Both abscisic acid and a nitrate transporter regulate the production of ROS and the expression of superoxide-generating NADPH oxidase enzymes to regulate root elongation.
Abstract
Abscisic acid (ABA) modulates root growth in plants grown under normal and stress conditions and can rescue the root growth defects of the Medicago truncatula lateral root-organ defective (latd) mutant. Here, we demonstrate that reactive oxygen species (ROS) function downstream of ABA in the regulation of root growth by controlling cell elongation. We also show that the MtLATD/NUMEROUS INFECTIONS AND POLYPHENOLICS (NIP) nitrate transporter is required for ROS homeostasis and cell elongation in roots and that this balance is perturbed in latd mutants, leading to an excess of superoxide and hydrogen peroxide and a corresponding decrease in cell elongation. We found that expression of the superoxide-generating NADPH oxidase genes, MtRbohA and MtRbohC (for respiratory burst oxidase homologs), is increased in latd roots and that inhibition of NADPH oxidase activity pharmacologically can both reduce latd root ROS levels and increase cell length, implicating NADPH oxidase function in latd root growth defects. Finally, we demonstrate that ABA treatment alleviates ectopic ROS accumulation in latd roots, restores MtRbohC expression to wild-type levels, and promotes an increase in cell length. Reducing the expression of MtRbohC using RNA interference leads to increased root elongation in both wild-type and latd roots. These results reveal a mechanism by which the MtLATD/NIP nitrate transporter and ABA modulate root elongation via superoxide generation by the MtRbohC NADPH oxidase.
Abscisic acid (ABA) is a key regulator of both plant development and stress responses. ABA is often thought of as a growth inhibitor, negatively regulating aspects of plant development such as root elongation (Pilet, 1975) and seed germination (Finkelstein et al., 2002), but in certain stress conditions, such as drought, ABA can also play a positive role, promoting root elongation and maintaining root growth (Sharp et al., 2004). Recently, ABA has been shown to also have a positive effect on root growth under nonstressed conditions, indicating that ABA can also act as a growth stimulator (Cheng et al., 2002). ABA regulates root growth by maintaining the Arabidopsis (Arabidopsis thaliana) root apical meristem, where it promotes the quiescence of the quiescent center as well as the local suppression of differentiation (Zhang et al., 2010). However, the role of ABA in cell elongation during root growth has not been investigated.
The LATERAL ROOT ORGAN DEFECTIVE/NUMEROUS INFECTIONS AND POLYPHENOLICS (LATD/NIP) gene, also known as NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER FAMILY1.7 (NPF1.7), encodes a nitrate transporter of Medicago truncatula that is required for root and nodule meristem development (Veereshlingam et al., 2004; Bright et al., 2005; Liang et al., 2007; Yendrek et al., 2010; Bagchi et al., 2012; Léran et al., 2014). In plants homozygous for the latd mutation, the strongest allele of the LATD/NIP locus, the root apical meristem (RAM) undergoes exhaustion as development proceeds, ultimately arresting by 21 d (Bright et al., 2005; Liang et al., 2007; Yendrek et al., 2010; Bagchi et al., 2012). latd mutants also exhibit severe defects in the elongation of primary and lateral roots and in the formation of symbiotic root nodules (Veereshlingam et al., 2004; Bright et al., 2005; Liang et al., 2007). Interestingly, application of ABA can maintain the viability of the latd RAM and prevent it from arresting (Liang et al., 2007). The mechanism by which ABA acts to rescue root elongation in latd mutants is unknown. latd mutants have normal levels of ABA (Liang et al., 2007), so the defect is likely to be in transport or signaling. The rescue of latd root growth by ABA indicates a positive role for ABA in RAM function but not the mechanism by which it regulates root elongation.
Root growth is a function both of cell division and cell elongation. Rapid root growth is due largely to rapid elongation of cells in the growing root. Cell division is restricted to the meristematic region at the root tip and is required for continued growth of the root. Newly formed cells at the edge of the meristem exit the cell cycle and begin the process of differentiation, first elongating, undergoing a rapid increase in size, then beginning to differentiate (Benfey et al., 2010). The transition between proliferative growth and differentiation in the root is controlled by the transcription factor UPBEAT1, which regulates reactive oxygen species (ROS) homeostasis (Tsukagoshi et al., 2010).
ROS are important signaling molecules that affect the response to environmental signals as well as many aspects of plant development. In Arabidopsis root development, ROS signaling is not only crucial for RAM organization and maintenance (De Tullio et al., 2010; Tsukagoshi et al., 2010) but also for root hair formation (Foreman et al., 2003). Disruption in ROS signaling leads to defects in these developmental processes. ROS are also integral parts of hormone signaling networks (Mittler et al., 2011) and play an important role in plant responses under stress conditions (Achard et al., 2006, 2008). In drought conditions, ABA induces stomatal closure by stimulating the production of the ROS molecule hydrogen peroxide (H2O2) in guard cells (Zhang et al., 2001; Wang and Song, 2008; Jannat et al., 2011) via a pathway involving guard cell-specific NADPH oxidases (Kwak et al., 2003). Plasma membrane-localized NADPH oxidases, named RESPIRATORY BURST OXIDASE HOMOLOGS (Rboh) in plants, are enzymes that play a key regulatory role in the ROS signaling network, catalyzing the production of extracellular superoxide (O2.−) from molecular oxygen (for review, see Suzuki et al., 2011). Subsequently, short-lived O2.− can be converted by superoxide dismutases (SODs) to H2O2, which can signal both intracellularly and extracellularly (Alscher et al., 2002).
Because of their role in O2.− production, Rboh genes play critical functions in processes modulating ROS homeostasis, both in stress responses and during development, including that of roots. In Arabidopsis, AtRbohC/ROOT HAIR DEFECTIVE2 is required for root hair tip growth (Foreman et al., 2003; Takeda et al., 2008) and AtRbohF is required for Casparian strip formation (Lee et al., 2013) and functions with AtRbohD to regulate root length in response to ABA (Kwak et al., 2003). In Phaseolus vulgaris, PvRbohB promotes lateral root elongation (Montiel et al., 2012, 2013) and the formation of symbiotic root nodules (Montiel et al., 2012; Arthikala et al., 2013). In M. truncatula, MtRbohA promotes nitrogen fixation in root nodules, but whether it functions in root growth has not been explored (Marino et al., 2011), and no other MtRboh genes have been functionally characterized.
Here, we report that ROS functions downstream of ABA in the regulation of root growth by controlling cell elongation. We find that latd mutant roots have higher levels of both O2.− and H2O2 than wild-type roots and that the expression of two O2.−-generating NADPH oxidase genes, MtRbohA and MtRbohC, is increased in latd mutant roots while that of the predicted apoplastic ROS-scavenging gene COPPER/ZINC SUPEROXIDE DISMUTASE (Cu/ZnSOD) is reduced. Additionally, we demonstrate that ABA can rescue the ectopic ROS accumulation and cell elongation defects in latd mutant roots, as can decreasing Rboh function with the pharmacological inhibitor diphenylene iodonium (DPI). We find that ABA treatment reduces MtRbohC expression in latd roots to wild-type levels and that inhibiting MtRbohC expression using RNA interference (RNAi) leads to increased root elongation in both the wild type and latd mutants. In summary, we find that ABA functions as a positive growth regulator stimulating cell elongation, and thus root elongation, via the inhibition of RbohC-mediated ROS signaling, rescuing the cell elongation defect of mutants lacking MtLATD/NIP.
RESULTS
The Balance of ROS Levels Is Disrupted in latd Mutant Roots
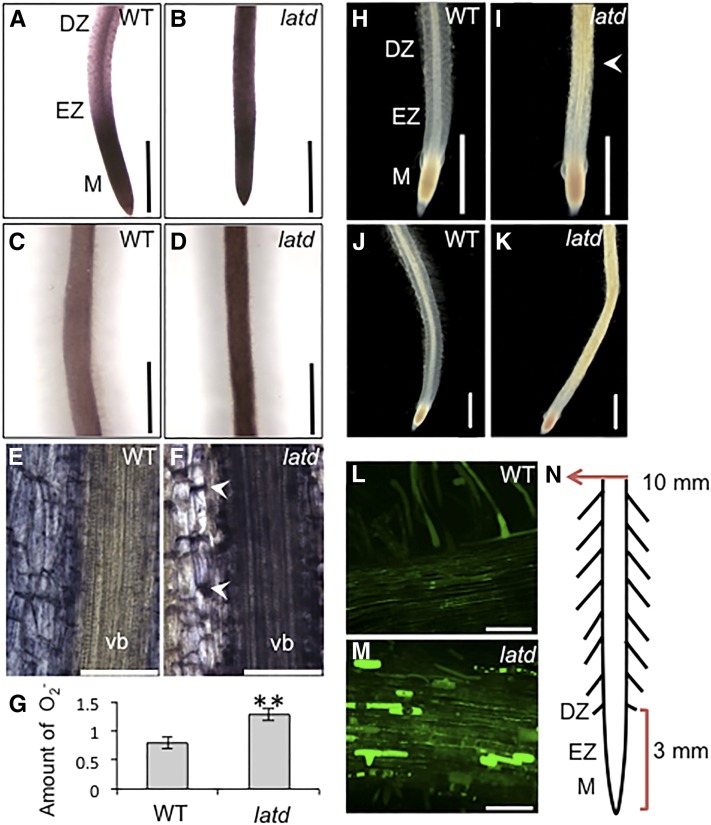
The phenotypes of latd mutants are pleiotropic, with defects in several aspects of root growth as well as in stomatal function (Bright et al., 2005; Liang et al., 2007). The reduced root length, as well as reduced root hair growth relative to the wild type, is reminiscent of the root hair phenotype of AtrbohC/rhd2 mutants and the root growth phenotype of AtrbohF mutants (Foreman et al., 2003; Kwak et al., 2003; Takeda et al., 2008). The RAM arrest of latd mutants is reminiscent of that of root meristemless mutants, which are defective in the synthesis of glutathione, a central component in the regulation of cellular redox homeostasis (Cheng et al., 1995; Vernoux et al., 2000); however, glutathione treatment does not rescue latd root growth (Bright et al., 2005). To test whether latd mutant roots have altered ROS levels, we used the ROS-reactive dyes nitroblue tetrazolium (NBT) and 3,3′-diaminobenzidine (DAB) to measure levels of the ROS molecules O2.− and H2O2, respectively, in different parts of the root (Ramu et al., 2002; Fester and Hause, 2005). We found that wild-type roots had strong NBT staining in the tip, with the strongest staining in the first 1 mm of primary roots, but only weak staining in the rest of the root (Fig. 1A). In wild-type roots, the intensity of NBT staining starts to decrease in the elongation zone and differentiation zone, behind the root meristem (Fig. 1A), with only light staining observed at a distance of 10 mm from the root tip (Fig. 1C). In longitudinal sections of the mature region of wild-type roots (taken approximately 5 mm from the tip), staining is found mostly in the cortex and endodermis, with almost none in the vasculature (Fig. 1E). Lateral roots have a similar O2.− accumulation pattern, with the darkest staining in the tips (Supplemental Fig. S1). Given the pattern of NBT staining in the wild type, we focused on two locations in the roots: the first 3 mm of the root tip containing approximately 1 mm of meristem, and a region 10 mm away from the tip, where cells have finished elongation and are mature (Fig. 1N). In latd mutants, the root tips showed strong NBT staining in the root tip, as in the wild type, but unlike the wild type, strong staining was also detected in the elongation zone, the differentiation zone (Fig. 1B), and the mature region of the roots, even as far as 10 mm from the root tip (Fig. 1D), leading to an overall increase of O2.− in the root (Fig. 1G). We observe a similar pattern in plants carrying the nip-1 mutation, a weaker allele of the LATD/NIP locus (Veereshlingam et al., 2004; Yendrek et al., 2010; Supplemental Fig. S2). The strength of NBT staining in the latd, nip-1, and nip-3 mutants reflects an allelic series corresponding to the severity of root and nodule defects (Veereshlingam et al., 2004; Bright et al., 2005; Teillet et al., 2008; Yendrek et al., 2010; Supplemental Fig. S2), although both latd and nip-1 have similar nitrate transport defects (Bagchi et al., 2012). Longitudinal sections of latd mutant roots revealed that mature regions of the latd root (5 mm from the root tip) exhibit ectopic accumulation of O2.−, especially in the inner cortex, endodermis, and vascular bundle (Fig. 1F). In latd roots, this ectopic NBT staining is detected particularly in the cross walls of cortical cells but not on the sides parallel to the main axis of the root (Fig. 1F, arrowheads).
Figure 1.
latd mutant roots have altered ROS levels and localization. A to D, Representative images of 5-d-old wild-type (WT) and latd mutant roots stained with NBT to indicate O2.−. latd mutants have increased O2.− levels at both the root tips (A and B) and the mature region of the roots (C and D), taken at 10 mm from root tips, as shown in N. DZ, Differentiation zone; EZ, elongation zone; M, meristem. Bars = 1 mm. E and F, Longitudinal thick sections of NBT-stained mature roots 5 mm from the tip. Ectopic O2.− accumulation in latd is observed in the cortical cell end walls (arrowheads) and in the vascular bundle (vb) at this location in the root. Sections are 90 μm thick. Bars = 100 μm. G, Quantification of NBT staining in wild-type and latd roots. The amount of O2.− represents nmol of reduced NBT per g of tissue. Data were collected from three biological replicates with 28 roots total for each genotype. **, One-way ANOVA was performed, giving P < 0.01. H to K, Whole-mount DAB staining of H2O2 accumulation in 5-d-old wild-type and latd roots. Root tip staining is similar in both wild-type and latd roots, with staining observed in the meristem. DAB staining is higher throughout the latd root, increasing strongly at the end of the differentiation zone (white arrowhead in I). Bars = 1 mm. L and M, Mature root 10 mm from the root tip (as shown in N) of 7-d-old roots stained with H2DCFDA to label H2O2. Note the occasional brightly labeled epidermal cells in latd. Bars = 100 μm. All staining above was from at least three biological replicates for a total of 10 to 30 plants for each condition. N, Diagram showing the locations of images in this figure. The red bracket indicates the position of root tips (3 mm, containing about 1 mm of meristem), and the red arrow indicates the position of the mature region photos (10 mm from the tips).
H2O2 detected by DAB is localized primarily in the root tip of wild-type plants (Fig. 1H). latd mutants also show root tip DAB staining (Fig. 1I); however, DAB staining starts to give a strong signal approximately 2 mm from the tip of latd roots in the differentiation zone (Fig. 1I, arrowhead), where wild-type plants exhibit only a very low signal (Fig. 1H). This increased H2O2 accumulation in latd roots persists throughout the mature region of the root, whereas wild-type roots display no significant DAB staining in these regions (Fig. 1, J and K). When dihydrofluorescein diacetate (H2DCFDA) was used to detect intracellular H2O2, we also observed increased levels of H2O2 in latd roots as compared with the wild type (Fig. 1, L and M). Curiously, occasionally entire cells in the epidermis of latd mutant roots were lit uniformly with a strong H2DCFDA signal (Fig. 1M). Together, these data show that latd mutant roots have higher levels of both O2.− and H2O2 than wild-type roots and that these are found ectopically in the elongation zone, the differentiation zone, and the mature region of the root.
Cell Elongation and Expansion Are Severely Reduced in latd Mutant Roots
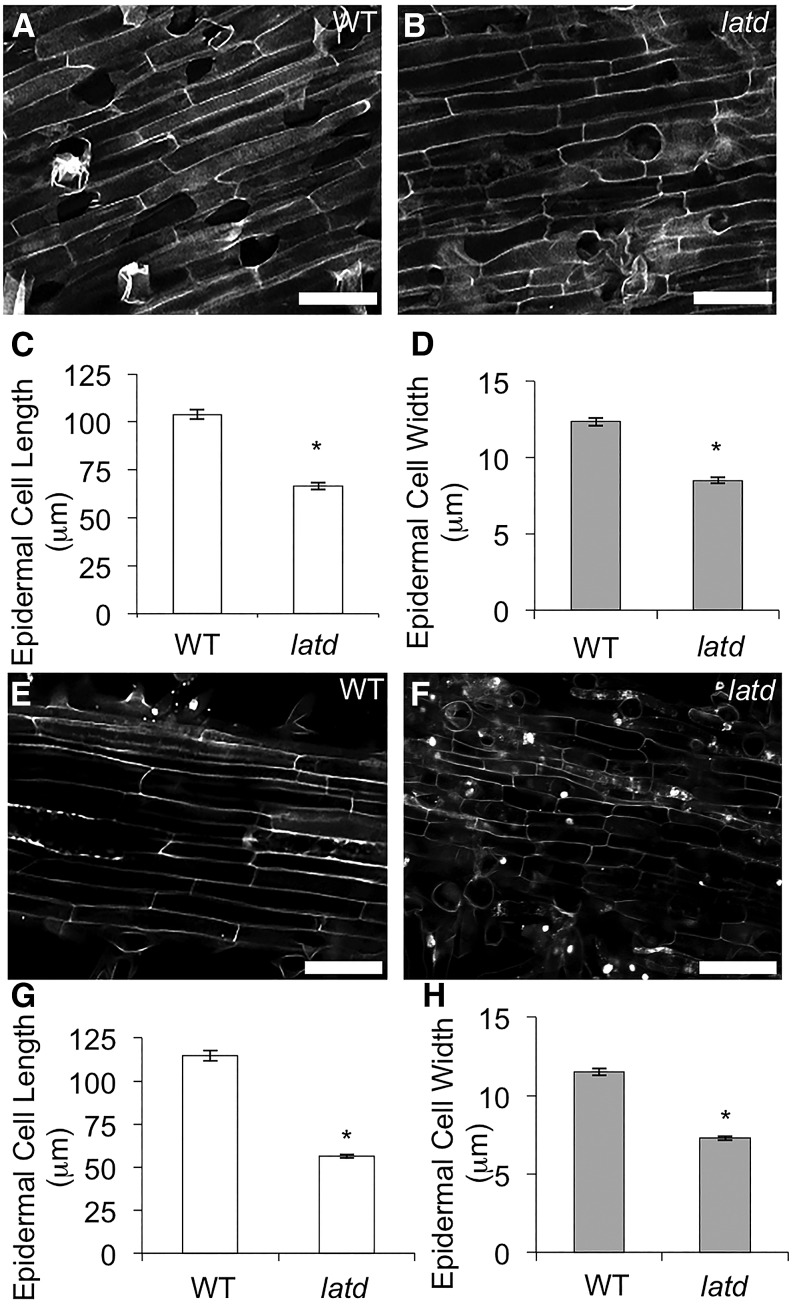
latd mutants have short primary roots as well as lateral roots that fail to elongate (Liang et al., 2007). ROS are important signaling molecules that regulate root growth and root hair elongation (Foreman et al., 2003; Monshausen et al., 2007). We wondered whether the short-root phenotype of latd mutants might be the result of defects in cell elongation. To test whether latd mutant roots have cell elongation defects, we measured epidermal cell length at 5 d, when latd roots grow at a rate only slightly less than that of the wild type (Bright et al., 2005). To ensure that we were measuring comparable cells in both the wild type and latd, we measured cells at the same distance, 10 mm from the root tip in the mature zone, where root hair growth was complete. At this developmental stage, latd mutant roots still have organized cell files as wild-type plants do (Fig. 2, A and B). We found that at the same distance from the root tip, latd mutant roots have significantly shortened epidermal cells that are nearly 40% shorter than wild-type roots (Fig. 2C). We also found that the width of latd root epidermal cells is significantly narrower than that of the wild type (Fig. 2D).
Figure 2.
latd mutant roots are defective in cell elongation. A, B, E, and F, Representative confocal images of propidium iodide-stained root epidermal cells from 5-d-old (A and B) and 21-d-old (E and F) wild-type (WT; A and E) and latd mutant (B and F) roots. All images were taken 10 mm from the root tip. Bars = 50 μm. C, D, G, and H, Average epidermal cell length (C and G) and width (D and H) from 5-d-old (C and D) and 21-d-old (G and H) wild-type and latd mutant roots. Graphs display means ± se of n > 50 cells (10 cells per root from at least five roots in each genotype) for length measurements and n > 100 cells (20 cells per root from at least five roots in each genotype) for width measurements. All measurements were taken at 10 mm from the root tip. Asterisks indicate statistically significant differences, with P < 0.001, by Student’s t test.
The Cell Elongation Defect in latd Mutant Roots Becomes Increasingly Severe as Meristem Arrest Progresses
The meristem of latd roots undergoes exhaustion as development proceeds and arrests completely by 3 weeks (Bright et al., 2005; Liang et al., 2007). We wondered whether cell elongation defects in latd mutant roots might also become more severe as development progressed. In order to test this, we quantified epidermal cell length in latd mutants at 21 d old, at which point root growth had completely arrested. As described above, we measured cells 10 mm from the root tip, in the mature zone, so that we are measuring fully differentiated cells. In this way, we could test whether cells that had recently completed differentiation at 5 or 21 d had differences in length. We found that at 21 d, cells at this position in latd mutant roots show more severe elongation defects, with epidermal cell length more than 50% shorter than in the wild type and significantly narrower (Fig. 2, E–H). Epidermal cell shape becomes more variable in latd mutant roots, varying from a standard rectangular shape by being more triangular, having pointed ends, or having more irregular shapes (Fig. 2F). Thus, our data suggest that both the cell elongation defects and cell shape variability in latd mutant epidermal cells become more severe as the meristem arrests.
ABA Rescues Both the latd ROS and Cell Elongation Defect
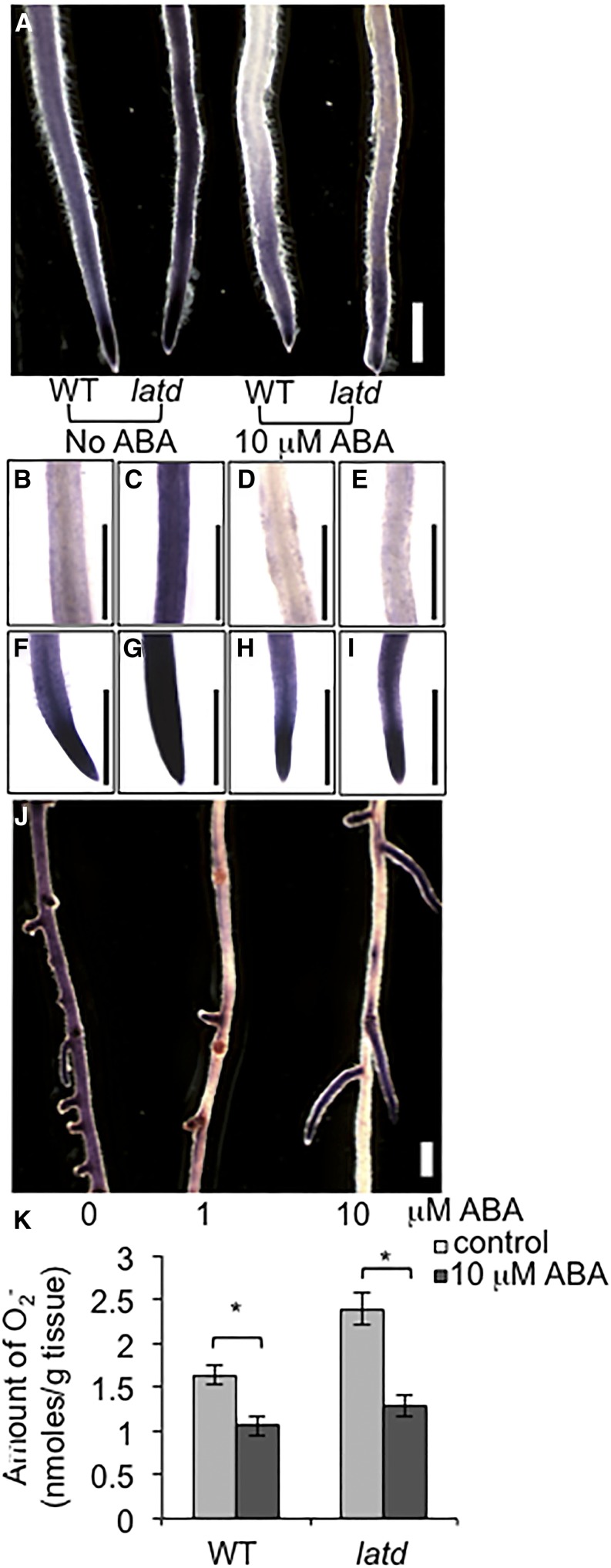
ABA can rescue the short-root phenotype as well as the lateral root elongation defects of latd mutants (Liang et al., 2007). Since latd roots have increased levels of ROS (Fig. 1), we asked whether ABA might rescue latd root defects by decreasing ROS levels. To test this hypothesis, we used NBT staining to detect O2.− in wild-type plants and latd mutant roots treated with or without ABA for 5 d. We found that 10 μm ABA decreases O2.− levels in wild-type 3-mm root tips (Fig. 3A), the mature region (10 mm from the tip; Fig. 3, B and D), and overall O2.− amount in the whole root (Fig. 3K). The same effect of ABA on O2.− levels is also observed in wild-type lateral roots (Supplemental Fig. S1). At this concentration, ABA also decreases O2.− levels in latd mutant root in both root regions (Fig. 3, A, C, and E). However, these ABA treatments did not obviously affect NBT staining in the meristem (approximately the first 1 mm of the root tips): the tips of both primary and lateral roots remain darkly stained with NBT in both wild-type and latd roots grown at both concentrations of ABA (Fig. 3, F–I; Supplemental Fig. S1). Again, we checked the roots of nip-1, an allele of latd, and found that ABA decreased O2.− levels in nip-1 roots in a similar manner (Supplemental Fig. S3). We also examined the effect of ABA on O2.− levels in latd roots at a later developmental stage, 21 d, when latd root growth arrests in plants grown in the absence of ABA (Liang et al., 2007). We found that continuous ABA treatment also decreases O2.− levels in a dose-dependent manner at this stage (Fig. 3J). The similar effect of ABA on O2.− levels in the wild type and latd may indicate that the response to ABA in latd is intact but that the endogenous factors that control the ROS balance are disrupted.
Figure 3.
ABA lowers O2.− levels in wild-type (WT) and latd mutant roots. A, Whole-mount NBT staining of 5-d-old wild-type A17 and latd mutant roots grown continuously with and without 10 μm ABA. All NBT staining was repeated four times with at least 30 roots for each condition. Bar = 1 mm. B to I, NBT staining of A17 (B, D, F, and H) and latd mutant (C, E, G, and I) root meristems grown with (D, E, H, and I) and without (B, C, F, and G) 10 μm ABA. B to E, Staining of 10 mm distance from the root tips. F to I, Root tips (1.5 mm). Bars = 1 mm. J, NBT staining of 21-d-old latd mutant roots grown continuously on 0, 1, or 10 μm ABA. Note the elongating lateral roots on latd plants grown on 10 μm ABA, a concentration that restores lateral root growth in the latd mutant. Bar = 1 mm. K, Quantification of NBT staining in wild-type and latd roots. Plants were grown on BNM with and without 10 μm ABA for 7 d. The amount of O2.− represents nmol of reduced NBT per g of tissue. Data were collected from two biological replicates for a total of n = 19 to 33 roots per treatment. Error bars represent se. Asterisks represent statistical significance between treatments (P < 0.05) using one-way ANOVA.
To test whether exogenous ABA also decreases H2O2 levels, we used DAB to stain for H2O2 in ABA-treated roots. We found that a 5-d continuous 10 μm ABA treatment did not significantly decrease DAB staining in wild-type roots (Supplemental Fig. S4A), indicating that ABA may regulate H2O2 differently in roots than O2.−, or alternatively, that ABA may regulate H2O2 at a different time point. However, in the mature region of latd roots, where ABA-rescued lateral roots develop, ABA decreased H2O2 accumulation (Supplemental Fig. S4, B and C). We found that, as the concentration of ABA applied increases, the lower boundary of H2O2 accumulation in the mature region of latd roots is shifted shootward, away from the root tip (Supplemental Fig. S4, D and E, arrowheads).
We then tested whether ABA could also rescue the cell elongation defect in latd mutant roots. We measured epidermal cell length and width in wild-type and latd roots treated with 10 μm ABA and found that 10 μm ABA increases both the length and width of epidermal cells in latd mutant roots but decreases them in the wild type (Fig. 4, A and B). Since LATD/NIP transports nitrate (Bagchi et al., 2012), we also tested whether 10 mm nitrate could regulate ROS levels in either wild-type or latd roots but found no significant effect (data not shown). Thus, our findings indicate that ABA can rescue both ROS levels and cell elongation defects in latd mutant roots and decrease O2.− levels in wild-type roots, suggesting that ROS may function in the signaling pathway downstream of ABA in regulating root elongation.
Figure 4.
Epidermal cell length (A) and width (B) of latd mutant roots can be increased by adding 10 μm ABA. For each genotype in each treatment, graphs display the mean of n = 50 to 60 cells from at least five roots for cell length and n = 102 to 126 cells from at least five roots for cell width. Error bars represent se, and letters indicate statistically significant differences between different treatments and genotypes, with P < 0.05, using Student’s t test. WT, Wild type.
Decreasing ROS Levels Can Increase Cell Elongation in latd Roots
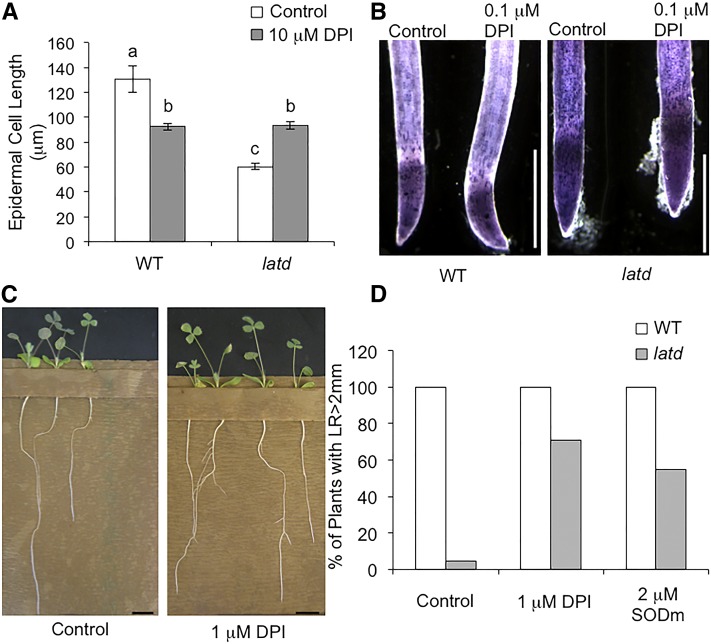
ROS molecules are important for regulating plant growth and development (Swanson and Gilroy, 2010). Thus, an imbalance of ROS levels can cause defects in root growth and differentiation. We wondered whether the high ROS levels found in latd mutants were an indirect effect of the latd mutation or rather a direct cause of the root elongation defects. To test this question, we directly manipulated ROS levels in the root and measured the length and width of newly mature epidermal cells as described above. To decrease O2.− levels, we used either DPI to inhibit the activity of NADPH oxidases, which catalyze the production of O2-, or EUK 134 (a SOD mimetic) to chemically mimic the activity of SODs (Rong et al., 1999), which scavenge O2.−, converting it to H2O2. We found that after 2 d of DPI treatment, latd mutant roots developed significantly longer epidermal cells (Fig. 5A). In contrast, the same DPI treatment decreases epidermal cell length in wild-type roots (Fig. 5A), suggesting that ROS levels must reach an optimal balance for normal root elongation and that the increased ROS in latd roots is mediated by NADPH oxidase activity. This 2-d DPI treatment is sufficient to reduce O2.− accumulation in wild-type and latd roots, especially at the root tip and elongation zone (Fig. 5B). In addition, latd mutants treated with DPI or EUK 134 showed an increase from 4.3% to 70.6% or 54.6%, respectively, in the percentage of plants that developed lateral roots longer than 2 mm (Fig. 5, C and D). Together, these data indicate that decreasing ROS levels in latd mutant roots, either by inhibiting production or increasing scavenging, can increase cell elongation, and thus root elongation, in both primary and lateral roots and that the increased ROS levels in latd roots are mediated, at least in part, by NADPH oxidase activity.
Figure 5.
Decreasing ROS levels in latd mutants rescues both cell and lateral root elongation defects. A, Epidermal cell length in wild-type (WT) and latd mutant roots treated with 0.1 μm DPI for 2 d. Plants were first grown in plain BNM for 8 d in growth pouches, then BNM supplemented with 0.1 μm DPI was added into the pouches. Graphs display means of n = 40 to 80 cells from at least four roots for each genotype. Cell length was measured in cells 1 cm from the root tip. Error bars represent se, and letters indicate statistically significant differences between genotypes and treatments, with P < 0.05, using Student’s t test. B, NBT staining in wild-type and latd roots grown on BNM with or without 0.1 μm DPI for 2 d. Note the decreased O2.− staining in DPI-treated roots. This experiment was repeated twice with at least five roots for each condition. Bars = 1 mm. C, Twenty-one-day-old latd mutant plants in growth pouches grown continuously with either control medium (left) or medium containing 1 μm DPI (right). Note the increased lateral root length on DPI-treated roots. Bars = 1 cm. D, Percentage of wild-type and latd mutant plants grown as in C that develop lateral roots (LR) longer than 2 mm. Plants were grown for 21 d with either control medium or medium supplemented with 1 μm DPI or 2 μm EUK 134 (a SOD mimetic [SODm]). Data are from two biological replicates with n > 10 for each column.
Regulation of Rboh Gene Expression by LATD/NIP and ABA
Our data show that latd mutants have increased levels of root O2.− (Fig. 1) and decreased length of epidermal cells (Fig. 2). Inhibiting the activity of NADPH oxidases with DPI decreases the level of O2.− and partially restores the cell elongation defect (Fig. 5). This observation indicates that the excess ROS production in latd mutants is mediated by NADPH oxidases. The fact that DPI mimics the effect of ABA on latd root ROS levels, cell length, and root elongation (Figs. 3 and 4; Liang et al., 2007) suggests that NADPH oxidases may be a target of ABA signaling as well. The Rboh family comprises 10 genes in Arabidopsis and at least seven in M. truncatula (Sagi and Fluhr, 2006; Lohar et al., 2007; Marino et al., 2011). Gene duplications occurred independently in the Brassicaceae and the Fabaceae; thus, similar gene names do not indicate orthologous genes (Marino et al., 2011; Montiel et al., 2012). To determine which MtRboh gene or genes were correlated with the latd root defects, we examined the expression of these genes using quantitative reverse transcription (qRT)-PCR to see whether the expression of any of these corresponded to latd root ROS levels and cell length phenotypes. We predicted that expression of the MtRboh gene responsible should be increased in latd mutants and decreased by ABA to approximately wild-type levels in latd and further in the wild type, since this is the pattern of O2.− staining that we observed (Figs. 1 and 3). We found that expression of MtRbohA and MtRbohC was increased in latd roots while that of MtRbohD was decreased (Fig. 6, A and B). Several of the MtRboh genes were ABA responsive, with MtRbohA, MtRbohB, and MtRbohF expression increased by ABA and MtRbohC and MtRbohG expression decreased (Fig. 6, A and B). However, only RbohC expression fulfilled our predictions, namely, that expression is increased in latd over the wild type and ABA treatments that rescue the latd root ROS and cell length defects (continuous treatment with 10 μm ABA) reduce the expression of RbohC to wild-type levels in latd and further in wild-type roots (Fig. 6A), suggesting that RbohC is likely the NADPH oxidase responsible for latd root ROS and cell elongation defects. The regulation of RbohC by ABA in both genetic backgrounds suggests that RbohC may also mediate ABA’s effect on root ROS and cell length in wild-type roots as well as those of latd mutants.
Figure 6.
latd mutant roots have altered expression of several ROS-related genes. A, Relative expression of MtRbohC in wild-type (WT) and latd mutant roots. B, Relative expression of six other MtRboh genes in wild-type and latd mutant roots. C, Relative expression of a putative apoplastic Cu/ZnSOD (ApoCu/ZnSOD) in wild-type and latd mutant roots. D, Relative expression of a putative cell wall-localized peroxidase (cwPRX2) in wild-type and latd mutant roots. E, Relative expression of RIP1 in wild-type and latd mutant roots. Whole-root tissue of 7-d-old plants grown continuously on BNM with or without 10 μm ABA was harvested and analyzed by qRT-PCR. Graphs represent means ± se from three biological replicates, with n = 20 for each genotype in each treatment per replicate. One-way ANOVA was performed to analyze statistical differences (P < 0.05) for each gene examined. Different letters indicate statistically significant differences between genotypes and treatments. Primers used for qRT-PCR are listed in Supplemental Table S1.
Other ROS-Related Genes Are Also Regulated by LATD/NIP and ABA
ROS are dynamic signaling molecules whose levels are under tight control, as a result of the balance between production and scavenging (Mittler et al., 2011). We wondered whether the high O2.− and H2O2 levels in latd mutant roots (Fig. 1) might be caused not just by increased expression of O2.−-generating Rboh genes (Fig. 6, A and B) but also by decreased expression of ROS-scavenging enzymes.
To further examine the regulation of the ROS-scavenging system in latd mutants, we examined the expression of SOD genes as well as peroxidases and other antioxidant genes. SOD enzymes act directly on O2.−, so we examined the expression of several SOD genes in both wild-type and latd plants. Since plants have multiple SOD proteins targeted to different subcellular regions, we used BLAST to identify genes for putative cell wall-localized, chloroplast-localized, and cytoplasm-localized SODs. The cotton (Gossypium hirsutum) Cu/ZnSOD, GhCSD3, is known to be targeted to the cell wall (Kim et al., 2008). We searched for an M. truncatula gene with high homology and found that TC183733 has 81% amino acid identity to GhCSD3. This putative apoplastic Cu/ZnSOD (ApoCu/ZnSOD; Medtr6g029200) has slightly but significantly decreased expression in latd mutant roots (Fig. 6C). ABA can increase this Cu/ZnSOD expression in wild-type roots and also in latd roots to restore its expression to wild-type levels (Fig. 6C), the inverse of the RbohC expression pattern. On the other hand, an FeSOD gene (GenBank accession no. AFK34552.1; Medtr1g048990) that has 95% identity to a Medicago sativa FeSOD (GenBank accession no. AAL32441.1), predicted to be localized to the chloroplast (Alscher et al., 2002; Rubio et al., 2004; Asensio et al., 2012), as well as a Cu/ZnSOD (CytoCu/ZnSOD; Medtr7g114240) predicted to be cytoplasmic (Macovei et al., 2011) did not show a difference in gene expression between latd and wild-type roots (Supplemental Fig. S5).
Because many of the LATD-regulated ROS genes we had identified are associated with cell wall ROS production (three MtRboh genes and the putatively cell wall-localized ApoCu/ZnSOD), we asked whether other putatively cell wall-localized ROS enzymes might be regulated by LATD. Peroxidases have been shown to catalyze cell wall cross-linking in response to various environmental stimuli (Almagro et al., 2009). We examined a putative cell wall peroxidase, cwPRX2 (GenBank accession no. AES66966.1; Medtr2g084000), with high expression levels in roots according to the M. truncatula Gene Expression Atlas (Benedito et al., 2008; He et al., 2009) and with 82% amino acid identity to the cell wall-localized PsPOX11 peroxidase from Pisum sativum (Kawahara, 2006). We found that the expression of cwPRX2 was signficantly up-regulated in latd mutant roots and up-regulated by ABA only in latd mutants (Fig. 6D). Another peroxidase, RHIZOBIUM-INDUCED PEROXIDASE (RIP1; Medtr5g074860), which is expressed in roots but induced to higher levels by nodulation and H2O2 (Ramu et al., 2002), has decreased expression in latd roots, and ABA can stimulate its expression in a pattern similar to that of ApoCu/ZnSOD, although RIP1 expression is more strongly affected by the latd mutation (Fig. 6E). Finally, we checked the expression of some antioxidant genes that are known to be altered in M. truncatula in response to drought stress (Filippou et al., 2011). We found that expression of the drought stress-responsive genes ALTERNATIVE OXIDASE (AOX; Medtr5g026620) and GLUTATHIONE S-TRANSFERASE (GST; Medtr1g026140) was also altered in latd roots, but not the expression of cytosolic ASCORBATE PEROXIDASE (cAPX; Medtr4g061140; Supplemental Fig. S6; Filippou et al., 2011).
In summary, latd mutant roots exhibit altered expression levels of many ROS-related genes, for both ROS-generating and scavenging systems, and many of these are also regulated by ABA. Regulation of RbohC and apoplastic Cu/ZnSOD by latd and ABA correlates with root O2.− levels: the presence of the latd mutation increases RbohC and decreases apoplastic Cu/ZnSOD expression and ABA treatment reverses this effect, returning expression to wild-type levels, suggesting that RbohC and apoplastic Cu/ZnSOD expression may be important targets of a common ABA, LATD/NIP signaling pathway.
Silencing MtRbohC Stimulates Root Elongation and Lateral Root Formation
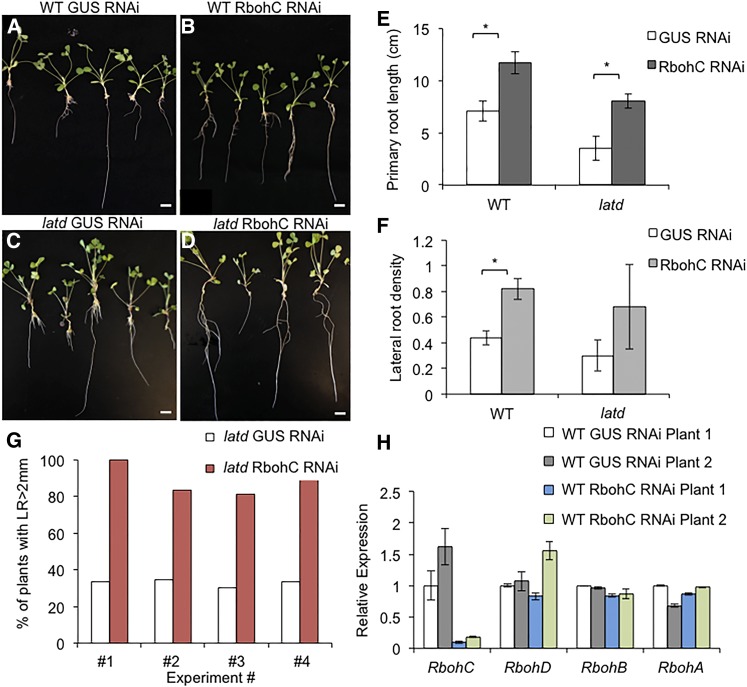
MtRbohC root expression correlates both with root length and with ROS levels: when RbohC expression is high (i.e. in untreated latd roots), roots have ectopic ROS accumulation and root length is short; when RbohC expression in latd roots is decreased by ABA treatment to wild-type untreated levels (Fig. 6A), ROS levels decrease and roots are longer. In order to test whether the decrease in RbohC expression is the cause of increased root length in ABA-treated latd roots, we used RNAi to reduce RbohC expression using Agrobacterium rhizogenes-mediated transformation. We confirmed that our RNAi approach efficiently reduced RbohC expression at least 5-fold, yet it did not affect the expression of RbohD, the most closely related MtRboh gene (Marino et al., 2011), or two other MtRboh genes, RbohB and RbohA, with strong root expression (Fig. 7H). We observed root elongation in RbohC RNAi-transformed roots for 10 d. Our results showed that in wild-type plants, roots transformed with the MtRbohC RNAi construct have longer primary roots and an increased lateral root density (Fig. 7, B and C). latd roots transformed with RbohC RNAi constructs showed similar results in both root elongation and lateral root density, although the latter one showed a big variance among four biological replicates, leading to a nonstatistical difference (Fig. 7, D and E). We think this is due to the variability in the nature of A. rhizogenes transformation, but latd roots transformed with MtRbohC RNAi vector showed increased lateral root density in three out of four biological replicates. Although decreasing RbohC expression in latd mutants did not show full rescue of its root defects, the percentage of roots that developed lateral roots longer than 2 mm increased to at least 80% from about 30% (Fig. 7G). Together, these findings indicate that down-regulation of MtRbohC expression using RNAi increases root elongation and lateral root formation in both the wild type and latd mutants, demonstrating an important role for MtRbohC as a negative regulator of root elongation.
Figure 7.
Silencing MtRbohC expression via RNAi increases root elongation and lateral root density. A to D, Representative images of the wild type (WT; A and B) and latd mutants (C and D) transformed with control vector (GUS RNAi; A and C) and MtRbohC RNAi vector (B and D) using A. rhizogenes transformation. Bars = 1 cm. E and F, Primary root length (E) and lateral root density (F) in wild-type and latd mutant roots transformed with the MtRbohC RNAi vector. Lateral root density is the lateral root number per cm of primary root. The graphs represent averages of four biological replicates, with n = 53 to 80 individual transformants total for each genotype and treatment. Error bars represent se, and asterisks indicate statistically significant differences using one-way ANOVA (P < 0.05). G, Percentage of latd transformants that developed lateral roots (LR) longer than 2 mm. The graph represents data from four biological replicates (experiments 1–4). For latd plants transformed with GUS RNAi, n = 6, 23, 13, and 11 for experiments 1 to 4, respectively; for latd plants transformed with MtRbohC RNAi, n = 7, 18, 16, and 12, respectively. H, Relative expression of MtRbohC, MtRbohD, MtRbohB, and MtRbohA in wild-type plants transformed with the control vector and the MtRbohC RNAi vector. The graph shows relative expression from two individual transformants for each vector. Note that only MtRbohC expression is knocked down in wild-type roots by the MtRbohC RNAi vector, and not the closely related MtRbohD gene, nor MtRbohA or MtRbohB. Error bars represent se from two technical replicates.
DISCUSSION
In this study, we demonstrate that the MtLATD/NIP/MtNPF1.7 transporter is required for ROS homeostasis and cell elongation in roots. In latd mutants, this balance is perturbed, leading to an excess of O2.− and H2O2 and a corresponding decrease in cell elongation (Figs. 1 and 2). We demonstrate that rescue of root growth by ABA in latd mutants is due to a decrease in ROS levels and an increase in cell elongation (Figs. 3 and 4). Directly manipulating ROS levels in latd mutant roots to reduce O2.− levels can also increase cell length and promote root elongation, indicating that it is the change in root ROS levels that increases cell elongation in ABA-treated latd roots (Fig. 5). We propose that ABA can regulate cell elongation via the manipulation of ROS levels in roots. Our data indicate that ABA reduces ROS levels and that this effect corresponds to changes in root length and cell length, suggesting that this is one mechanism by which ABA regulates root growth. The observation that the NADPH oxidase inhibitor, DPI, increases both cell and lateral root elongation in latd mutants indicates that the latd mutant root elongation phenotype is likely mediated by NADPH oxidase activity (Fig. 5). We also find that the expression of enzymes involved in ROS synthesis or degradation is regulated by ABA and LATD/NIP in a manner consistent with observed levels of ROS in roots (Fig. 6), suggesting transcriptional control of ROS-related enzymes by ABA and LATD/NIP. In particular, expression of the NADPH oxidase-encoding gene, MtRbohC, increases in latd roots and is decreased by ABA treatment, just as O2.− levels are (Fig. 6). Reducing the expression of MtRbohC leads to significant increases in root elongation in both wild-type and latd roots, indicating that MtRbohC is a negative regulator of root elongation and that its increased expression in latd roots is a major cause of the latd short-root phenotype. The change in MtRbohC expression in latd roots and in response to ABA and its correlation with root and cell length indicate that MtRbohC is a key target of LATD/NIP and ABA signaling in the root.
ABA Regulates Root Cell Elongation by Modulating ROS Levels
ABA plays an important role in regulating root length, both in drought conditions and during normal growth (Sharp et al., 1994). In well-watered plants, low ABA levels caused by mutations in ABA biosynthetic genes reduce root length, as do high levels of ABA caused by addition of the hormone to the growth medium (Sharp et al., 1994; Cheng et al., 2002; Lin et al., 2007), suggesting that under normal growth conditions there is an optimal ABA level for root elongation and that deviating above or below that level reduces the ability of the root to elongate. Root growth is driven by two interrelated processes: cell division and cell elongation. Root meristem length and cortical cell length in different Arabidopsis accessions are highly correlated, indicating the fundamental linkage between these two processes (Slovak et al., 2014). ABA has been shown to promote quiescent center quiescence and to suppress stem cell differentiation in the RAM, which indicates ABA’s positive role in regulating root growth (Zhang et al., 2010). In maize (Zea mays) plants, continued root elongation under conditions of low water potential is reduced in ABA-deficient mutants, and elongation can be restored by the addition of ABA (Sharp et al., 2004). Here, we show that ABA stimulates root cell elongation in latd mutants (Fig. 4) at the same concentrations that restore latd root tip morphology and meristem function (Liang et al., 2007). Interestingly, this concentration of ABA has the opposite effect on wild-type roots, where it decreases epidermal cell elongation (Fig. 4). This apparent contradiction is resolved by a model in which latd root tips experience either a low concentration of ABA or reduced responsiveness to ABA. As a result, addition of ABA to latd roots would bring them closer to a more optimal concentration of ABA. Wild-type roots, in contrast, already experience a concentration of ABA that is more optimal for root growth. Addition of ABA drives them farther from this optimum, and root elongation decreases.
ABA signals via ROS in multiple plant species and tissues, but whether ABA stimulates or inhibits ROS production differs in different species. In Arabidopsis and maize, ABA stimulates O2.− and H2O2 production in guard cells, leaves, and roots via NADPH oxidase activity (Guan et al., 2000; Jiang and Zhang, 2001, 2002; Kwak et al., 2003; He et al., 2012). In contrast, our findings indicate that ABA treatment reduces levels of O2.− in both wild-type and latd roots in M. truncatula (Fig. 3). The observation that addition of ABA increases ROS levels in Arabidopsis, but decreases it in M. truncatula, may be the reason for the opposite response of legume and nonlegume roots to ABA in the elaboration of root architecture (Liang and Harris, 2005). Briefly, ABA treatment stimulates lateral root formation in M. truncatula and several other legume species while simultaneously inhibiting primary root elongation, thus strongly increasing lateral root density. Conversely, ABA significantly decreases lateral root density in nonlegumes from several different plant families by inhibiting lateral root formation more strongly than it inhibits primary root elongation (Liang and Harris, 2005). The one exception is Casuarina glauca, a nonlegume that forms a nitrogen-fixing symbiosis with an actinomycete and that increases lateral root density in response to ABA just as most legumes tested do (Liang and Harris, 2005). Based on the observed diversity of root architecture patterns throughout the plant kingdom, it is not surprising that different plant species will have evolved different responsiveness to hormones or different hormone responses. Thus, our analysis of the ABA response pathway in Medicago roots may reveal insight into the mechanism by which different plant species elaborate different root architectures.
A LATD/NIP Signaling Pathway Interacts with ABA to Regulate MtRBOH Gene Expression
Here, we present evidence that the LATD/NIP nitrate transporter and the plant hormone ABA combine to regulate the expression of multiple Rboh genes in the M. truncatula root. Two genes, MtRbohA and MtRbohC, are regulated by both ABA and LATD/NIP, but in the case of RbohA, the effects of ABA and LATD/NIP are additive, and in the case of RbohC, they are antagonistic (Fig. 6). The expression of RbohC corresponds to the level of O2.− observed in latd roots. Under conditions that raise the expression of RbohC, ROS levels also rise, and under conditions that lower RbohC, ROS levels fall (Figs. 3 and 6). In addition, root and cell length correspond well with the levels both of O2.− and of RbohC expression (Fig. 4). Why might RbohC-mediated O2.− production be more important that that produced by other Rboh gene products? RbohC is expressed at lower levels in the root than any of the other Rboh genes (Marino et al., 2011), and the expression of many other genes is also regulated by LATD/NIP and ABA; thus, the regulation of RbohC expression may initially seem an unlikely mechanism. However, the tight correlation of RbohC expression with ROS levels and cell length suggests otherwise. The low expression of RbohC in root tissue may indicate that it is expressed only in a subset of cells. Ubeda-Tomás et al. (2008) have shown that endodermal cell expansion is rate limiting for elongation of the root as a whole, thus demonstrating that factors that affect a small subgroup of root cells may have effects at the level of the whole root. Perhaps the location of RbohC activity in the root is critical to its function in controlling cell and root length. It will be interesting to see which cells express RbohC in both the wild type and latd mutants and in this way add to our understanding of the control of root growth.
Could ABA-Regulated ROS Modulate Cell Elongation by Acting Directly on the Cell Wall?
Many studies have shown that ROS can interact with plant cell wall components in order to regulate growth (Gapper and Dolan, 2006). H2O2 has been shown to cross-link cell wall structural proteins during plant disease resistance in soybean (Glycine max; Brisson et al., 1994) or induce cell wall cross-linking between cell wall polymers in maize coleoptiles, thus stiffening cell walls and limiting growth (Schopfer, 1996). Conversely, hydroxyl radicals can induce cell wall loosening, thus allowing cell expansion (Fry, 1998). In our study, we noticed that O2.− accumulates ectopically at apical/basal cell junctions in latd mutant roots, which may lead to local cell wall stiffening (Fig. 1E). Although there is no direct evidence linking extracellular O2.− with cell wall modification, O2.− can be converted to H2O2 very rapidly either through nonenzymatic reactions or by enzymes such as SOD (Apel and Hirt, 2004). Thus, it is possible that an increased level of O2.− in the apoplast of latd mutant roots may cause cell wall cross-linking by generating more H2O2 in the cell wall and lead to premature growth cessation and, thus, shorter cells.
We find that ABA can decrease ROS levels in latd mutant roots and promote cell elongation. Since ROS can interact with cell wall components to either loosen or strengthen the cell wall to regulate growth, we hypothesize that ABA may function to regulate cell elongation by altering cell wall ROS levels. Alternatively, ABA regulation of cell elongation could be via a separate pathway. In M. truncatula seeds, it has been shown that the inhibition of germination by ABA is related to the reduced expression of genes involved in cell wall loosening and expansion (Gimeno-Gilles et al., 2009). It is likely that in order to restrict or promote growth, ABA may directly regulate cell wall stiffening or loosening by triggering changes in apoplastic ROS production and scavenging to modulate the balance of different ROS species in the cell wall.
The Nitrate Transporter MtLATD/NIP/MtNPF1.7 Regulates Root ROS Levels to Control Root Elongation
Uneven distribution of nitrate in the soil can locally stimulate root branching, thus altering the overall architecture of the root system (Zhang and Forde, 1998). The ability of the plant to respond to an inconsistent and changing environment is a reflection of the plasticity of root development. In order for the local environment to modulate root architecture, the root must first sense the change in environment, then coordinate development, most likely via a hormone, and ultimately regulate cell division and cell elongation. The LATD/NIP gene (MtNPF1.7) encodes a member of the NPF family of hormone, nitrate, and dipeptide or tripeptide transporters (Yendrek et al., 2010; Léran et al., 2014) and plays an important role in root architecture (Bright et al., 2005). The LATD/NIP protein has recently been shown to mediate the transport of nitrate into cells (Bagchi et al., 2012) and is required for both lateral root elongation as well as primary root growth (Bright et al., 2005). An attractive model is that LATD/NIP may function to coordinate root elongation with nitrate sensing, although its role in nitrate sensing is still unknown.
Loss of LATD/NIP function results in complete loss of lateral root elongation: lateral roots arrest immediately after emergence from the primary root (Bright et al., 2005). This block to lateral root elongation can be bypassed by adding ABA to the growth medium (Liang et al., 2007). ABA levels in whole latd mutant seedlings are indistinguishable from those in the wild type (Liang et al., 2007); thus, LATD/NIP must function after ABA synthesis. Since the latd root development defect can be rescued by adding ABA to the growth medium, the likeliest possibility is that latd mutants can produce ABA but are unable to transport it to the needed location. The fact that this problem can be circumvented by providing ABA in the medium suggests that the medium is in contact with the responding tissue, perhaps the root tip or epidermis. Alternatively, ABA and LATD/NIP could function in parallel to independently regulate both ROS levels and root elongation by different mechanisms. NRT1.1 (AtNPF6.3) transports both nitrate and auxin, thus directly linking nitrate sensing with auxin transport and the control of root growth (Krouk et al., 2010). It is interesting to speculate that LATD/NIP could perform a similar role in sensing nitrate and regulating ABA transport, either directly or indirectly. Other NPF family members have recently been shown to transport ABA (Kanno et al., 2012), suggesting that the link between LATD/NIP and ABA transport could be direct. We are currently testing this possibility.
Our findings that functioning of the LATD/NIP transporter is required for ROS homeostasis, which regulates cell elongation in the developing root, reveal a key role for LATD/NIP in the control of root growth and suggest a possible link to soil nitrate. Our data demonstrate that, together, LATD/NIP, in concert with ABA, control the expression of the O2.−-generating RbohC enzyme in the root in a way that corresponds to O2.− levels and to cell length. We do not yet know whether the effect of ROS and LATD/NIP on cell elongation is mediated by direct changes in cell wall loosening or rigidity or indirectly via cytoplasmic changes, perhaps to the cytoskeleton. The severe root defects caused by the loss of LATD/NIP function, despite the existence of several very close Medicago spp. homologs, indicate an essential role for this nitrate transporter in root development via the maintenance of ROS homeostasis and suggest an intimate relationship between LATD/NIP function and ABA signaling.
MATERIALS AND METHODS
Plant Growth Conditions
Medicago truncatula seeds were scarified with concentrated sulfuric acid for 10 min, rinsed six times with sterile water, and sterilized in 30% (v/v) bleach before imbibing for 5 to 6 h and shaking at room temperature. Seeds were cold treated for at least 24 h before germinating on a moistened, sealed petri plate overnight in the dark. The A17 line was used as the wild-type control in all experiments. Seedlings were grown on 25- × 25-cm petri dishes (Nunc; http://www.nuncbrand.com/) or in growth pouches (http://www.mega-international.com/) containing buffered nodulation medium (BNM) at pH 6.5 (Ehrhardt et al., 1992) and placed vertically in an MTR30 Conviron growth chamber at 20°C, 50% humidity, and a 16-h-light/8-h-dark cycle with an intensity of 100 μE m−2 s−1. Growth pouches were placed in a sealed Styrofoam box with a clear plastic dome as a lid. For ABA treatment, (±)-ABA (A1049; Sigma-Aldrich; http://www.sigmaaldrich.com/) was added to the medium after autoclaving to reach specified concentrations. DPI (Sigma-Aldrich) and EUK 134 (Caymen Chemicals) were dissolved in dimethyl sulfoxide (DMSO) and added to liquid medium in growth pouches to reach specified concentrations.
ROS Staining
For O2.− staining, NBT (Sigma-Aldrich) was used as described by Ramu et al. (2002) with modifications. Roots were incubated in 10 mm sodium phosphate buffer (pH 7.8) with 10 mm NaN3 and 1 mg mL−1 NBT for 30 min at 37°C. The reaction was stopped by removing the NBT staining solution and washing roots twice in 80% (v/v) ethanol. All NBT staining was repeated four times for a total of at least 30 roots per condition. Whole-mount images were made using a Leica dissecting microscope.
For longitudinal root sections, a minimum of five stained roots were cut into 1-cm segments and sectioned on a Lancer Vibratome Series 1000. All sections were 90 μm thick. The images of sections were made using an Olympus microscope.
For O2.− quantification, NBT-stained roots were first ground in liquid N2 into a fine powder. Then, the powder was dissolved in 2 m KOH:DMSO (1:1.16, v/v) followed by centrifugation at 12,000g for 10 min. Absorbance at 630 nm was immediately measured and then compared with a standard curve plotted from known amounts of NBT in the KOH:DMSO mix (Ramel et al., 2009). Experiments were from three biological replicates for a total of nine to 28 roots per condition and genotype.
For H2O2 staining, roots were incubated in 1 mg mL−1 DAB (Sigma-Aldrich) in sodium citrate buffer at room temperature overnight, then washed and cleared with 10% (v/v) lactic acid (Fester and Hause, 2005). Stained roots were imaged using a Leica dissecting microscope. For an alternative H2O2 probe, 25 μm H2DCFDA (Sigma-Aldrich) in sodium phosphate buffer, pH 7, was used to stain whole roots for 30 min. Stained roots were then washed with fresh sodium phosphate buffer three times and imaged with a Zeiss LSM 510 confocal microscope using the green emission filter. DAB staining was repeated three times for a total of 20 roots per condition.
Cell Length Measurement and Confocal Microscopy
To measure cell length, whole roots were first stained with propidium iodide (Sigma-Aldrich) followed by washing in sterile water twice, then imaged with a Zeiss LSM 510 confocal microscope. Epidermal cell length was averaged from at least 10 cells per root at a distance of 1 cm from the root tips from at least five roots examined for each treatment. Cell length was measured using Zeiss LSM 510 software.
qRT-PCR
Total RNA was extracted from whole roots using the RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s protocol. Each treatment in all experiments was from 15 to 20 pooled roots; all experiments were repeated at least three times. RNA was quantified with a nanodrop and then followed with DNase treatment (Turbo DNase-free kit; Ambion). RNA was subsequently cleaned and concentrated with the Qiagen RNA Cleanup Kit. All RNAs were checked with an Agilent 2100 Bioanalyzer for good quality and integrity. Complementary DNA was synthesized from 1 μg of total RNA using the SuperScript III first-strand synthesis system (Life Technologies, Invitrogen). qRT-PCR was performed using the ABI StepOnePlus Real-Time PCR system (Applied Biosystems) using SYBR Green reagent (VWR). Data were collected with SDS 2.2 software (Applied Biosystems). Target gene relative expression was determined by normalizing with the geometric mean of the expression of two endogenous controls: UBIQUITIN CARRIER PROTEIN9 and PLANT DEFENSIN2 (Kakar et al., 2008). Statistical analysis was performed using one-way ANOVA in SPSS software (version 20.0.0) on the relative gene expression from three biological replicates. Primers used in qRT-PCR were designed using Primer Express version 3.0.1 software and are listed in Supplemental Table S1.
RNAi Vector Construction and Agrobacterium rhizogenes-Mediated Transformation
A 231-bp region of MtRbohC with low similarity to other MtRboh genes was amplified from wild-type root complementary DNA (RbohC RNAi_for, 5′-CACCATGGGAATTGATGAAATTGAAGA-3′; RbohC RNAi_rev, 5′-GCCAAGCAAATCAAGTTTCTC-3′). The purified PCR fragment was then cloned into the Gateway pENTR Directional TOPO Cloning Kit (Invitrogen) following the manufacturer’s instructions and recombined into the destination vector pK7GWIWG2(II)-RedRoot (Vlaams Instituut voor Biotechnologie, Plant Systems Biology, University of Ghent; Karimi et al., 2002; Op den Camp et al., 2011) to produce the RbohC RNAi vector pCZ42. The pCZ42 vector was transformed into A. rhizogenes strain ArquaI (Quandt et al., 1993) and then used to transform both A17 and latd M. truncatula roots (Limpens et al., 2004). As a control, some plants of each genotype were transformed with a GUS RNAi construct. After 2 weeks, transgenic roots were identified by screening for DsRed fluorescence using a 525-nm long-pass emission filter on a Nikon TE200 microscope. All untransformed roots and all transformed roots but one were cut off the composite plants. Composite plants containing untransformed shoots and a single transformed root were then transferred to growth pouches containing BNM, placed in Styrofoam boxes covered with a plastic dome, and sealed with packing tape to maintain humidity. The boxes were placed in the growth chambers with settings as described above. Plants were grown for 10 d, and the medium was replenished once during this period. At 10 d, roots were examined with the fluorescence microscope again to confirm DsRed fluorescence before recording the final root growth phenotype. Experiments were done three times, for a total of 25 to 37 composite plants of each genotype/vector combination over the course of the four experiments.
The sequences for genes used in this article can be found under the following accession numbers: MtLATD/NIP (Medtr1g009200), MtRbohA (Medtr1g083290), MtRbohB (Medtr3g098380), MtRbohC (Medtr3g098350), MtRbohD (Medtr3g098320), MtRbohE (Medtr8g095520), MtRbohF (Medtr7g060540), MtRbohG (Medtr7g113130), ApoCu/ZnSOD (Medtr6g029200), CytoCu/ZnSOD (Medtr7g114240), cwPRX2 (Medtr2g084000), cAPX (Medtr4g061140), GST (Medtr1g026140), AOX (Medtr5g026620), FeSOD (Medtr1g048990), RIP1 (Medtr5g074860), UBC9 (Medtr7g116940), and PDF2 (Medtr6g084690).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Seven-day-old wild-type lateral roots at different developmental stages stained for O2− with NBT.
Supplemental Figure S2. The nip-1 allele of the LATD/NIP gene has increased O2.− levels.
Supplemental Figure S3. ABA decreases O2.− levels in nip-1 roots, as it does for latd roots.
Supplemental Figure S4. DAB staining of H2O2 in 5-d-old wild-type and latd roots grown with and without 10 μm ABA.
Supplemental Figure S5. Relative expression of FeSOD and a cytoplasmic SOD (cytoSOD) in 7-d-old wild-type and latd mutant roots with and without 10 μm ABA for 24 h.
Supplemental Figure S6. Gene expression analysis for other ROS-related antioxidant genes, AOX, cAPX, and GST, in 7-d-old wild-type and latd mutant roots with and without 10 μm ABA for 24 h.
Supplemental Table S1. Primers used in qRT-PCR.
Supplementary Material
Acknowledgments
We thank Marilyn Wadsworth (University of Vermont Microscopy Imaging Center) for advice and training on confocal microscopy; Alan Howard (University of Vermont Statistical Consulting Clinic) for advice on statistical analysis; Jill Preston (University of Vermont) for use of the qRT-PCR machine; and Mary Tierney, Philip Lintilhac, Nick Heintz (University of Vermont), Li Zuo (Ohio State University), and members of our laboratory for suggestions and comments on the project.
Glossary
- ABA
abscisic acid
- RAM
root apical meristem
- ROS
reactive oxygen species
- H2O2
hydrogen peroxide
- SOD
superoxide dismutase
- O2.−
superoxide
- DPI
diphenylene iodonium
- RNAi
RNA interference
- NBT
nitroblue tetrazolium
- DAB
3,3′-diaminobenzidine
- H2DCFDA
dihydrofluorescein diacetate
- qRT
quantitative reverse transcription
- BNM
buffered nodulation medium
- DMSO
dimethyl sulfoxide
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–0920096 to J.M.H.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Renou JP, Berthomé R, Harberd NP, Genschik P. (2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Almagro L, Gómez Ros LV, Belchi-Navarro S, Bru R, Ros Barceló A, Pedreño MA. (2009) Class III peroxidases in plant defence reactions. J Exp Bot 60: 377–390 [DOI] [PubMed] [Google Scholar]
- Alscher RG, Erturk N, Heath LS. (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53: 1331–1341 [PubMed] [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Arthikala MK, Montiel J, Nava N, Santana O, Sánchez-López R, Cárdenas L, Quinto C. (2013) PvRbohB negatively regulates Rhizophagus irregularis colonization in Phaseolus vulgaris. Plant Cell Physiol 54: 1391–1402 [DOI] [PubMed] [Google Scholar]
- Asensio AC, Gil-Monreal M, Pires L, Gogorcena Y, Aparicio-Tejo PM, Moran JF. (2012) Two Fe-superoxide dismutase families respond differently to stress and senescence in legumes. J Plant Physiol 169: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Bagchi R, Salehin M, Adeyemo OS, Salazar C, Shulaev V, Sherrier DJ, Dickstein R. (2012) Functional assessment of the Medicago truncatula NIP/LATD protein demonstrates that it is a high-affinity nitrate transporter. Plant Physiol 160: 906–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Benfey PN, Bennett M, Schiefelbein J. (2010) Getting to the root of plant biology: impact of the Arabidopsis genome sequence on root research. Plant J 61: 992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright LJ, Liang Y, Mitchell DM, Harris JM. (2005) The LATD gene of Medicago truncatula is required for both nodule and root development. Mol Plant Microbe Interact 18: 521–532 [DOI] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb C. (1994) Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell 6: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JC, Seeley KA, Sung ZR. (1995) RML1 and RML2, Arabidopsis genes required for cell proliferation at the root tip. Plant Physiol 107: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Tullio MC, Jiang K, Feldman LJ. (2010) Redox regulation of root apical meristem organization: connecting root development to its environment. Plant Physiol Biochem 48: 328–336 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR. (1992) Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256: 998–1000 [DOI] [PubMed] [Google Scholar]
- Fester T, Hause G. (2005) Accumulation of reactive oxygen species in arbuscular mycorrhizal roots. Mycorrhiza 15: 373–379 [DOI] [PubMed] [Google Scholar]
- Filippou P, Antoniou C, Fotopoulos V. (2011) Effect of drought and rewatering on the cellular status and antioxidant response of Medicago truncatula plants. Plant Signal Behav 6: 270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Fry SC. (1998) Oxidative scission of plant cell wall polysaccharides by ascorbate-induced hydroxyl radicals. Biochem J 332: 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper C, Dolan L. (2006) Control of plant development by reactive oxygen species. Plant Physiol 141: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno-Gilles C, Lelièvre E, Viau L, Malik-Ghulam M, Ricoult C, Niebel A, Leduc N, Limami AM. (2009) ABA-mediated inhibition of germination is related to the inhibition of genes encoding cell-wall biosynthetic and architecture: modifying enzymes and structural proteins in Medicago truncatula embryo axis. Mol Plant 2: 108–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan LM, Zhao J, Scandalios JG. (2000) Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J 22: 87–95 [DOI] [PubMed] [Google Scholar]
- He J, Benedito VA, Wang M, Murray JD, Zhao PX, Tang Y, Udvardi MK. (2009) The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10: 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Duan Y, Hua D, Fan G, Wang L, Liu Y, Chen Z, Han L, Qu LJ, Gong Z. (2012) DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 24: 1815–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannat R, Uraji M, Morofuji M, Islam MM, Bloom RE, Nakamura Y, McClung CR, Schroeder JI, Mori IC, Murata Y. (2011) Roles of intracellular hydrogen peroxide accumulation in abscisic acid signaling in Arabidopsis guard cells. J Plant Physiol 168: 1919–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Zhang J. (2001) Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol 42: 1265–1273 [DOI] [PubMed] [Google Scholar]
- Jiang M, Zhang J. (2002) Involvement of plasma-membrane NADPH oxidase in abscisic acid- and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 215: 1022–1030 [DOI] [PubMed] [Google Scholar]
- Kakar K, Wandrey M, Czechowski T, Gaertner T, Scheible WR, Stitt M, Torres-Jerez I, Xiao Y, Redman JC, Wu HC, et al. (2008) A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4: 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M. (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109: 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kawahara TM, Namba H, Toyoda K, Kasai T, Sugimoto M, Inagaki Y, Ichinose Y, Shiraishi T. (2006) Induction of defense responses in pea tissues by inorganic phosphate. J Gen Plant Pathol 72: 129–136 [Google Scholar]
- Kim HJ, Kato N, Kim S, Triplett B. (2008) Cu/Zn superoxide dismutases in developing cotton fibers: evidence for an extracellular form. Planta 228: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JDG, Schroeder JI. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N. (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412 [DOI] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, et al. (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Liang Y, Harris JM. (2005) Response of root branching to abscisic acid is correlated with nodule formation both in legumes and nonlegumes. Am J Bot 92: 1675–1683 [DOI] [PubMed] [Google Scholar]
- Liang Y, Mitchell DM, Harris JM. (2007) Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Dev Biol 304: 297–307 [DOI] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R. (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH. (2007) Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol 143: 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar DP, Haridas S, Gantt JS, VandenBosch KA. (2007) A transient decrease in reactive oxygen species in roots leads to root hair deformation in the legume-rhizobia symbiosis. New Phytol 173: 39–49 [DOI] [PubMed] [Google Scholar]
- Macovei A, Balestrazzi A, Confalonieri M, Buttafava A, Carbonera D. (2011) The TFIIS and TFIIS-like genes from Medicago truncatula are involved in oxidative stress response. Gene 470: 20–30 [DOI] [PubMed] [Google Scholar]
- Marino D, Andrio E, Danchin EG, Oger E, Gucciardo S, Lambert A, Puppo A, Pauly N. (2011) A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol 189: 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Messerli MA, Shi C, Gilroy S. (2007) Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc Natl Acad Sci USA 104: 20996–21001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel J, Arthikala MK, Quinto C. (2013) Phaseolus vulgaris RbohB functions in lateral root development. Plant Signal Behav 8: e22694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel J, Nava N, Cárdenas L, Sánchez-López R, Arthikala MK, Santana O, Sánchez F, Quinto C. (2012) A Phaseolus vulgaris NADPH oxidase gene is required for root infection by rhizobia. Plant Cell Physiol 53: 1751–1767 [DOI] [PubMed] [Google Scholar]
- Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, Ammiraju JSS, Kudrna D, Wing R, Untergasser A, et al. (2011) LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science 331: 909–912 [DOI] [PubMed] [Google Scholar]
- Pilet PE. (1975) Abscisic acid as a root growth inhibitor: physiological analyses. Planta 122: 299–302 [DOI] [PubMed] [Google Scholar]
- Quandt H, Pühler A, Broer I. (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Ramel F, Sulmon C, Bogard M, Couée I, Gouesbet G. (2009) Differential patterns of reactive oxygen species and antioxidative mechanisms during atrazine injury and sucrose-induced tolerance in Arabidopsis thaliana plantlets. BMC Plant Biol 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu SK, Peng HM, Cook DR. (2002) Nod factor induction of reactive oxygen species production is correlated with expression of the early nodulin gene rip1 in Medicago truncatula. Mol Plant Microbe Interact 15: 522–528 [DOI] [PubMed] [Google Scholar]
- Rong Y, Doctrow SR, Tocco G, Baudry M. (1999) EUK-134, a synthetic superoxide dismutase and catalase mimetic, prevents oxidative stress and attenuates kainate-induced neuropathology. Proc Natl Acad Sci USA 96: 9897–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MC, James EK, Clemente MR, Bucciarelli B, Fedorova M, Vance CP, Becana M. (2004) Localization of superoxide dismutases and hydrogen peroxide in legume root nodules. Mol Plant Microbe Interact 17: 1294–1305 [DOI] [PubMed] [Google Scholar]
- Sagi M, Fluhr R. (2006) Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol 141: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P. (1996) Hydrogen peroxide-mediated cell-wall stiffening in vitro in maize coleoptiles. Planta 199: 43–49 [Google Scholar]
- Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT. (2004) Root growth maintenance during water deficits: physiology to functional genomics. J Exp Bot 55: 2343–2351 [DOI] [PubMed] [Google Scholar]
- Sharp RE, Wu Y, Voetberg GS, Saab IN, LeNoble ME. (1994) Confirmation that abscisic acid accumulation is required for maize primary root elongation at low water potentials. J Exp Bot 45: 1743–1751 [Google Scholar]
- Slovak R, Göschl C, Su X, Shimotani K, Shiina T, Busch W. (2014) A scalable open-source pipeline for large-scale root phenotyping of Arabidopsis. Plant Cell 26: 2390–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. (2011) Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol 14: 691–699 [DOI] [PubMed] [Google Scholar]
- Swanson S, Gilroy S. (2010) ROS in plant development. Physiol Plant 138: 384–392 [DOI] [PubMed] [Google Scholar]
- Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L. (2008) Local positive feedback regulation determines cell shape in root hair cells. Science 319: 1241–1244 [DOI] [PubMed] [Google Scholar]
- Teillet A, Garcia J, de Billy F, Gherardi M, Huguet T, Barker DG, de Carvalho-Niebel F, Journet EP. (2008) api, a novel Medicago truncatula symbiotic mutant impaired in nodule primordium invasion. Mol Plant Microbe Interact 21: 535–546 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Swarup R, Coates J, Swarup K, Laplaze L, Beemster GT, Hedden P, Bhalerao R, Bennett MJ. (2008) Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol 10: 625–628 [DOI] [PubMed] [Google Scholar]
- Veereshlingam H, Haynes JG, Penmetsa RV, Cook DR, Sherrier DJ, Dickstein R. (2004) nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol 136: 3692–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inzé D, et al. (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Song CP. (2008) Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol 178: 703–718 [DOI] [PubMed] [Google Scholar]
- Yendrek CR, Lee YC, Morris V, Liang Y, Pislariu CI, Burkart G, Meckfessel MH, Salehin M, Kessler H, Wessler H, et al. (2010) A putative transporter is essential for integrating nutrient and hormone signaling with lateral root growth and nodule development in Medicago truncatula. Plant J 62: 100–112 [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG. (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang H, Han W, De Smet I, Talboys P, Loya R, Hassan A, Rong H, Jürgens G, Knox JP, Wang MH. (2010) ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J 64: 764–774 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang L, Dong F, Gao J, Galbraith DW, Song CP. (2001) Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol 126: 1438–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.