Pi deprivation shifts the subcellular distribution of two key brassinosteroid-related transcriptional effectors to trigger shallower root system architecture.
Abstract
Plants feature remarkable developmental plasticity, enabling them to respond to and cope with environmental cues, such as limited availability of phosphate, an essential macronutrient for all organisms. Under this condition, Arabidopsis (Arabidopsis thaliana) roots undergo striking morphological changes, including exhaustion of the primary meristem, impaired unidirectional cell expansion, and elevated density of lateral roots, resulting in shallow root architecture. Here, we show that the activity of two homologous brassinosteroid (BR) transcriptional effectors, BRASSINAZOLE RESISTANT1 (BZR1) and BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1 (BES1)/BZR2, blocks these responses, consequently maintaining normal root development under low phosphate conditions without impacting phosphate homeostasis. We show that phosphate deprivation shifts the intracellular localization of BES1/BZR2 to yield a lower nucleus-to-cytoplasm ratio, whereas replenishing the phosphate supply reverses this ratio within hours. Phosphate deprivation reduces the expression levels of BR biosynthesis genes and the accumulation of the bioactive BR 28-norcastasterone. In agreement, low and high BR levels sensitize and desensitize root response to this adverse condition, respectively. Hence, we propose that the environmentally controlled developmental switch from deep to shallow root architecture involves reductions in BZR1 and BES1/BZR2 levels in the nucleus, which likely play key roles in plant adaptation to phosphate-deficient environments.
Plant performance depends on its ability to remodel its growth and development in response to changes in nutrient availability in the rhizosphere. Phosphorus is an essential macronutrient that is acquired through the root system as inorganic phosphate (Pi; also known as orthophosphate) from the soil. In many soils, soluble Pi levels are suboptimal for plant growth and productivity, mainly because of its slow diffusion, its high chemical reactivity that subsequently leads to formation of insoluble complexes, and its conversion to organic forms by soil bacteria (Vance et al., 2003). In response to low Pi availability, plants reshape their root system architecture (RSA) from a deeper to a shallower and broader structure. This developmental switch is thought to maximize Pi exploitation, because Pi is mainly present in the upper layers of the soil (López-Arredondo et al., 2014). In Arabidopsis (Arabidopsis thaliana), developmental reprogramming in response to low Pi leads to inhibition of primary root growth by unknown mechanisms. Subsequently, meristem exhaustion and inhibition of unidirectional cell expansion are observed alongside increased lateral root (LR) and root hair density and length, which together result in a shallower RSA (Sánchez-Calderón et al., 2005; Abel, 2011; Chiou and Lin, 2011; Péret et al., 2011; Niu et al., 2013; Zhang et al., 2014). Other cellular and physiological changes include increased expression of Pi transporter genes, activation of secreted acid phosphatases (APases), and accumulation of anthocyanin in leaves.
Plants respond to changes in Pi levels by deploying interconnected local and systemic signaling pathways that are triggered by direct contact with external phosphate and the overall Pi homeostasis of the plant, respectively (Chiou and Lin, 2011). The systemic signaling pathway controls Pi homeostasis alongside specific aspects of the root development response, whereas the local pathway controls the aforementioned changes in RSA independent of the internal phosphate content. Hence, low Pi is locally sensed by the root tip; a potential sensor candidate has been recently proposed (Rouached et al., 2011). A number of downstream components have been shown to mediate the RSA alterations triggered by local Pi sensing. They include the interacting P5-type ATPase called Phosphate Deficiency Response2 (PDR2; Ticconi et al., 2004) and Low Phosphate Root1 (LPR1) and LPR2 (Svistoonoff et al., 2007; Ticconi et al., 2009). Loss of function of PDR2 confers enhanced meristem exhaustion in response to Pi deficiency, whereas the lpr1;lpr2 double mutant has longer roots compared with the wild type. Hormonal activity has also been implicated in different developmental aspects of this response with promotive (e.g. auxin and ethylene) and repressive (e.g. GAs) effects (Chiou and Lin, 2011; Zhang et al., 2014). Plant response to low Pi levels involves transcriptional activation of many phosphate starvation-induced genes that are controlled by transcription factors of different families. However, thus far, they have not been implicated in root meristem response to low Pi.
The brassinosteroid (BR) signaling pathway regulates numerous physiological and developmental processes. The cascade is triggered on BR binding to the BRASSINOSTEROID INSENSITIVE1 (BRI1) receptor at the cell surface followed by a series of signaling events, leading to inhibition of the Glycogen synthase kinase-3 BRASSINOSTEROID INSENSITIVE2 (BIN2; Clouse, 2011). When BR levels are low, BIN2 levels rise, and the protein phosphorylates and inactivates the two key transcription factor homologs BRASSINAZOLE RESISTANT1 (BZR1) and BRI1-ETHYL METHANESULFONATE-SUPPRESSOR1 (BES1)/BZR2 (Wang et al., 2002; Yin et al., 2002). In response to high BRs, BZR1 and BES1 are dephosphorylated by Protein phosphatase 2A (Tang et al., 2011), which renders them active; this enhances their stability and accumulation in the nucleus, where they homodimerize or heterodimerize and finally, bind DNA at defined known cis-elements, leading to regulation of the expression of many genes (He et al., 2005; Yin et al., 2005; Sun et al., 2010; Yu et al., 2011). BZR1 and BES1/BZR2 are plant-specific highly homologous transcription factors that bear both genetically redundant and unique roles (Wang et al., 2002; Yin et al., 2002, 2005). Dominant mutations (bes1-D and bzr1-D) promote a hypophosphorylated state, resulting in their constitutive activity and consequential suppression of different phenotypic abnormalities in BR-insensitive and BR-deficient mutants (Wang et al., 2002; Yin et al., 2002; Tang et al., 2011). In agreement, their constitutive activity confers resistance to the compound brassinazole (BRZ), a specific inhibitor of the BR biosynthesis enzyme DWARF4 (DWF4).
In roots, BRs have both promoting and inhibitory effects on growth depending on the intensity of the signal (Fridman and Savaldi-Goldstein, 2013); bri mutants (e.g. bri1) are dwarf, bearing extremely short roots, a result of longer cell cycle duration and reduced cell elongation (González-García et al., 2011; Hacham et al., 2011). Conversely, BR-treated roots are inhibited because of premature cell exit from the cell cycle, which accounts for the slightly shorter roots in bes1-D plants compared with the wild type (González-García et al., 2011). Enhanced BR signaling triggered by impaired spatial distribution of BRI1 also limits unidirectional cell expansion and whole-root growth (Fridman et al., 2014).
Here, we considered whether BR regulation of root growth is responsive to environmental cues and found that BES1/BZR2 and BZR1 block plant responses to Pi deprivation. Plants expressing bes1/bzr2-D or bzr1-D failed to modulate virtually all aspects of root adaptation to adverse Pi conditions, which manifested as largely indistinguishable root meristem, cell morphology, and LR density regardless of whether these plants were grown under adequate or low Pi conditions. Other known physiological responses, such as anthocyanin accumulation in the shoot and APase activity, were similarly largely indistinguishable. This dramatic arrest of a central developmental switch occurred despite normal sensing the Pi deficiency by bes1/bzr2-D and bzr1-D roots, which was measured by their Pi content and the response of Pi-starvation responsive genes. We show that low Pi conditions shift the localization of BES1 from the nucleus to the cytoplasm, whereas adequate Pi conditions trigger its relocalization in the nucleus. Thus, subcompartmentalization shifts of the key signaling factor dictate the plant response to environmental cues, providing unique insights into developmental plasticity in plants.
RESULTS
BZR1 and BES1 Activity Confers Resistance to Low Pi Availability
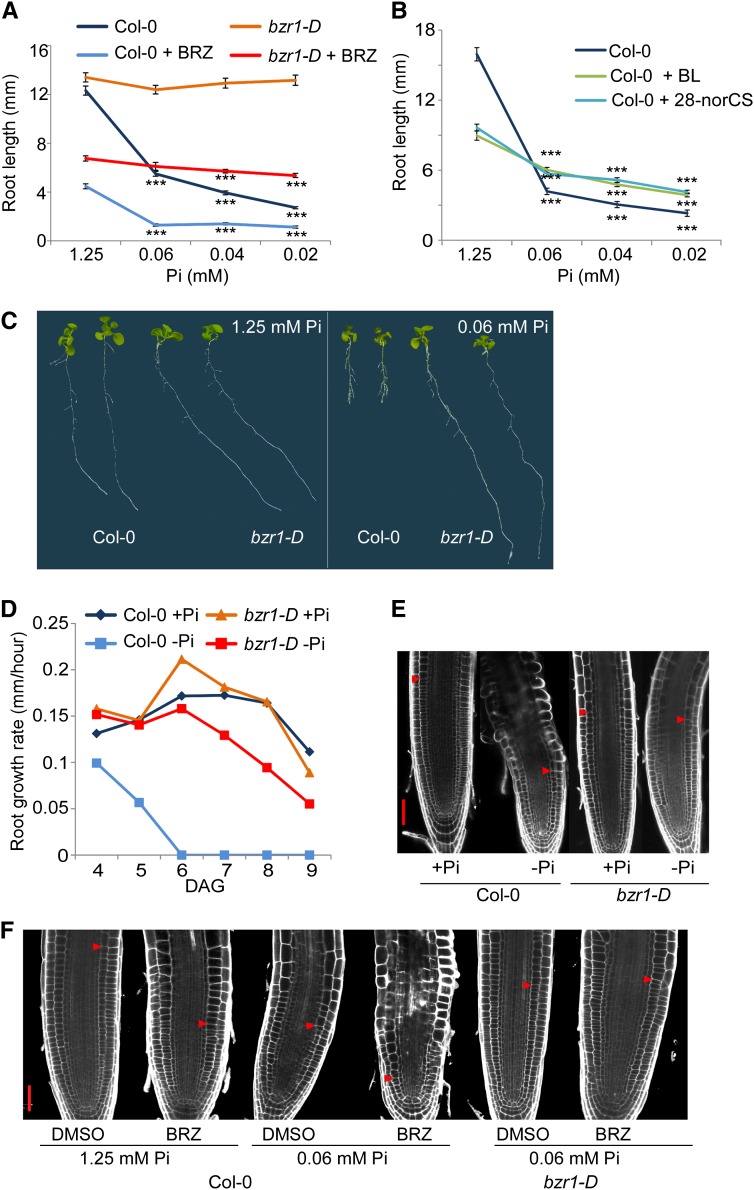
To assess whether BR-mediated root growth is modulated by environmental signals, we performed a root sensitivity assay in response to decreasing Pi availability in the medium and found a clear association with BES1 and BZR1 activity (Fig. 1A; Supplemental Fig. S1). In wild-type plants, primary root growth was severely inhibited when available Pi concentration was reduced to 60 μm (Fig. 1, A and B). Strikingly, roots of the bzr1-D mutants remained long at the lowest tested Pi concentration (20 μm; Fig. 1, A and C). By contrast, root response to increasing salt concentrations revealed no correlation with these genes, suggesting their specific involvement in response to low Pi stress only (Supplemental Fig. S1D). Application of the BR biosynthesis inhibitor BRZ to wild-type plants enhanced root growth inhibition, reaching full inhibition at 60 μm Pi. Under adequate Pi conditions, bzr1-D roots were inhibited by the drug but to a lesser extent compared with the wild type, which is in agreement with the constitutive activity of the mutant, even in the absence of BRs. Furthermore, BRZ treatment did not affect bzr1-D root insensitivity to low Pi (Fig. 1A).
Figure 1.
BZR1 activity confers root insensitivity to Pi deprivation. A, Root sensitivity to decreasing concentrations of Pi; root lengths of wild-type seedlings (Col-0) and bzr1-D grown in the presence or absence of the BR biosynthesis inhibitor BRZ are shown. Error bars represent se. ***, P < 0.001 with two-tailed Student's t test. B, Root sensitivity to decreasing concentrations of Pi; root length of wild-type (Col-0) seedlings grown in the absence and presence of 1 nm BL and 20 nm 28-norCS. Error bars represent se. ***, P < 0.001 with two-tailed Student's t test. C, Phenotype of Col-0 and bzr1-D plants grown in adequate versus low Pi (60 μm) conditions. D, Root growth rate of wild-type (Col-0) and bzr1-D plants grown in the presence or absence of Pi (1 μm). DAG, Days after germination. E and F, Confocal microscopy image of Col-0 and bzr1-D root meristems. E, Roots of seedlings grown as in D. Arrows indicate the transition zone. Bar = 20 μm. F, Roots of seedlings grown in 60 μm Pi as in C. Bar = 40 μm.
To test whether BES1/BZR2 (hereafter BES1) activity also blocks root response to low phosphate availability, we established plant lines expressing an equivalently stabilized, constitutively active variant of the protein (POLYUBIQUITIN10 [pUBQ10]-bes1-D). Like bzr1-D, pUBQ10-bes1-D roots remained remarkably longer, even when Pi was almost fully depleted (1 μm; Supplemental Fig. S1, B and C). Quantification of root growth and RSA in medium with depleted Pi showed a rapid deceleration in the growth rate of wild-type roots within 1 d of seedling transfer to low Pi medium and complete growth arrest within 3 d (Fig. 1D). By contrast, bzr1-D roots exhibited an only slightly lower growth rate after 4 d of seedling transfer to low Pi medium (Fig. 1D). Root growth arrest in response to low Pi is a result of reduced meristematic activity and unidirectional cell expansion. Cellular analysis revealed that both growth parameters remained largely unaffected in bzr1-D as opposed to wild-type plants (Fig. 1E). In agreement, the small meristem size observed in response to 60 μm Pi was even smaller by BRZ in the wild type but remained largely unaffected in the presence of bzr1-D (Fig. 1, A and F).
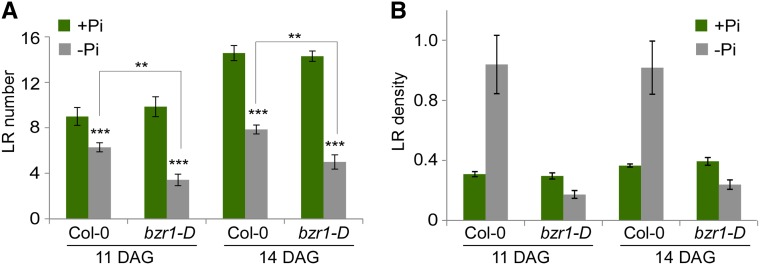
LR densities on days 8 and 11 from plant exposure to low Pi levels were calculated when seedlings were 11 and 14 d old, respectively, by dividing the total number of LRs by the root length (Fig. 2). Under high Pi conditions, no differences in LR density between the wild type and bzr1-D were observed. However, in response to low Pi levels, wild-type roots exhibited elevated LR density, whereas that of bzr1-D was reduced. Taken together, these data show that reduced BR levels promote root response to low Pi availability, whereas constitutive BZR1 and BES1/BZR2 activity confers significant insensitivity to this cue, resulting in plants with unaffected RSA, despite the severe conditions.
Figure 2.
BZR1 activity blocks RSA modulation in response to Pi deprivation. A, Total number of LRs in the wild type and bzr1-D on days 11 and 14 after germination when grown in the presence or absence of Pi (1 μm). B, LR density in these same lines. Note the high versus low LR density in the wild type and bzr1-D, respectively, in Pi deprivation conditions. Error bars represent se. **, P < 0.01 with two-tailed Student's t test. ***, P < 0.001 with two-tailed Student's t test. DAG, Days after germination.
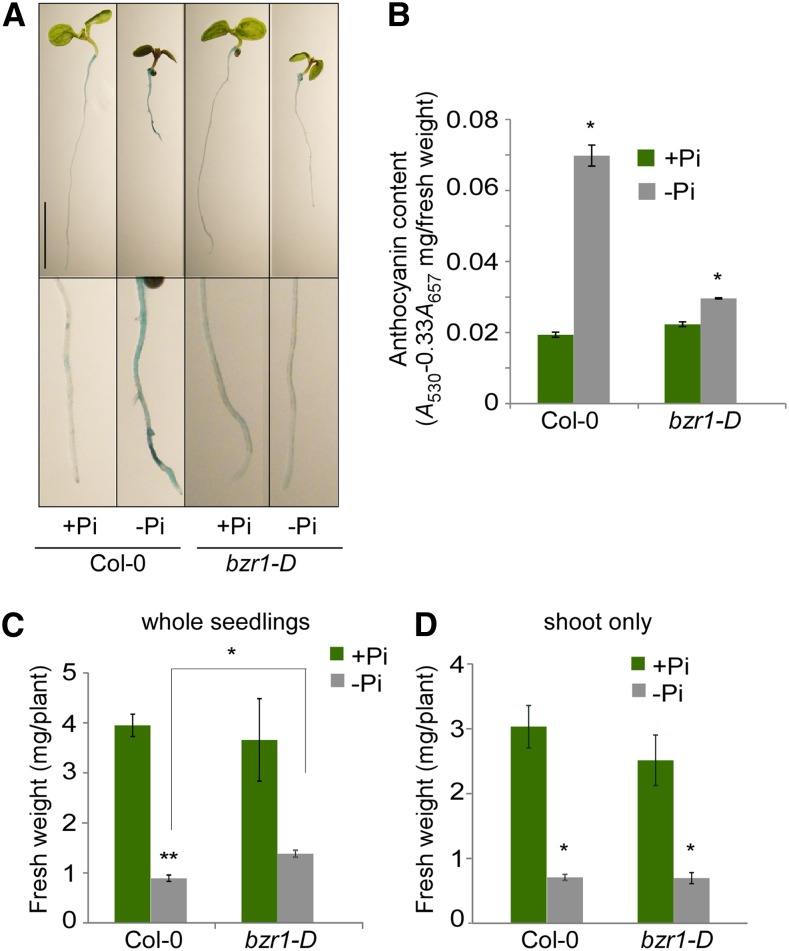
Pi deprivation stimulates additional physiological responses, including elevation of APase activity and anthocyanin accumulation in the shoot. Thus, we chose to evaluate the impact of constitutive BZR1 activity on these responses. In a qualitative in vivo staining assay performed to assess APase activity (Fig. 3A), high staining accumulation in wild-type roots grown under Pi deprivation (1 μm) conditions indicated that APase activity was triggered as expected. By contrast, no conditional stimulation of APase activity was observed in bzr1-D roots, which maintained low staining under both high and low Pi conditions (Fig. 3A). A similar lack of conditional APase activity stimulation was observed in pUBQ10-BES1-D plants, which was expected from their common RSA phenotype reported above, whereas pUBQ10-BES1 plants showed a normal response (Supplemental Fig. S2).
Figure 3.
Pi deprivation-triggered secreted APase activity, anthocyanin accumulation, and reduction of seedling fresh weight are impaired by BZR1. A, In vivo root APase staining. Blue precipitates on the root surface represent APase cleavage of exogenously applied 5-bromo-4-chloro-3-indolyl phosphate substrate. Note the low staining in bzr1-D roots compared with the wild type under Pi-deprived conditions (1 μm). Bar = 5 mm. B, Anthocyanin content. C and D, Fresh weight of the wild type and bzr1-D grown as under Pi-deprived conditions. C, Fresh weigh of shoots and roots. D, Fresh weigh of shoots of the same seedlings used in C. Note that, under depleted Pi conditions, roots contribute to the increase in fresh weight of bzr1-D in contrast to the wild type. Error bars represent se. *, P < 0.05 with two-tailed Student's t test. **, P < 0.01 with two-tailed Student's t test.
When grown on low Pi medium, the wild-type and pUBQ10-BES1 cotyledons had a dark shade, whereas those of bzr1-D remained fairly green, suggesting high accumulation of anthocyanin in the former (Fig. 3A; Supplemental Fig. S2). Quantification of anthocyanin content revealed that their levels were dramatically increased in response to low Pi conditions, whereas only a small elevation was detected in bzr1-D seedlings (Fig. 3B). Finally, the typical loss in fresh weight of wild-type seedlings grown under low Pi conditions was less severe in bzr1-D whole seedlings, which had an overall higher mean fresh weight compared with the wild type in the absence of Pi (1 μm) and under low (60 μm) Pi conditions (Fig. 3C; Supplemental Table S1). Comparing the fresh weight of shoots only revealed a similar reduction between wild-type and bzr1-D shoots, suggesting roots as the major unaffected organ under Pi-depleted conditions (Fig. 3D).
Transcriptional Activation of Phosphate Starvation-Induced Genes and Endogenous Phosphate Content Are Unaffected by BZR1 and BES1 Activity
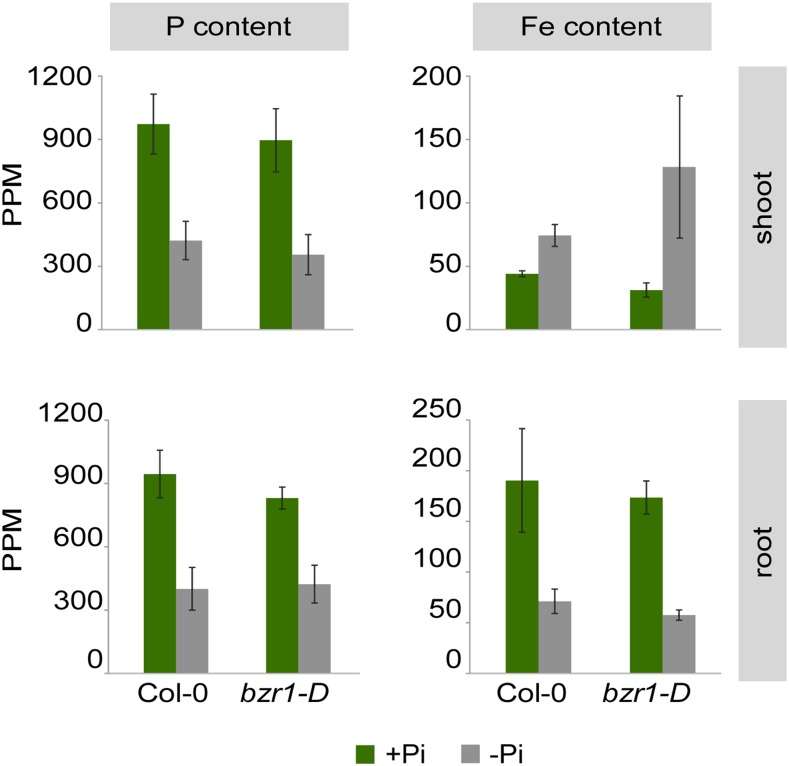
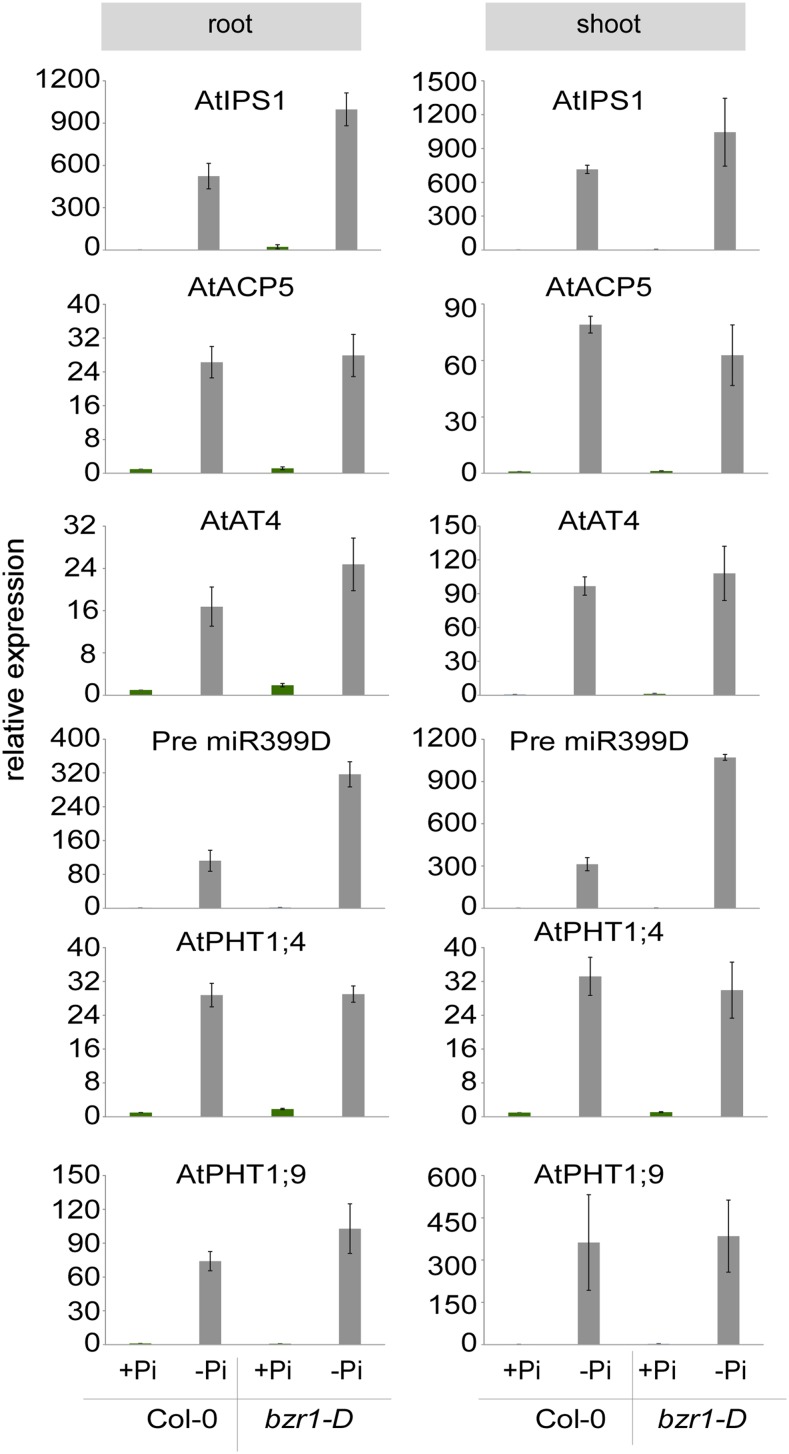
BZR1 and BES1 activity may block RSA response to low Pi by elevating endogenous phosphorus content and decreasing iron levels that would potentially compensate for low Pi (Svistoonoff et al., 2007). To test this hypothesis, we performed an Inductively Coupled Plasma (ICP) analysis and quantified and compared phosphorus and iron content in plants grown under adequate versus deprived Pi conditions (Fig. 4). No significant differences in phosphorus content were detected between wild-type and bzr1-D lines. We also noted a seemingly opposing change in iron content between root and shoot, with a reduction and increase in response to low Pi availability in both wild-type and bzr1-D plants, respectively. Thus, the internal change in iron content did not coincide with impaired developmental and physiological response to low Pi in bzr1-D plants. We next asked whether the normal drop in endogenous phosphorus content in bzr1-D plants is associated with a normal response of phosphate starvation-induced genes. To this end, we analyzed the relative expression of known phosphate starvation-induced genes in shoots and roots of wild-type and bzr1-D plants grown under adequate versus depleted Pi conditions (Fig. 5). In wild-type plants, transcripts corresponding to all phosphate starvation-induced genes tested were elevated as expected. Interestingly, normal stimulation of these genes was also observed in bzr1-D plants (Fig. 5). Taken together, the physiological and developmental insensitivity to low Pi levels is a result of BZR1 and BES1 acting independently or downstream of a low Pi-sensing pathway, which triggers phosphate starvation-induced gene activity.
Figure 4.
BZR1 activity does not impact phosphorus and iron homeostasis. ICP analysis to determine phosphorus and iron content in roots and shoots of the wild type and bzr1-D grown in the presence or absence of Pi (1 μm). Error bars represent se. Values are averages of two biological replicates.
Figure 5.
Pi deprivation-mediated induction of phosphate starvation-induced genes is not affected by BZR1 activity. Analysis of relative expression of different phosphate starvation-induced genes in roots and shoots of wild-type and bzr1-D plants grown in the presence or absence of Pi (1 μm). Error bars represent se. IPS1, Induced by phosphate starvation1; ACP5, ACID PHOSPHATE TYPE5; miR399, microRNA 399; PHT, Phosphate transporter.
Pi Deprivation Reduces the Accumulation of Specific BRs
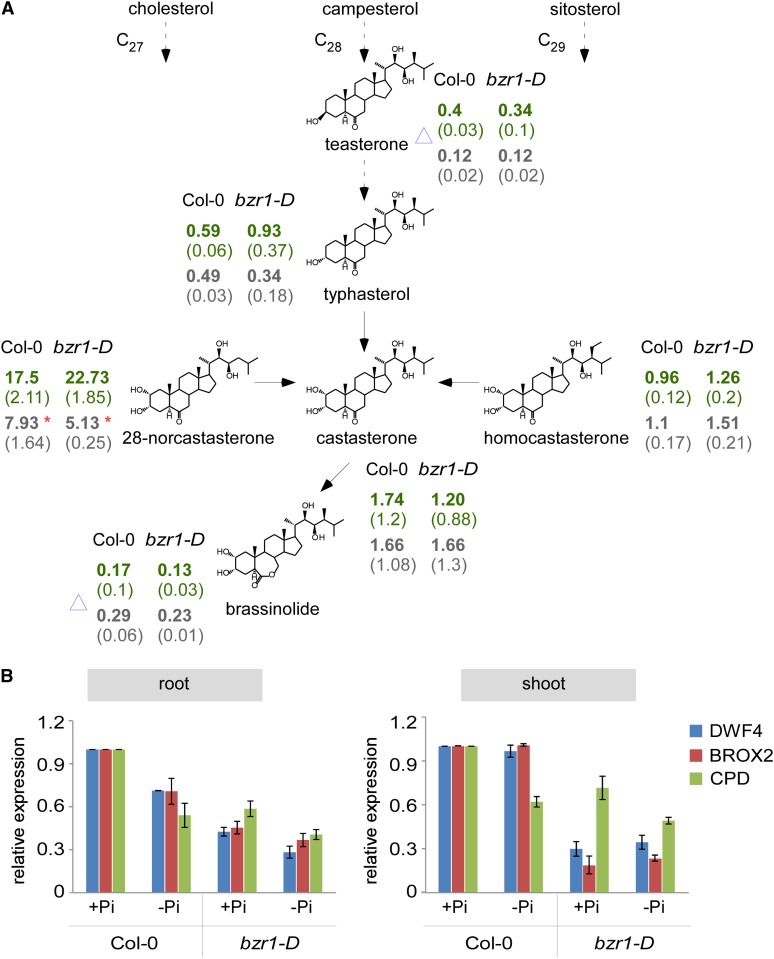
Based on the findings thus far, it was hypothesized that low Pi levels reduce BR activity and/or levels to promote the aforementioned developmental and physiological response. To address this hypothesis, the levels of various BR derivatives were analyzed. Detectable levels were measured for six molecules, including brassinolide (BL), the most active BR, and its immediate and indirect precursors castasterone (CS), 28-norCS, and homoCS (classified as C27, C28, and C29 BRs, respectively) in both wild-type and bzr1-D seedlings grown in the presence or absence of Pi (Fig. 6A; Supplemental Table S2). Mean 28-norCS levels were significantly reduced in both the wild type and the mutant, whereas no significant changes were observed in the levels of the other BR molecules (Fig. 6A). Teasterone and BL levels were marked as inconclusive (Fig. 6A, blue triangle), because some of the samples had values below detection and therefore, were considered noise (less than 0.1 pg mg−1 of tissue).
Figure 6.
Low Pi availability triggers a reduction of 28-norCS. A, Structures of detected BR molecules are shown and arranged according to their synthesis pathway. Numbers present the average levels (picograms per milligram) of BRs detected in whole seedlings of the wild type (Col-0) and bzr1-D grown under adequate (green) or deprived (gray) Pi (1 μm) conditions. Numbers in parentheses represent se. The blue triangles indicate that samples with measurements below detection (<0.1 pg mg−1) were also taken for average values (hence, considered noise). *, P < 0.05 with two-tailed Student's t test. B, Analysis of relative expression of BR biosynthesis genes in roots and shoots of wild-type and bzr1-D plants grown under adequate or deprived Pi (1 μm) conditions. Error bars represent se.
Both 28-norCS and CS are synthesized from their corresponding higher order precursors (cholesterol and campesterol, respectively) through mutual BR biosynthesis enzymes CONSTITUTIVE PHOTOMORPHOGENIC DWARF (CPD), DWF4, and BRASSINOSTEROID-6-OXIDASE2 (Yokota et al., 2001; Joo et al., 2012). The relative expression levels of these genes in wild-type shoot and roots (Fig. 6B) were moderately down-regulated in response to low Pi conditions, with all having lower expression in roots and only CPD changing in shoots. In bzr1-D plants, the expression levels of these genes were consistently lower compared with the wild type, which is in agreement with the known repressive activity of BZR1 on their expression; CPD and DWF4 (in roots) showed an even further reduction in response to low Pi conditions. Thus, low Pi availability reduces the levels of specific endogenous BR derivatives, likely by inhibiting the expression of BR biosynthesis genes.
To address whether 28-norCS desensitizes roots in response to low Pi availability, we performed a sensitivity assay in the absence and presence of this compound (Fig. 1B) and found that it desensitizes roots in a similar manner to BL, although at higher concentrations. Thus, 28-norCS is a bioactive BR or relevant BR precursor under these conditions.
Pi Deprivation Promotes Cytoplasmic Accumulation of BES1
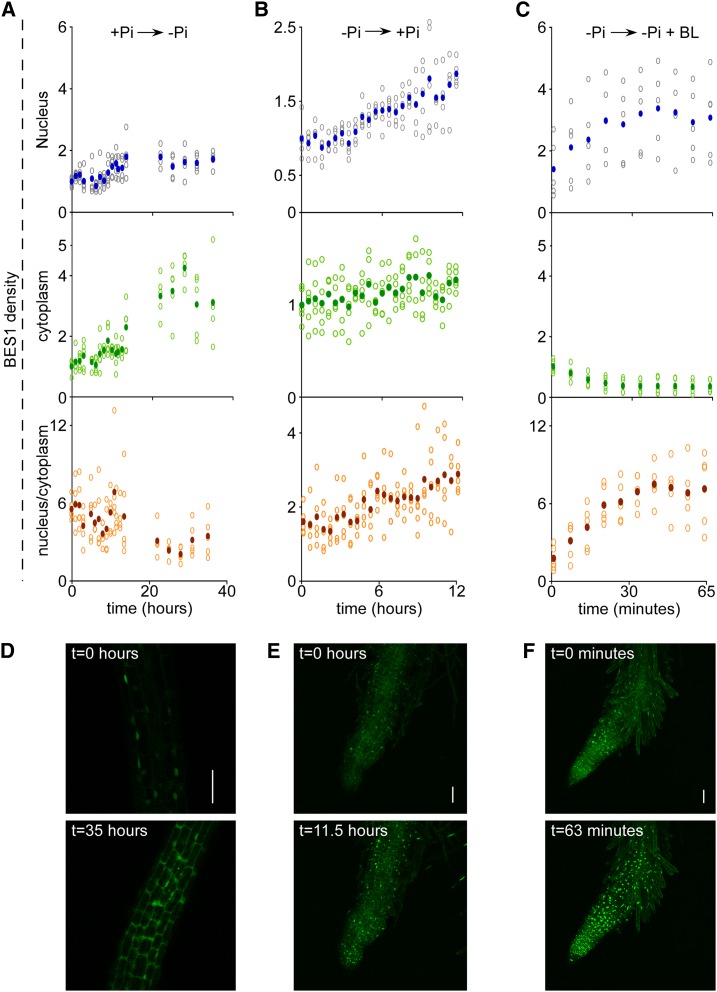
BR activates BZR1 and BES1 posttranslationally, enhancing their nuclear accumulation (Wang et al., 2002; Yin et al., 2002). To address whether low Pi-mediated reduction of BR levels is accompanied by a reduced nuclear-to-cytoplasmic ratio of the protein, we established pUBQ10-BES1-YPet (for Yellow fluorescent protein for energy transfer) lines for live imaging analysis and followed the BES1 signal in the same root immediately after its transfer from adequate Pi medium to Pi-deprived conditions for 35 h (Supplemental Movie S1). Quantification of average fluorescence signal in consecutive time intervals revealed initial overall elevation in both the nucleus and cytoplasm (Fig. 7A; Supplemental Fig. S3), likely a result of transcriptional activation driven by the promoter (data not shown). However, the cytoplasmic signal of BES1 was highly elevated compared with the nucleus, and it was noted after 11 h from transfer (Fig. 7, A and D). To address whether the shift in the subcellular localization of BES1 is reversible, pUBQ10-BES1-YPet seedlings were transferred from depleted to adequate phosphate levels (Fig. 7, B and E; Supplemental Fig. S3; Supplemental Movie S2), which resulted in a significant elevation in the nucleus and elevated nuclear-to-cytoplasmic ratio occurring within 7 h.
Figure 7.
Low Pi availability triggers cytoplasmic accumulation of BES1. A to C, Time course of the BES1 fluorescent signal measured in the nucleus and cytoplasm in the root elongation zone and the corresponding ratio (nucleus-cytoplasm) expressed as values relative to baseline. A, Fluorescent signal measured in the seedling transferred from adequate Pi medium to Pi-deprived conditions immediately before imaging. B, Fluorescent signal measured in the seedling transferred from Pi-deprived medium to adequate Pi conditions immediately before imaging. C, Fluorescent signal measured in the seedling transferred from Pi-deprived medium to the same medium supplemented with BL immediately before imaging. D to F, Snapshots of the BES1-YPet signal at selected time points (corresponding to A–C, respectively). D, A confocal section image. D and E, Maximal projection images to facilitate visualization of the aberrant root morphology. Bars = 50 μm.
To assess whether the cytoplasmic accumulation of the BES1 in response to deprived Pi conditions is BR regulated, we performed similar live-imaging analysis using seedlings that were transferred from Pi-deprived medium to Pi-deprived medium supplemented with 100 nm (Fig. 7, C and F; Supplemental Fig. S3; Supplemental Movie S3). Quantification of the fluorescence signal revealed rapid reversal of the low nuclear-to-cytoplasmic ratio within about 10 min that increased until reaching plateau within approximately 40 min.
DISCUSSION
Mechanisms controlling developmental plasticity in response to phosphate deprivation are poorly understood. Our study shows that the constitutive activity of two homologous transcription factors, BZR1 and BES1, impairs the developmental switch typically activated by low Pi availability. We propose that local sensing of Pi deprivation promotes their cytoplasmic accumulation, thus triggering the transcriptional reprogramming necessary to drive shallower RSA. Sensing adequate Pi concentrations restores their localization to the nucleus, thus providing an efficient modularity of development.
Local sensing of Pi availability is thought to trigger hormone-mediated modulation of RSA (Thibaud et al., 2010). In agreement with this model, constitutive BZR1 and BES1 activity blocks the RSA response to low Pi levels, despite the internal drop in phosphorus levels. This is further supported by a normal transcriptional response of phosphate starvation-induced genes associated with the systemic signal. Although the local sensing mechanism and its downstream signal transduction remain enigmatic, we hypothesize that a reduction in 28-norCS, mediated by a reduction in the BR biosynthesis gene expression shown here, could be one modality contributing to subcellular BZR1 changes. In agreement, our data show that low and high BR concentrations sensitize and desensitize root response, respectively, to low Pi. Accordingly, a previous transcriptome study comparing root response with local versus systemic Pi depletion revealed a 2-fold reduction in DWF4 transcript levels by the former pathway only (Thibaud et al., 2010). Interestingly, endogenous levels of CS and homoCS showed no change, but those of 28-norCS significantly dropped in response to low Pi conditions in both the wild type and bzr1-D. This suggests that the C27 branch of the BR biosynthesis pathway is targeted by defined environmental conditions. The complexity of the BR biosynthesis pathway may present a flexible means of modulating BR availability. Hence, both modulation of BR activity in response to low Pi availability and BR-mediated root growth inhibition might be context and tissue specific (Fridman and Savaldi-Goldstein, 2013). BL and CS are known to bind BRI1 at high and low affinities, respectively (Wang et al., 2001; Kinoshita et al., 2005; Hothorn et al., 2011; She et al., 2011). The binding affinity of 28-norCS to BRI1 is unknown. However, its high levels measured here, namely 20-fold higher than CS, likely compensate for a potentially lower affinity to its receptor under adequate Pi conditions.
Although high BR levels impaired root response to Pi depletion, they did not mimic the strong effect imposed by constitutively active BES1 and BZR1. Hence, we postulate that low Pi levels also trigger BR dose-independent mechanisms that control the activity of the transcription factors.
Various hormonal activities have been implicated in RSA modulation in response to low Pi availability. Of these, the GA-DELLA pathway is reminiscent of the BR-BZR1/BES1 response reported here (Jiang et al., 2007). In this model, low Pi conditions modulate the expression of GA biosynthesis genes, thereby leading to a reduction in internal GA content, which in turn, leads to DELLA accumulation and modulation of RSA without an apparent impact on phosphorus homeostasis (Jiang et al., 2007). It has been previously reported that DELLA proteins bind and inhibit BZR1/BES1 to promote photomorphogenesis (de Lucas et al., 2008; Bai et al., 2012; Gallego-Bartolomé et al., 2012; Li et al., 2012; Oh et al., 2012). Thus, the possibility that DELLA accumulation enhances plant response to low Pi availability by targeting BZR1/BES1 is an attractive hypothesis for future work.
In summary, the striking blockage of RSA modulation by BZR1 and BES1 in response to low Pi conditions uncovered here presents unique research avenues aimed to further understand how developmental reprogramming is achieved, and they can be leveraged toward potential biotechnology improvement of plant performance.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Chemical Treatments
All Arabidopsis (Arabidopsis thaliana) lines were in the Columbia-0 (Col-0) background. Seeds were sterilized as previously described (Fridman et al., 2014).
In all experiments, except for phosphorus and iron measurements, seedlings were initially germinated on one-half-strength Murashige and Skoog medium supplemented with 0.2% (w/v) Suc (Fridman et al., 2014), transferred after 3 d to either adequate (1.25 mm) or deficient (1 µm) Pi medium unless otherwise indicated, and grown at 22°C in continuous light (approximately 70 μmol m−2 s−1).
Phosphate-deficient medium was prepared by replacing KH2PO4 with equimolar KCl followed by pH adjustment to 5.8. Medium composition was modified from López-Bucio et al., 2002 and contained 10.2 mm NH4NO3, 9.4 mm KNO3, 2 mm CaCl2.2H2O, 0.625 mm MgSO4.7H2O, 0.0025 mm KI, 0.05 mm H3BO3, 0.0497 mm MnSO4_H2O, 0.024 mm ZnSO4_7H2O, 518 nm Na2MoO4_2H2O, 50.2 nm CuSO4_5H2O, 96.2 nm CoCl2_6H2O, 0.05 mm FeSO4.7H2O, 0.05 mm EDTA.2H2O, 0.2% (w/v) Suc, and vitamins (0.25 mg L−1 of nicotinic acid, 0.05 mg L−1 of thiamine HCl, 0.25 mg L−1 of pyridoxine HCl, and 50 mg L−1 of myo-inositol). For chemical and hormone treatments, 3-d-old seedlings were transferred to the relevant supplemented media and analyzed after 4 d. BRZ was dissolved in 100% dimethyl sulfoxide. BRZ was added to a final concentration of 3 μm.
Vector Constructs and Transgenic Lines
Plants were transformed by the standard floral dip method using Agrobacterium tumefaciens. containing the pMLBART. The pUBI10 promoter was cloned to the polylinker of BJ36 and the polylinker of BJ36 harboring YPet-HA by XhoI/KpnI. Untagged BES1-D was subcloned by KpnI/KpnI sites to the BJ36 harboring YPet-HA. For pUBQ10-BES1-YPet, BES1 was digested by ClaI/BamHI and subcloned to BJ36-YPet-HA digested by ClaI/BglII.
RNA Extraction and Expression Analysis
Total RNA was extracted from roots and shoots of 7-d-old seedlings using an RNA extraction kit (Sigma). Quantitative real-time PCR assays were performed in an ABI 7300 PCR (Applied Biosystems) using SYBR Green PCR Master Mix (Applied Biosystems; Life Technologies). Transcripts corresponding to At5g15400 were used as endogenous control. Relative expression values were calculated using the ΔΔ threshold cycle method. Two to three biological replicates, each with three technical repetitions, were performed for all reactions. Primers used for quantitative real-time PCR are listed in Supplemental Table S3.
Root Growth and RSA Analysis
For the root growth rate analysis, 3-d-old seedlings were transferred to adequate and deprived Pi (1 µm) media and scanned every 24 h. Root length was measured using the ImageJ software. For sensitivity assays, 3-d-old seedlings were transferred to medium containing varying low concentrations of Pi as indicated in the text. After 4 d, seedlings were analyzed for root length. For LR analysis, 3-d-old seedlings were transferred and grown on adequate or deprived Pi (1 µm) medium for an additional 8 and 11 d, and the total number of LRs was counted. LR density was calculated by dividing the average number of LRs by the average root length. Total fresh weight of seedlings grown for an additional 11 d after transfer to adequate or deprived Pi medium was measured.
Anthocyanin Content and in Vivo APase Activity
Anthocyanin measurements were performed 8 d after seedling transfer to adequate or deprived Pi medium. Anthocyanin content was measured as previously described (Kim et al., 2003). Briefly, 25 mg of seedlings were incubated overnight at 4°C in 300 µL of anthocyanin extraction buffer and 1% (w/v) HCl in methanol. After overnight incubation, 200 µL of water and 200 µL of chloroform were added. After centrifugation for 5 min at 12,000 rpm, the upper aqueous phase was used for spectrophotometric quantification of anthocyanin. Total anthocyanin per 1 mg of tissue was calculated as A530 − 0.33A657.
In vivo active APase staining was performed as previously described (Tomscha et al., 2004; Devaiah et al., 2007) with slight modifications; 3-d-old seedlings grown for an additional 4 d in adequate or deprived Pi medium were transferred to 0.1% (w/v) 5-bromo-4-chloro-3-indolyl phosphate solution and incubated at 37°C for 30 min. After clearance in 70% (w/v) alcohol for 4 h, roots were documented using the Olympus DP-72 camera connected to a stereomicroscope (M165 FC; Leica).
Phosphorus and Iron Measurements
Seedlings were germinated in Pi-adequate and Pi-depleted (1 μm) media for 7 d. Roots and shoots, harvested separately, were washed with distilled, deionized water and air dried on blotting paper. For each sample, 200 mg were dried at 100°C for 24 h, after which time the dry weight of the sample was measured again. The dried sample was mixed with 1 mL of concentrated HNO3 and maintained at 110°C for 2 h. After digestion, the final volume was adjusted to 8 mL with distilled deionized water and analyzed (iCAP 6000 Series of ICP-OES Spectrometer; Thermo Scientific).
Confocal Microscopy
Fluorescence signals were detected using a confocal laser-scanning microscope (LSM 510 META) with a 25× water immersion objective lens (numerical aperture 0.8) for snapshots and an LSM 700 META Inverted microscope (Zeiss) with a 20× air objective lens (NA 0.8) for live imaging.
For snapshots, roots were imaged in water supplemented with propidium iodide (10 μg mL−1). Propidium iodide was viewed at an excitation wavelength of 488 nm, and emission was collected at 575 nm. For live imaging, roots were placed on a dish with a number 1.5 glass bottom (catalog no. 81158; idibi), covered with Murashige and Skoog agar according to the tested conditions, and sealed to prevent drying by the plate lid. Z-stack images were obtained at 5-μm intervals. YPet was viewed at an excitation wavelength of 488 nm, and emission was collected at 518 nm. Fluorescence measurements were obtained using ImageJ software. Average cytoplasm and nucleus GFP intensity were measured using segmented line and polygon selection tools, respectively, from selected Z stacks.
BR Analysis
For BR content, samples of 30 to 40 mg dry weight were sonicated for 5 min and extracted overnight with stirring in ice-cold 60% (w/v) acetonitrile and 30 pmol of [2H3]BL, [2H3]CS, [2H3]24-epi-BL, and [2H3]24-epi-CS as internal standards (OlChemIm). After centrifugation, samples were further purified on polyamide SPE columns (Supelco) and then analyzed by ultra-HPLC tandem mass spectrometry (Micromass). The data were analyzed using Masslynx 4.1 software (Waters), and BR content was quantified using the standard isotope dilution method.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtBZR1 (AT1G75080), and AtBES1 (AT1G19350).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root length in response to adequate and deprived Pi conditions.
Supplemental Figure S2. APase activity in response to adequate and deprived Pi conditions.
Supplemental Figure S3. Low Pi availability triggers cytoplasmic accumulation of BES1.
Supplemental Table S1. Fresh weight of seedlings grown in adequate and deprived Pi conditions.
Supplemental Table S2. BR analysis in whole seedlings grown in adequate and deprived Pi conditions.
Supplemental Table S3. Primers used for qRT-PCR.
Supplementary Material
Acknowledgments
We thank Zhiyong Wang for the bzr1-D seeds and Jeff Long for the BJ36-Ypet-HA plasmid.
Glossary
- APase
acid phosphatase
- BL
brassinolide
- BR
brassinosteroid
- BRZ
brassinazole
- Col-0
Columbia-0
- CS
castasterone
- LR
lateral root
- Pi
inorganic phosphate
- RSA
root system architecture
Footnotes
This work was supported by the Israel Science Foundation (grant no. 592/13), by the Fund for Applied Research at Technion (to A.P.S.), and by the Israel Council for Higher Education (fellowship to A.P.S.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abel S. (2011) Phosphate sensing in root development. Curr Opin Plant Biol 14: 303–309 [DOI] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. (2007) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman Y, Elkouby L, Holland N, Vragović K, Elbaum R, Savaldi-Goldstein S. (2014) Root growth is modulated by differential hormonal sensitivity in neighboring cells. Genes Dev 28: 912–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman Y, Savaldi-Goldstein S. (2013) Brassinosteroids in growth control: how, when and where. Plant Sci 209: 24–31 [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadí D, Blázquez MA. (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 109: 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-García S, Russinova E, Caño-Delgado AI. (2011) Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J. (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Gao X, Liao L, Harberd NP, Fu X. (2007) Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol 145: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo SH, Kim TW, Son SH, Lee WS, Yokota T, Kim SK. (2012) Biosynthesis of a cholesterol-derived brassinosteroid, 28-norcastasterone, in Arabidopsis thaliana. J Exp Bot 63: 1823–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G. (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J. (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Li QF, Wang C, Jiang L, Li S, Sun SS, He JX. (2012) An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci Signal 5: ra72. [DOI] [PubMed] [Google Scholar]
- López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L. (2014) Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol 65: 95–123 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS. (2013) Responses of root architecture development to low phosphorus availability: a review. Ann Bot (Lond) 112: 391–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T. (2011) Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci 16: 442–450 [DOI] [PubMed] [Google Scholar]
- Rouached H, Stefanovic A, Secco D, Bulak Arpat A, Gout E, Bligny R, Poirier Y. (2011) Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J 65: 557–570 [DOI] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L. (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46: 174–184 [DOI] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang ZY, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S. (2004) Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J 37: 801–814 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Lucero RD, Sakhonwasee S, Adamson AW, Creff A, Nussaume L, Desnos T, Abel S. (2009) ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc Natl Acad Sci USA 106: 14174–14179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomscha JL, Trull MC, Deikman J, Lynch JP, Guiltinan MJ. (2004) Phosphatase under-producer mutants have altered phosphorus relations. Plant Physiol 135: 334–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yokota T, Sato T, Takeuchi Y, Nomura T, Uno K, Watanabe T, Takatsuto S. (2001) Roots and shoots of tomato produce 6-deoxo-28-norcathasterone, 6-deoxo-28-nortyphasterol and 6-deoxo-28-norcastasterone, possible precursors of 28-norcastasterone. Phytochemistry 58: 233–238 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liao H, Lucas WJ. (2014) Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J Integr Plant Biol 56: 192–220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.