Flavonols contribute to lateral root development through the regulation of auxin transport and root hair and they modulate the levels of reactive oxygen species.
Abstract
This study utilized tomato (Solanum lycopersicum) mutants with altered flavonoid biosynthesis to understand the impact of these metabolites on root development. The mutant anthocyanin reduced (are) has a mutation in the gene encoding FLAVONOID 3-HYDROXYLASE (F3H), the first step in flavonol synthesis, and accumulates higher concentrations of the F3H substrate, naringenin, and lower levels of the downstream products kaempferol, quercetin, myricetin, and anthocyanins, than the wild type. Complementation of are with the p35S:F3H transgene reduced naringenin and increased flavonols to wild-type levels. The initiation of lateral roots is reduced in are, and p35S:F3H complementation restores wild-type root formation. The flavonoid mutant anthocyanin without has a defect in the gene encoding DIHYDROFLAVONOL REDUCTASE, resulting in elevated flavonols and the absence of anthocyanins and displays increased lateral root formation. These results are consistent with a positive role of flavonols in lateral root formation. The are mutant has increased indole-3-acetic acid transport and greater sensitivity to the inhibitory effect of the auxin transport inhibitor naphthylphthalamic acid on lateral root formation. Expression of the auxin-induced reporter (DR5-β-glucuronidase) is reduced in initiating lateral roots and increased in primary root tips of are. Levels of reactive oxygen species are elevated in are root epidermal tissues and root hairs, and are forms more root hairs, consistent with a role of flavonols as antioxidants that modulate root hair formation. Together, these experiments identify positive roles of flavonols in the formation of lateral roots and negative roles in the formation of root hairs through the modulation of auxin transport and reactive oxygen species, respectively.
The phenylpropanoid pathway is one of the best-characterized systems of plant natural product biochemistry, despite its complexity (Winkel, 2006; Fraser and Chapple, 2011). This pathway produces lignins, sinapates, and flavonoids, which includes flavonones, flavonols, and anthocyanins, and a variety of more specialized secondary metabolites. In recent years, increasing interest in the roles of flavonoids in a variety of fundamental biological processes has spurred the characterization of both their biosynthesis and physiological and developmental activities (Taylor and Grotewold, 2005; Grotewold, 2006; Peer and Murphy, 2007). An intriguing feature of flavonoids is that there are rapid and profound changes in the abundance of these metabolites in response to changing environmental parameters and growth and developmental signals (Winkel-Shirley, 2002). Flavonoid biosynthesis is transcriptionally induced in response to elevated light and changing light quality (Broun, 2005; Azari et al., 2010), elevated auxin and ethylene (Buer et al., 2006; Lewis et al., 2011b; Watkins et al., 2014), oxidant stresses (Page et al., 2012), and drought stress (Fini et al., 2012). Genetic approaches have identified roles for the flavonols in an array of developmental and physiological responses in Arabidopsis (Arabidopsis thaliana; Taylor and Grotewold, 2005; Buer et al., 2010).
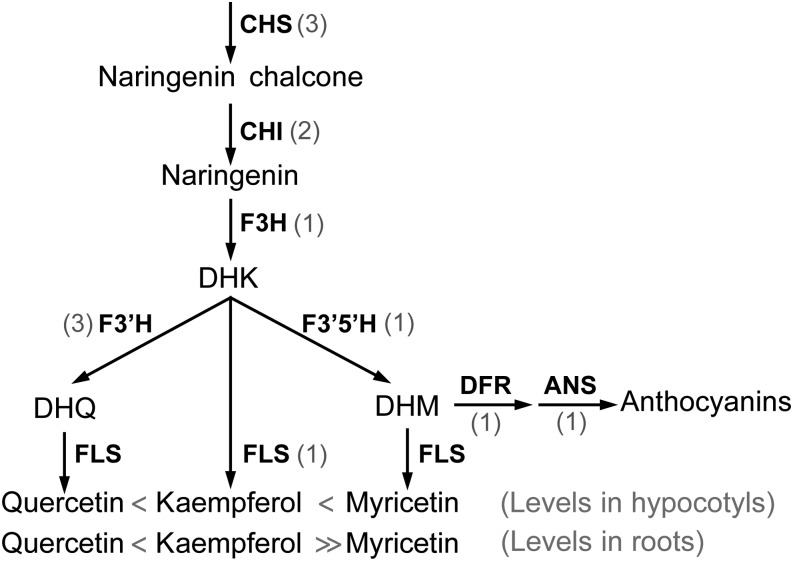
The genes encoding enzymes and regulatory factors controlling phenylpropanoid metabolism have been identified in Arabidopsis, and mutants in each step of the pathway have been isolated (Shirley et al., 1995; Winkel, 2006). This pioneering work led to the identification of transparent testa (tt) Arabidopsis mutants that have reduced pigmentation in the seed coat due to defects in genes encoding the central enzymes and regulatory molecules of this pathway (Shirley et al., 1995; Winkel-Shirley, 2001a). The biosynthetic pathway for flavonones, flavonols, and anthocyanins is illustrated in Figure 1 and highlights the order of enzymes and intermediates and defines the abbreviations that are used for both genes and protein products (Winkel-Shirley, 2001b; Koes et al., 2005). In addition to the metabolites and enzymes found in Arabidopsis, there are distinct enzymes that lead to unique metabolites in different species (Winkel-Shirley, 2001b). In tomato (Solanum lycopersicum), but not Arabidopsis, dihydrokaempferol (DHK) is also converted to dihydromyrectin (DHM) and then to either myricetin, by FLAVONOL SYNTHASE (FLS), or delphinidin-type anthocyanins, by DIHYDROFLAVONOL REDUCTASE (DFR) and ANTHOCYANIDIN SYNTHASE (ANS; Ballester et al., 2010). DHM is the primary substrate for anthocyanidin synthesis in tomato (Groenenboom et al., 2013). In addition to the synthesis of the core aglycone flavonols outlined in Figure 1, flavonols can be glycosylated to modulate structure and function (Winkel-Shirley, 2001a).
Figure 1.
The biosynthetic pathway leading to flavonols and anthocyanidins, showing enzymatic steps. Gray numbers indicate gene copies in tomato. Greater than/less than symbols describe relative levels of free quercetin, kaempferol, and myricetin in tomato hypocotyl and root tissues.
The presence of mutants in each step of the flavonoid pathway in Arabidopsis has provided strong evidence that flavonoids have important roles in modulating signaling pathways in plants. The tt4 mutant has a defect in the gene encoding CHALCONE SYNTHASE (CHS), the first committed step of the pathway, and has alterations in root elongation, gravitropism, and lateral root development (Brown et al., 2001; Buer and Muday, 2004; Peer et al., 2004; Buer et al., 2006; Lewis et al., 2011b). The root gravitropic delays and auxin transport defects in several tt4 alleles are reversed by chemical complementation with the flavonol precursor naringenin (Brown et al., 2001; Buer and Muday, 2004; Buer et al., 2006). Inflorescence branching and hypocotyl elongation are impaired in mutants in flavonoid pathway enzymes (Buer and Djordjevic, 2009). Aberrant accumulation of flavonoids has also been linked to hyponastic cotyledons, altered shape of pavement cells, and deformed trichomes in Arabidopsis (Kuhn et al., 2011). In tomato, a defect in a CHALCONE ISOMERASE (CHI) gene led to altered flavonoid and terpenoid accumulation in trichomes that resulted in a reduction in insect deterrence (Kang et al., 2014). In tomato and maize (Zea mays), fertilization and seed development are blocked in CHS RNA interference lines and mutants with reduced flavonoid synthesis, respectively (Mo et al., 1992; Schijlen et al., 2007). In contrast, tt4 mutants of Arabidopsis that make no flavonoids are fully fertile (Burbulis et al., 1996; Ylstra et al., 1996), suggesting that there is variation in the developmental processes regulated by flavonols between species.
One target of flavonols is auxin transport, which is elevated in inflorescences, hypocotyls, and roots of Arabidopsis plants with the tt4-2 mutation (Murphy et al., 2000; Brown et al., 2001; Buer and Muday, 2004; Lewis et al., 2011b), consistent with the absence of an endogenous inhibitor of auxin transport. The specific role of one flavonol, quercetin, in inhibiting indole-3-acetic acid (IAA) transport via ATP-BINDING CASSETTE PROTEIN B19 in vitro (Geisler et al., 2005) and inhibiting shootward auxin transport and gravitropism was shown using genetic approaches in Arabidopsis (Lewis et al., 2011b). In contrast, kaempferol is the active flavonoid in regulating rootward auxin transport in the Arabidopsis inflorescence (Yin et al., 2013). These results are consistent with specific flavonoids controlling distinct aspects of growth and development and auxin transport in a tissue-specific fashion.
Another mechanism by which flavonols may regulate growth and development is through antioxidant activity (Hernández et al., 2009; Pollastri and Tattini, 2011; Agati et al., 2012). Reactive oxygen species (ROS) can act as signaling molecules that regulate plant developmental processes (De Tullio et al., 2010; Swanson and Gilroy, 2010; Mittler et al., 2011), including guard cell physiology (Kwak et al., 2003; Watkins et al., 2014), root hair and primary root elongation (Monshausen et al., 2009; Ivanchenko et al., 2013), differentiation in the root apex (Bashandy et al., 2010; Tsukagoshi et al., 2010), and developing adventitious roots (Steffens et al., 2012). In both plants and animals, ROS can be generated through respiratory burst/NADPH oxidases (Suzuki et al., 2011), while ROS levels are reduced through antioxidant proteins, including thioredoxins, glutathione/glutaredoxins, and peroxidases (Mittler et al., 2011). Plants also produce many chemical antioxidants, including ascorbate, carotenoids, and flavonoids (Agati et al., 2012). The potential for flavonoids to act as antioxidants in both signaling and oxidative stress pathways has been debated (Hernández et al., 2009; Pollastri and Tattini, 2011; Agati et al., 2012). This idea has recently been tested in the tt4 mutant of Arabidopsis, which has a defect in the first enzyme of flavonoid synthesis. Flavonoids accumulate in guard cells on the surface of wild-type, but not tt4, leaves, and the tt4 mutant shows elevated ROS and altered stomatal closure, consistent with the flavonoids in these cells acting as antioxidants that modulate signaling (Watkins et al., 2014).
This study examines the regulation of tomato root architecture by flavonols and explores mechanisms by which flavonols may act to control development. The mutant anthocyanin reduced (are) has been suggested to have a defect in the gene encoding the enzyme FLAVONOID 3-HYDROXYLASE (F3H; Yoder et al., 1994), thereby reducing the levels of anthocyanins and their flavonol precursors. This study provides a more detailed molecular and biochemical analysis of this mutant and explores the developmental impact of flavonols in this important crop species. We also test the hypotheses that flavonols function to modulate lateral root development through the alteration of auxin transport capacity and root hair development through the modulation of ROS levels in planta, acting as antioxidants in vivo.
RESULTS
Examination of Genes Encoding Flavonoid Biosynthetic Pathway Enzymes in the are Mutant
The are tomato mutant was identified by its greatly reduced anthocyanin levels in vegetative tissues. Yet, as the name implies, the mutant has a reduction, but not an absence, of anthocyanins. Total anthocyanin extracts from hypocotyls of 5-d-old wild-type and are seedlings showed that are produces 13% of the amount of anthocyanin in the wild type (Supplemental Table S1), while anthocyanins are not detected in tomato roots in either genotype. A previous report indicated that are has a point mutation in the gene encoding F3H (Yoder et al., 1994). Yet, it has been suggested that there is only one tomato F3H gene, detected by Southern blot (De Jong et al., 2004), which was difficult to reconcile with the presence of detectable anthocyanins in this mutant.
Therefore, we examined the genes encoding enzymes in the flavonol and anthocyanidin biosynthetic pathway (Fig. 1) using the recently released tomato genome from the Sol Genomics Network (solgenomics.net). The flavonoid enzymes in Arabidopsis are encoded by single genes (Shirley et al., 1995; Winkel, 2006), but in tomato, several of the enzymes are encoded by multiple genes. The Sol Genomics Network database contains four putative copies of CHS (SGN-U579222, SGN-U580856, SGN-U581366, and SGN-U580262), with CHS2 (SGN-U580856) being the most highly expressed (Løvdal et al., 2010). There are two and three putative copies of CHI (SGN-U577427 and SGN-U579009) and FLAVONOID 3′-HYDROXYLASE (F3′H; SGN-U576659, SGN-U573255, and SGN-U570072), respectively. The lack of known tomato mutants defective in these genes may be due to redundant activity of the different copies of these genes. F3H, FLS, FLAVONOID 3′,5′-HYDROXYLASE (F3′5′H), DFR, and ANS have only one apparent copy (SGN-U563669, SGN-U569889, SGN-U602093, SGN-U569072, and SGN-U602582, respectively).
F3H was found on chromosome 2 and encodes a protein that has 93% amino acid identity to Arabidopsis F3H. Aligning F3H amino acid sequences from distantly related plant species, including tomato, Arabidopsis, rice (Oryza sativa), maize, and pea (Pisum sativum; Supplemental Fig. S1), showed that this sequence is highly conserved, especially in the dioxygenase N-terminal and 2 oxoglutarate (2OG)-Fe(II) oxygenase C-terminal domains, which characterize the class of enzyme to which F3H belongs (http://www.ncbi.nlm.nih.gov/cdd). Consistent with a single gene encoding F3H, the next closest match, SGN-U579904, had only 57% identity to the F3H protein and is annotated as a flavonone 3-hydroxylase-like/oxidoreductase protein and shares 67% amino acid identity to Arabidopsis DOWNY MILDEW RESISTANT6. This gene contained an Fe(II)-dependent 2OG oxygenase C-terminal domain but lacked the provisional flavonone 3-hydroxylase N-terminal domain found in F3H genes in tomato and other species. The point mutation, S117N, identified in the F3H gene of are resided within a highly conserved region of the dioxygenase N-terminal domain (De Jong et al., 2004). The absence of detectable F3H enzyme activity in are (Yoder et al., 1994) was consistent with this molecular defect, but it does not explain the ability of this mutant to make low levels of anthocyanin. One hypothesis is that low amounts of naringenin can be converted to downstream flavonoids by an enzyme other than F3H, such as FLS or ANS, as has been shown in vitro with Arabidopsis enzymes (Turnbull et al., 2004; Owens et al., 2008), both of which share the Fe(II)-dependent 2OG oxygenase domain with F3H, as determined using the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/cdd).
Flavonol Levels in are Seedlings
To provide insight into the metabolic phenotype of the are mutant, we used HPLC-mass spectrometry to determine flavonoid metabolite levels in tomato seedling hypocotyls and roots. Hydrolyzed flavonol samples from roots and hypocotyls of 6-d-old plants were extracted to quantify the total aglycone pools of naringenin, a flavonol precursor, and the flavonols quercetin, kaempferol, and myricetin using tandem mass spectrometry by comparison of the resulting spectra and retention times with those of authentic standards and a public spectral database (www.massbank.jp).
In wild-type hypocotyls, myricetin was the most abundant of the four compounds analyzed (Table I), consistent with previously published data (Groenenboom et al., 2013). Naringenin accumulated at much lower levels than the three flavonols, at 20% of quercetin, 6% of kaempferol, and 2% of myricetin concentrations, consistent with the efficient conversion of this precursor to later pathway intermediates. Kaempferol accumulated to greater than 3-fold higher levels than quercetin in this tissue. In roots, the most abundant flavonol was kaempferol, while quercetin was 10% of the levels of kaempferol, and myricetin was not detectable. Naringenin levels were 7% of the levels of kaempferol and 75% of the levels of quercetin.
Table I. Quantification of flavonoids from hypocotyl and root tissue from 6-d-old VF36 and are seedlings by liquid chromatography-mass spectrometry.
Averages and sd are presented. Asterisks indicate significant differences from the wild type with P ≤ 0.05, determined by Student’s t test (n = 10). ND indicates that the compound was not detectable because it was below the limit of detection, which was 0.1 ng g−1 fresh weight for kaempferol and 10 ng g−1 fresh weight for myricetin.
| Flavonoid | Hypocotyl |
Root |
||||
|---|---|---|---|---|---|---|
| VF36 | are | Ratioa | VF36 | are | Ratioa | |
| ng g−1 fresh wt | ng g−1 fresh wt | |||||
| Naringenin | 13.8 ± 0.7 | 89.1 ± 4.3* | 7.4 | 0.4 ± 0.1 | 1.2 ± 0.4 | 3.0 |
| Kaempferol | 230.5 ± 20.1 | 7.5 ± 0.3* | 0.1 | 6.1 ± 1.2 | ND | <0.02 |
| Quercetin | 72.2 ± 16.1 | 7.9 ± 1.8* | 0.1 | 0.6 ± 0.2 | 0.3 ± 0.2 | 0.5 |
| Myricetin | 942.2 ± 58.9 | 673.2 ± 29.8* | 0.7 | ND | ND | ND |
Ratio represents values from are divided by VF36.
If the primary lesion in are is a defect in F3H, the levels of the substrate of this enzyme, naringenin, should be higher in the mutant, while downstream flavonoids are expected to be lower. As hypothesized, naringenin was increased in are, with 7- and 3-fold increases relative to the wild type in hypocotyls and roots, respectively (Table I). In hypocotyls, kaempferol and quercetin were at 5% and 12% of wild-type levels in are, respectively (Table I). In are roots, kaempferol was below the threshold of detection of 0.1 ng g−1 fresh weight in are, corresponding to less than 2% of wild-type levels. Surprisingly, the levels of myricetin were much less perturbed in are hypocotyls, accumulating at 70% of wild-type levels.
To investigate the regulatory role of F3H in the flavonoid pathway in a situation of increased upstream enzymatic activity, we performed flavonoid analysis in seedlings (containing root and shoot tissues) of a tomato p35S:CHS-CHALCONE REDUCTASE (CHR) overexpression line (Schijlen et al., 2006). Kaempferol, quercetin, and myricetin levels were equivalent in the transgenic and the untransformed parental genotype; however, naringenin accumulated at much higher levels in the transgenic line than in the wild type (Supplemental Table S2). This result suggested that F3H was a bottleneck in the flavonoid pathway in this transgenic line, limiting the accumulation of downstream metabolites.
Analysis of the Expression of Genes Encoding Flavonoid Biosynthetic Enzymes
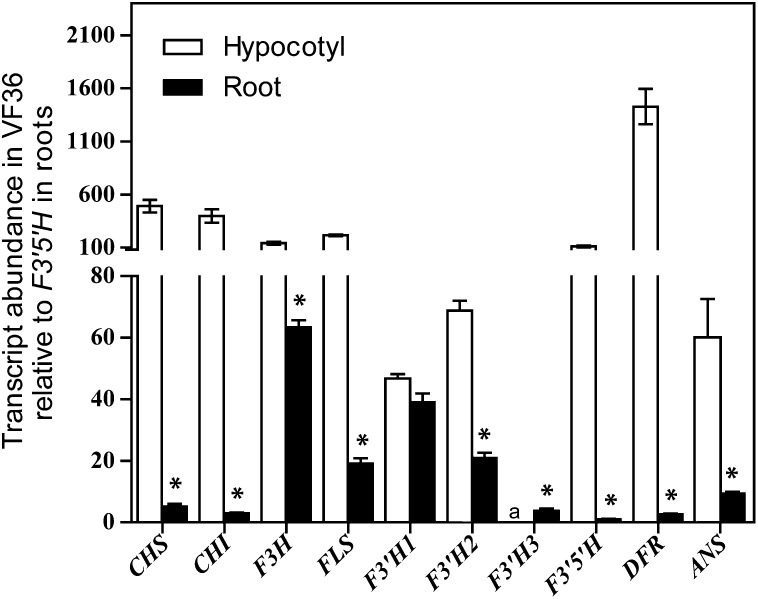
The accumulation of transcripts encoding flavonoid biosynthetic enzymes was examined in the wild type to look for relationships between transcriptional controls of the pathway and metabolite accumulation. The transcript abundance of each flavonoid biosynthetic gene was analyzed by quantitative real-time (qRT)-PCR in hypocotyls and roots of 6-d-old wild-type seedlings. Transcript levels were reported relative to the transcript with the lowest detectable abundance, which was F3′5′H in roots (Fig. 2). The statistical significance of the differences in transcript levels of flavonoid biosynthetic genes between hypocotyl and root tissue was examined by two-way ANOVA, performed for each gene. The F ratios and P values for each test are reported in Supplemental Table S3. The differences between tissues were significant for all genes.
Figure 2.
Transcript abundance of flavonoid biosynthetic genes in hypocotyls and roots of VF36. Quantities are reported normalized to F3′5′H in roots. Scales differ above and below the y axis break, to clearly show levels of low-abundance transcripts. Averages and se are reported for nine biological replicates, each with three technical replicates. Asterisks indicate differences between tissue types. Significance was determined by Student’s t test with P ≤ 0.05. a, Transcript level was below the threshold of detection.
An illustration is provided in Supplemental Figure S2C to facilitate visualization of the qualitative differences in flavonol transcript and metabolite levels between tissue types. The presence of high levels of CHS and CHI transcripts in hypocotyls was consistent with greater carbon flow from primary metabolism into the flavonoid pathway in hypocotyls than in roots. High levels of F3′5′H and DFR transcripts in hypocotyls were consistent with high levels of myricetin and anthocyanin in this tissue. In roots, the near-background levels of F3′5′H, DFR, and ANS transcripts reflected the absence of detectable myricetin and anthocyanins in these tissues. The relatively high levels of transcripts encoding F3H, FLS, and F3′H in roots suggested the efficient conversion of naringenin to downstream intermediates, leading to substantially higher levels of kaempferol and quercetin than naringenin. We asked whether the relatively high level of F3H transcript abundance in roots, compared with other flavonoid gene transcripts, was specific to the VF36 genotype. We performed qRT-PCR of F3H in Ailsa Craig (AC) and found that F3H transcript abundance in roots was 42% of the level in hypocotyls (Supplemental Fig. S3). The consistency of high levels of F3H transcripts among genotypes in roots compared with other flavonoid biosynthetic genes suggested an important regulatory role for the F3H protein in roots.
These data allowed us to ask whether the conversion of DHK to kaempferol, DHQ, or DHM was correlated with the levels of transcripts encoding the branch point enzymes FLS, F3′H, and F3′5′H that controlled their synthesis, respectively. The transcripts encoding FLS and F3′5′H were at equivalent levels, while the transcripts encoding three different F3′H isoenzymes were at individually lower levels but had a similar combined total. These results were not consistent with a model in which transcript and protein levels defined the metabolite partitioning at the DHK branch point. Rather, these results are consistent with a previous report that FLS had a higher affinity for DHK than DHQ when the activity of purified Arabidopsis FLS is examined in vitro (Owens et al., 2008). In roots, the higher levels of kaempferol than quercetin could have resulted from F3′H being at lower levels, but this was not consistent with the transcript levels, in which the combined F3′H transcripts were at much higher levels than FLS transcripts. These results suggested that posttranscriptional regulation of the F3′H gene may limit protein synthesis or enzyme activity. Extensive regulation of this activity was interesting considering the presence of three isoenzymes, which may allow the differential regulation of enzyme activity.
Analysis of the Expression of Genes Encoding Flavonoid Biosynthetic Enzymes in are
The levels of these transcripts were also examined in 6-d-old root and hypocotyl samples from the are mutant and were reported relative to F3′5′H in roots (Supplemental Fig. S2A) and the level of each transcript in the VF36 wild type (Supplemental Fig. S2B). There were no significant differences in F3H transcript abundance between are and VF36 in hypocotyls or roots, suggesting that the point mutation in are did not affect the expression of this gene. In roots of are, there were subtle changes in CHS, CHI, F3′5′H, DFR, and ANS compared with the wild type. The statistical significance of the differences in transcript levels of flavonoid biosynthetic genes between are and the wild type was also examined by two-way ANOVA, performed for each gene (Supplemental Table S3). The differences between genotype were significant for CHS, FLS, F3′H-1, F3′H-3, F3′5′H, and DFR transcripts. The two-way interactions of genotype and tissue were significant for CHS, FLS, F3′H-3, F3′5′H, and DFR. These statistical tests indicate that there are small, but significant differences in the transcript abundance of many of the flavonoid biosynthetic genes due to the are mutation and between hypocotyl and root tissues. Interestingly, transcripts of F3′H3, encoding an enzyme that may control the level of quercetin synthesis, were increased most significantly in both are roots and hypocotyls compared with the wild type. In are hypocotyls, F3′H3 and DFR transcripts were elevated 2- and 1.5-fold, respectively, compared with the wild type. This observation may have reflected a feed-forward effect of naringenin, causing downstream enzymes to accumulate to process the excess precursor.
FLS and ANS structure and enzyme activity measurements in other species suggested that they may allow naringenin to bypass F3H to produce the low levels of myricetin and anthocyanins in this mutant (Turnbull et al., 2004; Owens et al., 2008). Therefore, we asked whether transcript levels suggest a compensatory synthesis of these enzymes. In are hypocotyls, FLS transcripts decreased while ANS transcripts were at wild-type levels (Supplemental Fig. S2). The lack of increased abundance of the transcripts encoding ANS or FLS in are suggested that the synthesis of these two enzymes was not increased to compensate for a loss of F3H activity in are, although the normal abundance of these enzymes may be sufficient to convert low levels of naringenin to downstream anthocyanins.
Complementation of are with F3H Restored Flavonol Synthesis and Anthocyanin Pigmentation
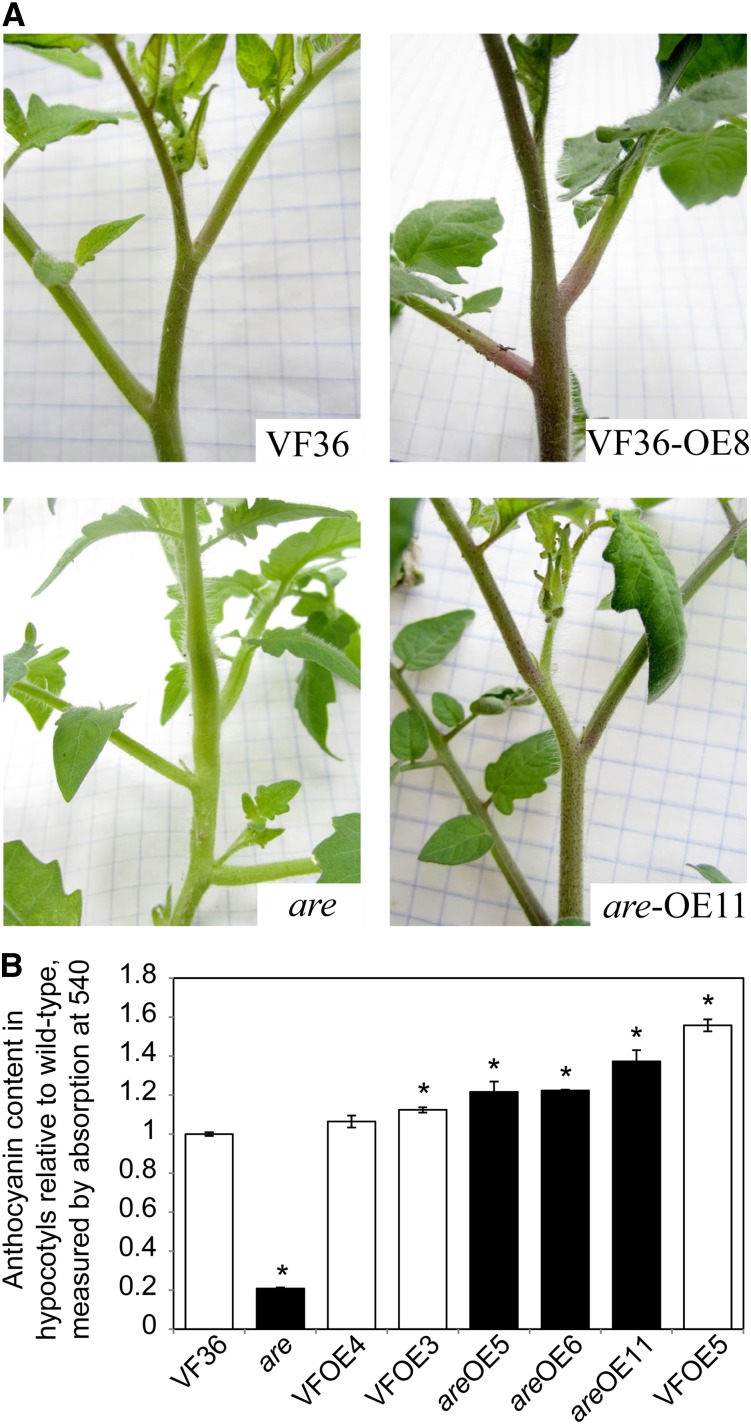
Although there is compelling evidence that the are mutant phenotype was associated with a mutation in the F3H gene (Yoder et al., 1994), the presence of residual anthocyanin synthesis requires an additional test of this conclusion. To provide direct evidence of an F3H mutation causing the are phenotype, the wild-type F3H sequence was cloned downstream of a 35S promoter and transformed into both are and VF36. All positive transformants in the are genetic background showed visible purple coloration due to anthocyanin production in stems and petioles, while untransformed are plants at this same growth stage did not show anthocyanin-linked coloration in any parts of the plant (Fig. 3A). Expanding leaves of similar sizes from all T0 plants were used to quantify F3H transcript abundance, illustrating as much as 50-fold increases in this transcript in some lines (Supplemental Fig. S4). Anthocyanins from a subset of are-35S:F3H and VF36-35S:F3H lines in the T1 generation were quantified in hypocotyls (Fig. 3B). In all are transgenic lines, the levels of anthocyanins were increased more than 5-fold, which resulted in levels equivalent to or greater than those in the VF36 parental line. In the VF36-35S:F3H and are-35S:F3H lines, two of three lines had anthocyanin contents that were significantly greater than untransformed VF36, but at a maximal 1.5-fold increase, suggesting that F3H overexpression was not sufficient for whole-pathway overproduction.
Figure 3.
Overexpression of a wild-type F3H gene increases anthocyanin production in wild-type plants and restores anthocyanin production in the are mutant. A, Images show the apical regions of untransformed parents and representative plants from the T0 generation. B, Anthocyanin content from hypocotyls was measured by spectrophotometry. Averages and se are presented. Asterisks indicate significant differences from untransformed VF36 with P ≤ 0.05 (n = 3–9), determined by Student’s t test. White and black bars signify lines in the VF36 and are backgrounds, respectively.
The levels of naringenin, kaempferol, quercetin, and myricetin were quantified in hypocotyls of individual are-35S:F3H and VF36-35S:F3H lines (Supplemental Table S4), and the averages of transgenic lines are compared with those of untransformed are and the wild type in Table II. The levels of naringenin in are-35S:F3H hypocotyls were dramatically decreased, to 17% of the levels in untransformed are, which were equivalent to the untransformed wild-type levels. Similarly in roots, naringenin levels dropped to 16% of the untransformed levels. The changes in kaempferol included a striking 29-fold increase in are-35S:F3H relative to the untransformed are, yielding levels that were less than 2-fold different from wild-type kaempferol levels. In are roots, kaempferol was below the threshold of detection of 0.1 ng g−1 fresh weight and reached 10.1 ± 0.7 ng g−1 fresh weight in are-35S:F3H, a 22-fold increase.
Table II. Quantification of flavonoids from 6-d-old seedlings of VF36, are, and averaged p35S:F3H transgenic lines.
Averages and se are reported (n = 10–12). Asterisks indicates significant differences from the untransformed genotype.
| Flavonoid | VF36 | VF36-OE | Ratioa | are | are-OE | Ratiob |
|---|---|---|---|---|---|---|
| ng g−1 fresh wt | ng g−1 fresh wt | |||||
| Naringenin | 12.3 ± 0.6 | 8.6 ± 0.9 | 0.7 | 81.9 ± 2.0 | 14.2 ± 0.7* | 0.2 |
| Kaempferol | 205.9 ± 27.0 | 59.3 ± 15.8* | 0.3 | 5.5 ± 1.0 | 160.5 ± 33.7* | 29.2 |
| Quercetin | 17.5 ± 7.1 | 2.1 ± 0.9* | 0.1 | 3.2 ± 0.6 | 0.3 ± 0.1* | 0.1 |
| Myricetin | 681.0 ± 45.5 | 673.5 ± 57.5* | 1.0 | 289.5 ± 22.0 | 778.1 ± 24.0* | 2.7 |
Ratio represents values from VF36-overexpression (OE) hypocotyls divided by VF36. bRatio represents values from are-OE hypocotyls divided by are.
In VF36-35S:F3H, the changes in metabolite accumulation were much more subtle. As in are, the levels of naringenin decreased, to 70% of the untransformed wild type. The levels of myricetin increased by 2.7-fold in are-35S:F3H and were unchanged in VF36-35S:F3H relative to untransformed genotypes. Surprisingly kaempferol and quercetin levels were lower in the transgenic line than in the untransformed wild type. Quercetin decreased to 11% or 14% of wild-type values in are-35S:F3H and VF36-35S:F3H, respectively. These transgenic lines performed greater conversion of naringenin to downstream metabolites, but with uneven distribution among the three flavonols. As enzymes of this pathway have been shown to be part of enzyme complexes (Burbulis and Winkel-Shirley, 1999; Dana et al., 2006; Crosby et al., 2011), it may be that the endogenous F3H complexes with other enzymes, such as F3′H, needed for quercetin synthesis and that the transgenic F3H enzyme did not assemble into these complexes.
Lateral Root Formation Was Modulated by the Level of F3H Expression
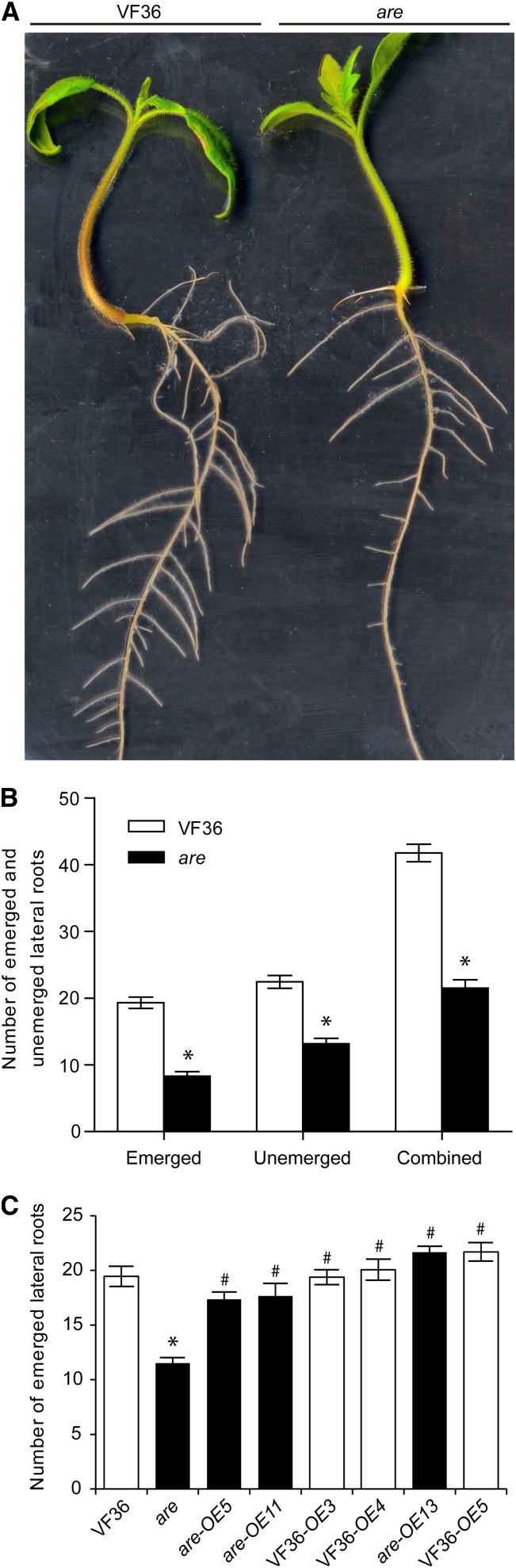
The effects of flavonols on rooting in tomato have not been studied previously, and the are mutant allowed us to ask if flavonol levels influenced root growth and branching, as they do in Arabidopsis. The number of unemerged lateral root primordia at stage 2 or later, the number of emerged lateral roots (Fig. 4), and length of primary roots (Supplemental Fig. S5B) were measured 6 d after germination in are and VF36. Seedlings of are produced fewer than half of the number of emerged lateral roots than the wild type. Unemerged lateral root primordia were observed in cleared roots and found to be greatly reduced in are compared with the wild type, indicating that the lateral root phenotype was not due to a defect in root emergence but a defect in root formation. Total combined emerged and unemerged lateral roots in are were 50% of that in the wild type. We also examined the root growth of are and the wild type in Turface, a soil substitute from which it is easy to remove roots. We found that lateral root numbers of are grown in Turface for 7 d were 55% of the number in wild-type roots (Supplemental Fig. S5A), which mirrored our observation of seedlings grown on agar medium. The average primary root length of are seedlings grown on agar medium was 86% of the length of the wild type (Supplemental Fig. S5B). Although this reduction in length was significant (P < 0.046, determined by Student’s t test), it was of much lower magnitude than the reduction in lateral root numbers (fewer than 50% of wild-type levels).
Figure 4.
Lateral root numbers are reduced in the are mutant. A, Image showing 6-d-old seedlings of the wild type and are displaying differences in lateral rooting and hypocotyl pigmentation. B, Numbers of emerged lateral roots, unemerged lateral root primordia, and combined totals of emerged and primordial roots. Averages and se are presented. Asterisks indicate differences between genotypes with P ≤ 0.05 (n = 15). C, Lateral root numbers are reported for untransformed VF36, are, and three F3H overexpression lines in each background genotype. Asterisks indicate differences from VF36, and number symbols indicate differences from are with P ≤ 0.05 (n = 16). Black bars indicate lines in the are background.
Lateral root quantification was performed as above on 6-d-old seedlings of the T1 generation in three independent transgenic lines. Transgenic are seedlings overexpressing wild-type F3H produced numbers of lateral roots that were significantly different from untransformed are, indicating that the rooting phenotype of are was reversed by this transgene and caused by a defect in F3H (Fig. 4C). The restoration of root formation in are-35S:F3H was paralleled by the accumulation of naringenin and kaempferol to wild-type levels. VF36-35S:F3H seedlings did not produce significantly more roots than nontransformed wild-type seedlings, consistent with more subtle changes in flavonol levels in these lines.
Flavonol Content Rather Than Naringenin Accumulation Was Linked to Changes in Root Development
The root developmental defect in are may be due to either increased naringenin concentration or decreases in the levels of flavonols or other metabolites downstream of F3H. We asked if genotypes with increased levels of flavonols show enhanced lateral root formation. The mutant anthocyanin without (aw) has a mutation in its DFR gene (Goldsbrough et al., 1994), encoding an enzyme that converts DHM to leucodelphinidin, an anthocyanin precursor. We found that kaempferol concentrations in roots and hypocotyls of aw were greater than 100-fold higher than in its wild-type parent, AC (Table III). Similarly, the levels of quercetin were higher in aw roots, at 42.3 ng g−1 fresh weight, while below the threshold of detection of 0.1 ng g−1 fresh weight in AC roots. The numbers of lateral roots in 6-d-old seedlings of aw were significantly higher than in the wild type (Supplemental Fig. S5C). These results were consistent with a positive correlation between flavonol concentrations in seedlings and lateral root formation in tomato.
Table III. Quantification of flavonoids from hypocotyl and root tissue from 6-d-old AC and aw seedlings.
Averages and se are reported (n = 6). Asterisks represent significant differences between genotypes with P ≤ 0.05, determined by Student’s t test. ND indicates that the compound was not detectable due to being absent or below the limit of detection, which was 0.1 ng g−1 fresh weight for naringenin and quercetin and 10 ng g−1 fresh weight for myricetin.
| Flavonoid | Hypocotyl |
Root |
||||
|---|---|---|---|---|---|---|
| AC | aw | Ratioa | AC | aw | Ratiob | |
| ng g−1 fresh wt | ng g−1 fresh wt | |||||
| Naringenin | 4.7 ± 1.2 | 12.2 ± 1.8* | 2.6 | ND | ND | ND |
| Kaempferol | 204.5 ± 12.3 | 20,533.6 ± 733.9* | 100.4 | 3.7 ± 1.2 | 451.9 ± 68.3* | 122.1 |
| Quercetin | 164.8 ± 21.4 | 1,049.5 ± 196.6* | 6.4 | ND | 42.3 ± 15.4 | >423.0 |
| Myricetin | 3,529.8 ± 246.6 | 2,627.8 ± 246.5 | 0.7 | ND | ND | ND |
To provide evidence that the rooting defect in are was not due to elevated levels of naringenin, we measured rooting in seedlings of p35S:CHS-CHR (Schijlen et al., 2006), which accumulated naringenin but contained wild-type levels of the flavonols kaempferol, quercetin, and myricetin (Supplemental Table S2). The numbers of emerged lateral roots in seedlings of p35S:CHS-CHR and its corresponding wild type were not significantly different, as reported in Supplemental Figure S6. These data suggested that the increased levels of naringenin observed in this transgenic line had no adverse effect on lateral root production.
Rootward IAA Transport Was Altered in the are Mutant
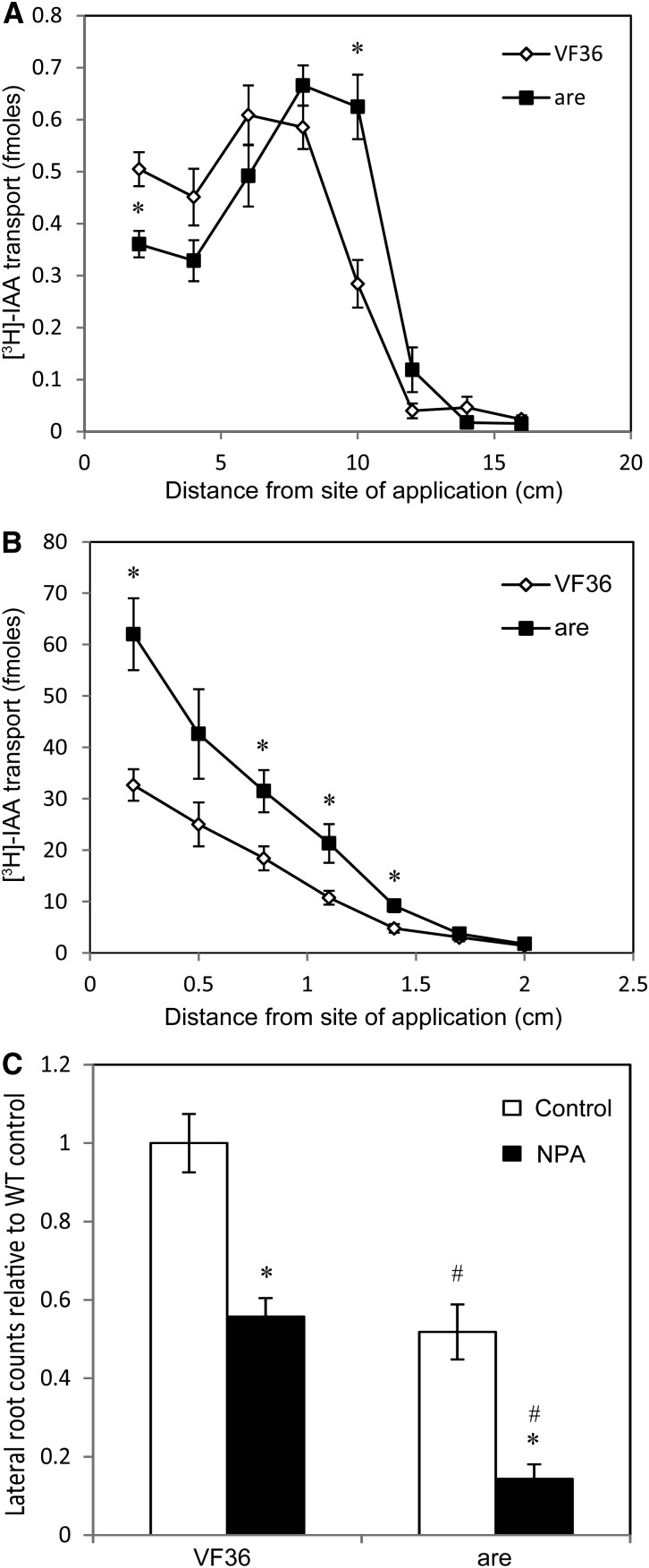
In Arabidopsis roots, flavonols act as negative regulators of auxin transport (Buer and Muday, 2004; Santelia et al., 2008; Kuhn et al., 2011; Lewis et al., 2011b). To ask if flavonols had similar activity in tomato roots, rootward IAA transport was measured in seedlings of are and the wild type to determine if IAA transport was altered in this mutant. A pulse-chase method of IAA transport was employed in which [3H]IAA is transiently applied to the hypocotyl apex or the root-shoot junction and, after 30 min, is chased by unlabeled IAA. In hypocotyl transport assays, the peak of radioactivity in are, corresponding to the pulse of [3H]IAA, occurred at a greater distance from the point of application than in the wild type, suggesting that auxin moved more rapidly in are hypocotyls than in the wild type (Fig. 5A). When the radioactivity in this peak was summed, it was also observed that the flux of auxin was greater in are hypocotyls (1.9 ± 0.2 versus 1.5 ± 0.2 fmol in the wild type), suggesting that more auxin moved from the shoot apex to the primary root in are than in the wild type. In root transport assays (Fig. 5B), all sections measured in are had a greater amount of radioactivity than corresponding wild-type sections. This suggested that roots of are transported more IAA to the primary root tip than the wild type, reducing IAA accumulation in the mature region of the root where lateral roots are elongating.
Figure 5.
Auxin transport and the response to an auxin transport inhibitor are altered in are. A and B, [3H]IAA pulse-chase transport in wild-type and are mutant seedlings. A, [3H]IAA rootward transport through hypocotyls from the shoot apex in are compared with the wild type. B, [3H]IAA rootward transport through roots from the root-shoot junction in are compared with the wild type. Averages and se are reported for n = 16. For A and B, asterisks indicate differences between are and the wild type with P ≤ 0.05. C, NPA or control droplets were placed 2 cm below the root-shoot junction, and roots were quantified below the application site after 3 d. Averages and se are reported for 15 seedlings. Asterisks indicate significant differences from the control treatment within genotypes with P ≤ 0.05, and number symbols indicate significant differences within treatments with P ≤ 0.05. WT, Wild type.
To test the model that enhanced IAA transport in are depleted IAA in regions where roots form, we treated roots with the auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA). Agar droplets containing 100 µm NPA were placed on the middle of the primary root of 2-d-old seedlings of the wild type and are. The number of emerged lateral roots below and above the NPA applications was quantified and compared with seedlings treated with control agar droplets 3 d later, as shown in Figure 5C and Supplemental Figure S7, respectively. In the control treatments, are formed 50% of the number of emerged lateral roots found in the wild type, consistent with untreated samples. We did not observe differences in root length between control and NPA treatment in either genotype. The number of lateral roots forming below the site of NPA application in the wild type was significantly reduced to 52% of the control value. However, the magnitude of reduction in are below the site of NPA application was even greater than in the wild type, to 32% of the control value. In the region of the root above the treatment, no difference in root number was observed in the wild type, while are treated with NPA had 73% of the number of lateral roots of control-treated roots, with a P value of 0.028 (Supplemental Fig. S7). This greater magnitude in reduction in are suggested that root formation in the mutant was more sensitive to the inhibition of auxin transport than in the wild type. The statistical significance of the differences in emerged lateral root numbers below the site of control or NPA treatment in are and the wild type was examined by a general linear model specifying a Poisson distribution to account for nonnormal distribution of integer data, with the output from that analysis included in Supplemental Table S5. The differences between genotype were significant (P < 0.00001), as were the differences between treatments (P < 0.00001). More importantly, the interaction of genotype and treatment was significant (P = 0.00062), suggesting that the inhibitory effect of NPA is enhanced in are. These results support the hypothesis that auxin transport changes in are roots are linked to the altered root development in this mutant line.
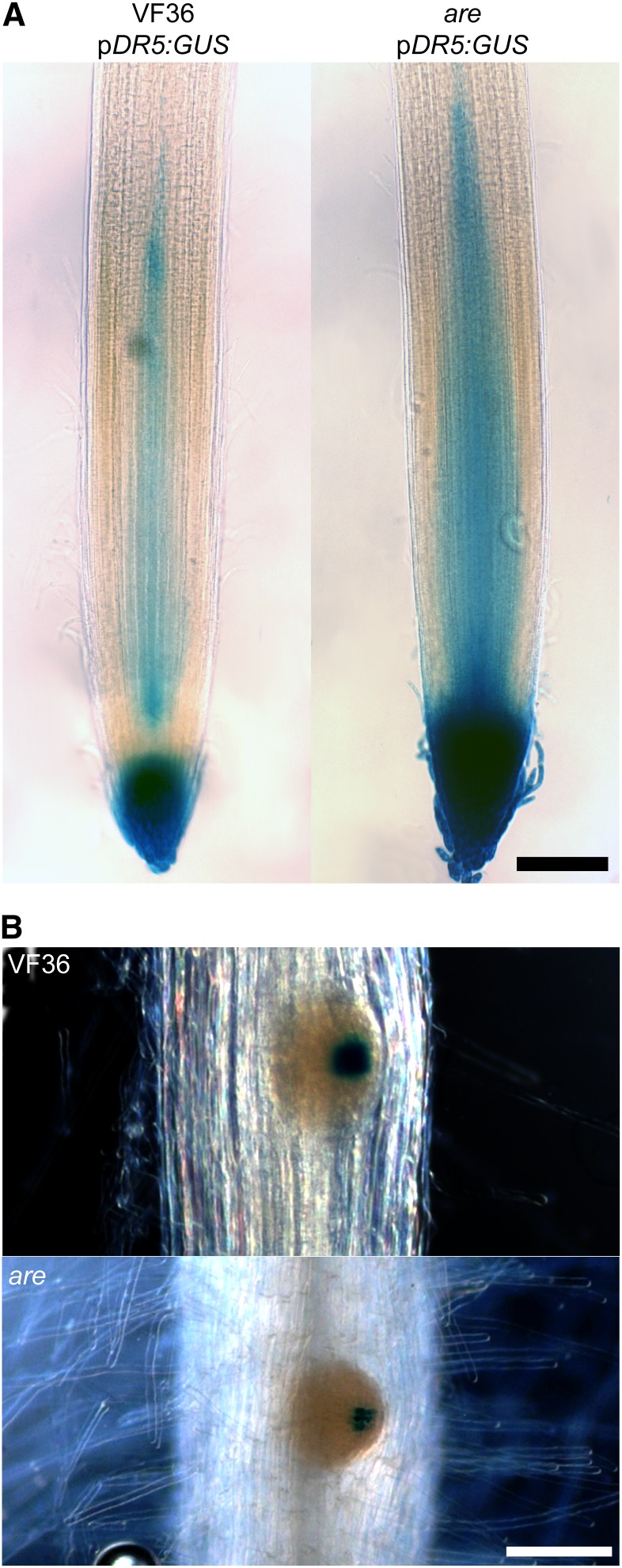
To test the model that the flux of IAA through are roots was altered, a pDR5:GUS auxin-responsive reporter was crossed into are and VF36. This reporter shows regions of elevated auxin-induced gene expression, which may be due to increased auxin accumulation or signaling. The levels of GUS product accumulation were examined 3 d after germination, the time at which lateral roots begin forming. GUS activity was visualized in cleared roots (Fig. 6). Primary root tips of are displayed more intense GUS staining than the wild type (Fig. 6A; Supplemental Fig. S8A). This is consistent with enhanced rootward IAA transport, described above, resulting in greater accumulation of auxin and the resulting auxin signaling in the primary root tip. In addition, are lateral root primordia displayed less intense GUS staining than the wild type (Fig. 6B; Supplemental Fig. S8B), consistent with less IAA accumulation in developing lateral root primordia due to enhanced rootward IAA transport toward the root tip
Figure 6.
The DR5:GUS auxin reporter indicates that more auxin accumulates in the root tip of the are mutant than in the wild type. A, GUS-stained primary root tips of wild-type or are seedlings containing the pDR5:GUS transgene. Bar = 200 µm. B, Lateral root primordia with GUS staining in wild-type or are seedlings. Bar = 100 µm. Seedlings were 3 d old, the stage at which lateral root primordia start forming and before the emergence of lateral roots.
ROS Levels and Root Hairs Were Increased in Roots of are
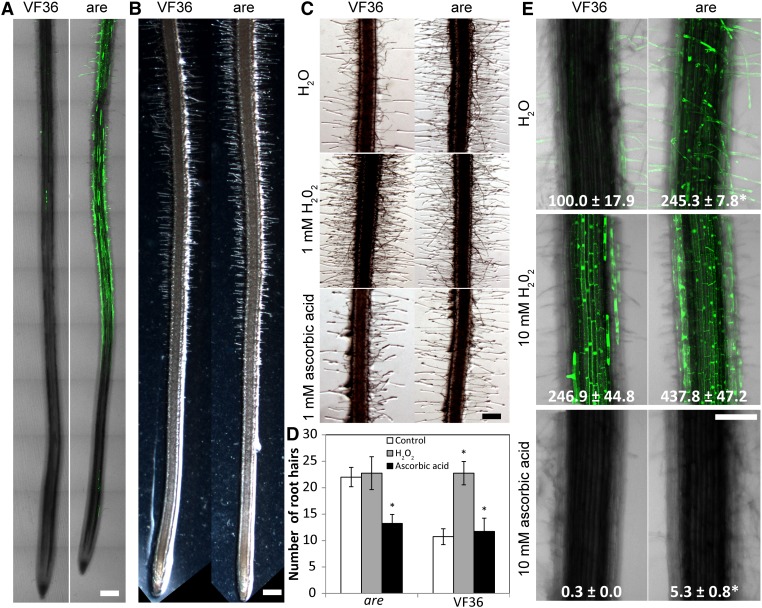
As flavonols can act as antioxidants in vitro (Hernández et al., 2009; Agati et al., 2012), we asked whether reduced flavonol accumulation in are roots led to the increased accumulation of ROS. Roots of are and the wild type were stained with a fluorescent ROS reporter, dihydrodichloro-fluorescein diacetate, which forms fluorescent 2′,7′-dichlorofluorescein (DCF) when oxidized by ROS, such as hydrogen peroxide (H2O2). Roots of 3-d-old seedlings were stained with DCF and examined using laser scanning confocal microscopy to capture multiple focal planes and tile scans, in which multiple images were digitally stitched together to allow the visualization of ROS distribution along the root (Fig. 7A). Most epidermal cells and root hairs of are showed bright DCF fluorescence, while little DCF fluorescence was observed in these cells of wild-type roots. ROS accumulated in the maturation zone, where root hair growth was most evident, but not in the elongation zone in tomato roots.
Figure 7.
ROS levels and root hair numbers are greater in roots of the are mutant compared with the wild type. A, Tile scan confocal images of mutant and wild-type roots showing the distribution of DCF staining along the root. Bar = 300 µm. B, Bright-field images of primary roots showing root hair density in mutant and the wild type. Bar = 200 µm. C, Bright-field images of mature regions of primary roots grown on control medium or medium containing H2O2 or ascorbic acid. Bar = 150 µm. D, Quantification of root hair numbers in mature regions of primary roots grown on control medium or medium containing H2O2 or ascorbic acid. Asterisks indicate significant differences from the control treatment within genotypes with P < 0.05. E, Confocal images of mature regions of 3-d-old roots treated with water, H2O2, or ascorbic acid and stained with DCF to observe ROS levels. Fluorescence was quantified, and numbers in each image represent percentages of wild-type fluorescence and se. Asterisks indicate significant differences from the wild type within treatments with P < 0.05. Bar = 200 µm.
DCF fluorescence accumulated in the root hairs of both the wild type and are, with much greater fluorescence detected in the are roots. These images also suggested that the number of root hairs differed between genotypes. As prior studies suggested roles for ROS in root hair development (Duan et al., 2010; Sundaravelpandian et al., 2013) and elongation (Jones et al., 2007; Monshausen et al., 2007; Swanson and Gilroy, 2010; Shin et al., 2011), we asked if are had increased root hair development. It was evident that there was a greater density of root hairs in are than in the wild type in the light micrographs of the apical tips of primary roots (Fig. 7B). This enhancement was also evident in the are images in Figure 6B, which were focused on the regions surrounding lateral root primordia. This localization of ROS in the maturation zone and accumulation in epidermal cells (Fig. 7A), from which root hairs form, suggested that increased ROS is positively correlated with root hair growth. Images of DCF fluorescence in VF36 and are were also collected at higher magnification in the root hair-forming region of the root (Fig. 7E, top frame). This fluorescent signal was quantified using a consistent area of interest focused on the primary root for all treatments, and the values normalized to untreated VF36 controls are noted in Figure 7E. There was a 2.5-fold increase in DCF signal in untreated are as compared with the wild type.
To confirm that the greater DCF fluorescence in are was due to a defect in F3H, mature root regions of three independent lines of are-35S:F3H and VF36-35S:F3H were stained with DCF and imaged. The levels of fluorescence detected in all transgenic lines in both genotypes were not significantly different from those in the untransformed wild type (Supplemental Fig. S9), consistent with elevated DCF being linked to reduced flavonol levels. Untransformed are had over 3-fold greater fluorescence than the untransformed wild type. The three are-35S:F3H lines examined had between 17% and 30% of the fluorescence of untransformed are, with levels reduced below those found in untransformed VF36. The effects were more subtle in the three VF36-35S:F3H lines examined, which had between 80% and 140% of the fluorescence of the untransformed wild type.
To test the hypothesis that ROS positively affects root hair growth, we manipulated the levels of ROS by treatment with the antioxidant ascorbic acid and the oxidant H2O2 (Fig. 7C). We quantified the number of root hairs forming in regions along the primary root that are equidistant from the root apex (Fig. 7D). In untreated control roots, we observed 2-fold more root hairs in are as compared to the number of root hairs along an equivalent length of primary root in the wild type. Consistent with the elevated ROS driving root hair formation, are roots treated with the antioxidant ascorbic acid show reduced levels of root hairs, to the levels found in the wild type. In contrast, treatment of the wild type with H2O2 led to increased numbers of root hairs, to levels that were equivalent to are. The reciprocal treatments of are with H2O2 and the wild type with ascorbic acid led to no significant changes, consistent with these genotypes having already elevated ROS or sufficient antioxidants to reduce ROS, respectively. The effects of these treatments on DCF fluorescence were also examined (Fig. 7E). This fluorescent signal was quantified using a consistent area of interest focused on the primary root for all treatments, and the values are noted in Figure 7E. In roots of both genotypes treated with H2O2, very bright DCF fluorescence was observed, with levels in VF36 reaching untreated are and the are roots showing less than 2-fold increase over the control treatment of this mutant. Very little DCF fluorescence was observed in roots of either genotype treated with ascorbic acid, suggesting that this treatment reduced ROS levels.
DISCUSSION
The roles of flavonols in plant development have been demonstrated using plants with genetic defects altering the synthesis of flavonoid metabolites (Brown et al., 2001; Buer and Muday, 2004; Peer et al., 2004; Buer et al., 2006; Lewis et al., 2011b). The presence of well-characterized mutants at each step in flavonoid biosynthesis in Arabidopsis has facilitated these studies (Winkel-Shirley, 2001b). To expand these studies to crop plants, additional characterization of genetic resources is needed. In tomato, mutants with altered anthocyanin synthesis have been isolated, but only a few have mapped mutations (Al-Sane et al., 2011). The recent release of completely sequenced genomes in several crops has also illustrated that the flavonoid biosynthetic enzymes are frequently encoded by multiple genes, as described here for tomato, further complicating mutant analyses. We focused on one tomato mutant, are, which has been reported to have a defective F3H gene (De Jong et al., 2004). Our analysis of the recently released tomato genome is consistent with the F3H enzyme being encoded by a single gene. This study uncovers a role for flavonols in tomato root development by examining mutant and transgenic lines, including the are mutant with and without complementation by the tomato F3H gene.
To conclusively demonstrate that anthocyanin reductions in the are mutant are linked to the F3H gene and no other secondary mutations, we complemented this mutant and transformed the parental line with the F3H gene under the control of the cauliflower mosaic virus 35S promoter. The are mutant has elevated naringenin and reduced levels of flavonols and anthocyanin compared with the wild type. The levels of naringenin, kaempferol, and anthocyanins were restored to wild-type levels in are containing the 35S:F3H transgene, consistent with a defective F3H gene. The wild-type plants transformed with this construct showed subtle changes in metabolite levels, consistent with the coordination of pathway enzymes that limited end product accumulation even in the presence of high levels of F3H. These results, combined with the absence of detectable F3H activity in the are mutant (De Jong et al., 2004), indicate that this plant is defective in the F3H gene and provides a genetic resource to ask if altered flavonol levels affect root development.
We examined the elongation and lateral root phenotypes of wild-type and are plants. There was a statistically significant, greater than 2-fold reduction in the formation of lateral roots in are when seedlings were grown on agar medium or in artificial soil (Turface). We were able to visualize lateral root primordia of stage 2 or later in Malachite Green-stained and cleared roots, indicating that the are mutation affects early stages of lateral root initiation. The similar decrease in emerged lateral roots suggests that emergence is not altered in are, but the reduced number of emerged lateral roots reflects reduced initiation events. The effect on root elongation is subtle, with a 14% difference in the length of primary roots. The ability of the 35S:F3H transgene to complement the are root phenotype mirrors the restoration in flavonol synthesis. The overexpression of this gene in the wild type has no significant effect on root formation, consistent with the smaller effects of this transgene on flavonol synthesis. The root phenotype of are could be due to either reduced levels of flavonols or downstream metabolites or elevated levels of naringenin. We observed elevated numbers of lateral roots in the aw mutant, which has elevated flavonols due to a DFR mutation, and a wild-type number of roots in a 35S:CHS:CHR transgenic line (Schijlen et al., 2006), which has elevated levels of naringenin but wild-type levels of flavonols. These results suggest a positive role of flavonols in lateral root initiation and are inconsistent with a negative role of naringenin.
Flavonoids have been shown to be negative regulators of auxin transport in Arabidopsis inflorescences and roots (Brown et al., 2001; Buer and Muday, 2004; Lewis et al., 2011b; Yin et al., 2013), so we asked if there is a similar regulation in tomato, which may, in turn, regulate lateral root formation. Transport of the radiolabeled IAA was measured using pulse-chase assays through both the hypocotyl and roots of the wild type and the are mutant. These assays provide information on both the rate and flux of IAA transport. In the hypocotyls and roots of are, there is an enhanced rate of IAA transport and a greater flux of IAA through the tissue and into the root. To test the functional significance of the altered auxin transport on root development, we treated young roots before they had detectable lateral root primordia with the auxin transport inhibitor NPA, applied mid root, and measured the effect of this treatment on the number of emerged lateral roots. Previous studies in Arabidopsis have demonstrated that such treatment blocks lateral root formation in sites that are on the rootward site of NPA treatment (Reed et al., 1998). We found fewer lateral roots in are after treatment with control agar, as observed previously. The magnitude of the reduction in roots forming on the rootward site of NPA treatment is 3-fold in are, as compared with the 2-fold effect in the wild type. This result is consistent with the more rapid depletion of auxin needed for lateral root formation from the zone below NPA application. In contrast, in the region above this treatment, NPA had no effect in the wild type and had a 27% reduction in emerged lateral roots in are. We asked how the are mutation affected the distribution of IAA throughout the root, as judged by examination of the DR5:GUS transgene, which reports auxin-induced gene expression that reflects both IAA distribution and responsiveness. We found enhanced auxin-induced gene expression at the primary root tip of this mutant, with reduced expression in developing lateral root primordia, consistent with elevated flux through the root in the are mutant. This elevated auxin flux and reduced lateral root formation parallel the positive effects of ethylene on auxin transport and the negative effects on lateral root development in both Arabidopsis and tomato (Negi et al., 2008, 2010; Lewis et al., 2011a).
The role of auxin in most aspects of lateral root initiation has been well studied in Arabidopsis, including priming pericycle cells in the basal meristem, controlling cell cycle progression and asymmetric division, and integrating responses to a number of hormones modulating this process, as described in recent reviews (Overvoorde et al., 2010; Muday et al., 2012; Lavenus et al., 2013; Van Norman et al., 2013). Of particular relevance to this study is a role of auxin transport in root development (Petrásek and Friml, 2009; Grunewald and Friml, 2010; Laskowski, 2013). Arabidopsis mutants that have defects in rootward IAA transport (such as auxin insensitive1, pinformed3 [pin3], and pin7) have reduced lateral root formation, consistent with limited levels of auxin available to drive lateral root formation (Benková et al., 2003; Laskowski et al., 2008; Lewis et al., 2011a). Similarly, both mechanical and chemical treatments that block auxin movement from the shoot into the root reduce lateral root development (Reed et al., 1998). In tomato, although the positive role of auxin and auxin transport in lateral root development was reported decades ago (Muday and Haworth, 1994; Muday et al., 1995), limited numbers of studies have explored the mechanisms for this regulation (Ivanchenko et al., 2006; de Jong et al., 2009; Negi et al., 2010; Dubrovsky et al., 2011; Bassa et al., 2012; Gupta et al., 2013). One consistent finding in both Arabidopsis and tomato is that treatments or mutations (such as those resulting in elevated levels of ethylene) that enhanced long-distance IAA transport at the expense of local accumulation of auxin can impair lateral root initiation (Negi et al., 2008, 2010). Auxin transport enhancement, alterations in auxin-induced gene expression, and reduced lateral root initiation in are are consistent with flavonols reducing auxin flux through wild-type roots, enhancing the accumulation of auxin at sites of root primordia formation. In are, the elevated flux through the root reduced auxin accumulation at sites of root initiation.
Another biochemical activity of flavonoids in roots may be to function as antioxidants. The ability of flavonols to reduce the amount of ROS has been demonstrated in vitro, but the effect of these molecules on ROS levels in vivo has been debated (Rice-Evans, 2001; Hernández et al., 2009; Pollastri and Tattini, 2011). In wild-type roots, the fluorescence of DCF, a nonspecific ROS sensor, was visible in root hairs, suggesting that wild-type levels of flavonols are sufficient to keep ROS at low levels in other root tissues. In are roots, DCF fluorescence is elevated in epidermal and cortical cells of the primary root, concentrated in the maturation zone as well as in root hairs. This ROS accumulation pattern is intriguing, based on a previously reported role of ROS in root hair development (Duan et al., 2010; Sundaravelpandian et al., 2013) and elongation (Jones et al., 2007; Monshausen et al., 2007; Swanson and Gilroy, 2010; Shin et al., 2011). We quantified the number of root hairs in the wild type and are and found a significant 2-fold enhancement in are. We asked whether this increased root hair formation is tied to the elevated ROS in are root hairs, by treating with the antioxidant ascorbic acid. This compound reduced ROS in are to background levels, as indicated by DCF imaging and reduced root hair formation to levels equivalent to the wild type. In contrast, treatment of the wild type with exogenous H2O2 increased root hair formation to levels equivalent to those in are. Together, these results suggest a role of flavonols as antioxidants in root hairs, with the ratio of flavonols to ROS modulating the number of root hairs. The altered ROS may also contribute to the auxin transport and lateral root phenotypes of are, as prior experiments have suggested such a linkage, particularly in Arabidopsis (Fernández-Marcos et al., 2013). We were unable to detect changes in auxin transport or lateral root formation by exogenous application of ROS or antioxidants in tomato roots, but the greater thickness of tomato roots, relative to Arabidopsis, may limit the penetration of these compounds to internal tissues that transport auxin and initiate roots.
In addition to using the are mutant to explore the developmental impact of reduced flavonol levels, we also gained new insight into the biochemistry of flavonol synthesis in tomato. A more complete understanding of the pathway of flavonoid synthesis will enable the metabolic engineering of tomato and other species, which are under way (Schijlen et al., 2006), to produce elevated levels of flavonoids that have important health-promoting activities in animals (Butelli et al., 2008). As outlined in Figure 1, this pathway has several branch point enzymes whose activity defines the metabolite partitioning in plant tissues. The absence of myricetin and anthocyanins in root tissues is clearly linked to developmental controls that limit the expression of the genes required for anthocyanin synthesis in these tissues, F3′5′H, DFR, and ANS, as illustrated in Supplemental Figure S3. This finding is consistent with another study that showed that metabolite differences in tomato cotyledons and hypocotyls were best predicted by the difference in F3′5′H transcript abundance affecting the relative accumulation of myricetin and anthocyanins (Groenenboom et al., 2013). Yet, the FLS and F3′H transcripts encoding the earlier branch point enzymes, in which DHK can be converted to either DHQ or kaempferol, do not show dramatic differences between hypocotyl and root tissues. Therefore, the observed metabolite levels cannot be explained by transcriptional controls of the genes encoding the enzymes that mediate this branch point, as FLS and F3′H are expressed in both tissues at roughly equivalent levels. These data suggest that there are posttranscriptional controls of these branch points. One of these controls is likely to confer a higher affinity of FLS for DHK than for DHQ, as reported previously for Arabidopsis FLS (Owens et al., 2008).
An additional complexity to this pathway is the ability of the are mutant with a defect in the only F3H gene producing no catalytically active F3H in tomato to synthesize some flavonols and anthocyanins (De Jong et al., 2004). Clearly, there is another reaction that must bypass this step, which may be mediated by FLS, which can have higher affinity for naringenin than for DHQ (Owens et al., 2008), or by ANS, which also bypasses this reaction (Turnbull et al., 2004). We explored the idea that there was a transcriptional change in these two enzymes in are that might facilitate this pathway bypass, but we detected no significant increases in these transcripts in are that might drive the synthesis of downstream metabolites. Our data suggest that F3H is an important point controlling flux through the flavonoid pathway. We have found in multiple genotypes a high abundance of F3H transcript in both hypocotyls and roots, leading to high flux from naringenin to DHK to kaempferol, the aglycone of which is the highest abundance flavonol in tomato roots. The absence of increased kaempferol and the presence of increased naringenin in p35S:CHS-CHR seedlings also supports the flux-regulating role of F3H.
The importance of studying the physiological effects of flavonoids in different species is apparent in comparing our findings in tomato with those in Arabidopsis. Previous reports show that flavonoid deficiencies in the Arabidopsis tt4 mutant result in greater numbers of lateral roots (Brown et al., 2001; Grunewald et al., 2012), opposite to the phenotype in are. However, auxin transport in tt4 roots is increased, as it is in are. That both instances of flavonoid deficiency result in similar auxin transport phenotypes but contrasting lateral root phenotypes suggests that increased flux of auxin through the root does not necessarily result in similar changes in auxin accumulation in the mature root region in tomato and Arabidopsis, possibly owing to the presence of many more cell files in tomato compared with Arabidopsis. In another example of species-specific flavonoid function, flavonoid deficiencies in many crop species result in male sterility (Mo et al., 1992; Schijlen et al., 2007), while the tt4 Arabidopsis mutant forms healthy pollen and displays normal seed production (Burbulis et al., 1996; Ylstra et al., 1996).
CONCLUSION
We have uncovered a positive role for flavonols in lateral root development in tomato through the use of transgenes and mutants with altered synthesis or activity of flavonol biosynthetic enzymes. This analysis has provided insight into the flavonoid biochemical pathway in tomato. We confirmed through transgene complementation that the are mutant harbors a defective F3H gene. Our findings show that F3H contributes to the regulation of concentrations of middle and late flavonoid pathway metabolites, particularly flavonols and anthocyanins. In addition, we have shown a positive correlation between lateral root production and flavonol concentrations in tomato, using other mutants and transgenes, which is opposite to that of Arabidopsis. We show that the influence of flavonols on lateral root production may be facilitated by their regulation of auxin transport, with decreased flavonol content resulting in greater auxin flux away from the maturation zone and an increased sensitivity to the auxin transport inhibitor NPA. Finally, we strengthen the evidence for the flavonols’ role as antioxidants in roots, showing that low flavonol concentrations result in greater ROS accumulation in the maturation zone, which is linked to increased numbers of elongated root hairs. Treatments with oxidants and antioxidants to manipulate ROS show a direct relationship between ROS levels and root hairs that are linked to the absence of flavonol antioxidants in are. These data provide a greater understanding of the role of F3H and flavonol synthesis in root growth and development in tomato.
MATERIALS AND METHODS
Reagents
Unless otherwise noted, chemicals and reagents were purchased from either Sigma-Aldrich (sigmaaldrich.com) or Thermo-Fisher Scientific (thermofisher.com).
Plant Materials and Seedling Growth
All mutant and wild-type seeds were obtained from the Tomato Genetic Resource Center (http://tgrc.ucdavis.edu/). The tomato (Solanum lycopersicum) CHS-CHR overexpression line and AC transformed with pDR5-GUS were generously provided by Arnaud Bovy (Schijlen et al., 2006) and Maria Ivanchenko (Dubrovsky et al., 2008), respectively. The DR5-GUS-expressing plants were crossed with are and VF36, and F2 seedlings were selected by screening for GUS staining and green hypocotyls. F3 seeds were generated from multiple F2 pDR5:GUS plants. The are mutant was backcrossed one time into VF36 to eliminate any unlinked mutations, and these progeny were used for most experiments. Seeds were surface sterilized by soaking in 95% (v/v) ethanol for 5 min and 20% (v/v) bleach for 30 min, autoclaved in distilled water for 5 min, followed by four more washes in water and placing in petri dishes lined with filter paper. Seeds were left in darkness for 3 to 4 d until radicles emerged. Unless otherwise noted, seeds with radicles of similar length (about 4 mm) were placed on 15- × 15-cm square petri dishes (Grenier Bio-One; greinerbioone.com) containing growth medium (1% [w/v] Suc, 0.5× Murashige and Skoog [MS] salts [Caisson Labs; caissonlabs.com], vitamin solution [1 µg mL−1 thiamine, 0.5 µg mL−1 pyridoxine, and 0.5 µg mL−1 nicotinic acid], 0.05% [w/v] MES, and 0.8% [w/v] agar, pH 5.6), with seven seeds per plate, and grown until 6 d after germination under continuous light at approximately 100 µmol m−2 s−1. For lateral root quantification in soil-like conditions, seeds were planted in Turface (turface.com) and kept watered for 14 d in our greenhouse. For quantification, Turface was poured out of pots, and roots were blotted dry before lateral roots were quantified by counting with a dissecting microscope.
Measurement of Transcript Abundance
For seedling experiments, entire roots or hypocotyls were collected 6 d after germination, with three seedlings per sample, and frozen in liquid nitrogen. Three independent experiments were performed with three biological and three technical replicates each. Tissues were pulverized using sterile micropestles (Thermo-Fisher) affixed to a Ryobi electric drill. RNA was then extracted from samples using the RNeasy plant mini kit from Qiagen (qiagen.com). RNA was quantified using a Nanodrop spectrophotometer (Thermo-Fisher) and treated with DNase (Promega; promega.com). DNase-treated RNA was converted to complementary DNA (cDNA) using SuperScript II reverse transcriptase and the cDNA first-strand synthesis kit, both from Life Technologies (lifetechnologies.com). cDNA synthesis utilized a mixture of poly(T) and random hexamer primers, both from Life Technologies. cDNA was treated with RNase H (Promega).
qRT-PCR was performed using SYBR Green Mastermix (Life Technologies) on an Applied Biosystems 7500 Fast Cycler. Primers used are listed in Supplemental Table S6 and were obtained using sequence from the appropriate genes from the Sol Genomics Network tomato database (http://solgenomics.net/) and designed using Primer3 software (http://frodo.wi.mit.edu/). Primers were purchased from Integrated DNA Technologies (idtdna.com) and tested for efficiency using a previously described protocol (Pfaffl, 2001). Analysis of transcript abundance was performed using the ΔΔCt method and normalization to actin transcript using efficiency-corrected calculations (Pfaffl, 2001).
Flavonol and Anthocyanin Extraction
For flavonol extraction, samples of roots or hypocotyls from six seedlings were collected in 1.5-mL microcentrifuge tubes (Thermo-Fisher) and frozen in liquid nitrogen (AirGas; airgas.com), then ground with a drill and micropestle. Extraction buffer (100% [v/v] acetone containing 5 µm formononetin [Indofine Chemicals; indofinechemical.com] for an internal standard) was added to samples at 3 µL mg−1 tissue. Hydrolysis to form aglycone flavonoids was carried out by adding an equal volume of 2 n hydrochloric acid and incubating at 70°C for 45 min. An equal volume of ethyl acetate was added to samples, which were vigorously mixed by shaking for 5 min. Samples were centrifuged at 14,000 rpm for 10 min. The top organic layers were transferred to HPLC vials (National Scientific; nationalscientific.com) and dried to completion in a vacuum centrifuge in line with a cold trap (Labconco; labconco.com). To each dried sample, 300 µL of acetone was added to resuspend the hydrolyzed residue, immediately followed by analysis.
For anthocyanin extraction, hypocotyls from four seedlings per sample were frozen in liquid nitrogen and ground with drill and pestle. Buffer consisting of 1% (v/v) hydrochloric acid in methanol was added to tissue at a rate of 2 µL mg−1. Extracts were incubated at 70°C for 1 h with shaking, then centrifuged at 14,000 rpm for 5 min. Supernatant was taken and added to 0.75-fold volume water. An equal volume of chloroform was added, and samples were shaken vigorously for 5 min before centrifuging at 14,000 rpm for 5 min. The aqueous phase was transferred to a cuvette and A540 was measured, with A675 subtracted, using a Beckman-Coulter (beckmancoulter.com) DU 730 spectrophotometer.
Quantification of Flavonols by Liquid Chromatography-Mass Spectrometry
Samples were run on a Thermo LTQ Orbitrap XL with electrospray ionization source, coupled to a Thermo Accela 1250 liquid chromatograph and autosampler (Thermo-Fisher), using a SecurityGuard guard column in line with a Luna 150 × 3 C18 column, both from Phenomenex (phenomenex.com). For flavonol analysis, 10 µL of each sample was injected with a solvent of water-acetonitrile, both containing 0.1% (v/v) formic acid in the following gradients: 95%:40% (v/v) water from 0 to 5 min, 40%:20% (v/v) water from 5 to 20 min, and 20%:95% (v/v) water from 20 to 23 min to recondition the column. Tandem mass spectrometry fragmentation spectra of flavonols were induced using 30 kV of collision-induced dissociation. Spectra of samples were compared with those of standards and those listed on the MassBank database (www.massbank.jp). Data were analyzed by quantifying peak areas using Thermo Xcalibur software and with normalization to the internal standard formononetin (Indofine Chemicals). Absolute quantities of metabolites were found by comparing peak area data with standard curves created using pure standards of naringenin, quercetin, kaempferol, and myricetin (Indofine Chemicals).
Microscopy
For counting emerged lateral roots, seedlings were grown on MS nutrient agar for 5 d. Plates were put on a stereomicroscope, and seedlings were left on the medium while roots were counted. Any root at or past stage VIII was counted as emerged. Roots of seedlings grown in Turface were placed in a shallow dish of water to spread out the roots while being counted. To quantify the number of lateral root primordia, seedlings were grown for 5 d on MS nutrient agar. Roots were then excised and submerged in an aqueous solution of 0.5% (w/v) Malachite Green for 24 h, followed by a brief wash in water and submerging in water for 24 h. Destained roots were placed on a Zeiss AxioObserver 3-D (zeiss.com) inverted microscope, and primordia were counted. Stage II and later primordia were detectable due to their darker green staining density compared with surrounding root tissue.
GUS staining was performed by incubating tissue for 24 h in GUS staining solution (500 μm ferrocyanide, 500 μm ferricyanide, 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid [Gold Biotechnology; goldbio.com], 100 mm sodium phosphate, pH 7, buffer, and 1% [v/v] Triton X-100). Staining solution was washed off by rinsing tissue in 100 mm sodium phosphate, pH 7, buffer for 1 h. Tissues were then fixed in 95% ethanol for 24 h. Tissues were cleared in a solution of chloral hydrate:water:glycerol (8:2:1, w/v/v) for 24 h before imaging. Differential interference contrast images were taken with a Zeiss AxioObserver 3-D inverted microscope with a 10× objective using a Hamamatsu Orca-3CCD (hamamatsu.com) digital color camera and Volocity software (Improvision; perkinelmer.com). Images were processed using ImageJ (http://rsbweb.nih.gov/ij/).
For imaging of root hairs, seedlings were grown on agar medium for 2 d after germination and imaged on a Leica MZ16 FA stereomicroscope at 35.7× zoom while leaving the roots undisturbed on the medium surface. Images were captured using an Infinity 2-2 CCD camera (lumenera.com) and Infinity Capture and Analyze software. Root hair images were captured focusing on the region of the root tip where root hair length equaled the width of the primary root and included 6.4 mm of root in the shootward direction. Light settings on the microscope were adjusted to provide high contrast between root hairs and background, to aid the identification of individual root hairs. For quantification, root hairs were counted in a 1-mm segment of each image chosen to have the most straight and intact root hairs, using the ImageJ Cell Counter plugin. To examine the effect of ROS on root hair formation, roots were transferred to medium containing either control medium or medium containing 1 mm H2O2 or 1 mm ascorbic acid for 2 d, and then images were captured, and root hair number was quantified.
Dried 2′,7′-dichlorodihydrofluorescein diacetate stock (Life Technologies) was resuspended in 20 µL of dimethyl sulfoxide and then diluted 1:1,000 in water to a concentration of 4.3 µm. Tomato roots were submerged in either distilled water, 10 mm H2O2, or 10 mm ascorbic acid for 15 min, followed by a 10-s rinse in distilled water. Roots were then submerged in DCF diacetate for 30 min, followed by a 10-s rinse in distilled water before imaging. Confocal microscopy was performed on a Zeiss LSM 710 confocal microscope with a 10× objective. DCF fluorescence was excited at 488 nm using a 35-MW argon laser with an emission band of 500 to 550 nm. Tile scan images were created by merging sequential images taken with a 10× objective, using the Zeiss Zen software. For Figure 7E, z-stack images were collected and combined. From these combined z-stacks, fluorescence intensity in a region of interest of 250 µm2 and in a consistent location in each image was quantified using Zeiss Zen 2011 software.
Transgenic Tomato Construction
The F3H gene was amplified from hypocotyl cDNA of VF36 and are using Phusion High-Fidelity polymerase (New England Biolabs; neb.com) with primers listed in Supplemental Table S6 to produce F3H with attB extensions. BP Clonase (Life Technologies) was used to clone the F3H cDNA into the pDONR-221 plasmid (Life Technologies) according to the manufacturer’s directions. The pDONR-221 plasmid containing the F3H cDNA was recombined into the pK7WG2 (http://gateway.psb.ugent.be/; Karimi et al., 2002) binary expression vector using LR Clonase (Life Technologies). Binary expression constructs were transformed into Agrobacterium tumefaciens LBA4404 electrocompetent cells (Takara Bio; clontech.com) via electroporation. A. tumefaciens-mediated transformation of tomato cotyledons was performed by the Transformation Facility at the North Carolina State University (http://projects.cals.ncsu.edu/planttransformation/index.html). This facility provided transformed tomato seedlings that were propagated in our greenhouse facility. Seedlings were screened for the NEOMYCIN PHOSPHOTRANSFERASE II transgene using primers listed in Supplemental Table S6 to confirm successful transformation and to confirm the presence of the transgene in subsequent generations.
IAA Transport
Tomato seedlings were grown on medium plates under the above conditions for 3 d after germination. Agar droplets (10 µL) containing 500 nm [3H]IAA (American Radiolabeled Chemicals) were used for precise placement of labeled IAA and were produced according to the previously published protocol (Lewis and Muday, 2009). For pulse-chase acropetal, or rootward, assays in roots, a droplet was placed just below the root-shoot junction and pressed so that it came in full contact with the root. For pulse-chase basipetal, or rootward, assays in hypocotyls, cotyledons, and apices were excised, and droplets were abutted to the site of excision. Seedlings were oriented vertically under yellow light. After 30 min, droplets containing [3H]IAA were replaced with new droplets containing 500 nm nonradioactive IAA. After 2 h, seedlings were sectioned. Starting 2 mm below the site of IAA application, sequential 3-mm sections were excised, and radioactivity was quantified in a scintillation counter (Beckman LS6500).
NPA Treatment of Roots
Tomato seedlings were grown on nutrient agar medium in a growth chamber under standard conditions for 2 d. Seedlings at this age did not have any lateral root primordia at stage 2 or later. Agar droplets of 30 µL were produced as detailed by Lewis and Muday (2009) with or without 100 µm NPA. This high dose was used as a localized treatment and at this concentration was shown to reduce the formation of lateral roots on the rootward side of the application in Arabidopsis (Reed et al., 1998). Control or NPA droplets were then placed over the primary root at a position 2 cm from the root-shoot junction, which, at that age, is approximately in the middle of the primary root. Seedlings were left in the growth chamber for another 3 d, and then the number of emerged lateral roots both above and below the site of agar application and root length were quantified.
Statistical Analyses
Student’s t tests were performed using Microsoft Excel software (microsoft.com). Two-way ANOVA was performed on the transcript data using GraphPad Prism 5 software (graphpad.com). To test if NPA root count data displayed a non-Gaussian distribution, a Pearson’s χ2 normality test was performed using R software (r-project.org). This test gave a P value of 0.0154, suggesting that the data were not normally distributed. Therefore, general linear modeling for a Poisson distribution was calculated using R software.
Sequence data from this article can be found on the solgenomics.net Web site under the following accession numbers: CHS (SGN-U579222, SGN-U580856, SGN-U581366, and SGN-U580262), CHS2 (SGN-U580856), CHI (SGN-U577427 and SGN-U579009), F3′H (SGN-U576659, SGN-U573255, and SGN-U570072), F3H (SGN-U563669), FLS (SGN-U569889), F3′5′H (SGN-U602093), DFR (SGN-U569072), and ANS (SGN-U602582).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multispecies F3H peptide sequence alignment.
Supplemental Figure S2. Transcript abundance of flavonoid biosynthetic genes in are.
Supplemental Figure S3. Transcript abundance of F3H in AC.
Supplemental Figure S4. Transcript abundance of F3H in 35S:F3H lines.
Supplemental Figure S5. Number of lateral roots of are in Turface, root length of are, and number of lateral roots in aw.
Supplemental Figure S6. Number of lateral roots in 35S:CHR-CHS seedlings.
Supplemental Figure S7. Number of lateral roots above the site of NPA application.
Supplemental Figure S8. GUS staining in root tips and lateral root primordia.
Supplemental Figure S9. DCF fluorescence in roots of 35S:F3H lines.
Supplemental Table S1. Anthocyanin levels in are hypocotyls.
Supplemental Table S2. Flavonoid levels in 35S:CHR-CHS seedlings.
Supplemental Table S3. ANOVA results of transcript abundance analysis.
Supplemental Table S4. Flavonoid levels in 35S:F3H hypocotyls and roots.
Supplemental Table S5. Results from a general linear model with a Poisson distribution for lateral root numbers.
Supplemental Table S6. Oligonucleotide primer sequences.
Supplementary Material
Acknowledgments
We thank Marcus Wright for Orbitrap operation assistance and advice; Kathryn Tully for liquid chromatography-mass spectrometry protocol optimization assistance; Anita McCauley, Glenn Mars, and Daniel Lewis for microscopy assistance and advice; T. Michael Anderson for advice on statistical analyses; Sergei Krasnyanski and the North Carolina State University Plant Transformation Laboratory for providing plant transformation services; and the Tomato Genetics Resource Center for providing tomato germplasm.
Glossary
- DHK
dihydrokaempferol
- DHM
dihydromyrectin
- IAA
indole-3-acetic acid
- ROS
reactive oxygen species
- qRT
quantitative real-time
- AC
Ailsa Craig
- NPA
1-N-naphthylphthalamic acid
- DCF
2′,7′-dichlorofluorescein
- H2O2
hydrogen peroxide
- MS
Murashige and Skoog
- cDNA
complementary DNA
Footnotes
This work was supported by the U.S. Department of Agriculture National Institute of Food and Agriculture program (grant no. 2009–65116–20436 to G.K.M.) and the National Science Foundation Major Research Instrumentation Program (grant nos. DBI–0500702, MRI–0722926, MRI–1039755, and 947028).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Agati G, Azzarello E, Pollastri S, Tattini M. (2012) Flavonoids as antioxidants in plants: location and functional significance. Plant Sci 196: 67–76 [DOI] [PubMed] [Google Scholar]
- Al-Sane K, Povero G, Perata P. (2011) Anthocyanin tomato mutants: overview and characterization of an anthocyanin-less somaclonal mutant. Plant Biosyst 145: 436–444 [Google Scholar]
- Azari R, Tadmor Y, Meir A, Reuveni M, Evenor D, Nahon S, Shlomo H, Chen L, Levin I. (2010) Light signaling genes and their manipulation towards modulation of phytonutrient content in tomato fruits. Biotechnol Adv 28: 108–118 [DOI] [PubMed] [Google Scholar]
- Ballester AR, Molthoff J, de Vos R, Hekkert B, Orzaez D, Fernández-Moreno JP, Tripodi P, Grandillo S, Martin C, Heldens J, et al. (2010) Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol 152: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, Reichheld JP. (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassa C, Mila I, Bouzayen M, Audran-Delalande C. (2012) Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato. Plant Cell Physiol 53: 1583–1595 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Broun P. (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8: 272–279 [DOI] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK. (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Djordjevic MA. (2009) Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J Exp Bot 60: 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Imin N, Djordjevic MA. (2010) Flavonoids: new roles for old molecules. J Integr Plant Biol 52: 98–111 [DOI] [PubMed] [Google Scholar]
- Buer CS, Muday GK. (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16: 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Sukumar P, Muday GK. (2006) Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol 140: 1384–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis IE, Iacobucci M, Shirley BW. (1996) A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell 8: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis IE, Winkel-Shirley B. (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96: 12929–12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EG, Hall RD, Bovy AG, Luo J, et al. (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Crosby KC, Pietraszewska-Bogiel A, Gadella TW, Jr, Winkel BS. (2011) Förster resonance energy transfer demonstrates a flavonoid metabolon in living plant cells that displays competitive interactions between enzymes. FEBS Lett 585: 2193–2198 [DOI] [PubMed] [Google Scholar]
- Dana CD, Bevan DR, Winkel BS. (2006) Molecular modeling of the effects of mutant alleles on chalcone synthase protein structure. J Mol Model 12: 905–914 [DOI] [PubMed] [Google Scholar]
- de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH. (2009) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57: 160–170 [DOI] [PubMed] [Google Scholar]
- De Jong WS, Eannetta NT, De Jong DM, Bodis M. (2004) Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor Appl Genet 108: 423–432 [DOI] [PubMed] [Google Scholar]
- De Tullio MC, Jiang K, Feldman LJ. (2010) Redox regulation of root apical meristem organization: connecting root development to its environment. Plant Physiol Biochem 48: 328–336 [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM. (2010) FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Napsucialy-Mendivil S, Duclercq J, Cheng Y, Shishkova S, Ivanchenko MG, Friml J, Murphy AS, Benková E. (2011) Auxin minimum defines a developmental window for lateral root initiation. New Phytol 191: 970–983 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E. (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marcos M, Sanz L, Lewis D, Muday G, Lorenzo O (2013) Control of auxin transport by reactive oxygen and nitrogen species. In R Chen, F Baluska, eds, Polar Auxin Transport (Signaling and Communication in Plants), Vol 17. Springer, Berlin, pp 103–117 [Google Scholar]
- Fini A, Guidi L, Ferrini F, Brunetti C, Di Ferdinando M, Biricolti S, Pollastri S, Calamai L, Tattini M. (2012) Drought stress has contrasting effects on antioxidant enzymes activity and phenylpropanoid biosynthesis in Fraxinus ornus leaves: an excess light stress affair? J Plant Physiol 169: 929–939 [DOI] [PubMed] [Google Scholar]
- Fraser CM, Chapple C. (2011) The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book 9: e0152 doi/10.1199/tab.0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, et al. (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Goldsbrough A, Belzile F, Yoder JI. (1994) Complementation of the tomato anthocyanin without (aw) mutant using the dihydroflavonol 4-reductase gene. Plant Physiol 105: 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenenboom M, Gomez-Roldan V, Stigter H, Astola L, van Daelen R, Beekwilder J, Bovy A, Hall R, Molenaar J. (2013) The flavonoid pathway in tomato seedlings: transcript abundance and the modeling of metabolite dynamics. PLoS ONE 8: e68960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, editor (2006) The Science of Flavonoids. Springer Science, New York [Google Scholar]
- Grunewald W, De Smet I, Lewis DR, Löfke C, Jansen L, Goeminne G, Vanden Bossche R, Karimi M, De Rybel B, Vanholme B, et al. (2012) Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci USA 109: 1554–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Friml J. (2010) The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J 29: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Shi X, Lindquist IE, Devitt N, Mudge J, Rashotte AM. (2013) Transcriptome profiling of cytokinin and auxin regulation in tomato root. J Exp Bot 64: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández I, Alegre L, Van Breusegem F, Munné-Bosch S. (2009) How relevant are flavonoids as antioxidants in plants? Trends Plant Sci 14: 125–132 [DOI] [PubMed] [Google Scholar]
- Ivanchenko MG, Coffeen WC, Lomax TL, Dubrovsky JG. (2006) Mutations in the Diageotropica (Dgt) gene uncouple patterned cell division during lateral root initiation from proliferative cell division in the pericycle. Plant J 46: 436–447 [DOI] [PubMed] [Google Scholar]
- Ivanchenko MG, den Os D, Monshausen GB, Dubrovsky JG, Bednárová A, Krishnan N. (2013) Auxin increases the hydrogen peroxide (H2O2) concentration in tomato (Solanum lycopersicum) root tips while inhibiting root growth. Ann Bot (Lond) 112: 1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Yang Z, Smirnoff N. (2007) NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J Exp Bot 58: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Kang JH, McRoberts J, Shi F, Moreno JE, Jones AD, Howe GA. (2014) The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol 164: 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Kuhn BM, Geisler M, Bigler L, Ringli C. (2011) Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiol 156: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M. (2013) Lateral root initiation is a probabilistic event whose frequency is set by fluctuating levels of auxin response. J Exp Bot 64: 2609–2617 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AF, Scheres B. (2008) Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18: 450–458 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Muday GK. (2009) Measurement of auxin transport in Arabidopsis thaliana. Nat Protoc 4: 437–451 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Negi S, Sukumar P, Muday GK. (2011a) Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138: 3485–3495 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Ramirez MV, Miller ND, Vallabhaneni P, Ray WK, Helm RF, Winkel BS, Muday GK. (2011b) Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol 156: 144–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løvdal T, Olsen KM, Slimestad R, Verheul M, Lillo C. (2010) Synergetic effects of nitrogen depletion, temperature, and light on the content of phenolic compounds and gene expression in leaves of tomato. Phytochemistry 71: 605–613 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP. (1992) Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA 89: 7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Messerli MA, Shi C, Gilroy S. (2007) Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc Natl Acad Sci USA 104: 20996–21001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Weisenseel MH, Gilroy S. (2009) Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21: 2341–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday GK, Haworth P. (1994) Tomato root growth, gravitropism, and lateral development: correlation with auxin transport. Plant Physiol Biochem 32: 193–203 [PubMed] [Google Scholar]
- Muday GK, Lomax TL, Rayle DL. (1995) Characterization of the growth and auxin physiology of roots of the tomato mutant, diageotropica. Planta 195: 548–553 [DOI] [PubMed] [Google Scholar]
- Muday GK, Rahman A, Binder BM. (2012) Auxin and ethylene: collaborators or competitors? Trends Plant Sci 17: 181–195 [DOI] [PubMed] [Google Scholar]
- Murphy A, Peer WA, Taiz L. (2000) Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211: 315–324 [DOI] [PubMed] [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK. (2008) Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J 55: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Sukumar P, Liu X, Cohen JD, Muday GK. (2010) Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J 61: 3–15 [DOI] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T. (2010) Auxin control of root development. Cold Spring Harb Perspect Biol 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DK, Alerding AB, Crosby KC, Bandara AB, Westwood JH, Winkel BS. (2008) Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol 147: 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M, Sultana N, Paszkiewicz K, Florance H, Smirnoff N. (2012) The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: further evidence for redox control of anthocyanin synthesis. Plant Cell Environ 35: 388–404 [DOI] [PubMed] [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS. (2004) Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS. (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Friml J. (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri S, Tattini M. (2011) Flavonols: old compounds for old roles. Ann Bot (Lond) 108: 1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK. (1998) Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice-Evans C. (2001) Flavonoid antioxidants. Curr Med Chem 8: 797–807 [DOI] [PubMed] [Google Scholar]
- Santelia D, Henrichs S, Vincenzetti V, Sauer M, Bigler L, Klein M, Bailly A, Lee Y, Friml J, Geisler M, et al. (2008) Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J Biol Chem 283: 31218–31226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schijlen E, Ric de Vos CH, Jonker H, van den Broeck H, Molthoff J, van Tunen A, Martens S, Bovy A. (2006) Pathway engineering for healthy phytochemicals leading to the production of novel flavonoids in tomato fruit. Plant Biotechnol J 4: 433–444 [DOI] [PubMed] [Google Scholar]
- Schijlen EG, de Vos CH, Martens S, Jonker HH, Rosin FM, Molthoff JW, Tikunov YM, Angenent GC, van Tunen AJ, Bovy AG. (2007) RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol 144: 1520–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin LJ, Huang HE, Chang H, Lin YH, Feng TY, Ger MJ. (2011) Ectopic ferredoxin I protein promotes root hair growth through induction of reactive oxygen species in Arabidopsis thaliana. J Plant Physiol 168: 434–440 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Steffens B, Kovalev A, Gorb SN, Sauter M. (2012) Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling. Plant Cell 24: 3296–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaravelpandian K, Chandrika NNP, Schmidt W. (2013) PFT1, a transcriptional Mediator complex subunit, controls root hair differentiation through reactive oxygen species (ROS) distribution in Arabidopsis. New Phytol 197: 151–161 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. (2011) Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol 14: 691–699 [DOI] [PubMed] [Google Scholar]
- Swanson S, Gilroy S. (2010) ROS in plant development. Physiol Plant 138: 384–392 [DOI] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8: 317–323 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Nakajima J, Welford RW, Yamazaki M, Saito K, Schofield CJ. (2004) Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis: anthocyanidin synthase, flavonol synthase, and flavanone 3beta-hydroxylase. J Biol Chem 279: 1206–1216 [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Xuan W, Beeckman T, Benfey PN. (2013) To branch or not to branch: the role of pre-patterning in lateral root formation. Development 140: 4301–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JM, Hechler PJ, Muday GK. (2014) Ethylene-induced flavonol accumulation in guard cells suppresses reactive oxygen species and moderates stomatal aperture. Plant Physiol 164: 1707–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel BS (2006) The biosynthesis of flavonoids. In E Gotewold, ed, The Science of Flavonoids. Springer, New York, pp 71–96 [Google Scholar]
- Winkel-Shirley B. (2001a) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001b) It takes a garden: how work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol 127: 1399–1404 [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Yin R, Han K, Heller W, Albert A, Dobrev P, Zazimalova E, Schaffner A (2013) Kaempferol 3-O-rhamnoside-7-O rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol 201: 466–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylstra B, Muskens M, Van Tunen AJ. (1996) Flavonols are not essential for fertilization in Arabidopsis thaliana. Plant Mol Biol 32: 1155–1158 [DOI] [PubMed] [Google Scholar]
- Yoder JI, Belzile F, Tong Y, Goldsbrough A. (1994) Visual markers for tomato derived from the anthocyanin biosynthetic pathway. Euphytica 79: 163–167 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.