Cellular and molecular events are inherent to the formation and development of the root cap in rice.
Abstract
The tip of the root is covered by a thimble-shaped root cap that is the site of perception and transduction for many environmental stimuli. Until now, little was known about how the root cap of rice (Oryza sativa) develops and functions to regulate the adaptive behavior of the root. To address this, we examined the formation of the rice root cap during embryogenesis and characterized the anatomy and structure of the rice radicle root cap. We further investigated the role of the quiescent center in the de novo origin of the root cap. At the molecular level, we found that shoot-derived auxin was absolutely needed to trigger root cap regeneration when the quiescent center was removed. Our time-course analysis of transcriptomic dynamics during the early phases of root cap regeneration indicated that changes in auxin signaling and appropriate levels of cytokinin are critical for root cap regeneration after the removal of the root cap. Moreover, we identified 152 genes that produce root cap-specific transcripts in the rice root tip. These findings together offer, to our knowledge, new mechanistic insights into the cellular and molecular events inherent in the formation and development of the root cap in rice and provide a basis for future research on the developmental and physiological function of the root cap of monocot crops.
The root cap is the terminal-most tissue of the root of most plants. Accumulated evidence over the last 50 years has shown that the root cap not only has a role in the protection of the proximal root meristem (PRM), but also directs root growth in response to various environmental stimuli including gravity (gravitropism), unilateral light (phototropism), touch (thigmotropism), gradients in temperature (thermotropism), humidity (hydrotropism), and ions and other chemicals (chemotropism; Ponce et al., 2000; Barlow, 2003). Moreover, recent findings suggest that border cells, produced by detachment of differentiated root cap cells, play a key role in plant defense and the regulation of rhizosphere microbial populations (Hawes et al., 2012; Driouich et al., 2013).
The root cap originates at the opposite end of the embryo to the shoot apex and consists of the columella and lateral root cap regions. In the model dicot plant species Arabidopsis (Arabidopsis thaliana), the columella root cap is originated by an asymmetric division of the hypophysis derived from the basal cell (Scheres et al., 1994; Jenik et al., 2007), whereas the formation of the lateral root cap is initiated in the embryo proper, which in turn is formed from the apical cell (Scheres et al., 1994). In monocots such as maize (Zea mays) and rice (Oryza sativa), however, the embryonic origin of root cap remains unknown because cell divisions after the first asymmetric zygotic division are highly variable and unpredictable (Suzuki et al., 1992, 1993; Chandler et al., 2008).
During postembryonic development, root cap cells are continuously renewed by the stem cells. Lateral root cap cells in the Arabidopsis root are produced by the periclinal division of the epidermis-lateral root cap stem cells independent of columella root cap stem cells, which give rise to columella root cap cells through an anticlinal division (Dolan et al., 1993). In maize and rice, histological analysis of sections of the radicle and primary root tips suggests that the columella and lateral root cap may originate from the same type of stem cells independent of those of the epidermis (Williams, 1947; Iijima et al., 2008; Rebouillat et al., 2009). However, this hypothesis remains to be proven by cell lineage tracing (Scheres et al., 1994; Kidner et al., 2000; Kurup et al., 2005).
Unlike in Arabidopsis, the root cap in maize and rice is structurally separated from the PRM by a thick cell wall boundary called the root cap junction. The presence of such a boundary makes it possible to detach the intact root cap from the rest of the root tip and use decapped roots to study the function of the root cap in controlling root growth and development (Juniper et al., 1966; Hahn et al., 2008). Earlier studies in maize also revealed that the root cap regenerates from reprogrammed distal quiescent center (QC) cells within 72 h after its removal (Barlow, 1974; Feldman, 1976), suggesting that the QC plays an important role in the de novo origin of a new root cap in decapped maize roots. However, in both maize and Arabidopsis, as early as 24 h after removal of the root cap and the QC, a new set of cells expressed the root cap markers although the QC was not yet reestablished (Ponce et al., 2000; Sena et al., 2009), indicating that root cap regeneration could occur in the absence of a functional QC. Auxin, which positions the new stem cell niche of the Arabidopsis root after laser ablation of the QC (Xu et al., 2006; Grieneisen et al., 2007), has been implicated to play a critical role in the de novo origin and development of the root cap. However, the sequence of molecular events leading to the formation of the root cap remains uncertain.
In this work, we examined the formation of the rice root cap during embryogenesis, and characterized the anatomy and structure of the postembryonic radicle root cap. We further investigated the role of auxin and QC in the de novo origin and development of the root cap, and analyzed global transcriptional changes during the early phases of root cap regeneration. Collectively, our data offer new mechanistic insights into the cellular and molecular events inherent to the formation and development of the root cap.
RESULTS
Root Cap Formation during Embryogenesis
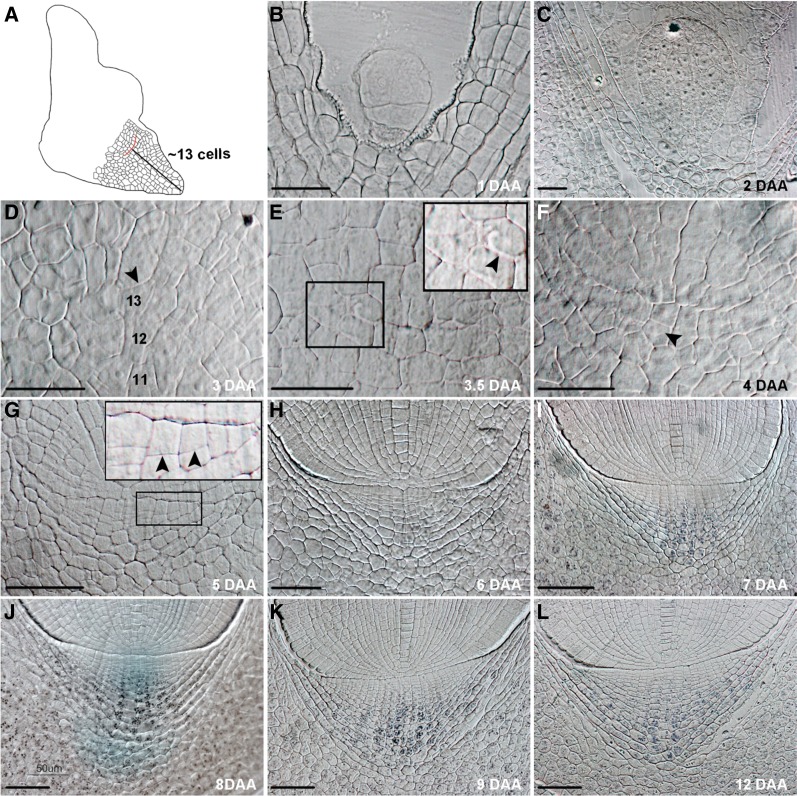
To study the formation of the root cap during embryogenesis, semithin sections of embryos at different developmental stages were cut and examined with a light microscope (Fig. 1). We found that embryonic roots of rice have a closed meristem with a thick cell wall forming the boundary between the root cap and the rest of the root apex (Fig. 1, H–L), similar to that reported for maize and other grasses (Clowes and Juniper, 1964; Sievers et al., 2002). This boundary was referred to as the cap junction (Clowes and Juniper, 1964) and could be unambiguously recognized at 4 d after anthesis (DAA; Fig. 1F). To reveal the exact position where the cap junction emerges, we determined the number of cells between the cell at the basal end of the suspensor and the cap junction in sections of embryos at 4 DAA. We found that the cap junction appeared at an approximately 13-cell distance (mean 12.8 ± 1.3, n = 5; Fig. 1, A and D) from the cell at the basal end of the suspensor. We thus postulated that cells at this position at around 3 DAA (Fig. 1D) are the progenitor cells that would produce the cap junction. Indeed, cells at this position divided anticlinally at around 3.5 DAA and the cap junction appeared (Fig. 1E; Supplemental Fig. S1A). Cells above the cap junction form the QC and PRM, whereas cells below the cap junction develop as the root cap stem cell, which will give rise to the root cap. At 4 DAA, the junction extended after the division of more progenitor cells (Fig. 1F; Supplemental Fig. S1B), and some of the root cap stem cells divided anticlinally to produce a daughter cell (Fig. 1F). More root cap stem cells divided at 5 DAA (Fig. 1G) and a thimble-shaped root cap appeared at 6 DAA with up to eight layers of root cap cells (Fig. 1H). At 7 and 8 DAA, 13 layers of root cap cells could be observed. Starch granules appeared at 7 DAA in the lower 10 layers of columella root cap cells (Fig. 1I), whereas starch granules in lateral root cap cells could only be readily seen at 8 DAA (Fig. 1J). Starch granule formation in the columella and lateral root cap cells indicates that these cells are fully differentiated and thus the root cap might be fully functional. Approximately 16 layers of root cap cells, of which the lower 13 layers could be stained with Lugol solution, were observed from 9 to 12 DAA (Fig. 1, K and L), indicating that the root cap is fully developed at 9 DAA and no further cell division occurred in the top three layers (Fig. 1, K and L), which will give rise to the root cap meristem during postembryonic root development.
Figure 1.
Origin and development of the root cap during rice embryogenesis. A, Schematic view of a medial longitudinal section through a rice embryo at around 4 DAA. The red line denotes the developing cap junction. The black line shows an average 13-cell distance between the cell at the basal end of the suspensor and the developing cap junction. B, Embryo at 1 DAA. C, Embryo at 2 DAA. D to F, Cells at the region where the radicle and cap junction originate. The 11, 12, and 13 in D denote the cell counts at the distance from the cell at the basal end of the suspensor of an embryo at 3.5 DAA. The inset in E is an enlarged view of the boxed region. Arrowheads in D to F point to the putative initiation position of the cap junction (D), an extending part of the cap junction formed by an anticlinal cell division (E), and a daughter cell produced by an anticlinal division of the root cap stem cell (F), respectively. G to L, Development of the root cap in the embryo at 5 DAA (G), 6 DAA (H), 7 DAA (I), 8 DAA (J), 9 DAA (K), or 12 DAA (L). The inset in G is an enlarged view of the boxed region. Arrowheads in G indicate that more root cap stem cells divided at 5 DAA. Lugol staining showed that starch granules appeared from 7 DAA in the lower 10 layers of columella root cap cells (I) and starch granules in lateral root cap cells could only be readily seen from 8 DAA (J–L). Scale bar = 25 μm for B to F and 50 μm for G to L.
Cell Fate and Cell Lineage in the Radicle Root Cap
We found that during the seedling stage, the radicle root cap contains 13 to 14 layers of columella root cap cells and has a similar anatomy and structure as the embryonic root cap (compare Figs. 1, J–L, and 2, A and B; Supplemental Fig. S2, A–C). However, the tip angle of the root cap only stabilized when the root length was longer than 10 mm (Supplemental Fig. S2D). Therefore, we used roots slightly longer than 10 mm for further analysis of cellular organization of the radicle root cap.
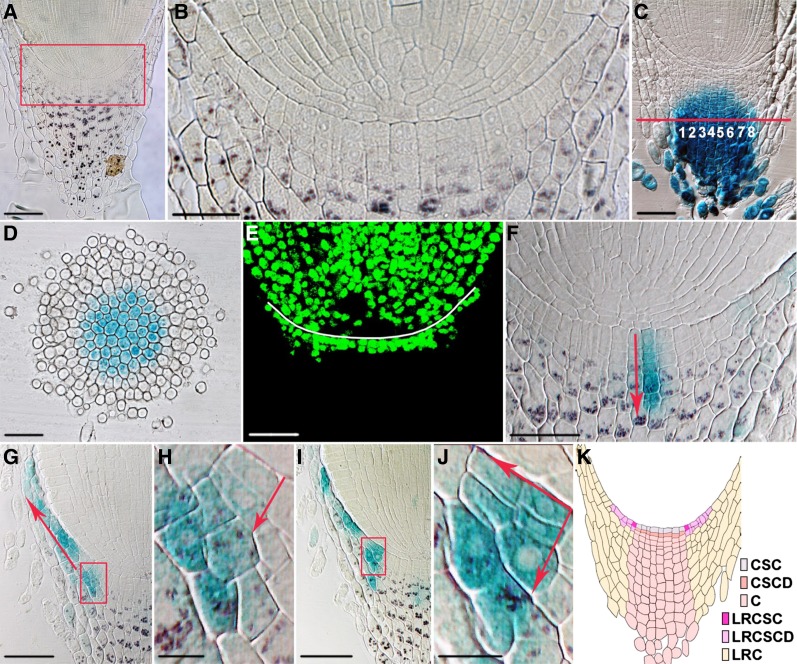
Figure 2.
Cell fate and cell lineage in the radicle root cap. A, Medial longitudinal section of the radicle root tip. Starch granules in the columella root cap were revealed by Lugol staining. B, Enlarged view of the boxed region in A. C, Medial longitudinal section of the radicle root tip of the columella root cap-specific GAL4/UAS::GUS enhancer trap line A788. Note that GUS staining could be observed in eight columns of columella root cap cells. D, Cross section at the position indicated by the red line in C. Cells with GUS staining are columella root cap cells, surrounded by three to five layers of GUS-negative lateral root cap cells. E, EdU cell proliferation assay revealing stem cells and their daughters in the root cap. The white line denotes the position of the cap junction. F to J, Medial longitudinal sections of the radicle root tips of the 35S::Spm-GUS lines, stained with both GUS and Lugol solution. GUS-marked clonal sectors suggest the presence of both columella (F) and lateral root cap lineages (G–J). Arrows in F to H and J show the direction of cell division. H and J are enlarged views of the boxed regions in G and I, respectively. K, Cartoon showing the medial longitudinal view of the root cap. Cell types are color coded. C, Columella; CSC, columella stem cell; CSCD, columella stem cell daughter; LRC, lateral root cap; LRCSC, lateral root cap stem cell; LRCSCD, lateral root cap stem cell daughter. All images were taken at the same magnification except B, H, and J. Bar = 50 μm for A, C to G, and I; 25 μm for B; and 10 μm for H and J.
To distinguish the columella root cap from the lateral root cap, we screened transfer DNA enhancer trap lines from the Rice Mutant Database (http://rmd.ncpgr.cn; Wu et al., 2003) and identified a GAL4/UAS::GUS enhancer trap line A788 as a columella root cap-specific marker (Supplemental Fig. S2C). Longitudinal semithin sections of the radicle root tips of A788 revealed that the columella root cap comprised eight columns of cells (Fig. 2C). Transverse semithin sections showed that the columella root cap cells are located in the center of the radicle root cap, surrounded by three to five layers of lateral root cap cells. Along the central radial axis, eight cells with GUS staining could be found (Fig. 2D), confirming the number of columns observed with longitudinal sections. Moreover, the number of GUS-positive cells on the cross section was approximately 50, in agreement with the estimated columella root cap cell number (π × 42 = 50.3). Between the columella root cap and the root cap junction is the columella root cap meristem, which consists of eight columns and one to two layers of cells that had no Lugol staining but had 5-ethynyl-2′-deoxyuridine (EdU) staining (Fig. 2, B and E). The cells in the upper tier are stem cells of the columella root cap that, as shown by GUS-marked clonal sectors generated by the transposition of a nonautonomous defective Suppressor-mutator (Spm) transposable element from maize (Tissier et al., 1999; Barkoulas et al., 2008; Supplemental Fig. S3), could produce the columella root cap stem cell daughter cell at the lower tier through an anticlinal division and eventually form the entire column of columella root cap cells (Fig. 2F). Lateral root cap cells, on the other hand, appeared to be originated from stem cells unrelated to the columella root cap lineage, because we could only find GUS-marked clonal sectors containing cells of the lateral root cap alone but not in combination with cells of the columella root cap (Fig. 2, G–J). Analysis of these clonal sectors, together with the EdU incorporation assay (Fig. 2E) and Lugol staining (Fig. 2, F–J), revealed the existence of a lateral root cap meristem, which consists of lateral root cap stem cells and lateral root cap stem cell daughter cells produced by both periclinal and anticlinal divisions of lateral root cap stem cells (Fig. 2, E and G–J). These daughter cells could undergo anticlinal divisions and eventually differentiate into starch granule-containing lateral root cap cells (Fig. 2, G–J). Together, these data allowed us to propose a cell lineage map for the rice root cap (Fig. 2K), which underpins future studies on the root cap in rice and other monocot crop species.
Root Cap Regeneration Is Regulated by Auxin and the QC
Although the role of the root cap in root growth and environmental perception and response has been extensively studied (Juniper et al., 1966; Tsugeki and Fedoroff, 1999; Hahn et al., 2008), little attention has been given to the determination of how root cap formation and development is controlled. To address this, we performed surgical removal of the root cap (deCAP; Fig. 3A), excision of root cap and quiescent center (deQC; Fig. 3E), and excision of root cap, quiescent center, and part of proximal root meristem (dePRM; Fig. 3I) as described in the “Materials and Methods.” We then examined the de novo origin and development of the root cap during regeneration of excised tissues.
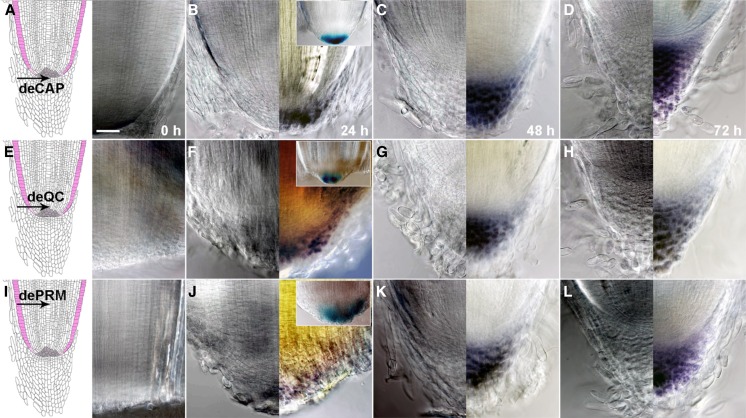
Figure 3.
Root cap regeneration after deCAP, deQC, or dePRM. A to D, Morphology of the radicle root tips at 0 h (A, right), 24 h (B), 48 h (C), or 72 h (D) after deCAP, without (A, right; and B–D, left) or with (B–D, right) Lugol staining. The left image in A is a cartoon showing the medial longitudinal view of the radicle root tip. The arrow points to the position of deCAP. The inset in B shows that GUS expression was detected in A788 24 h after deCAP. E to H, Morphology of the radicle root tips at 0 h (E, right), 24 h (F), 48 h (G), or 72 h (H) after deQC, without (E, right; and F–H, left) or with (F–H, right) Lugol staining. The arrow in E (left) points to the position of deQC. The inset in F shows that GUS expression was detected in A788 24 h after deQC. I to L, Morphology of the radicle root tips at 0 h (I, right), 24 h (J), 48 h (K), or 72 h (L) after dePRM, without (I, right; and J–L, left) or with (J–L, right) Lugol staining. The arrow in I (left) points to the position of dePRM. The inset in J shows that GUS expression was detected in A788 24 h after dePRM. All images were taken at the same magnification. Bar = 50 μm.
We found that the timing and sequence of regenerative events that occurred after deCAP (Fig. 3, A–D), deQC (Fig. 3, E–H), or dePRM (Fig. 3, I–L) in rice were essentially the same as previously described in maize (Barlow, 1974; Feldman, 1976; Ponce et al., 2000). Within about 24 h, Lugol staining and expression of columella root cap-specific marker A788 reappeared in the outer layers of the regenerating root cap (Fig. 3, B, F, and J, and insets), indicating the formation of new columella root cap layers. More root cap layers formed at 48 h after deCAP (Fig. 3C), deQC (Fig. 3G), or dePRM (Fig. 3K) and a complete new root cap regenerated at 72 h (Fig. 3, D, H, and L).
Recent studies in Arabidopsis show that the accumulation of auxin in the root tip is the earliest sign of tissue regeneration triggered by laser ablation of the QC (Xu et al., 2006) or dePRM (Sena et al., 2009). We thus monitored the expression of the auxin-responsive DR5::GUS reporter (Zhao et al., 2009) in the rice root tip over the period of regeneration. DR5::GUS expression was found to be reduced in the QC but was upregulated in the stele of decapped root tips at 1 h after deCAP (compare Fig. 4, A and B), and its expression was further enhanced in the stele and became accumulated at the position of the QC and regenerating root cap at 24 h (Fig. 4C). A strong DR5::GUS expression was observed in the regenerating root cap at 48 h (Fig. 4D) and an expression pattern similar to that in intact roots was seen at 72 h (Fig. 4E), a stage at which a new root cap was fully regenerated. These observations indicate that the initiation of the regeneration process in decapped rice roots involves dynamic changes of auxin levels in the QC and stele. The expression dynamics of DR5::GUS over the first 72 h after deQC and dePRM was remarkably similar to that observed after deCAP (compare Fig. 4, A–E with F–O) except for at 24 h, when a broader DR5::GUS expression domain was observed after deQC and dePRM (compare Fig. 4, C, H, and M).
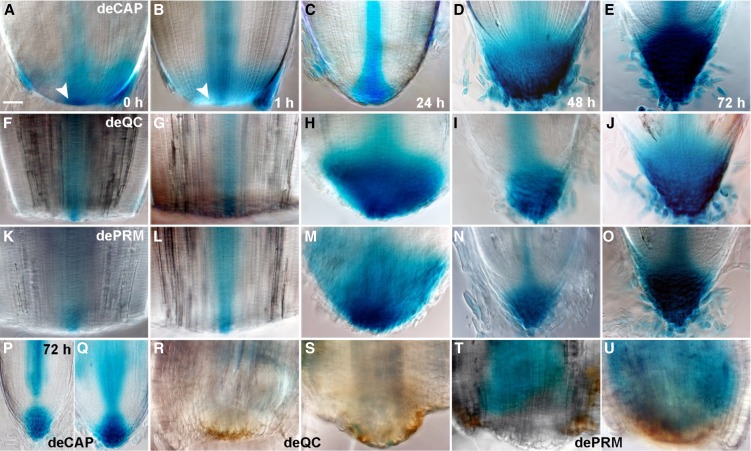
Figure 4.
Auxin distribution in the regenerating root tip. A to E, Expression of the auxin-responsive reporter DR5::GUS at 0 h (A), 1 h (B), 24 h (C), 48 h (D), or 72 h (E) after deCAP. Arrowheads in A and B indicate that DR5::GUS expression was reduced in the QC at 1 h (B) compared with 0 h (A) after deCAP. F to J, DR5::GUS expression at 0 h (F), 1 h (G), 24 h (H), 48 h (I), or 72 h (J) after deQC. K to O, DR5::GUS expression at 0 h (K), 1 h (L), 24 h (M), 48 h (N), or 72 h (O) after dePRM. P and Q, DR5::GUS expression in regenerating root tips at 72 h after deCAP and removal of the shoot part (P) or the shoot part together with the maturation zone of the radicle root (Q). R to U, DR5::GUS expression in the root tips at 72 h after deQC (R and S) or dePRM (T and U) and removal of the shoot part (R and T) or the shoot part together with the maturation zone of the radicle root (S and U). Note that auxin accumulation in the tip of damaged roots and regeneration of the root tip was not observed. All images were taken at the same magnification. Bar = 50 μm.
Root development is controlled by both shoot-derived and root-generated auxin (Overvoorde et al., 2010). To examine the role of shoot-derived auxin during root cap regeneration, we next removed the shoot part, either alone or together with the maturation zone of the radicle root, and performed the deCAP experiment (Fig. 4, P and Q). We found that the expression dynamics of DR5::GUS and the regeneration processes were not affected, although the expression level of DR5::GUS decreased and the root cap had fewer cell layers (Fig. 4, P and Q). These results suggested that when the auxin supply from the shoot was removed, auxin accumulation and regeneration could occur after deCAP in a root-autonomous manner, and that shoot-derived auxin has a role in root cap development. By contrast, auxin accumulation in the damaged root apex and regeneration of the root cap were not observed at 72 h after deQC (Fig. 4, R and S) or dePRM (Fig. 4, T and U) when we simultaneously removed the auxin supply from the shoot, indicating an essential role for the QC in root cap regeneration after the removal of shoot auxin supply. Taken together, we conclude that root cap regeneration in rice requires the presence of the QC or shoot-derived auxin.
Factors Involved in the Regeneration of the Root Cap
To gain further insight into the mechanisms that control root cap formation and development in rice, we next utilized an RNA sequencing (RNAseq) approach to identify transcripts involved in the regeneration of the root cap. Given that changes in DR5::GUS expression were observed in the QC and stele at 1 h after deCAP (Fig. 4B) and that starch granules but not A788 expression started to appear in the terminal-most cells at around 12 h (Fig. 5H; data not shown), we generated gene expression profiles with RNAs isolated from the root apex at 0, 1, or 12 h after deCAP, respectively. We hypothesized that these two time points (1 and 12 h) represent early regeneration events that lead to the formation of a new root cap.
Figure 5.
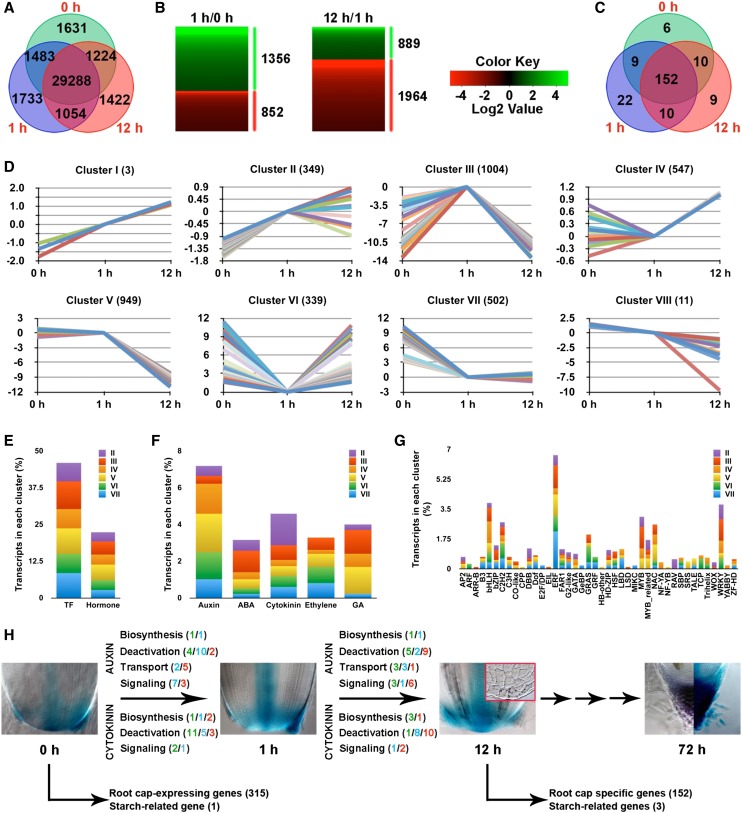
Transcriptome dynamics during early phases of root cap regeneration and identification of root cap-specific genes. A, Venn diagram illustrating the number of unique (nonoverlapping circles) and common (overlapping circles) transcripts identified in the regenerating root tip at 0, 1, and 12 h after deCAP. B, Heat map showing differentially induced (in green) and repressed (in red) transcripts identified between consecutive time points (1 h/0 h and 12 h/1 h). Transcripts with an absolute threshold value of a log2 ratio ≥ 1 and a false discovery rate ≤ 0.001 were identified as differentially expressed. Details of the transcripts are presented in Supplemental Table S1. C, Venn diagram depicting the number of root cap genes whose transcripts were not detected at 0, 1, and 12 h after deCAP. Details of the transcripts are presented in Supplemental Table S4. D, Clusters (I–VIII) of 3,704 transcripts with common expression changes during early phases of root cap regeneration, identified by k-means clustering of differentially expressed transcripts shown in B and listed in Supplemental Table S3. E to G, Percentage of transcripts encoding transcription factors (TFs) and genes involved in the biosynthesis, deactivation, and signaling of five major classes of plant hormones in each cluster. E, All classes. F and G, Individual classes. Details of the transcripts are presented in Supplemental Table S3. H, Summary for the presented data. Transcripts associated with metabolism and signaling of auxin and cytokinin exhibited dynamic transcriptional changes in response to deCAP. The numbers are color coded to indicate the following: green, induced; red, repressed; and blue, no change. Transcripts of 315 root cap genes were identified as common transcripts for 0, 1, and 12 h (Supplemental Table S4), suggesting that they are also expressed in the root meristem. Transcripts of 152 root cap genes, including three of four starch-related genes identified previously in the rice root cap, had no expression within 12 h (Supplemental Table S4), indicating that they are root cap specific in the rice root tip (Supplemental Fig. S5). DR5::GUS expression at 0 h (Fig. 4A), 1 h (Fig. 4B), 12 h, and 72 h (Fig. 4E) and Lugol staining at 12 h (inset) and 72 h after deCAP are shown.
A total of 37,835 transcripts were identified by mapping the clean reads (>40 million for each time point) to the annotated rice genes, of which 29,288 transcripts had detectable expression at 0, 1, and 12 h (Fig. 5A), suggesting that these transcripts have expression in the PRM. A comparison of reads per kilobase transcriptome per million mapped reads (RPKM) values between consecutive time points (1 h/0 h and 12 h/1 h) revealed significant transcriptional changes during the regeneration processes. A list of 3,704 transcripts was identified as differentially expressed transcripts for either or both 1 h/0 h and 12 h/1 h (Fig. 5B; Supplemental Table S1) and a subset of these transcripts was selected and validated by quantitative real-time (qRT)-PCR (Supplemental Fig. S4; Supplemental Table S2). Based on their expression dynamics over the three successive time points, these transcripts were classified into eight clusters (Fig. 5D; Supplemental Table S3), including two categories of transcripts that upregulated (cluster I, 3 transcripts) or downregulated (cluster VIII, 11 transcripts) continuously at 1 h and 12 h after deCAP, two categories of transcripts that were upregulated (cluster II, 349 transcripts) or downregulated (cluster VII, 502 transcripts) at 1 h and then remained largely unchanged at 12 h, two categories of transcripts that were either transiently upregulated (cluster III; 1,004 transcripts) or downregulated (cluster VI; 339 transcripts), and two categories of transcripts that were upregulated (cluster IV, 547 transcripts) or downregulated (cluster V, 949 transcripts) late.
Given the importance of auxin in rice root cap regeneration, transcripts encoding genes known or implicated to play a role in the metabolic and signaling processes of auxin were identified in these clusters, along with transcripts associated with other four major classes of phytohormones: abscisic acid (ABA), cytokinin, ethylene, and GA, which may interact with auxin to regulate rice root cap regeneration. We found that the percentage of auxin- and GA-related transcripts is highest in cluster V (Fig. 5F), making the percentage of total hormone-related transcripts in this cluster highest among all clusters (Fig. 5E). The second highest cluster for auxin is cluster IV (Fig. 5F), which, like cluster V, consists of differentially expressed transcripts for 12 h/1 h but not 1 h/0 h (Fig. 5D). These results suggest that a large portion of auxin-related transcripts exhibited altered expression only after 1 h. Functional classification of auxin-related transcripts further revealed that genes responsible for auxin deactivation, auxin signaling, and auxin transport, but not for auxin biosynthesis, were highly enriched (Fig. 5H; Supplemental Table S3). Significantly more transcripts encoding genes that might deactivate auxin were found to be downregulated at 12 h than 1 h, in agreement with increased expression of DR5::GUS in the regenerating root tip (Fig. 5H). Moreover, auxin transport-related transcripts were found to be largely downregulated at 1 h but upregulated at 12 h (Fig. 5H), suggesting that auxin transport in the root tip was impaired by deCAP but recovered during root cap regeneration. Intriguingly, we found that a transcript encoding OsIAA23, a QC-specific AUX/IAA gene in the rice root tip (Ni et al., 2011), was downregulated at 1 h and then remained lowly expressed at 12 h (Supplemental Table S3). Stabilizing mutations in domain II of OsIAA23 were known to cause the progressive loss of the QC and thus terminal differentiation of the root cap (Ni et al., 2011, 2014), suggesting that auxin signaling in the QC is essential for the formation and development of the root cap. By contrast, cytokinin-related transcripts were strikingly enriched in cluster II compared with other clusters (Fig. 5F). Functional classification of cytokinin-related transcripts showed that deCAP had no significant effects on cytokinin signaling and biosynthesis. However, transcripts encoding genes that might deactivate cytokinin were significantly overrepresented in the list (Supplemental Table S3). Eleven of 19 transcripts were upregulated at 1 h, whereas 10 transcripts were downregulated at 12 h (Fig. 5H), suggesting that a significant portion of bioactive cytokinins was deactivated shortly after deCAP and that cytokinin deactivation was markedly reduced at 12 h. The highest cluster for ABA and the second highest cluster for GA is cluster III (Fig. 5D). Functional classification of ABA- and GA-related transcripts in this cluster indicated that ABA biosynthesis and GA deactivation were transiently induced by deCAP. No significant preference of ethylene-related transcripts to any of the clusters was found (Fig. 5F).
Because transcription factors are ultimately responsible for the differential expression of transcripts, we next identified transcripts that encode putative transcription factors of various families in each of these clusters and found that APETALA2/ERF, WRKY, BHLH, MYB, NAC, and various types of zinc finger family genes are abundantly presented in the list (Fig. 5G; Supplemental Table S3). The percentages of transcription factor-encoding transcripts in clusters III, V, and VII are relatively higher than in other clusters (Fig. 5E), suggesting that a higher portion of transcription factors were transiently induced by deCAP and/or had reduced transcripts at 12 h after deCAP. Notably, among 25 ARF genes identified in rice (Wang et al., 2007), only OsARF16 showed differential expression after deCAP (Supplemental Fig. S4; Supplemental Table S3). OsARF16 is a close homolog of ARF7 and ARF19 in Arabidopsis (Wang et al., 2007). Roots of arf7 arf19 double mutants are agravitropic (Okushima et al., 2005), suggesting a role for OsARF16 in root cap regeneration and recovery of gravitropic response. In addition, transcripts encoding OsWOX10 and OsWOX11, two close homologs of WOX11 and WOX12 in Arabidopsis, were found to be induced within 1 h by deCAP and then either stayed unchanged (OsWOX10) or reduced significantly (OsWOX11) at 12 h (Supplemental Table S3). WOX11 and WOX12 were recently shown to control cell fate transition during de novo root organogenesis in Arabidopsis (Liu et al., 2014) and overexpression of OsWOX11 could induce ectopic root formation in the rice shoot (Zhao et al., 2009), indicating a role for OsWOX10 and OsWOX11 in the specification of new root cap stem cell fate after deCAP.
Genes with Root Cap-Specific Transcripts in the Rice Root Tip
A major advantage of the RNAseq approach over other transcript profiling methods is that it uses absolute rather than relative values, allowing a discrete measurement for each transcript. This enabled us to identify a list of genes whose transcripts were likely not expressed in the decapped root apex at 0 h after deCAP based on a cutoff value of RPKM < 1. A comparison between this list of genes and a list of 521 genes (Supplemental Table S4) identified previously by microarray analysis as preferentially expressed in the rice root cap (Takehisa et al., 2012) revealed an overlap of 177 genes, of which 152 genes had transcripts remained inactive at 1 h and 12 h (Fig. 5, C and H; Supplemental Table S4), suggesting that these genes could generate transcripts that are specifically expressed in the root cap in the rice root tip. This idea was largely supported by the root gene expression profile (Supplemental Fig. S5) compiled from the RiceXPro database (Sato et al., 2013). Moreover, three of four starch-related genes identified previously as root cap-expressing genes (Takehisa et al., 2012) appeared to be root cap specific in the root tip (Fig. 5H; Supplemental Fig. S5), suggesting that these genes have a role in the formation and function of starch granules in the root cap.
DISCUSSION
Our anatomical analysis during rice embryogenesis shows that the columella root cap and lateral root cap arise from a distinct cell layer with an approximately 13-cell distance from the cell at the basal end of the suspensor at around 3.5 DAA. Our cell lineage analysis in the postembryonic rice radicle root cap revealed that the lateral root cap cells are produced by both periclinal and anticlinal divisions of the lateral root cap stem cells, independent of the stem cells that give rise to the epidermis or columella root cap cells. Although the molecular mechanisms controlling the origin and development of the root cap in rice are still largely unknown, studies on the AUX/IAA family gene OsIAA23 suggested that the auxin signaling in the QC plays a critical role in the maintenance of the QC identity and root cap development (Ni et al., 2011, 2014). It will be interesting to see whether stabilizing mutations in OsIAA23 completely abolish root cap regeneration after deCAP at the seedling stage when the root cap is still present in the mutant root. In addition, a rice Glu receptor-like gene (GLR3;1) has been implicated to play a role in lateral root cap development (Li et al., 2006). Compared with the wild-type control, a diminution in the QC size and a large number of cell layers in the lateral root cap were found in the glr3;1 mutant, whereas columella root cap development was not affected by loss of GLR3;1 function. Whether and how GLR3;1 coordinates the maintenance of the QC identity and lateral root cap development remains to be addressed. One possibility is that it involves a cross talk between Glu signaling and auxin signaling in the root tip (Walch-Liu et al., 2006).
deCAP led to a transient reduction of DR5::GUS expression in the QC at 1 h, suggesting that the level of auxin in the QC has to be transiently reduced to permit cell division in the QC and subsequent generation of new root cap stem cells. In the presence of shoot-derived auxin, deQC or dePRM was not able to abolish the ability of root cells to regenerate the damaged tissue, but this was not the case when the auxin supply from the shoot was removed. This observation suggests that, in the absence of a functional stem cell niche, shoot-derived auxin is absolutely needed to activate stem cell-like properties dispersed in the root meristem to mediate complete organ regeneration. The precise identity of cells with stem cell-like properties remains to be established.
With the RNAseq approach, we further expand our knowledge of organ regeneration to the molecular level. Our time-course transcriptomics profile substantiates a role for auxin in the early phases of root cap generation. The identification of transcripts encoding OsIAA23, OsARF16, OsWOX11, and OsWOX12 in the list of differentially expressed transcripts is in good agreement with the findings of previous studies on these genes or their close homologs in Arabidopsis (Okushima et al., 2007; Zhao et al., 2009; Ni et al., 2011; Liu et al., 2014; Ni et al., 2014). These genes are likely key components of the regeneration machinery in plants. Other components of this machinery may include LBD/ASL genes, which are regulated by both ARFs and WOXs (Inukai et al., 2005; Okushima et al., 2007; Lee et al., 2009; Fan et al., 2012; Liu et al., 2014) and are present in the list of differentially expressed transcripts identified by this study (Supplemental Table S3).
Besides auxin-related transcripts, transcripts related to the metabolism and signaling of ABA, cytokinin, ethylene, and GA were identified as differentially regulated by deCAP (Supplemental Table S3). Although it remains a challenge to interpret the changes of these transcripts in a biological context, our analysis indicates that cytokinin has a crucial role in root cap regeneration. Fifteen of 40 putative cytokinin-O-glucosyltransferases identified from the rice genome (Rice GT Database, http://ricephylogenomics.ucdavis.edu/cellwalls/gt/; Cao et al., 2008) were found to be differentially regulated by deCAP (Supplemental Table S3). Cytokinin-O-glycosides represent inactive, stable storage forms of cytokinins and can be rapidly converted back into active cytokinins (Frébort et al., 2011), suggesting that appropriate levels of active cytokinins are critical in root cap regeneration.
By cleanly removing the root cap from the root apex of rice, we were able to use the RNAseq approach, which allows a better discrimination of transcripts with low and no expression than the microarray method, to identify a list of transcripts that were not present in the decapped apex and encode 152 of 521 genes that are preferentially expressed in the rice root cap (Takehisa et al., 2012). We believe that these transcripts are root cap specific in the rice root tip because none of them were detected in the three time points analyzed. Future studies on these root cap-specific transcripts will help to elucidate molecular and cellular mechanisms controlling the development and function of the root cap in rice and other agriculturally important monocot species.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa ssp. japonica) lines used in this study are in the Zhonghua11 background. The DR5::GUS line was described in Zhao et al. (2009). A788 was isolated from a rice GAL4/UAS::GUS enhancer trap collection (Wu et al., 2003). The 35S::Spm-GUS line was generated using the SLJ8313 construct (Tissier et al., 1999). For study of root cap formation during rice embryogenesis, seeds were planted under natural long-day conditions in the experimental field. For regeneration studies of the radicle root, rice seeds were surface sterilized and incubated vertically on petri plates containing one-half-strength Murashige and Skoog medium (Duchefa Biochemie) and were then germinated for 2 to 3 d in the dark at 28°C.
Microscopy Analyses of Root Cap Anatomy and Structure
Root cap development during embryogenesis and in the seedling stage was analyzed with differential interference contrast microscopy (Nikon Eclipse 80i) over time, beginning prior to 1 DAA. In brief, caryopses or radicle roots tips were excised from the rest of the embryos or roots and were immediately fixed in 5% (v/v) formaldehyde, 5% (v/v) acetic acid, 45% (v/v) ethanol, and 45% (v/v) distilled, deionized water at 4°C, followed by vacuum infiltration until the samples sank to the bottom of container. Fixed samples were embedded with Technovit 7100 (Heraeus Kulzer) and cut to semithin sections of 2- to 5-μm thickness with a Leica RM2265 microtome for imaging. The tip angle of the radicle root cap was measured by ImageJ software (National Institutes of Health). For visualization of starch granules, sections were stained for 1 min in Lugol solution (Fluka) and were then imaged with the differential interference contrast microscope.
Root Tip Excision and Regeneration Assays
deCAP was performed according to the method described by Barlow and Hines (1982). For dePRM and deQC experiments, the root cap and QC were excised together with (dePRM) or without (deQC) part of the PRM (with a size equal to the length of the root cap). Excised tips were placed onto the square petri plates containing one-half-strength Murashige and Skoog medium and were cultured in the dark at 28°C. The regeneration of the root cap was analyzed with Lugol staining and GUS staining of marker lines.
Histochemical Analysis of GUS Activity
GUS activity was assayed in the staining solution at 37°C. For DR5::GUS and A788 enhancer trap lines, 30 min and 1 h of staining were performed, respectively. For 35S::Spm-GUS lines, root tips were stained for various time periods depending on the transposition activity of the Spm element in the root cap.
EdU Incorporation Assay
The EdU incorporation assay was performed using an EdU kit from RiboBio, according to the manufacturer’s protocol. Two-day-old rice seedlings were immersed for 24 h in 50 μm EdU solution at 28°C in the dark. EdU images were then captured with a Leica TCS SP2 confocal laser-scanning microscope equipped with an ×40 water immersion objective and were analyzed with Leica LAS AF software.
Whole-Transcriptome RNAseq Analysis
For transcriptome sequencing, total RNA was extracted from the root tip region (with a size equal to the length of the root cap) of the rice seedlings at 0, 1, and 12 h after deCAP, using the RNeasy Plant Mini Kit (Qiagen). RNA integrity and quantity were determined with the Agilent 2100 Bioanalyzer per the manufacturer’s recommendation. Enrichment of mRNA from the total RNA, complementary DNA synthesis, and construction of the library were performed at the Beijing Genome Institute. A total of three libraries were sequenced using an Illumina HiSeq 2000 sequencing system. The raw reads were filtered by removing the adapter sequences and low-quality sequences (e.g. those containing more than 5% unknown bases or more than 30% nucleotides with sequence quality value below 10). The clean reads were then aligned to the rice genome with SOAPaligner/SOAP2 (Li et al., 2009) with no more than five mismatches allowed in the alignment. The level of gene expression was calculated by using the RPKM method (Mortazavi et al., 2008). An absolute threshold value of a log2 ratio ≥ 1 was used to select differentially expressed transcripts. A false discovery rate ≤ 0.001 (Benjamini and Yekutieli, 2001) was further used to estimate the correction for false positive and false negative errors.
qRT-PCR
For qRT-PCR, total RNA was extracted as described for the RNAseq analysis. Primers were designed with PRIMER EXPRESS 2.0 software (PE Applied Biosystems) to amplify 87- to 204-bp products. Primer sequences and information on the 16 selected genes are listed in Supplemental Table S2. The rice ACTIN1 gene was used for data normalization. Three technical replicates were generated per sample type.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AB047313 (ACTIN1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Formation of the cap junction during rice embryogenesis.
Supplemental Figure S2. The tip angle of the radicle root cap and the number of columella root cap layers in the radicle.
Supplemental Figure S3. Schematic diagram of the cell lineage tracing system.
Supplemental Figure S4. Validation of 16 differentially expressed transcripts by qRT-PCR.
Supplemental Figure S5. Expression patterns of 152 root cap genes in the rice root.
Supplemental Table S1. Differentially expressed transcripts identified between consecutive time points (1/0 h and 12/1 h).
Supplemental Table S2. Primer sequences used for the validation of 16 differentially expressed transcripts by qRT-PCR.
Supplemental Table S3. k-Means clustering of differentially expressed transcripts.
Supplemental Table S4. Genes/transcripts expressed in the rice root cap.
Supplementary Material
Acknowledgments
We thank Jonathan D.G. Jones, Yu Zhao, and the Rice Mutant Database for providing DNA constructs and rice materials and Chen Li and Liang Bao for helpful discussions.
Glossary
- PRM
proximal root meristem
- QC
quiescent center
- DAA
d after anthesis
- EdU
5-ethynyl-2′-deoxyuridine
- Spm
Suppressor-mutator
- deCAP
surgical removal of the root cap
- deQC
excision of root cap and quiescent center
- dePRM
excision of root cap, quiescent center, and proximal root meristem
- RNAseq
RNA sequencing
- RPKM
reads per kilobase transcriptome per million mapped reads
- qRT
quantitative real-time
- ABA
abscisic acid
Footnotes
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. (2008) A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet 40: 1136–1141 [DOI] [PubMed] [Google Scholar]
- Barlow P. (1974) Regeneration of the cap of primary roots of Zea mays. New Phytol 73: 937–954 [Google Scholar]
- Barlow P. (2003) The root cap: cell dynamics, cell differentiation and cap function. J Plant Growth Regul 21: 261–286 [Google Scholar]
- Barlow PW, Hines ER. (1982) Regeneration of the root cap of Zea mays L. and Pisum sativum L.: a study with the scanning electron microscope. Ann Bot (Lond) 49: 521–529 [Google Scholar]
- Benjamini Y, Yekutieli D. (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188 [Google Scholar]
- Cao PJ, Bartley LE, Jung KH, Ronald PC. (2008) Construction of a rice glycosyltransferase phylogenomic database and identification of rice-diverged glycosyltransferases. Mol Plant 1: 858–877 [DOI] [PubMed] [Google Scholar]
- Chandler J, Nardmann J, Werr W. (2008) Plant development revolves around axes. Trends Plant Sci 13: 78–84 [DOI] [PubMed] [Google Scholar]
- Clowes FAL, Juniper BE. (1964) The fine structure of the quiescent centre and neighbouring tissues3. J Exp Bot 15: 622–623 [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Driouich A, Follet-Gueye M-L, Vicré-Gibouin M, Hawes M. (2013) Root border cells and secretions as critical elements in plant host defense. Curr Opin Plant Biol 16: 489–495 [DOI] [PubMed] [Google Scholar]
- Fan M, Xu C, Xu K, Hu Y. (2012) LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res 22: 1169–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman LJ. (1976) The de novo origin of the quiescent center regenerating root apices of Zea mays. Planta 128: 207–212 [DOI] [PubMed] [Google Scholar]
- Frébort I, Kowalska M, Hluska T, Frébortová J, Galuszka P. (2011) Evolution of cytokinin biosynthesis and degradation. J Exp Bot 62: 2431–2452 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B. (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Hahn A, Zimmermann R, Wanke D, Harter K, Edelmann HG. (2008) The root cap determines ethylene-dependent growth and development in maize roots. Mol Plant 1: 359–367 [DOI] [PubMed] [Google Scholar]
- Hawes MC, Curlango-Rivera G, Xiong Z, Kessler JO. (2012) Roles of root border cells in plant defense and regulation of rhizosphere microbial populations by extracellular DNA ‘trapping’. Plant Soil 355: 1–16 [Google Scholar]
- Iijima M, Morita S, Barlow PW. (2008) Structure and function of the root cap. Plant Prod Sci 11: 17–27 [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W. (2007) Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol 23: 207–236 [DOI] [PubMed] [Google Scholar]
- Juniper BE, Groves S, Landau-Schachar B, Audus LJ. (1966) Root Cap and the Perception of Gravity. Nature 209: 93–94 [Google Scholar]
- Kidner C, Sundaresan V, Roberts K, Dolan L. (2000) Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta 211: 191–199 [DOI] [PubMed] [Google Scholar]
- Kurup S, Runions J, Köhler U, Laplaze L, Hodge S, Haseloff J. (2005) Marking cell lineages in living tissues. Plant J 42: 444–453 [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J. (2009) LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol 151: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhu S, Song X, Shen Y, Chen H, Yu J, Yi K, Liu Y, Karplus VJ, Wu P, et al (2006) A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem. Plant Cell 18: 340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam T-W, Yiu SM, Kristiansen K, Wang J. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L. (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Ni J, Wang G, Zhu Z, Zhang H, Wu Y, Wu P. (2011) OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J 68: 433–442 [DOI] [PubMed] [Google Scholar]
- Ni J, Zhu Z, Wang G, Shen Y, Zhang Y, Wu P. (2014) Intragenic suppressor of Osiaa23 revealed a conserved tryptophan residue crucial for protein-protein interactions. PLoS ONE 9: e85358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T. (2010) Auxin control of root development. Cold Spring Harb Perspect Biol 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce G, Luján R, Campos ME, Reyes A, Nieto-Sotelo J, Feldman LJ, Cassab GI. (2000) Three maize root-specific genes are not correctly expressed in regenerated caps in the absence of the quiescent center. Planta 211: 23–33 [DOI] [PubMed] [Google Scholar]
- Rebouillat J, Dievart A, Verdeil JL, Escoute J, Giese G, Breitler JC, Gantet P, Espeout S, Guiderdoni E, Périn C. (2009) Molecular Genetics of Rice Root Development. Rice 2: 15–34 [Google Scholar]
- Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y. (2013) RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res 41: D1206–D1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P. (1994) Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120: 2475–2487 [Google Scholar]
- Sena G, Wang X, Liu HY, Hofhuis H, Birnbaum KD. (2009) Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers A, Braun M, Monshausen GB (2002) The root cap: structure and function. In Y Waisel, A Eshek, U Kafkafi, eds, Plant Roots: The Hidden Half, Ed 3, Marcel Dekker Inc., New York, pp 51–74 [Google Scholar]
- Suzuki K, Miyake H, Taniguchi T, Maeda E. (1993) Ultrastructural studies on rice globular embryos with emphasis on epidermis initiation. Jpn J Crop Sci 62: 116–125 [Google Scholar]
- Suzuki K, Taniguchi T, Maeda E. (1992) Ultrastructure and cleavage pattern of rice proembryos. Jpn J Crop Sci 61: 292–303 [Google Scholar]
- Takehisa H, Sato Y, Igarashi M, Abiko T, Antonio BA, Kamatsuki K, Minami H, Namiki N, Inukai Y, Nakazono M, et al (2012) Genome-wide transcriptome dissection of the rice root system: implications for developmental and physiological functions. Plant J 69: 126–140 [DOI] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JD. (1999) Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki R, Fedoroff NV. (1999) Genetic ablation of root cap cells in Arabidopsis. Proc Natl Acad Sci USA 96: 12941–12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Liu LH, Remans T, Tester M, Forde BG. (2006) Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant Cell Physiol 47: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y. (2007) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394: 13–24 [DOI] [PubMed] [Google Scholar]
- Williams BC. (1947) The structure of the meristematic root tip and origin of the primary tissues in the roots of vascular plants. Am J Bot 34: 455–462 [Google Scholar]
- Wu C, Li X, Yuan W, Chen G, Kilian A, Li J, Xu C, Li X, Zhou DX, Wang S, et al (2003) Development of enhancer trap lines for functional analysis of the rice genome. Plant J 35: 418–427 [DOI] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B. (2006) A molecular framework for plant regeneration. Science 311: 385–388 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hu Y, Dai M, Huang L, Zhou DX. (2009) The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.