The diversity of postembryonic root forms and their functions add to our understanding of the genes, signals and mechanisms regulating lateral and adventitious root branching in the plant models Arabidopsis and rice.
Abstract
Root branching is critical for plants to secure anchorage and ensure the supply of water, minerals, and nutrients. To date, research on root branching has focused on lateral root development in young seedlings. However, many other programs of postembryonic root organogenesis exist in angiosperms. In cereal crops, the majority of the mature root system is composed of several classes of adventitious roots that include crown roots and brace roots. In this Update, we initially describe the diversity of postembryonic root forms. Next, we review recent advances in our understanding of the genes, signals, and mechanisms regulating lateral root and adventitious root branching in the plant models Arabidopsis (Arabidopsis thaliana), maize (Zea mays), and rice (Oryza sativa). While many common signals, regulatory components, and mechanisms have been identified that control the initiation, morphogenesis, and emergence of new lateral and adventitious root organs, much more remains to be done. We conclude by discussing the challenges and opportunities facing root branching research.
Branching through lateral and adventitious root formation represents an important element of the adaptability of the root system to its environment. Both are regulated by nutrient and hormonal signals (Bellini et al., 2014; Giehl and von Wirén, 2014), which act locally to induce or inhibit root branching. The net effect of these adaptive responses is to increase the surface area of the plant root system in the most important region of the soil matrix for resource capture (e.g. surface layers for phosphorus uptake and deeper layers for nitrate uptake) or to secure anchorage. Different species use different combinations of lateral or adventitious roots to achieve this, with lateral roots dominating the root system of eudicots while adventitious (crown and brace) roots dominate the root system of monocots, including in cereal crops.
Our understanding of the mechanisms controlling lateral and adventitious root development has accelerated in recent years, primarily through research on model plants. The simple anatomy and the wealth of genetic resources in Arabidopsis (Arabidopsis thaliana) have greatly aided embryonic and postembryonic root developmental studies (De Smet et al., 2007; Péret et al., 2009a; Fig. 1, A and E). Nevertheless, impressive recent progress has been made studying root branching in other crop species, notably cereals such as maize (Zea mays) and rice (Oryza sativa).
Figure 1.
A to D, Schematics showing diversity in root system architecture at both seedling (left) and mature (right) stages in eudicots (A and C) and monocots (B and D). A, Arabidopsis root system. B, Maize root system. C, Tomato root system (for clarity, stem-derived adventitious roots are only shown in the labeled region). D, Wheat root system. E and F, Cross sections of emerging lateral root primordia in Arabidopsis (E) and rice (F). E and F are adapted from Péret et al. (2009b).
In this Update, we initially describe the diversity of postembryonic root forms in eudicots and monocots (Fig. 1). Next, we highlight recent advances in our understanding of the genes, signals, and mechanisms regulating lateral root and adventitious root branching in Arabidopsis, rice, and maize. Due to space limits, we cannot provide an exhaustive review of this subject area, focusing instead on recent research advances. However, we direct readers to several recent in-depth reviews on lateral root (Lavenus et al., 2013; Orman-Ligeza et al., 2013) and adventitious root development (Bellini et al., 2014).
ROOT BRANCHING CLASSES AND NOMENCLATURE
Higher plant roots can be classified into roots derived from the embryo (embryonic roots) and those formed after germination from existing roots or nonroot tissues (postembryonic roots). Postembryonic roots arising from tissues other than roots are termed adventitious. Root systems based on development of the primary embryonic root, such as those of dicots (Figs. 1, A and C, and 2A) are also known as taproot or allorhizic systems; those in monocots, composed mostly of adventitious roots, are termed fibrous or homorhizic root systems (Figs. 1, B and D, and 2B).
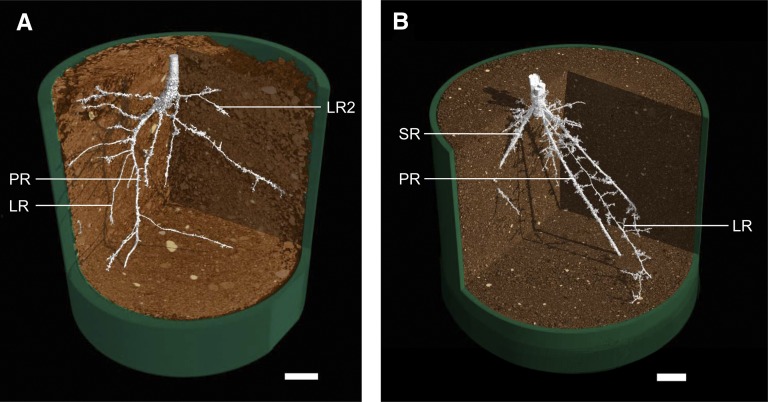
Figure 2.
Taproot (A) and fibrous (B) root systems. Root systems of tomato (A) and wheat (B) were imaged using x-ray microscale computed tomography. LR, Lateral root; LR2, second order lateral root; PR, primary root; SR, seminal root. Scans were taken at 12 d (A) and 3 weeks (B) after germination. Roots are false colored in white, and soil is false-colored in brown. Bars = 0.5 cm (A) and 1 cm (B).
Embryonic Roots
In many eudicots, the embryonic root (radicle) develops into a single primary or tap root that remains active throughout the life cycle and develops several orders of lateral root. Examples are the primary root of Arabidopsis (Fig. 1A) and the taproot in tomato (Solanum lycopersicum; Figs. 1C and 2A). In contrast, the embryonic root system of monocots, which consists of both primary and seminal roots (Figs. 1, B and D, and 2B), is of importance only during the seedling stage, with postembryonic adventitious roots making up the bulk of the root system (Hochholdinger and Zimmermann, 2008).
Postembryonic Roots
Postembryonic roots are classified according to time of emergence as early or late (Hochholdinger et al., 2004b). Early postembryonic roots in eudicots (Fig. 1, A and C; Table I) include lateral roots derived from the primary root and junction roots: adventitious roots that form at the root-hypocotyl junction (Falasca and Altamura, 2003). Late postembryonic roots include other forms of adventitious root (e.g. those arising from the hypocotyl) and adventitious-derived lateral roots.
Table I. Mutant phenotypes for embryonic and postembryonic roots in dicots.
Dashes indicate unstudied phenotypes, italic text denotes complete absence of root type/growth rate, boldface text indicates reduced root number/growth, and boldface italic text denotes increased root number/growth. This table is based on supplemental table 1 from Bellini et al. (2014) and formatting based on Hochholdinger et al. (2004b). The original description of the mutant phenotypes detailed in the table can be found in: Boerjan et al. (1995), Delarue et al. (1998), Fukaki et al. (2002), Sorin et al. (2005), Sibout et al. (2006), Dello Ioio et al. (2007), and Negi et al. (2010).
| Locus | Embryonic Roots |
Early Postembryonic Roots | Late Postembryonic Roots | |||
|---|---|---|---|---|---|---|
| Primary Roots | Lateral Roots on the Primary Roots | Junction Roots | Adventitious Roots (Hypocotyl) | Lateral Roots | ||
| On the Junction Roots | On the Adventitious Roots | |||||
| Arabidopsis thaliana argonaute1 | Shorter | (With auxin) normal | – | (With auxin) none, etiolated less | – | – |
| Arabidopsis thaliana superroot1 (Atsur1) | Shorter | More | More | Many more | – | – |
| Atsur2 | Shorter | More | More | Many more | – | – |
| Strigolactone mutants (pea [Pisum sativum]; Arabidopsis) | Shorter | More | – | Many more (but shorter in pea) | – | In pea mutants normal |
| Arabidopsis thaliana YUCCA1 | – | – | – | Many more | – | – |
| Arabidopsis thaliana cytokinin oxidase mutants | Longer | More | – | More | – | – |
| Arabidopsis thaliana Arabidopsis histidine kinase mutants | Longer | No lateral roots elongation | – | More adventitious roots, normal elongation | – | – |
| Arabidopsis thaliana isopentenyl transferase | Longer | More | – | – | ||
| Epinastic (tomato) | Shorter | Less | More | More | – | – |
| Never ripe (tomato) | Longer | More | More | Less | – | – |
| Ethylene response factor3 (Populus spp.) | – | More | – | More | – | – |
| Arabidopsis thaliana coronatine insensitive1 | – | Less | – | More | – | – |
| Arabidopsis thaliana elongated hypocotyl5 (Athy5) | Normal | More | – | More | – | – |
| Athy5 hy5-homolog (double mutant) | Shorter | Delayed and less | – | Delayed | – | – |
| Arabidopsis thaliana solitary root | Normal | None | – | – | – | – |
| Arabidopsis thaliana auxin response2 (Ataxr2) | Normal growth rate | More | – | Less | – | – |
| Ataxr3 | Reduced growth rate | – | – | More | – | – |
| Arabidopsis thaliana short hypocotyl2 | Reduced growth rate | Less | – | Less | – | – |
In monocots, the majority of the root system is usually composed of adventitious postembryonic roots derived from the shoot. In cereals, these include two classes of nodal root, the crown and brace roots, which develop from belowground and aboveground nodes, respectively (Fig. 1, B and D). Crown roots in wheat (Fig. 1D) form at the lower three to seven nodes (Kirby, 2002), and in maize they are organized on average in six underground whorls (Fig. 1B). Maize also has two to three whorls of aboveground brace roots (also termed stilt or prop roots), which are functional for water and nutrient uptake and also important for support (Hoppe et al., 1986).
ROOT BRANCHING IN ARABIDOPSIS
Eudicot lateral root organogenesis has primarily been studied in the model Arabidopsis (Malamy and Benfey, 1997; Péret et al., 2009a), originally in two dimensions (Malamy and Benfey, 1997) and very recently via three-dimensional imaging studies (Lucas et al., 2013). Arabidopsis roots comprise single layers of epidermal, cortex, and endodermal tissues that surround the pericycle and vasculature (Fig. 1E). Lateral roots initiate from pairs of pericycle cells that originate from three cell files adjacent to the xylem pole (Fig. 3; De Smet et al., 2007; Lucas et al., 2008). These cells undergo several rounds of anticlinal, periclinal, and tangential cell divisions to form a new lateral root primordium (Fig. 3; Malamy and Benfey, 1997), ultimately resulting in a fully emerged new organ mimicking the primary root in terms of cellular organization. The major developmental stages (Fig. 3) and important new regulatory insights are described below.
Figure 3.
Gene regulatory networks regulating adventitious root (top) and lateral root (bottom) development in Arabidopsis. While knowledge of early steps in adventitious root prebranch site formation is very limited, initiation and emergence rely on gene networks regulated by auxin (IAA), jasmonic acid (JA), and strigolactones (top). GH3, GRETCHEN HAGEN3; HAE/HSL2, HAESA RECEPTOR-LIKE PROTEIN KINASE/HAESA-LIKE 2; IDA, INFLORESCENCE DEFICIENT IN ABSCISSION. In contrast, lateral root formation and development have been studied in more detail. Developing lateral root primordia have been formally divided into eight stages (I–VIII; Malamy and Benfey, 1997). Different auxin response gene networks regulate early steps of prebranch site formation, initiation, and emergence (bottom). Different colors denote different tissues.
Prebranch Site Formation
The very earliest expressed marker during lateral root development is luciferase (LUC) under the control of the synthetic auxin response element DR5 (Ulmasov et al., 1997) that is expressed in an oscillatory manner in xylem pole cells (underlying xylem pole pericycle cells) in a region termed the oscillation zone, located just behind the primary root tip. Prebranch site formation occurs when the auxin response oscillation has reached a maximum in this region, demarking the position of the future lateral root primordium. DR5::LUC expression in the oscillation zone is dependent on AUXIN RESPONSE FACTOR7 (ARF7; Moreno-Risueno et al., 2010), ARF6, ARF8, and ARF19 (De Rybel et al., 2010) and on the auxin repressor protein INDOLE-3-ACETIC ACID INDUCIBLE28 (IAA28; as well as IAA8 and IAA19). IAA28, ARF7, and ARF19 control the expression of downstream target genes such as the transcription factor GATA23 (Fig. 3) that, until recently, was one of the only regulators identified to have a role in prebranch site formation (De Rybel et al., 2010). AtMYB93, a member of the R2R3 MYB class of transcription factor, is expressed exclusively and transiently in root endodermal cells overlying new primordia, from where it negatively regulates lateral root development (Gibbs et al., 2014). AtMYB93 expression is auxin inducible, and Atmyb93 mutants exhibit reduced sensitivity to this key signal during lateral root development. PIN-FORMED3 (PIN3) is also transiently expressed in endodermal cells overlying prebranch sites, where it transports auxin into underlying pericycle cells to create a local auxin maxima that triggers pericycle cell division (Marhavý et al., 2013).
The regulation of the mechanical and/or hydraulic properties of endodermal cells overlying prebranch sites appears critical for lateral root initiation. Endodermal cells overlying prebranch sites must initially undergo a loss of volume so that pericycle cells can enlarge prior to their first anticlinal division (Vermeer et al., 2014). This spatial accommodation appears to be an auxin-regulated process. Disrupting the auxin response specifically in overlying endodermal cells completely blocks asymmetric pericycle cell divisions required for lateral root initiation (Vermeer et al., 2014). Likely targets for regulation by auxin include aquaporins (water channels) that are globally repressed by this signal during lateral root development (Péret et al., 2012).
Prebranch sites are also regulated by a newly discovered adaptive mechanism termed lateral root hydropatterning (Bao et al., 2014; Fig. 4). This adaptive mechanism involves suppressing lateral root initiation on the dry side of a root and maintaining lateral root development in tissues directly in contact with soil moisture, thereby ensuring that roots are better able to allocate their resources and maximize water and nutrient foraging. Hydropatterning appears to be auxin regulated, as mutations in auxin synthesis (tryptophan aminotransferase of Arabidopsis1), transport (pin2/pin3/pin7), or the disruption of auxin responses in endodermal and cortical cells overlying prebranch sites disrupt this adaptive response. In the light of recent work by Vermeer et al. (2014), hydropatterning may regulate lateral root initiation through blocking spatial accommodation in overlying tissues.
Figure 4.
Hydropatterning in maize roots. Microscale computed tomography-generated images show maize seedlings grown through a continuous volume of soil (A) and a macropore of air (B). Root tissue is false colored in white, and soil is false-colored in brown. Bars = 5 mm.
Cytokinin has been proposed to act antagonistically to auxin during lateral root development (Aloni et al., 2006; Fukaki and Tasaka, 2009); exogenous application inhibits initiation of lateral root primordia (Li et al., 2006; Laplaze et al., 2007). Cytokinin regulates the transcription of a subset of PINs (Fukaki et al., 2002; Laskowski et al., 2008; Lewis et al., 2011; Marhavý et al., 2013) and acts as a polarizing cue to determine the directionality of auxin flow during lateral root morphogenesis (Marhavý et al., 2013). Abscisic acid (ABA) inhibits lateral root formation by increasing the expression of microRNAs (miR393; Chen et al., 2012) and associated transcription factors (ABA INSENSITIVE4), which reduce TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX expression and consequently downstream auxin responses.
Lateral Root Initiation and Morphogenesis
The first visible cellular change leading to lateral root primordia involves several pairs of xylem pole pericycle cells, in which the nuclei of adjacent cells migrate together followed by division to form a pair of small daughter cells flanked by two larger cells (Fig. 3). Auxin transport and response components are necessary for this process (Casimiro et al., 2001; Dubrovsky et al., 2008). AUX1 and PIN auxin influx and efflux carriers optimize auxin supply and promote organ initiation (Laskowski et al., 2008; Lewis et al., 2011; Marhavý et al., 2013), while the SOLITARY ROOT (SLR)/IAA14-ARF7-ARF19 and BODENLOS/IAA12-ARF5 auxin response pathways are also required (Fukaki et al., 2002, 2005; Okushima et al., 2005; Vanneste et al., 2005; De Smet et al., 2010; Vernoux et al., 2011; Fig. 3).
Only a few downstream targets of the SLR/IAA14-ARF7-ARF19 auxin response pathway have been identified. These include members of the LATERAL ORGAN BOUNDARIES DOMAIN (LBD)/ASYMMETRIC LEAVES2-LIKE family of transcription factors, such as LBD16, LBD18, and LBD29, that positively regulate lateral root formation (De Smet et al., 2010). LBD16 is expressed in pericycle cell pairs prior to lateral root initiation (Goh et al., 2012). The authors demonstrated that by engineering the LBD16 transcriptional activator to become a repressor, the initial nuclear migration event in pericycle cell pairs could be blocked. This reveals that induction of LBD16 and possibly several related family members triggers the initial nuclear migration event.
Following lateral root initiation, primed pericycle cells undergo anticlinal cell division to generate a stage I primordium (Fig. 3). Further rounds of tangential and periclinal divisions produce a dome-shaped primordium (Lucas et al., 2013). Intriguingly, the pattern of formative cell divisions in new lateral root primordia appears not to be highly stereotypical, since disrupting new cell wall positioning using aurora mutants (aur1/aur2) has no effect on the shape of the primordium. However, blocking the auxin response in tissues overlying newly initiated lateral root primordia results in major changes in dome shape. This suggests that lateral root morphogenesis is largely controlled by the mechanical properties of the overlying tissues rather than the precise pattern of formative divisions (Lucas et al., 2013).
Lateral Root Emergence
New Arabidopsis lateral root primordia must emerge through the three outer cell layers of the parental root (Figs. 1E and 3). This process is promoted by shoot-derived auxin (Bhalerao et al., 2002; Swarup et al., 2008) channeled by newly initiated primordia via the sequential induction of PIN3 and LIKE AUX1 3 (LAX3) auxin efflux and influx carrier genes in overlying cortical cells (Swarup et al., 2008; Péret et al., 2013). This dynamic pattern of auxin carrier expression functions to first mobilize, and then accumulate, auxin in a subset of cells directly overlying new lateral root primordia. This serves to delimit the expression of auxin-inducible genes that triggers cell separation and facilitates lateral root emergence.
The process of auxin-regulated cell separation in overlying cells requires cell wall-remodeling enzymes such as polygalacturonase (Neuteboom et al., 1999; Laskowski et al., 2006; Swarup et al., 2008; Lee et al., 2013) that promote cell separation in front of emerging lateral root primordia. This process also uses key components of the floral abscission response machinery (Kumpf et al., 2013). Auxin also facilitates organ emergence by controlling the hydraulic properties of the primordia and overlying tissues by repressing aquaporin expression (Péret et al., 2012).
ABA also functions as a promoter of postemergence lateral root growth by enhancing auxin-dependent transcription through the action of the ABA receptor PYR1-LIKE8 (PYL8; Zhao et al., 2014). PYL8 has been shown to interact directly with the transcription factor MYB77 (also MYB44 and MYB73) ,which, by means of interactions with ARF7, can increase the expression of genes, including LBD16 and LBD29, that are known to promote lateral root formation and elongation (Okushima et al., 2005, 2007; Wilmoth et al., 2005).
ADVENTITIOUS ROOTING IN ARABIDOPSIS
Adventitious roots can form on the Arabidopsis hypocotyl (typically after etiolation) or at the root-hypocotyl junction (junction roots). Junction roots can be induced to emerge after the primary root is damaged or its growth terminates (Lucas et al., 2011). Despite their distinct regulation, in both cases the visible stages of adventitious root formation follow an equivalent pattern to lateral roots, including pericycle asymmetric cell divisions (Fig. 3; Falasca and Altamura, 2003; Della Rovere et al., 2013).
Despite the anatomical similarities between adventitious and lateral root morphogenesis, their regulation exhibits some clear differences (summarized in Bellini et al., 2014). The most obvious overlap is the central role of auxin signaling controlling initiation as well as later primordia development and emergence. The major players mediating auxin signaling during adventitious rooting are the activating ARF6 and ARF8 and the repressing ARF17 (Fig. 3; Gutierrez et al., 2009). The expression of ARF6 and ARF8 is controlled by MIR167, while ARF17 expression is regulated by MIR160. ARF6, ARF8, and ARF17 oppositely regulate jasmonic acid homeostasis via regulating jasmonic acid-modifying GRETCHEN HAGEN3 (GH3) enzymes (Gutierrez et al., 2009, 2012; Bellini et al., 2014). Auxin transporters are also important for adventitious root formation, with both influx (LAX3) and efflux (PIN1) carriers required for auxin accumulation in the early stages of adventitious root organogenesis (Della Rovere et al., 2013). An ATP-binding cassette transporter mediating IAA efflux (ABCB19) has also been linked to adventitious root formation (Sukumar et al., 2013). Other members of the same family have been shown to transport the auxin precursor indole-3-butyric acid (Růžička et al., 2010; Strader and Bartel, 2011), which, among all tested auxin isoforms, has the highest competence to induce adventitious root formation (Nordström et al., 1991). Additional hormone signals regulate adventitious roots (Bellini et al., 2014; Fig. 3). For example, ethylene regulates the positioning of adventitious root primordia (A. Rasmussen, Y. Hu, D. Van Der Straeten, and D. Geelen, unpublished data), while strigolactones block adventitious root formation in Arabidopsis (Kohlen et al., 2012; Rasmussen et al., 2012; Bellini et al., 2014), most likely by interfering with auxin transport and independent of the inhibitory effect of cytokinin (Kohlen et al., 2012; Rasmussen et al., 2012).
ROOT BRANCHING IN CEREALS
Lateral Root Development in Cereals
Lateral root formation in cereals has been classified into three developmental phases: organ initiation, growth through the cortex, and emergence through the epidermis (Orman-Ligeza et al., 2013). In contrast to Arabidopsis, which has a central vascular cylinder and a single layer of cortical cells, maize, wheat (Triticum spp.), and rice have 10 to 15 cortical cell layers surrounding the xylem and phloem poles, which alternate within the stele (Fig. 1, E and F; Smith and De Smet, 2012). In maize, wheat, rice, and barley (Hordeum vulgare), lateral roots originate from pericycle and endodermal cells located opposite the phloem poles (Casero et al., 1995; Demchenko and Demchenko, 2001; Hochholdinger et al., 2004a), contrasting with eudicots, in which primordia originate from pericycle cells opposite protoxylem poles (Casimiro et al., 2003). Due to the higher number of phloem poles in cereals (usually 10 or more; Smith and De Smet, 2012), lateral roots initiate in several longitudinal files, with the number of files roughly proportional to the stele diameter (Bell and McCully, 1970; Draye et al., 1999; Draye, 2002).
As in Arabidopsis, the establishment of auxin response maxima is crucial in lateral root initiation in maize. Marker lines expressing red fluorescent protein under control of the DR5 promoter have indicated the presence of auxin response maxima in differentiating xylem cells and in cells surrounding the protophloem vessels (Jansen et al., 2012). Furthermore, targeted genome-wide transcriptome analysis of lateral root initiation has highlighted common transcriptional regulation between Arabidopsis and maize, suggesting the conservation of genes involved in lateral root initiation across angiosperms (Jansen et al., 2013).
A key difference in lateral root emergence compared with Arabidopsis is that in cereals cortical cells overlying lateral root primordia reenter mitosis and divide (Bell and McCully, 1970). In barley, these divisions can be observed as early as 10 to 12 h after the first asymmetric division in the pericycle. Many of the resulting small cells enter hydrogen peroxide-mediated cell death following penetration by the primordium (Orman-Ligeza et al., 2013). Loosening of the cell walls via hydrolytic enzymes also occurs in cereals (Bell and McCully, 1970). Unlike Arabidopsis, cell division and programmed cell death are required in cereals to allow the emerging lateral organ to penetrate the larger number of cortical cell layers (Fig. 1F). A number of lateral root mutants have been described in cereals (Table II), the majority of which are auxin related (Hochholdinger and Tuberosa, 2009). For example, in maize, mutation of rootless with undetectable meristems1 (RUM1), which encodes a member of the Auxin/IAA (Aux/IAA) family of transcription factors, leads to the failure of lateral root initiation and disruptions in vascular patterning (Woll et al., 2005; von Behrens et al., 2011; Zhang et al., 2014).
Table II. Mutant phenotypes for embryonic and postembryonic roots in monocots.
Dashes indicate unstudied phenotypes, italic text denotes complete absence of root type/growth rate, boldface text indicates reduced root number/growth, and boldface italic text increased root number/growth. This table is based on supplemental table 1 from Bellini et al. (2014) and formatting from Hochholdinger et al. (2004b).
| Locus | Embryonic Roots |
Early Postembryonic Roots |
Late Postembryonic Roots |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Primary Roots | Seminal Roots | Lateral Roots |
Crown Roots (Node 1) | Crown Roots (Node 2) | Brace Roots | Lateral Roots |
|||
| On the Primary Roots | On the Seminal Roots | On the Crown Roots | On the Brace Roots | ||||||
| Osiaa11 | – | – | None | Normal | – | – | – | – | |
| Zea mays rootless with undetectable meristems1 | – | None | None | None | Normal | Normal | Normal | Normal | Normal |
| Oryza sativa crown rootless1 (Oscrl1)/adventitious rootless1 (arl1), Oscrl2 | – | Less | None | None | None | None | None | None | |
| Zea mays rootless concerning crown and seminal roots (Zmrtcs) | – | None | – | None | None | None | None | None | None |
| Oscrl4/Oryza sativa guanine nucleotide exchange factor for ADP-ribosylation factor | – | – | Less | – | None | None | None | None | None |
| Oryza sativa YUCCA1 | – | – | – | More | – | – | – | – | |
| Oscrl5 | – | – | Normal | Normal | Less | Less | Less | Less | Less |
| Oryza sativa wuschel-related homeobox11 | – | – | – | – | Delayed | ||||
| Zea mays lateral rootless1 | – | – | None | None | None | None | None | None | None |
| Oryza sativa lateral rootless1 (Oslrt1) | – | – | None | None | – | – | – | – | – |
| Oslrt2 | |||||||||
| Zea mays rootless1 | – | – | None | None | None | None | None | ||
| Zea mays short lateral roots1 (Zmslr1) | – | – | Shorter | Shorter | Normal | Normal | Normal | Normal | Normal |
| Zmslr2 | |||||||||
| Zmslr1, Zmslr2 | |||||||||
| Oryza sativa cytokinin oxidase4 | Longer | – | – | – | More and faster growth | More and faster growth | – | – | – |
| Rice strigolactone mutants (d10, d17, d3, d14, d27) | Shorter | Shorter | – | More | Shorter but normal numbers | Shorter but normal numbers | – | – | – |
In rice, mutation of the Aux/IAA gene Osiaa13 causes lateral root defects by blocking the auxin-mediated degradation of this auxin response repressor protein (Kitomi et al., 2008). Transcriptome analysis using laser microdissection and microarray profiling has identified 71 genes associated with lateral root initiation in rice (Takehisa et al., 2012), nine of which are differentially regulated in the Osiaa13 mutant compared with the wild type (Kitomi et al., 2008), with a further seven genes containing an auxin response element in their promoter sequence.
Aux/IAA protein degradation leads to the release of ARF transcription factors that regulate a set of downstream target genes. These sequences include LBDs such as CRL1. Mutations in CRL1 have been shown to reduce lateral root number by 70% (Inukai et al., 2005). OsARF16 is orthologous to Arabidopsis ARF7 and ARF19 sequences and binds auxin response elements in the CRL1 promoter. Hence, there appears to be strong conservation of the regulatory network components controlling lateral root initiation between Arabidopsis and rice.
Adventitious Root Development in Cereals
Cereal crops have several classes of adventitious roots, including crown roots and brace roots (Fig. 1, B and D; Table II). Both crown and brace roots develop at nodes; belowground in the case of crown roots and from aboveground nodes for brace roots. Maize forms approximately 70 stem-formed roots with an average six whorls of crown roots and three whorls of brace roots (Hochholdinger et al., 2004b).
Crown Roots in Cereals
Crown roots are postembryonic stem-formed roots that develop in the lower nodes of monocots (Table II; Hochholdinger et al., 2004a, 2004b). These roots exist below the soil surface and are similar to aboveground brace roots (Hochholdinger et al., 2004b). It has been reported that the seminal and primary roots do not persist in some monocot species, but even when they do, the crown and brace root systems dominate the functional root system (Hochholdinger et al., 2004a, 2004b). The Zmrtcs mutant, which has no crown roots and no seminal roots but produces a normal primary root with laterals, exhibits up-regulation of stress-related genes, probably due to the inadequacy of the primary and lateral roots to support the developing plant (Muthreich et al., 2013).
Crown roots of rice and maize originate from cells next to the peripheral cylinder of vascular bundles in the stem that undergo one to two periclinal divisions (Fig. 5; Ito et al., 2006; Kitomi et al., 2011b; Muthreich et al., 2013). Cells in the inner layers of the primordia divide to form epidermis/endodermis and a central cylinder initial (Itoh et al., 2005). The cells in the outer layer divide anticlinally to form the root cap initial. Endodermal cells then undergo several rounds of asymmetric periclinal divisions to produce the cortical cell layers. Root cap cells form the columella through periclinal divisions, while the cells of the central cylinder continue division to become dome shaped. Primordia only become connected to the vascular bundle system at the time of organ emergence (Itoh et al., 2005).
Figure 5.
Gene regulatory networks regulating crown root (adventitious root) development in rice. Different colors denote different tissues. ARR, ARABIDOPSIS RESPONSE REGULATOR; NBD, 2,5-norbornadiene (ethylene inhibitor); PAC, paclobutrazol (GA inhibitor); WOX, WUSCHEL-related homeobox.
The regulation of crown root formation has been best studied in rice (Fig. 5). Auxin is a central regulator of crown root formation. CRL4 controls PIN-regulated auxin transport (Cho et al., 2014). In rice, the crl4/Osgnom1 mutants are devoid of crown roots but also show reduced numbers of lateral roots (Liu et al., 2009; Coudert et al., 2010). Auxin signals via AUX/IAA and ARF1 proteins to induce the expression of CRL1/ARL1, encoding an LBD transcription factor (Kitomi et al., 2011b). Mutants lacking CRL1/ARL1 produce no crown roots and have reduced lateral root formation (Coudert et al., 2010).
Auxin regulation of crown root formation also requires cytokinin signaling (Fig. 5). ARF1 induction of the APETALA2/ETHYLENE RESPONSE FACTOR class of transcription factor CRL5 up-regulates ARABIDOPSIS RESPONSE REGULATOR1, a type A cytokinin response regulator inhibiting a cytokinin response (Zhao et al., 2009; Kitomi et al., 2011a). The cytokinin oxidase OsCKX4 is up-regulated by both auxin and cytokinin signals via binding of ARF25, RICE TYPE-A RESPONSE REGULATOR2 (OsRR2), and OsRR3 to its promoter and is expressed in the base of the stem where crown roots are initiated, where it regulates cytokinin abundance (Gao et al., 2014). Later stages of crown root development are dominated by other hormone interactions, in particular ethylene, GA, and ABA (Steffens et al., 2006). Programmed cell death of the epidermal layers adjacent to crown root primordia and crown root elongation are induced by the cooperative effect of ethylene and GAs, while ABA antagonizes these two signals (Mergemann and Sauter, 2000; Steffens et al., 2006). A number of signals, mutants, and genes that control root branching have also been described in maize (summarized in Table II).
Brace Roots in Cereals
Brace roots, also known as buttress, stilt, or prop roots, are adventitious roots that form from aboveground nodes in a variety of monocots. Brace roots in maize (Fig. 1B) are important for lodging resistance and for water and nutrient uptake during the later stages of plant growth (Varney and Canny, 1993; Wang et al., 1994). They also substantially affect grain yield under soil flooding, presumably by improving gas exchange and/or topsoil foraging (Hochholdinger and Tuberosa, 2009). Brace roots form endogenously (like crown roots), but not all penetrate the soil. Lateral roots only form on the brace roots that penetrate the soil, and they provide additional lodging resistance plus facilitate water and nutrient update (Hochholdinger et al., 2004a, 2004b). Little is known about the underlying molecular mechanisms specific to brace root initiation; however, the existence of maize mutants such as rtcs-1 and rtcs-2, which lack adventitious and seminal lateral roots, indicates common genetic and/or hormonal control pathways of adventitious root initiation (Hetz et al., 1996). Other adaptive traits are also likely to be under common hormonal and genetic control. For example, the angle of emergence of brace and other adventitious root growth has been shown to become steeper under water- and nitrogen-limiting conditions in certain maize lines (Trachsel et al., 2013).
Maps of gene expression specific to maize brace roots indicate that many common pathways are shared between brace, lateral, and adventitious root morphogenesis and emergence, including genes participating in auxin transport and signaling plus cell wall synthesis and degradation (Li et al., 2011). Several genes were identified as specifically expressed in brace root tissues, including sugar transporters, potassium transport channels, and genes involved in wound and pathogen responses. A study using microRNA transcript profiling uncovered 14 conserved and 16 novel microRNAs differentially expressed in brace roots (Li et al., 2014). Other genetic studies have found multiple quantitative trait loci specific for brace root tier number (Ku et al., 2012), many of which have multiple epistatic interactions with each other, highlighting the complexity of the gene regulatory network controlling brace root initiation and growth.
CONCLUSION AND FUTURE CHALLENGES
In summary, impressive steps have been made dissecting the genetic, genomic, and cellular basis of root branching in model plants and crops such as Arabidopsis, rice, and maize in the last decade (for review, see Lavenus et al., 2013; Orman-Ligeza et al., 2013). Common and specific signals, regulatory components, and mechanisms have been identified that control the initiation, morphogenesis, and emergence of new lateral and adventitious root organs. Despite this progress, much more remains to be done.
Major challenges facing researchers include the following.
(1) Integrating the avalanche of root genetic and genomic information into a coherent framework of regulatory interactions in order to generate new insights into the underlying mechanisms controlling root branching. Two distinct approaches have been employed to construct root regulatory networks. The first approach is direct and involves a systematic experimental screen of pairwise interactions among a carefully selected subset of potentially interacting root genes and/or proteins. This approach is exemplified by the elegant work of Siobhan Brady (University of California, Davis) and her collaborators using root tissue-specifically expressed transcription factors in conjunction with the yeast one-hybrid technique to build several tiers of a root gene regulatory network (Brady et al., 2011). The second approach employs a more indirect approach that exploits the availability of root omics (predominantly transcriptomic) data sets to infer gene regulatory networks through statistical analysis tools and identifying regulatory relationships between genes. Although this latter approach is able to rapidly generate models of gene regulatory networks, they ultimately require experimental validation. Nevertheless, mathematical and computational modeling are becoming much more important as our knowledge of root regulatory pathways becomes increasingly complex.
(2) Bridging the genotype-to-phenotype gap by relating key regulatory genes to organ-scale lateral and adventitious root branching traits. Researchers are bridging this gap by integrating quantitative genetic or genome-wide association studies with root phenotyping approaches. For example, genetic diversity panels of crops such as maize are being screened for variation in root branching traits using root phenotyping techniques such as “shovelomics” (Trachsel et al., 2011). By comparing the root phenotypic data sets with genotyping data available for diversity panel lines, researchers are then able to pinpoint DNA polymorphisms that control the root branching trait of interest. This provides a powerful approach to identify new root branching regulatory genes. Nevertheless, the interplay between these scales is complex, and an integrative approach is essential to understand the underlying biological mechanisms. Band et al. (2012) argued that, in order to relate root genotype to phenotype, we must move beyond the gene and organ scales and employ multiscale modeling approaches to predict emergent properties at the tissue, organ, organism, and rhizosphere levels. In reality, both approaches are likely to prove of great value in the coming years.
(3) Understanding how root branching interacts with other root traits and environmental conditions to optimize crop performance and resource capture. Functional structural plant models provide a powerful approach to probe these relationships. For example, the functional structural plant model program SimRoot links root growth and root system architecture with the nutrient acquisition process (Postma and Lynch, 2011). Postma et al. (2014) recently used SimRoot to assess the optimal lateral root branching density for maize for capturing nitrate (a mobile soil resource) and phosphorus (an immobile soil resource). Model simulations revealed that the optimal lateral root branching density in the maize root system depends on the relative availability of these different macronutrients, with the optimum shifting to more branches when the nitrate-to-phosphorus ratio is high. By employing a modeling approach, Postma et al. (2014) were able to simulate many millions of permutations of the different root and nutrient parameters, something that is obviously beyond the logistics of direct measurements, highlighting the benefit of this approach in parallel with traditional experimental studies.
(4) Studying the regulation of root branching under more realistic growth conditions. There are many advantages to analyzing the regulation of root branching under highly controlled aseptic growth conditions such as agar plates. However, by not studying roots in their soil environment, we are likely missing novel adaptive responses to their microenvironment. Techniques to noninvasively visualize roots growing in soil such as microscale computed tomography and magnetic resonance imaging (for review, see Mooney et al., 2012) show great promise. Nevertheless, additional approaches need to be developed to simultaneously noninvasively visualize dynamic changes in root gene expression in a soil environment. Such technical advances would help uncover the hidden half of plants and reveal further new insights into the control of root branching.
Glossary
- ABA
abscisic acid
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council and the Engineering and Physical Sciences Research Council Centres for Integrative Systems Biology Program (to the Centre for Plant Integrative Biology), the European Research Council (advanced investigator grant FUTUREROOTS to M.J.B and J.A.A.), the Biotechnology and Biological Sciences Research Council (Professorial Fellowship to M.J.B., R.T., and J.A.A.), the Distinguished Scientist Fellowship Program at King Saud University (to M.J.B.), the Belgian Science Policy Office (grant no. IAP7/29 to D.M.W., U.V., and M.J.B.), and a Newton International Fellowship (to A.R).
References
- Aloni R, Aloni E, Langhans M, Ullrich CI. (2006) Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann Bot (Lond) 97: 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Fozard JA, Godin C, Jensen OE, Pridmore T, Bennett MJ, King JR. (2012) Multiscale systems analysis of root growth and development: modeling beyond the network and cellular scales. Plant Cell 24: 3892–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Aggarwal P, Robbins NE, II, Sturrock CJ, Thompson MC, Tan HQ, Tham C, Duan L, Rodriguez PL, Vernoux T, et al. (2014) Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc Natl Acad Sci USA 111: 9319–9324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JK, McCully M. (1970) A histological study of lateral root initiation and development in Zea mays. Protoplasma 70: 179–205 [Google Scholar]
- Bellini C, Pacurar DI, Perrone I. (2014) Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 65: 639–666 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Eklöf J, Ljung K, Marchant A, Bennett M, Sandberg G. (2002) Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Onckelen HV, Van Montagu M, Inze D. (1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Zhang L, Megraw M, Martinez NJ, Jiang E, Yi CS, Liu W, Zeng A, Taylor-Teeples M, Kim D, et al. (2011) A stele-enriched gene regulatory network in the Arabidopsis root. Mol Syst Biol 7: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero PJ, Casimiro I, Lloret PG. (1995) Lateral root initiation by asymmetrical transverse divisions of pericycle cells in four plant species: Raphanus sativus, Helianthus annuus, Zea mays, and Daucus carota. Protoplasma 188: 49–58 [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ. (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, et al. (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li Z, Xiong L. (2012) A plant microRNA regulates the adaptation of roots to drought stress. FEBS Lett 586: 1742–1747 [DOI] [PubMed] [Google Scholar]
- Cho SH, Yoo SC, Zhang H, Lim JH, Paek NC. (2014) Rice NARROW LEAF1 regulates leaf and adventitious root development. Plant Mol Biol Rep 32: 270–281 [Google Scholar]
- Coudert Y, Périn C, Courtois B, Khong NG, Gantet P. (2010) Genetic control of root development in rice, the model cereal. Trends Plant Sci 15: 219–226 [DOI] [PubMed] [Google Scholar]
- Delarue M, Prinsen E, Onckelen HV, Caboche M, Bellini C. (1998) Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J 14: 603–611 [DOI] [PubMed] [Google Scholar]
- Della Rovere F, Fattorini L, D’Angeli S, Veloccia A, Falasca G, Altamura MM. (2013) Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of Arabidopsis. Ann Bot (Lond) 112: 1395–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Demchenko NP, Demchenko KN. (2001) Resumption of DNA synthesis and cell division in wheat roots as related to lateral root initiation. Russ J Plant Physiol 48: 755–764 [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. (2010) A novel AUX/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voss U, Vanneste S, Benjamins R, Rademacher EH, Schlereth A, De Rybel B, Vassileva V, Grunewald W, et al. (2010) Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci USA 107: 2705–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- Draye X. (2002) Consequences of root growth kinetics and vascular structure on the distribution of lateral roots. Plant Cell Environ 25: 1463–1474 [Google Scholar]
- Draye X, Delvaux B, Swennen R. (1999) Distribution of lateral root primordia in root tips of Musa. Ann Bot (Lond) 84: 393–400 [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E. (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falasca G, Altamura MM. (2003) Histological analysis of adventitious rooting in Arabidopsis thaliana (L.) Heynh seedlings. Plant Biosystems 137: 265–273 [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. (2005) Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44: 382–395 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tasaka M. (2009) Hormone interactions during lateral root formation. Plant Mol Biol 69: 437–449 [DOI] [PubMed] [Google Scholar]
- Gao S, Fang J, Xu F, Wang W, Sun X, Chu J, Cai B, Feng Y, Chu C. (2014) CYTOKININ OXIDASE/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol 165: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Voß U, Harding SA, Fannon J, Moody LA, Yamada E, Swarup K, Nibau C, Bassel GW, Choudhary A, et al. (2014) AtMYB93 is a novel negative regulator of lateral root development in Arabidopsis. New Phytol 203: 1194–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RFH, von Wirén N. (2014) Root nutrient foraging. Plant Physiol 166: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H. (2012) The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139: 883–893 [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C. (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mongelard G, Floková K, Pacurar DI, Novák O, Staswick P, Kowalczyk M, Pacurar M, Demailly H, Geiss G, et al. (2012) Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 24: 2515–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz W, Hochholdinger F, Schwall M, Feix G. (1996) Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J 10: 845–857 [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K. (2004a) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9: 42–48 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Tuberosa R. (2009) Genetic and genomic dissection of maize root development and architecture. Curr Opin Plant Biol 12: 172–177 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Woll K, Sauer M, Dembinsky D. (2004b) Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programmes. Ann Bot (Lond) 93: 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Zimmermann R. (2008) Conserved and diverse mechanisms in root development. Curr Opin Plant Biol 11: 70–74 [DOI] [PubMed] [Google Scholar]
- Hoppe DC, McCully ME, Wenzel CL. (1986) The nodal roots of Zea: their development in relation to structural features of the stem. Can J Bot 64: 2524–2537 [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Tanakamaru K, Morita S, Abe J, Inanaga S. (2006) Lateral root development, including responses to soil drying, of maize (Zea mays) and wheat (Triticum aestivum) seminal roots. Physiol Plant 127: 260–267 [Google Scholar]
- Jansen L, Hollunder J, Roberts I, Forestan C, Fonteyne P, Van Quickenborne C, Zhen RG, McKersie B, Parizot B, Beeckman T. (2013) Comparative transcriptomics as a tool for the identification of root branching genes in maize. Plant Biotechnol J 11: 1092–1102 [DOI] [PubMed] [Google Scholar]
- Jansen L, Roberts I, De Rycke R, Beeckman T. (2012) Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos Trans R Soc Lond B Biol Sci 367: 1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Kirby EJM. (2002) Botany of the wheat plant. In BC Curtis, S Rajaram, HG Macpherson, eds, Bread Wheat Improvement and Production. FAO Plant Production and Protection Series No. 30. FAO, Rome, pp 30–52 [Google Scholar]
- Kitomi Y, Ito H, Hobo T, Aya K, Kitano H, Inukai Y. (2011a) The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J 67: 472–484 [DOI] [PubMed] [Google Scholar]
- Kitomi Y, Kitano H, Inukai Y. (2011b) Molecular mechanism of crown root initiation and the different mechanisms between crown root and radicle in rice. Plant Signal Behav 6: 1270–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi Y, Ogawa A, Kitano H, Inukai Y. (2008) CRL4 regulates crown root formation through auxin transport in rice. Plant Root 2: 19–28 [Google Scholar]
- Kohlen W, Charnikhova T, Lammers M, Pollina T, Tóth P, Haider I, Pozo MJ, de Maagd RA, Ruyter-Spira C, Bouwmeester HJ, et al. (2012) The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol 196: 535–547 [DOI] [PubMed] [Google Scholar]
- Ku LX, Sun ZH, Wang CL, Zhang J, Zhao RF, Liu HY, Tai GQ, Chen YH. (2012) QTL mapping and epistasis analysis of brace root traits in maize. Mol Breed 30: 697–708 [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Stø IM, Butenko MA, Péret B, Riiser ES, Bennett MJ, Aalen RB. (2013) Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc Natl Acad Sci USA 110: 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al. 2007) Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Biller S, Stanley K, Kajstura T, Prusty R. (2006) Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol 47: 788–792 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AF, Scheres B. (2008) Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18: 450–458 [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim MJ, Kim NY, Lee SH, Kim J. (2013) LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J 73: 212–224 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Negi S, Sukumar P, Muday GK. (2011) Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138: 3485–3495 [DOI] [PubMed] [Google Scholar]
- Li R, Li J, Li S, Qin G, Novák O, Pěnčík A, Ljung K, Aoyama T, Liu J, Murphy A, et al. (2014) ADP1 affects plant architecture by regulating local auxin biosynthesis. PLoS Genet 10: e1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mo X, Shou H, Wu P. (2006) Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol 47: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Li YJ, Fu YR, Huang JG, Wu CA, Zheng CC. (2011) Transcript profiling during the early development of the maize brace root via Solexa sequencing. FEBS J 278: 156–166 [DOI] [PubMed] [Google Scholar]
- Liu S, Wang J, Wang L, Wang X, Xue Y, Wu P, Shou H. (2009) Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res 19: 1110–1119 [DOI] [PubMed] [Google Scholar]
- Lucas M, Guédon Y, Jay-Allemand C, Godin C, Laplaze L. (2008) An auxin transport-based model of root branching in Arabidopsis thaliana. PLoS ONE 3: e3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, Voß U, Swarup K, De Smet I, Van Damme D, Lawrence T, Peret B, Moscardi E, et al. (2013) Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci USA 110: 5229–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, et al. (2011) SHORT-ROOT regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 155: 384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Marhavý P, Vanstraelen M, De Rybel B, Zhaojun D, Bennett MJ, Beeckman T, Benková E. (2013) Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO J 32: 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergemann H, Sauter M. (2000) Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol 124: 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney S, Pridmore T, Helliwell J, Bennett M. (2012) Developing x-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil 352: 1–22 [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthreich N, Majer C, Beatty M, Paschold A, Schützenmeister A, Fu Y, Malik WA, Schnable PS, Piepho HP, Sakai H, et al. (2013) Comparative transcriptome profiling of maize coleoptilar nodes during shoot-borne root initiation. Plant Physiol 163: 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Sukumar P, Liu X, Cohen JD, Muday GK. (2010) Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J 61: 3–15 [DOI] [PubMed] [Google Scholar]
- Neuteboom LW, Ng JM, Kuyper M, Clijdesdale OR, Hooykaas PJ, van der Zaal BJ. (1999) Isolation and characterization of cDNA clones corresponding with mRNAs that accumulate during auxin-induced lateral root formation. Plant Mol Biol 39: 273–287 [DOI] [PubMed] [Google Scholar]
- Nordström AC, Jacobs FA, Eliasson L. (1991) Effect of exogenous indole-3-acetic acid and indole-3-butyric acid on internal levels of the respective auxins and their conjugation with aspartic acid during adventitious root formation in pea cuttings. Plant Physiol 96: 856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman-Ligeza B, Parizot B, Gantet PP, Beeckman T, Bennett MJ, Draye X. (2013) Post-embryonic root organogenesis in cereals: branching out from model plants. Trends Plant Sci 18: 459–467 [DOI] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. (2009a) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Péret B, Larrieu A, Bennett MJ. (2009b) Lateral root emergence: a difficult birth. J Exp Bot 60: 3637–3643 [DOI] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, Band LR, Voß U, Postaire O, Luu DT, Da Ines O, Casimiro I, Lucas M, et al. (2012) Auxin regulates aquaporin function to facilitate lateral root emergence. Nat Cell Biol 14: 991–998 [DOI] [PubMed] [Google Scholar]
- Péret B, Middleton AM, French AP, Larrieu A, Bishopp A, Njo M, Wells DM, Porco S, Mellor N, Band LR, et al. (2013) Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Mol Syst Biol 9: 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma JA, Dathe A, Lynch J. (2014) The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiol 21: 233916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma JA, Lynch JP. (2011) Root cortical aerenchyma enhances the growth of maize on soils with suboptimal availability of nitrogen, phosphorus, and potassium. Plant Physiol 156: 1190–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A, Mason MG, De Cuyper C, Brewer PB, Herold S, Agusti J, Geelen D, Greb T, Goormachtig S, Beeckman T, et al. (2012) Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol 158: 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Strader LC, Bailly A, Yang H, Blakeslee J, Langowski L, Nejedla´ E, Fujita H, Itoh H, Syono K, et al. (2010) Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc Natl Acad Sci USA 107: 10749–10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS. (2006) Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet 2: e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, De Smet I. (2012) Root system architecture: insights from Arabidopsis and cereal crops. Philos Trans R Soc Lond B Biol Sci 367: 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, Geiss G, McKhann H, Garcion C, Vaucheret H, Sandberg G, et al. (2005) Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17: 1343–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Wang J, Sauter M. (2006) Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta 223: 604–612 [DOI] [PubMed] [Google Scholar]
- Strader LC, Bartel B. (2011) Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant 4: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar P, Maloney GS, Muday GK. (2013) Localized induction of the ATP-binding cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol 162: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Takehisa H, Sato Y, Igarashi M, Abiko T, Antonio BA, Kamatsuki K, Minami H, Namiki N, Inukai Y, Nakazono M, et al. (2012) Genome-wide transcriptome dissection of the rice root system: implications for developmental and physiological functions. Plant J 69: 126–140 [DOI] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP. (2011) Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341: 75–87 [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP. (2013) Maize root growth angles become steeper under low N conditions. Field Crops Res 140: 18–31 [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, De Rybel B, Beemster GT, Ljung K, De Smet I, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M, et al. (2005) Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varney GT, Canny MJ. (1993) Rates of water uptake into the mature root system of maize plants. New Phytol 123: 775–786 [Google Scholar]
- Vermeer JE, von Wangenheim D, Barberon M, Lee Y, Stelzer EH, Maizel A, Geldner N. (2014) A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343: 178–183 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Behrens I, Komatsu M, Zhang Y, Berendzen KW, Niu X, Sakai H, Taramino G, Hochholdinger F. (2011) Rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. Plant J 66: 341–353 [DOI] [PubMed] [Google Scholar]
- Wang XL, McCully ME, Canny MJ. (1994) The branch roots of Zea. New Phytol 126: 21–29 [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW. (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43: 118–130 [DOI] [PubMed] [Google Scholar]
- Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS, Hochholdinger F. (2005) Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol 139: 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Paschold A, Marcon C, Liu S, Tai H, Nestler J, Yeh CT, Opitz N, Lanz C, Schnable PS, et al. (2014) The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. J Exp Bot 65: 4919–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hu Y, Dai M, Huang L, Zhou DX. (2009) The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xing L, Wang X, Hou YJ, Gao J, Wang P, Duan CG, Zhu X, Zhu JK. (2014) The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci Signal 7: ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]