhabiting diverse environments that include beaches, rocky slopes, riverbanks, roadsides, and areas surrounding agriculture fields (Horton et al., 2012). A large number of accessions has been collected over the past decades from locations all over the world and made available to the scientific community. Importantly, these accessions of Arabidopsis exhibit a striking diversity of phenotypic variation of morphology and physiology (Koornneef et al., 2004) and can be used to understand the genetic and molecular bases of traits using quantitative genetics. Variations of traits are measured in a panel of genetically distinct plant strains and then correlated with the occurrence of genetic markers in these plants. Linked or associated genome regions can eventually be identified, and additional analysis can be conducted to find the causal genes. Self-fertilizing species, such as Arabidopsis, are particularly suited for such approaches, because they can be maintained as inbred lines and therefore, need to be genotyped only one time, after which they can be phenotyped multiple times. In the past, natural variation has been used to map causal genes mainly by using recombinant inbred lines (RILs) approaches; these are very powerful but lack a high mapping resolution, and they can only capture a very small subset of the allelic diversity (Korte and Farlow, 2013). However, the advent of new and cheap large-scale genotyping and sequencing technologies has enabled large-scale, high-resolution genotyping (Horton et al., 2012) and even the complete sequencing of a large number of plant strains (http://1001genomes.org; 3,000 Rice Genomes Project, 2014). With these data, genome-wide association studies (GWASs) for identification of alleles responsible for many different quantitative traits have become feasible (Weigel, 2012). In these studies, traits of a large number of accessions are measured and subsequently associated with genotyped markers, most frequently single-nucleotide polymorphism. Although GWASs are a very powerful tool and in principle, allow for a high mapping accuracy, a notable disadvantage is that the complexity of the population structure can confound these studies. However, there has been remarkable progress addressing this issue (Atwell et al., 2010; Segura et al., 2012).
In this review, we highlight recent progress in understanding the genetic bases of natural variation of growth, development, and physiology of the root system. After briefly explaining how root growth and development give rise to the root system architecture (RSA), we highlight natural variation and what has been learned from it for fundamental processes in root growth and development, root growth responses to nutrient availability, and ion uptake and homeostasis.
ROOT GROWTH AND RSA
Growth and development of the primary root (PR) and higher-order roots lead to a three-dimensional root system. Such a root system represents a highly complex and integrated organ that is important for plant productivity, because many soil resources are unevenly distributed and the spatial organization of the root system and the ability to perform optimal foraging, therefore, determine the capacity of a plant to exploit available resources (Lynch, 1995). The spatial configuration of this root system in its environment is termed RSA (Lynch, 1995). RSA is shaped by a complex interplay of different growth traits.
Much of the molecular basis of root traits has been learned using the root of Arabidopsis. This is not only because of the readily available genetic tools and resources of this model plant but also, because roots of Arabidopsis can be easily cultured on agar plates and these small and simply structured transparent roots allow for live microscopy. Like other dicotyledonous plants, Arabidopsis has one main or PR and many LRs. Only the PR is present as the radicle in the plant embryo, although the PR undergoes intensive postembryonic growth and development (Grunewald et al., 2007; Fig. 1A). The longitudinal axis represents a developmental timeline, because cell division, cell elongation, and differentiation occur in specific zones of the root system (Birnbaum et al., 2003). After the PR has grown for multiple days, LRs start to form and emerge. In Arabidopsis, LRs develop from special pericycle cells called xylem pole pericycles, which display meristematic characteristics such as a dense cytoplasm and small vacuoles (Himanen et al., 2004; Parizot et al., 2008). Formation of LRs includes three main stages: initiation, primordial formation, and emergence/elongation (Malamy and Benfey, 1997).
Figure 1.
Tissue architecture and natural variation of the root of Arabidopsis. A, The Arabidopsis root and its developing tissues. Schematic of a young Arabidopsis root (center). The developmental zones of the PR tip and its tissue architecture (left) and LR primordium (right). Different tissue types are indicated by different colors. B, Natural variation of root growth. Graphical depiction of 10-d-old seedlings of 13 divergent Arabidopsis accessions grown at the same time on 0.2-strength MS.
RSA is shaped by not only growth rates of the PR and LRs and branching patterns but also, the growth direction of each root of the root system. Gravity represents an important stimulus for the young roots to grow downward in the soil and thus, be able to acquire water and nutrients for the plant (Kutschera and Briggs, 2012). In Arabidopsis, whereas the PR grows along the vector of gravity (positive gravitropism), LRs initially grow in a more horizontal orientation and only later curve and display positive gravitropism (Kiss et al., 2002).
All the aforementioned developmental processes can be adjusted in response to environmental cues, eventually leading to different RSA configurations (Fig. 1B). The RSA, thus, represents a prime example of phenotypic plasticity, which is the ability of an organism to adopt different phenotypes in response to environmental changes.
USING NATURAL VARIATION TO IDENTIFY REGULATORS OF ROOT GROWTH AND DEVELOPMENT
Natural accessions of Arabidopsis exhibit great variation in different morphological and physiological traits, and this has been used to identify genes and their alleles that regulate these traits (Alonso-Blanco et al., 2009). However, such efforts were largely restricted to processes that could be observed in above-ground tissues, and for almost 10 years, allelic variation of only one gene, BREVIS RADIX (BRX), was known to be specifically involved in natural variation of root development (Mouchel et al., 2004). The BRX gene was identified by quantitative trait loci (QTL) analysis and shown to be responsible for about 80% of the variation of the PR length in an RIL population derived from crossing an accession with a long PR with one with a very short PR. BRX controls the cell proliferation and elongation (Mouchel et al., 2004). Subsequent analysis showed that BRX mediates the interaction between brassinosteroids and auxin pathways, which is required for root growth (Mouchel et al., 2006). Later analysis suggested that BRX is also involved in cytokinin-mediated inhibition of LR development (Li et al., 2009) and furthermore, the auxin-cytokinin cross talk that coordinates cell division and differentiation in the root tip (Scacchi et al., 2010). Combining high-throughput confocal microscopy imaging, GWAS, and expression analysis, a then-uncharacterized gene encoding an F-box protein was found that underlies the natural variation of root development in a broad set of accessions (Meijón et al., 2014). Allelic variation of this gene, coined KURZ UND KLEIN, determines a large fraction of variance for two highly correlated traits: meristem length and mature cell length (Meijón et al., 2014). The major portion of this effect was caused by sequence variation in the coding region of the KURZ UND KLEIN gene (Meijón et al., 2014). Using GWAS for 16 root growth traits on each day of a 5-d time course, 35 highly significant associations could be detected. The most significant association led to the identification of a condition-dependent regulator of root growth. Interestingly, the minor allele of this Calcium Sensor Receptor (CaS) gene was found to be enriched in populations in coastal regions, suggesting that variation in CaS could have adaptive significance for growth in coastal environments (Slovak et al., 2014). A chemical genetics approach combined with mapping using an RIL population that used different sensitivity to a small molecule ([5-(3,4-dichlorophenyl)furan-2-yl]-piperidine-1-ylmethanethione) that inhibits abscisic acid (ABA) -induced gene expression and signaling and eventually leads to a reduction of PR growth identified a Toll-Interleukin1 Receptor nucleotide binding-leucine-rich repeat coding gene and its natural variants underlying these responses (Kim et al., 2012). These findings provided a glimpse of the links between plant immune signaling components and the regulation of root growth.
Recently, light has also been shed on the genetic mechanisms that control root plasticity responses. Using allometric (size to shape) root traits represented in principal component values (Ristova et al., 2013), it was shown that root plasticity of the control accession (ecotype Columbia of Arabidopsis-0 [Col-0]) generated by different hormonal and nitrogen treatments broadly recapitulates the variation found in natural Arabidopsis accessions (Rosas et al., 2013). One particular allometric root phenotype that captures the proportion of LRs along the PR length was used for GWASs, and two genes underlying this trait were identified (PHOSPHATE1 [PHO1] and ROOT SYSTEMS ARCHITECTURE1 [RSA1]; Rosas et al., 2013). PHO1 was previously characterized as a phosphate transporter (Poirier et al., 1991; Hamburger et al., 2002), whereas RSA1 was a novel gene identified in the study to be involved in regulating root allometry (Rosas et al., 2013). Interestingly, these allometric phenotypes were conditional to specific environments, and RSA1 and PHO1 mediated these responses in the context of auxin, ABA, and nitrate signaling pathways (Rosas et al., 2013).
The genetic basis of natural variation of root growth and development was approached in not only Arabidopsis but also, other species. In an interspecies cross of a wild tomato (Solanum peruvianum) and a domesticated tomato (Solanum lycopersicum) species, numerous loci for multiple traits could be mapped, some of which cannot be found in the simple Arabidopsis root (Ron et al., 2013). Multiple studies for traits related to RSA were also conducted in rice (Oryza sativa). Using RIL-based QTL and subsequent near isogenic lines-based fine mapping for a trait for deep rooting in rice, allelic variation of the gene DEEPER ROOTING1 was determined to cause a deep rooting phenotype by affecting the growth angle of roots, which over time, translates into deeper rooting and confers enhanced drought tolerance (Uga et al., 2013). Furthermore, by phenotyping three-dimensional traits of rice RSA, a large number of QTL could be identified in an RIL population (Topp et al., 2013). Finally, GWAS identified 37 loci associated with root traits, of which several genes were already implicated in root development (Courtois et al., 2013).
NATURAL VARIATION OF NUTRIENT AVAILABILITY-DEPENDENT ROOT GROWTH RESPONSES
A hallmark of the plasticity of the root system is that it finely tunes its topology to nutrient availability (Fig. 1B; for comprehensive review, see Giehl and von Wirén, 2014). In particular, changes in nutrient availability lead to effects on root growth rate, LR emergence, and root hair density as well as variations in growth directions. Over time, these nutrient availability-induced changes can have tremendous impact on RSA (for review, see Malamy, 2005; Osmont et al., 2007; Giehl and von Wirén, 2014). In fact, RSA is tuned quantitatively to environmental parameters, which was recently shown by two comprehensive studies evaluating the effect of deficiencies of 12 nutrients at four different levels of concentrations (Gruber et al., 2013) and looking at 32 binary combinations of nutrient deficiencies and light parameters (Kellermeier et al., 2014). Importantly, this adjustment of RSA is achieved by the independent regulation of individual root traits, suggesting an involvement of multiple regulatory components, some of which act in a tissue-specific manner (Gruber et al., 2013). These environmental responses are subject to substantial natural variation within species (Fig. 1B). For instance, Arabidopsis accessions that originated from geographically different areas show very different responses to a variety of stresses (Gifford et al., 2013; Rosas et al., 2013), suggesting an adaptive value in adjusting root growth to different soil conditions. In this section, we focus on a subset of nutrients for which studies of natural variation led to the identification of novel regulators of RSA or provided other mechanistic insight.
Natural Variation of Nitrogen Availability in the Root
In most natural environments, nitrogen is the limiting factor for plant growth and development (Vidal and Gutiérrez, 2008). Because of the significance of nitrogen in survival and growth, plants have evolved many different mechanisms to adjust to various levels of nitrogen availability (Zhang et al., 2007). Four morphological responses to different nitrogen levels have been described in the Arabidopsis root (Zhang et al., 2007), indicating a high level of root developmental plasticity relating to nitrogen availability (Nibau et al., 2008). These responses include (1) stimulation of LR elongation by localized application of nitrate, (2) systemic inhibitory effects of high nitrate concentrations on LR growth, (3) suppression of LR initiation in high Suc to nitrogen (carbon-nitrogen ratio) environments, and (4) simultaneous inhibition of PR and stimulation of LR initiation and outgrowth by l-Glu, an organic form of nitrogen (Zhang et al., 2007). There is increasing evidence that nitrogen signals modulate hormonal status and that hormonal signals interplay with nitrogen nutrition, controlling plant growth and development (for review, see Kiba et al., 2011; Krouk et al., 2011). For example, plants of Arabidopsis and soybean (Glycine max) grown on high NO−3 concentrations showed decreased levels of auxin in their roots (Caba et al., 2000; Walch-Liu et al., 2006). Several molecular links between nitrogen and auxin signaling cross talk were identified (Gifford et al., 2008; Krouk et al., 2010; Vidal et al., 2010). Similarly, nitrogen supply controls cytokinin status, and nitrate application, for instance, induces the expression in both shoots and roots of a gene that is involved in cytokinin biosynthesis (Sakakibara et al., 2006). Furthermore, ABA is implicated in nitrogen signaling (Signora et al., 2001).
There has been ample documentation of natural variation of root growth responses to nitrogen, primarily by QTL mapping in RIL. Using such an approach, genomic regions controlling root growth and root mass on treatments with nitrogen could be identified (Rauh et al., 2002). However, the mapped regions were not shared across different nitrogen sources, suggesting that different interactions exist between genotypes and nitrogen treatments (Rauh et al., 2002). Using a set of 23 Arabidopsis accessions, four different growth response strategies to nitrate limitation and starvation were identified, suggesting that distinct patterns of adaptation exist within a species (Ikram et al., 2012). Although one group of accessions showed a tolerance to nitrate limitation (low nitrogen), a second group had the highest resistance to complete nitrogen starvation; the other two groups displayed low tolerance to nitrogen limitation and nitrogen starvation (Ikram et al., 2012). Similarly, another study found substantial variation within 24 accessions of root traits in nitrogen-limited environments compared with conditions with moderate levels of nitrogen (De Pessemier et al., 2013). Nevertheless, specific genes underlying the natural variation of nitrogen availability-dependent root growth responses remained elusive. The combination of genome-wide association mapping with gene expression analysis could close this gap. Root phenotypes were quantified across 96 Arabidopsis accessions in two distinct nitrogen environments (low versus high nitrogen). Interestingly, root traits were modulated independently of each other in these two nitrogen environments. For instance, two accessions (NFA-8 and Sq-8) that showed very similar root architecture phenotypes in low-nitrogen conditions displayed very different root architectures in high-nitrogen environments (Gifford et al., 2013). Genome-wide association mapping combined with expression data analysis among these accessions resulted in 13 genes that were high-confidence candidates for modulating root growth in response to nitrogen. The analyses of transfer DNA insertion mutant lines could confirm 3 of 13 genes to be involved in nitrogen availability-dependent root growth responses (Gifford et al., 2013). Notably, an important role for jasmonic acid signaling in nitrogen-dependent growth response and its natural variation was established, with mutants of JASMONATE RESPONSIVE1 and Phenazine biosynthesis PhzC showing altered LR length in low-nitrogen but not high-nitrogen content environments, which was in strong accordance with predictions derived from GWAS and expression data.
Natural Variation of Phosphorous Availability in the Root
Phosphorus is an essential macronutrient for plants and plays an important role in numerous biological processes, including metabolism, nucleic acid synthesis, photosynthesis, glycolysis, respiration, membrane synthesis and stability, enzyme activation/inactivation, redox reactions, signaling, carbohydrate metabolism, and nitrogen fixation (Vance et al., 2003). Similar to the case of nitrogen, plants have evolved diverse morphological responses to different levels of phosphorus availability in the environment. Initial studies in the Arabidopsis Col-0 reference accession found that low phosphorus causes a reduction of the PR length, whereas LR number and growth are increased (Williamson et al., 2001; Linkohr et al., 2002; Reymond et al., 2006); there is, however, substantial natural variation of this response (see below). Many genes involved in the regulation of root development in response to different phosphorus levels have been identified (for review, see Niu et al., 2013).
Similar to nitrogen, diverse root morphological and physiological responses of Arabidopsis accessions grown in environments with different phosphorus availability were reported (Narang et al., 2000). When a set of 73 accessions was screened for root responses to low phosphorus levels, only 50% of the accessions showed a reduction of PR length and LR number, about 25% did not show a significant response, and the remaining accessions responded by changes in either the PR or LRs (Chevalier et al., 2003). Similarly, root growth in rice distinctively responds to different phosphorus environments. Like in Arabidopsis, diverse root elongation response patterns to phosphorus deficiency could be observed among 62 rice varieties, with some varieties showing increased root growth rates and others showing reduced root growth rates (Shimizu et al., 2004). Using two contrasting strains to generate a hybrid population and subsequent QTL mapping, a single QTL could be found to explain 20% of the variation between the two parental accessions. Analysis of root traits of a mapping population of Brassica napus for response to low versus high phosphorus levels identified multiple genomic regions (Shi et al., 2013).
Natural variation of the phosphorus root growth response could be used to map causal genes for this response. Using an RIL population, the genetics underlying PR growth in response to low phosphorus and three QTL were mapped in Arabidopsis (Reymond et al., 2006). The causal gene of the major QTL, coined LOW PHOSPHATE ROOT1 (a multicopper oxidase), was subsequently mapped (Svistoonoff et al., 2007). Interestingly, additional studies showed that inhibition of root growth in phosphorus-deficient medium is, in fact, caused by iron toxicity (Ward et al., 2008; see below).
In soybean, an acid phosphatase (GmACP1) was identified that regulates tolerance to phosphorus deficiency by combining RIL-based linkage analysis with genome-wide and candidate gene association analyses to identify the causal gene in the mapping interval (Zhang et al., 2014).
Regulation of Root Development in Response to Potassium and Iron Availability
In the Col-0 reference strain of Arabidopsis, mild potassium (K+) deficiency leads to a decrease in PR length, whereas severe potassium deficiency additionally leads to a decrease in LR length and an increase in the density of second-order LRs (Gruber et al., 2013). In Arabidopsis accessions, however, two contrasting strategies for adaptation to potassium deficiency have been observed (Kellermeier et al., 2013). One strategy is that, in response to potassium starvation, the growth rate of the PR is maintained, whereas the elongation of the LRs is significantly reduced. In contrast, other accessions use a strategy that drastically reduces the growth of the PR, leading to a complete arrest on longer potassium starvation while maintaining the LR elongation (Kellermeier et al., 2013).
Roots also respond strongly to iron deprivation. The best characterized response is an increase in surface area by the formation of ectopic root hairs (Schmidt et al., 2000) and the development of hairs with bifurcated tips (two tips; Müller and Schmidt, 2004; Perry et al., 2007). Root growth rates are affected by iron deficiency as well, and whereas mild iron deficiency causes increased PR length, severe deficiency causes a strong reduction of PR and LR lengths (Gruber et al., 2013). Conversely, local supply of iron mainly stimulates the emergence and elongation of the LRs because of an increased rootward auxin transport into iron-exposed LRs, whereas LR initiation itself is less affected (Giehl et al., 2012). Although natural variation of root morphological responses to different iron availability has not been reported so far, transcriptome profiling of roots showed that there are crucial differences in the early responses and adaptation to iron deprivation among natural variants (Stein and Waters, 2012). Most notably, iron homeostasis-related processes are also of remarkable importance in the context of the other nutrient-dependent root growth responses. For instance, it was shown that the inhibition of root growth under phosphate-limited growth conditions is largely caused by iron toxicity (excess of iron), and on removal of iron in phosphorus-deficient medium, the PR continued to grow (Ward et al., 2008). Moreover, it was shown that this iron-dependent effect is subject to natural variation (Ward et al., 2008). Finally, natural variation in the iron homeostasis-related transporter FERRIC REDUCTASE DEFECTIVE3 gene underlies altered root growth responses on high zinc levels in Arabidopsis (Pineau et al., 2012), providing another example of the tight coupling of nutrient homeostasis systems.
Natural Variation of Ion Uptake, Transport, and Homeostasis
Not only root growth and development but also, the molecular transport systems that are active in the roots and determine ion levels in the plant are subject to natural variation. Studying these phenomena is crucial to understand how some plants cope with levels of nutrients that are toxic to others and thus, might pave the way to generating plants that can grow on a wider variety of soils.
One of the most prominent cases is high salinity. It plays an important role in agricultural productivity, because excessive soluble salts in the soil are toxic for the plants and restrict plant growth (Xiong and Zhu, 2002). Most crop plants are glycophytes and therefore, cannot tolerate high salinity (in contrast to haplophytes, which can grow in high-salinity environments), and high salinity causes osmotic, ionic, and oxidative stress for these plants (Zhu, 2011). The major ionic stress component in high-salinity environments is sodium toxicity (sodium chloride), which causes rapid and dramatic changes in gene expression, much of it corresponding to ABA-induced gene expression responses (Zhu, 2002; Fujita et al., 2011). At the level of root growth, high salinity elicits a strong inhibitory effect on LR length, whereas the PR was found to be much less sensitive to salt treatments (Duan et al., 2013). Additional analysis of mutants impaired in ABA signaling and an ABA-responsive reporter line showed that ABA signaling plays a major role in LR inhibition in response to high salinity (Duan et al., 2013). QTL analysis in rice also showed that genomic variation exists between salt-tolerant and salt-susceptible varieties for sodium ion (Na+) and K+ uptake in shoots and roots (Lin et al., 2004). There were no overlaps of the QTL between shoot and root traits, suggesting that distinct regulation of Na+ and K+ transport exists in shoot and roots (Lin et al., 2004). Interestingly, natural variation of Arabidopsis was also reported in root hydraulic profiles (water transport capacity) in salt-stressed conditions (Sutka et al., 2011). Using GWAS to identify associations with the Na+ accumulation capacity, it was found that a sodium ion transporter Arabidopsis High-Affinity Potassium Transporter1;1 (AtHKT1;1) has a major role in regulation of this trait (Baxter et al., 2010). This is consistent with the function of AtHKT1;1, which controls unloading of Na+ from the xylem in the root to control the accumulation of Na+ in the shoots (Møller et al., 2009). Potentially causal polymorphism in the coding as well as upstream and downstream regions of the AtHKT1 gene were identified in the accessions with contrasting Na+ accumulation patterns (Rus et al., 2006). A strong indication for the adaptive relevance of allelic variation at the AtHKT1;1 was identified by analyzing distribution patterns of accessions containing weak and strong alleles of AtHKT1;1. In particular, the weak allele of AtHKT1;1 is strongly enriched in populations that grow close to coastal regions or inland regions known to have saline soils, suggesting a role for AtHKT1;1 in local adaptation to saline environments (Baxter et al., 2010). Another locus important for Na+ transport was identified by an RIL-based approach, in which an Na+ exclusion QTL was fine mapped to a protein kinase Calcineurin B-Like-Interacting Protein Kinase16 (CIPK16). Sequence variation in the promoter of CIPK16 was shown to be responsible for different Na+ exclusion levels, and higher expression of the kinase in the root was associated with higher Na+ exclusion in the shoot, which in turn, leads to higher salinity tolerance (Roy et al., 2013).
The overabundance of other elements can also cause toxicity symptoms in plants. In acidic soils, aluminum is released by clay minerals in the soil and has a negative effect on root growth, thus leading to poor yield (Kochian et al., 2004). A study in rice combining an RIL-based QTL approach and GWAS for root growth of plants in high-aluminum conditions identified a significant association in proximity to a known transporter for aluminum expressed in the root (Famoso et al., 2011). Using different haplotypes of this rice Nramp Aluminium Transporter1 (OsNRAT1) gene locus, it could subsequently be shown that sequence variations of promoter and coding regions both play an important role in regulating aluminum tolerance in rice (Li et al., 2014). At the molecular level, this tolerance conferred by natural alleles of OsNRAT1 seems to be caused by higher expression of the transporter and more efficient translocation of aluminum from the root cell wall to the root cell and subsequently, in the vacuole, thus decreasing aluminum toxicity (Li et al., 2014). Several different natural variation approaches identified aluminum-activated malate transporter1 to underlie aluminum tolerance in wheat (Triticum aestivum; Sasaki et al., 2004; Raman et al., 2005; Zhou et al., 2007; Cai et al., 2008). More recently, GWASs in 1,055 wheat accessions identified known and many novel regions associated with aluminum resistance (Raman et al., 2010), supporting the notion of the power of the GWAS technique.
Natural sequence variation in the promoter of another transporter coding gene Mitochondrial Molybdenum Transporter1 (MOT1) determines the levels of molybdenum accumulation in Arabidopsis accessions. Although MOT1 is expressed in the root and the shoot, grafting experiments showed that the molybdenum phenotype can be attributed to MOT1 function in the root. Interestingly, MOT1 does not facilitate the transport at the plasma membrane, because it is located in the membrane of mitochondria and thus, not directly involved in the uptake of molybdenum into the root (Baxter et al., 2008). A large fraction of the variation in copper tolerance in Arabidopsis, which was evaluated by measurements of root growth, was attributed to the heavy metal-transporting P-type adenosine triphosphatase5 (HMA5) adenosine triphosphatase (Kobayashi et al., 2008). This is consistent with the involvement of HMA5 in root-to-shoot transport (Andrés-Colás et al., 2006).
CONCLUSION
Rapid progress in assessing the extent of natural variation and using it to identify novel regulators of root growth and RSA has been made in the past few years. Although much of the initial progress was made using RIL populations, GWASs have recently emerged as an excellent tool in various plant species. Developments in sequencing technology will certainly facilitate the sequencing of large numbers of strains of multiple species, which is a prerequisite for GWAS approaches. Using GWAS, it will be possible to dissect the genetic basis of traits in plant species that could so far not be studied genetically or where genetic approaches have been very difficult.
A traditional bottleneck for both RIL and GWAS approaches is phenotyping. However, for root phenotyping, many solutions are available (http://www.plant-image-analysis.org), and root phenotyping pipelines have recently been developed that specifically aim at the quantification of a large number of root traits at a high throughput (Ristova et al., 2013; Slovak et al., 2014).
Because of practical considerations, such as image acquisition and throughput, almost all studies that involve quantitative phenotyping of roots are conducted on plates containing nutrient solutions solidified by agar or other gelling agents. There is no generally accepted standard medium for growing seedlings for root studies, and different laboratories use full-strength Murashige and Skoog medium (MS), dilutions of MS, or custom-made media (Dubrovsky and Forde, 2012). Although this principally allows for very well-defined growth conditions, even such a reductionistic setup can create unexpected complexity. For instance, the use of different types of agar, agarose, and gel media introduces significant variations of nutrients concentrations into the medium, which was recently reported (Gruber et al., 2013). This can potentially be a source of significant variation and most importantly, might confound results for nutrient level-dependent root growth responses. Another potential caveat with regard to this is the ion confounding that occurs when the ions of interest are covaried with the associated ions in the salts used to reconstitute the nutrient solution (Niedz and Evens, 2007). Therefore, a risk exists that, when dropping out an ion in a growth condition, the observed consequences are not entirely because of the ion that was removed. Despite these caveats, no simple solution presents itself, because even greater complexities and experimental variation can be expected in any soil-based system. In particular, in approaches using quantitative genetics, the aforementioned issues do not cause fundamental problems, because different genotypes are compared under the same nutrient and agar conditions, therefore allowing trait quantification and mapping of genes under almost identical conditions at least within one experimental series.
Overall, the enormous progress in the last years clearly shows not only that the use of natural variation facilitates analysis of the genetic basis of complex traits and the discovery of new regulators but also, that such approaches can identify alleles that can be further used to study the molecular mechanisms of the genes underlying the trait variation or be easily used for breeding purposes. It can, thus, be expected that much will still be learned from the study of natural variation of root growth.
Acknowledgments
We thank Hatem Rouached and members of the laboratory of W.B. for critically reading the article and Thomas Friese for editing the article.
Glossary
- ABA
abscisic acid
- Col-0
ecotype Columbia of Arabidopsis-0
- GWAS
genome-wide association study
- LR
lateral root
- MS
Murashige and Skoog medium
- PR
primary root
- QTL
quantitative trait loci
- RIL
recombinant inbred line
- RSA
root system architecture
Footnotes
This work is supported by the Austrian Academy of Science through the Gregor Mendel Institute (to D.R. and W.B.) and the European Union (FP7 COFUND PLANT FELLOWS Grant to D.R.).
References
- Algeo TJ, Scheckler SE. (1998) Terrestrial-marine teleconnections in the Devonian: links between the evolution of land plants, weathering processes, and marine anoxic events. Philos Trans R Soc Lond B Biol Sci 353: 113–130 [Google Scholar]
- Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M. (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Colás N, Sancenón V, Rodríguez-Navarro S, Mayo S, Thiele DJ, Ecker JR, Puig S, Peñarrubia L. (2006) The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J 45: 225–236 [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M, et al. (2010) A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet 6: e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Muthukumar B, Park HC, Buchner P, Lahner B, Danku J, Zhao K, Lee J, Hawkesford MJ, Guerinot ML, et al. (2008) Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1). PLoS Genet 4: e1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Caba JM, Centeno ML, Fernández B, Gresshoff PM, Ligero F. (2000) Inoculation and nitrate alter phytohormone levels in soybean roots: differences between a supernodulating mutant and the wild type. Planta 211: 98–104 [DOI] [PubMed] [Google Scholar]
- Cai S, Bai GH, Zhang D. (2008) Quantitative trait loci for aluminum resistance in Chinese wheat landrace FSW. Theor Appl Genet 117: 49–56 [DOI] [PubMed] [Google Scholar]
- Chevalier F, Pata M, Nacry P, Doumas P, Rossignol M. (2003) Effects of phosphate availability on the root system architecture: large-scale analysis of the natural variation between Arabidopsis accessions. Plant Cell Environ 26: 1839–1850 [Google Scholar]
- Courtois B, Audebert A, Dardou A, Roques S, Ghneim-Herrera T, Droc G, Frouin J, Rouan L, Gozé E, Kilian A, et al. (2013) Genome-wide association mapping of root traits in a japonica rice panel. PLoS ONE 8: e78037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Herder G, Van Isterdael G, Beeckman T, De Smet I. (2010) The roots of a new green revolution. Trends Plant Sci 15: 600–607 [DOI] [PubMed] [Google Scholar]
- De Pessemier J, Chardon F, Juraniec M, Delaplace P, Hermans C. (2013) Natural variation of the root morphological response to nitrate supply in Arabidopsis thaliana. Mech Dev 130: 45–53 [DOI] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR. (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Forde BG. (2012) Quantitative analysis of lateral root development: pitfalls and how to avoid them. Plant Cell 24: 4–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, Bustamante C, Kochian LV, McCouch SR. (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7: e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124: 509–525 [DOI] [PubMed] [Google Scholar]
- Gibling MR, Davies NS. (2012) Palaeozoic landscapes shaped by plant evolution. Nat Geosci 5: 99–105 [Google Scholar]
- Giehl RF, Lima JE, von Wirén N. (2012) Localized iron supply triggers lateral root elongation in Arabidopsis by altering the AUX1-mediated auxin distribution. Plant Cell 24: 33–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RF, von Wirén N. (2014) Root nutrient foraging. Plant Physiol 166: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Banta JA, Katari MS, Hulsmans J, Chen L, Ristova D, Tranchina D, Purugganan MD, Coruzzi GM, Birnbaum KD. (2013) Plasticity regulators modulate specific root traits in discrete nitrogen environments. PLoS Genet 9: e1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber BD, Giehl RF, Friedel S, von Wirén N. (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Parizot B, Inze D, Gheysen G, Beeckman T. (2007) developmental biology of roots- one common pathway for all angiosperms. Int J Plant Dev Biol 1: 212–225 [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y. (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Van Montagu M, Inzé D, Beeckman T. (2004) Transcript profiling of early lateral root initiation. Proc Natl Acad Sci USA 101: 5146–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. (2012) Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram S, Bedu M, Daniel-Vedele F, Chaillou S, Chardon F. (2012) Natural variation of Arabidopsis response to nitrogen availability. J Exp Bot 63: 91–105 [DOI] [PubMed] [Google Scholar]
- Kellermeier F, Armengaud P, Seditas TJ, Danku J, Salt DE, Amtmann A. (2014) Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26: 1480–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F, Chardon F, Amtmann A. (2013) Natural variation of Arabidopsis root architecture reveals complementing adaptive strategies to potassium starvation. Plant Physiol 161: 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Kudo T, Kojima M, Sakakibara H. (2011) Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J Exp Bot 62: 1399–1409 [DOI] [PubMed] [Google Scholar]
- Kim TH, Kunz HH, Bhattacharjee S, Hauser F, Park J, Engineer C, Liu A, Ha T, Parker JE, Gassmann W, et al. (2012) Natural variation in small molecule-induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in Arabidopsis. Plant Cell 24: 5177–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss JZ, Miller KM, Ogden LA, Roth KK. (2002) Phototropism and gravitropism in lateral roots of Arabidopsis. Plant Cell Physiol 43: 35–43 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kuroda K, Kimura K, Southron-Francis JL, Furuzawa A, Kimura K, Iuchi S, Kobayashi M, Taylor GJ, Koyama H. (2008) Amino acid polymorphisms in strictly conserved domains of a P-type ATPase HMA5 are involved in the mechanism of copper tolerance variation in Arabidopsis. Plant Physiol 148: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV, Hoekenga OA, Pineros MA. (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55: 459–493 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Korte A, Farlow A. (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B. (2011) A framework integrating plant growth with hormones and nutrients. Trends Plant Sci 16: 178–182 [DOI] [PubMed] [Google Scholar]
- Kutschera U, Briggs WR. (2012) Root phototropism: from dogma to the mechanism of blue light perception. Planta 235: 443–452 [DOI] [PubMed] [Google Scholar]
- Li J, Mo X, Wang J, Chen N, Fan H, Dai C, Wu P. (2009) BREVIS RADIX is involved in cytokinin-mediated inhibition of lateral root initiation in Arabidopsis. Planta 229: 593–603 [DOI] [PubMed] [Google Scholar]
- Li JY, Liu J, Dong D, Jia X, McCouch SR, Kochian LV. (2014) Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc Natl Acad Sci USA 111: 6503–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY. (2004) QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet 108: 253–260 [DOI] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM. (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29: 751–760 [DOI] [PubMed] [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE. (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28: 67–77 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Down and out in Arabidopsis: the formation of lateral roots. Trends Plant Sci 2: 390–396 [Google Scholar]
- Malamy JE, Ryan KS. (2001) Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol 127: 899–909 [PMC free article] [PubMed] [Google Scholar]
- Meijón M, Satbhai SB, Tsuchimatsu T, Busch W. (2014) Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat Genet 46: 77–81 [DOI] [PubMed] [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M. (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS. (2004) Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 18: 700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Osmont KS, Hardtke CS. (2006) BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443: 458–461 [DOI] [PubMed] [Google Scholar]
- Müller M, Schmidt W. (2004) Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol 134: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang RA, Bruene A, Altmann T. (2000) Analysis of phosphate acquisition efficiency in different Arabidopsis accessions. Plant Physiol 124: 1786–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Gibbs DJ, Coates JC. (2008) Branching out in new directions: the control of root architecture by lateral root formation. New Phytol 179: 595–614 [DOI] [PubMed] [Google Scholar]
- Niedz RP, Evens TJ. (2007) Regulating plant tissue growth by mineral nutrition. In Vitro Cell Dev Biol Plant 43: 370–381 [Google Scholar]
- Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS. (2013) Responses of root architecture development to low phosphorus availability: a review. Ann Bot (Lond) 112: 391–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. (2007) Hidden branches: developments in root system architecture. Annu Rev Plant Biol 58: 93–113 [DOI] [PubMed] [Google Scholar]
- Parizot B, Laplaze L, Ricaud L, Boucheron-Dubuisson E, Bayle V, Bonke M, De Smet I, Poethig SR, Helariutta Y, Haseloff J, et al. (2008) Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiol 146: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry P, Linke B, Schmidt W. (2007) Reprogramming of root epidermal cells in response to nutrient deficiency. Biochem Soc Trans 35: 161–163 [DOI] [PubMed] [Google Scholar]
- Pigliucci M. (2005) Evolution of phenotypic plasticity: where are we going now? Trends Ecol Evol 20: 481–486 [DOI] [PubMed] [Google Scholar]
- Pineau C, Loubet S, Lefoulon C, Chalies C, Fizames C, Lacombe B, Ferrand M, Loudet O, Berthomieu P, Richard O. (2012) Natural variation at the FRD3 MATE transporter locus reveals cross-talk between Fe homeostasis and Zn tolerance in Arabidopsis thaliana. PLoS Genet 8: e1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. (1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman H, Stodart B, Ryan PR, Delhaize E, Emebiri L, Raman R, Coombes N, Milgate A. (2010) Genome-wide association analyses of common wheat (Triticum aestivum L.) germplasm identifies multiple loci for aluminium resistance. Genome 53: 957–966 [DOI] [PubMed] [Google Scholar]
- Raman H, Zhang K, Cakir M, Appels R, Garvin DF, Maron LG, Kochian LV, Moroni JS, Raman R, Imtiaz M, et al. (2005) Molecular characterization and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48: 781–791 [DOI] [PubMed] [Google Scholar]
- Rauh L, Basten C, Buckler S., IV (2002) Quantitative trait loci analysis of growth response to varying nitrogen sources in Arabidopsis thaliana. Theor Appl Genet 104: 743–750 [DOI] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- 3,000 Rice Genomes Project (2014) The 3,000 rice genomes project. Gigascience 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristova D, Rosas U, Krouk G, Ruffel S, Birnbaum KD, Coruzzi GM. (2013) RootScape: a landmark-based system for rapid screening of root architecture in Arabidopsis. Plant Physiol 161: 1086–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV. (2012) The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Dorrity MW, de Lucas M, Toal T, Hernandez RI, Little SA, Maloof JN, Kliebenstein DJ, Brady SM. (2013) Identification of novel loci regulating interspecific variation in root morphology and cellular development in tomato. Plant Physiol 162: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas U, Cibrian-Jaramillo A, Ristova D, Banta JA, Gifford ML, Fan AH, Zhou RW, Kim GJ, Krouk G, Birnbaum KD, et al. (2013) Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root systems architecture. Proc Natl Acad Sci USA 110: 15133–15138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SJ, Huang W, Wang XJ, Evrard A, Schmöckel SM, Zafar ZU, Tester M. (2013) A novel protein kinase involved in Na(+) exclusion revealed from positional cloning. Plant Cell Environ 36: 553–568 [DOI] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE. (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N. (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11: 440–448 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37: 645–653 [DOI] [PubMed] [Google Scholar]
- Savolainen O, Lascoux M, Merilä J. (2013) Ecological genomics of local adaptation. Nat Rev Genet 14: 807–820 [DOI] [PubMed] [Google Scholar]
- Scacchi E, Salinas P, Gujas B, Santuari L, Krogan N, Ragni L, Berleth T, Hardtke CS. (2010) Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proc Natl Acad Sci USA 107: 22734–22739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W, Tittel J, Schikora A. (2000) Role of hormones in the induction of iron deficiency responses in Arabidopsis roots. Plant Physiol 122: 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, Nordborg M. (2012) An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet 44: 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Shi T, Broadley MR, White PJ, Long Y, Meng J, Xu F, Hammond JP. (2013) High-throughput root phenotyping screens identify genetic loci associated with root architectural traits in Brassica napus under contrasting phosphate availabilities. Ann Bot (Lond) 112: 381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu A, Yanagihara S, Kawasaki S, Ikehashi H. (2004) Phosphorus deficiency-induced root elongation and its QTL in rice (Oryza sativa L.). Theor Appl Genet 109: 1361–1368 [DOI] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H. (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Slovak R, Göschl C, Su X, Shimotani K, Shiina T, Busch W. (2014) A scalable open-source pipeline for large-scale root phenotyping of Arabidopsis. Plant Cell 26: 2390–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RJ, Waters BM. (2012) Use of natural variation reveals core genes in the transcriptome of iron-deficient Arabidopsis thaliana roots. J Exp Bot 63: 1039–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutka M, Li G, Boudet J, Boursiac Y, Doumas P, Maurel C. (2011) Natural variation of root hydraulics in Arabidopsis grown in normal and salt-stressed conditions. Plant Physiol 155: 1264–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee CR, Zurek PR, Symonova O, Zheng Y, Bucksch A, Mileyko Y, Galkovskyi T, et al. (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci USA 110: E1695–E1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trontin C, Tisné S, Bach L, Loudet O. (2011) What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants? Curr Opin Plant Biol 14: 225–231 [DOI] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45: 1097–1102 [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA. (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA. (2008) A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol 11: 521–529 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov II, Filleur S, Gan Y, Remans T, Forde BG. (2006) Nitrogen regulation of root branching. Ann Bot (Lond) 97: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG. (2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol 147: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Nordborg M. (2005) Natural variation in Arabidopsis. How do we find the causal genes? Plant Physiol 138: 567–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM. (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. (2002) Salt tolerance. Arabidopsis Book 1: e0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Song H, Cheng H, Hao D, Wang H, Kan G, Jin H, Yu D. (2014) The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genet 10: e1004061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rong H, Pilbeam D. (2007) Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J Exp Bot 58: 2329–2338 [DOI] [PubMed] [Google Scholar]
- Zhou LL, Bai GH, Ma HX, Carver BF. (2007) Quantitative trait loci for aluminum resistance in wheat. Mol Breed 19: 153–161 [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2011) Plant salt tolerance. Trends Plant Sci 6: 66–71 [DOI] [PubMed] [Google Scholar]