The functional soil microbiome is an important parameter in developing a sustainable and effective strategy to increase crop yield and food security.
Abstract
There is considerable evidence in the literature that beneficial rhizospheric microbes can alter plant morphology, enhance plant growth, and increase mineral content. Of late, there is a surge to understand the impact of the microbiome on plant health. Recent research shows the utilization of novel sequencing techniques to identify the microbiome in model systems such as Arabidopsis (Arabidopsis thaliana) and maize (Zea mays). However, it is not known how the community of microbes identified may play a role to improve plant health and fitness. There are very few detailed studies with isolated beneficial microbes showing the importance of the functional microbiome in plant fitness and disease protection. Some recent work on the cultivated microbiome in rice (Oryza sativa) shows that a wide diversity of bacterial species is associated with the roots of field-grown rice plants. However, the biological significance and potential effects of the microbiome on the host plants are completely unknown. Work performed with isolated strains showed various genetic pathways that are involved in the recognition of host-specific factors that play roles in beneficial host-microbe interactions. The composition of the microbiome in plants is dynamic and controlled by multiple factors. In the case of the rhizosphere, temperature, pH, and the presence of chemical signals from bacteria, plants, and nematodes all shape the environment and influence which organisms will flourish. This provides a basis for plants and their microbiomes to selectively associate with one another. This Update addresses the importance of the functional microbiome to identify phenotypes that may provide a sustainable and effective strategy to increase crop yield and food security.
In recent years, the term plant microbiome has received substantial attention, since it influences both plant health and productivity. The plant microbiome encompasses the diverse functional gene pool, originating from viruses, prokaryotes, and eukaryotes, associated with various habitats of a plant host. Such plant habitats range from the whole organism (individual plants) to specific organs (e.g. roots, leaves, shoots, flowers, and seeds, including zones of interaction between roots and the surrounding soil, the rhizosphere; Rout and Southworth, 2013). The rhizosphere is the region of the soil being continuously influenced by plant roots through the rhizodeposition of exudates, mucilages, and sloughed cells (Uren, 2001; Bais et al., 2006; Moe, 2013). Thus, plant roots can influence the surrounding soil and inhabiting organisms. Mutually, the rhizosphere organisms can influence the plant by producing regulatory compounds. Thus, the rhizospheric microbiome acts as a highly evolved external functional milieu for plants (for review, see Bais et al., 2006; Badri et al., 2009b; Pineda et al., 2010; Shi et al., 2011; Philippot et al., 2013; Spence and Bais, 2013; Turner et al., 2013a; Spence et al., 2014). In another sense, it is considered as a second genome to a plant (Berendsen et al., 2012). Plant rhizospheric microbiomes have positive or negative influence on plant growth and fitness. It is influenced directly by beneficial mutualistic microbes or pathogens and indirectly through decomposition, nutrient solubilization, nutrient cycling (Glick 1995), secretion of plant growth hormones (Narula et al., 2006; Ortíz-Castro et al., 2008; Ali et al., 2009; Mishra et al., 2009), antagonism of pathogens (Kloepper et al., 2004), and induction of the plant immune system (Pieterse et al., 2001; Ramamoorthy et al., 2001; Vessey, 2003; Rudrappa et al., 2008, 2010). The establishment of plant and rhizospheric microbiome interaction is a highly coordinated event influenced by the plant host and soil. Recent studies show that plant host and developmental stage has a significant influence on shaping the rhizospheric microbiome (Peiffer et al., 2013; Chaparro et al., 2014).
There are various factors involved in the establishment of the rhizospheric and endophytic microbiome. They are greatly affected by soil and host type (Bulgarelli et al., 2012; Lundberg et al., 2012). Apart from these factors, other external factors such as biotic/abiotic stress, climatic conditions, and anthropogenic effects also can impact the microbial population dynamics in particular plant species. Plant host species differences can mainly be perceived from the secretory exudates by microbes. The root exudates act as a crucial driving force for multitrophic interactions in the rhizosphere involving microbes, neighboring plants, and nematodes (Bais et al., 2006). Thus, it is important to understand root exudate-shaped microbial community profiling in establishing phenotypes involved in plant health. Microbial components associated with plant hosts have to respond to these exudates along with utilizing them in order to grow competitively in a complex interactive root environment. Commonly, there are three groups of microbes present in the rhizosphere, commensal, beneficial, and pathogenic microbes, and their competition for plant nutrition and interactions confer the overall soil suppressiveness against pathogens and insects (Berendsen et al., 2012).
Traditionally, the components of the plant microbiome were characterized by isolating and culturing microbes on different media and growth conditions. These culture-based techniques missed the vast majority of microbial diversity in an environment or in plant-associated habitats, which is now detectable by modern culture-independent molecular techniques for analyzing whole environmental metagenomes (comprising all organisms’ genomes). Over the last 5 years, these culture-independent techniques have dramatically changed our view of the microbial diversity in a particular environment, from which only less than 1% are culturable (Hugenholtz et al., 1998). After discovering the importance of the conserved 16S ribosomal RNA (rRNA) sequence (Woese and Fox, 1977) and the first use of denaturing gradient gel electrophoresis (DGGE) of the amplified 16S rRNA gene in the analysis of a microbial community (Muyzer et al., 1993), there was a sudden explosion of research toward microbial ecology using various molecular fingerprinting techniques. Apart from DGGE, thermal gradient gel electrophoresis, and fluorescence in situ hybridization, clone library construction of microbial community-amplified products and sequencing emerged as other supporting techniques for better understanding of microbial ecology (Muyzer, 1999). Furthermore, there are many newer techniques used to understand the microbiome, from metagenomics to metaproteomics (Friedrich, 2006; Mendes et al., 2011; Knief et al., 2012; Rincon-Florez et al., 2013; Schlaeppi et al., 2014; Yergeau et al., 2014). These techniques cover the whole microbiome, instead of selecting particular species, unlike conventional microbial analysis. However, their presence was not yet correlated well with the phenotypic manifestation (phenome) they establish in the host plant.
As a consequence of population growth, food consumption is also increasing. On the other hand, cultivable agricultural land and productivity are significantly reduced due to global industrialization, drought, salinity, and global warming (Gamalero et al., 2009). This problem is only addressed by practicing the sustainable agriculture that protects the health of the ecosystem. The basic principle of sustainable agriculture is to significantly reduce the chemical input, such as fertilizers, insecticides, and herbicides, while reducing the emission of greenhouse gas. Manipulation of the plant microbiome has great potential in reducing the incidence of pests and diseases (van Loon et al., 1998; Kloepper et al., 2004; Van Oosten et al., 2008), promoting plant growth and plant fitness, and increasing productivity (Kloepper and Schroth 1978; Lugtenberg and Kamilova, 2009; Vessey, 2003). Single strains or mixed inoculum treatments induced resistance to multiple plant diseases (Jetiyanon and Kloepper, 2002). In recent years, several microbial biofertilizers and inoculants were formulated, produced, marketed, and successfully used by farmers worldwide (Bhardwaj et al., 2014). Although plants are being considered as a metaorganism (East, 2013), our understanding of the exact manifestation of this microbiome on plant health in terms of phenotypes is insufficient. Of late, there is a surge to understand and explore the genomic wealth of rhizosphere microbes. Hence, this Update will focus mainly on existing knowledge based on the root microbiome, its functional importance, and its potential relationship to the establishment of a host phenome, toward achieving sustainable agriculture.
CHARACTERIZATION OF THE SOIL MICROBIOME
Plant microbiomes are diverse, consisting of detrimental pathogens, potential endophytes, and beneficial symbionts (Beattie and Lindow, 1995; Rosenblueth and Martínez-Romero, 2006). As the soil matrix is considered a favorable niche (Lavelle and Spain, 2001), the bacterial density can reach up to 106 to 107 cells cm−2 (Hirano and Upper, 2000; Lindow and Brandl, 2003). But, classically, the microbial diversity was evaluated by isolating and culturing on different nutrient media and growth conditions. Various plant nutritional and regulatory requirements are fulfilled by microbial activities inside, on the surface, and in proximate soil surroundings (Vessey, 2003; Lugtenberg and Kamilova, 2009). These nutritional requirements mainly include nitrogen, phosphorous, and iron. In addition, elements such as nitrogen, phosphorous, and iron also have the ability to regulate plant growth by stimulating the production of plant growth regulators. To screen the potential bacteria involved in plant growth promotion, culture-based methods were routinely used to date (Forchetti et al., 2007; Beneduzi et al., 2008; Taulé et al., 2012). These techniques generally include simple plate assays or growing the microbes in broth to find plant beneficial activities. They are also used to find the genetic components behind these beneficial phenotypes and their characterization. However, the culture-dependent approaches miss the majority of the nonculturable microbial diversity in the microbiome. In addition, there are very few studies that suggest the involvement of plant-mediated recruitment of the rhizospheric microbiome.
The exact revelation of the microbial population in the rhizosphere and on the root surface is solely dependent on the sampling and sequencing techniques used, and this poses a difficult challenge. Furthermore, the ways and representative numbers for extracting microbes from rhizospheric samples are also crucial in this analysis. There are several factors, including plant host genotype, soil type, and cultivation practices (discussed in detail in the following sections), that are key drivers in shaping the rhizospheric microbial community (Philippot et al., 2013; Turner et al., 2013a; Huang et al., 2014). By using the stable isotope probing (SIP) technique, Haichar et al. (2008) found that the root exudates of various plants (wheat [Triticum aestivum], maize [Zea mays], rape [Brassica napus], and Medicago truncatula grown in similar soil) are involved in shaping the rhizospheric bacterial community. The rhizospheric microbes assimilating root exudates were separated by analyzing only the DGGE profile of [13C]DNA (fixed by plants from a controlled 13CO2 environment), whereas the organisms utilizing soil organic matter were analyzed using [12C]DNA. This study revealed that certain groups of bacteria, such as order Sphingobacteriales and genus Myxococcus, can exclusively utilize root exudates from all plants, while bacteria from the order Sphingomonadales are found to utilize carbon sources from both root exudates and soil organic matter. Importantly, some of the bacterial groups (genus Enterobacter and order Rhizobiales as generalists) are commonly present in all four plant species. This result implies that some bacteria can have wide host survival ability, overcoming host specificity limitations, although some bacteria can fall into the stringent nutritional requirement category, which are attracted to and supported by specific host root exudate compounds.
In the analysis of the whole microbiome, the initial effort was started with the discovery of a conserved 16S rRNA gene sequence and its application and PCR in the identification of microorganisms (Woese and Fox, 1977; Mullis et al., 1987). To date, there are rigorous improvements achieved with these techniques, yielding to metagenomics, in order to study and understand the microbiome in a holistic perspective in a short period. Very recently, these technological advancements were extensively reviewed and analyzed in terms of potentials, drawbacks, and demands (Whiteley et al., 2006; Berlec, 2012; Dini-Andreote and van Elsas, 2013; Rincon-Florez et al., 2013). These methods include starting with whole metagenome sampling, followed by purification, separation, and sequencing, and finally data analysis and interpretation. Especially, the sequencing technology is going through rapid development, as it provides wide and in-depth views of metagenomics, and today is broadly named high-throughput sequencing (HTS) or next-generation sequencing. These HTS techniques include use of the 454 Genome Sequencer (Roche Diagnostics), the HiSeq 2000 (Illumina), and the AB SOLiD System (Life Technologies; Lundberg et al., 2012; Peiffer et al., 2013; Rincon-Florez et al., 2013; Yergeau et al., 2014). Furthermore, other advanced techniques, such as DNA/RNA-SIP and DNA arrays (PhyloChip and functional gene arrays), also have promising features in the analysis of microbiomes, especially their functional parts (Friedrich, 2006; Mendes et al., 2011; Rincon-Florez et al., 2013; Uhlik et al., 2013). At present, there is a transition from metagenomics to metatranscriptomics, as the latter answers the diversity and functional part of the microbiome, rather than only showing the diversity, like the former. It was also recently perceived that the functional versatility and function-based diversity of the microbiome are likely to be dominant factors in niches rather than mere diversity (Barret et al., 2011; Chaparro et al., 2012; Turner et al., 2013b).
In metatranscriptomics approaches, RNA-SIP, quantitative reverse transcription-PCR, and complementary DNA analysis coupled with pyrosequencing provide advanced functional insights into microbiome activities in the soil and rhizosphere (Leininger et al., 2006; Whiteley et al., 2006; Uhlik et al., 2013). Particularly, the importance of RNA-SIP was emphasized in future studies for temporal analysis of the flow of root-derived carbon and the differentiation of primary and secondary microbial utilizers, which have higher rates of labeling than their genes and need not depend on cell division, unlike DNA-SIP (Haichar et al., 2008; Bressan et al., 2009; Uhlik et al., 2013). By overcoming the general constraints of quantitative PCR and microarray technology in analyzing the gene expression of a complex community, these advanced technologies still face enormous challenges. These challenges include selecting either mRNA or rRNA alone according to a study objective, achieving wider coverage of an ecological RNA pool, increasing the sensitivity of sequencing to reach ecologically important data. Peiffer et al. (2013) have shown the significant community differences among 27 maize inbred lines (genetic variation in a single species) with common enriched populations in the maize rhizosphere. They also included a pilot study to select suitable primer sets (out of four previously reported sets) and found the V3-V4 region of the 16S rRNA gene (primers 515F and 806R) to be most suitable, due to its enrichment of classifiable sequences as well as its reduced amplification of maize plastid-related sequences. This attempt implies the vital consideration of primer selection before performing any wide metagenomics or metatranscriptomics studies. However, metaproteomics has an entirely different approach, targeting the active functional part of the microbiome, and involves extracting the metaproteome from samples and using mass spectrometry for peptide fingerprinting (Keiblinger et al., 2012; Kolmeder and de Vos, 2014). Both metatranscriptomics and metaproteomics are in the early stages of development and face many challenges due to sampling constraints (removing humics and clays) and data acquisition (Keiblinger et al., 2012). Analyzing and assigning clusters of orthologous groups of proteins from metagenomic and metaproteomic (existing and future) data are another important process-centric approach and need to be complemented by other techniques to determine the diversity and functional relatedness of the rhizospheric microbiome (Barret et al., 2011; Keiblinger et al., 2012).
After the introduction of molecular techniques to analyze the whole community of bacteria, the great plate count anomaly (Amann et al., 1995) surfaced to the scientific community. Henceforth, almost all microbial community studies will involve molecular fingerprinting techniques, along with the culture-dependent techniques that are still valuable for deeper understanding of individual characterizations and are useful for studying interactions with host plants. Although there are many technical innovations in HTS that lead to insightful and wider understanding of the microbiome phylotypes and functions, Dini-Andreote and van Elsas (2013) have emphasized its hindrance in testing ecological hypotheses and the current need of a paradigm shift from HTS (or inclusive efforts) to studies of fundamental questions about yet-unexplored plant-soil-microbiota systems, especially toward phenotypic diversity of the rhizospheric microbiome on both spatial and temporal levels.
FACTORS DETERMINING THE SOIL MICROBIOME STRUCTURE AND FUNCTION
In the complex and dynamic plant root interaction with the microbiome, both biotic and abiotic factors play critical roles for microbiome composition, richness, and diversity. Biotic factors, such as host genotypes, cultivars, developmental stages, proximity to root, and root architecture, and abiotic factors, such as temperature, soil pH, seasonal variation, and the presence of rhizospheric deposits, act as chemical signals for microbes and influence the microbiome community structure and function (for review, see Berg and Smalla, 2009; Dennis et al., 2010; Berendsen et al., 2012; Chaparro et al., 2012; Minz et al., 2013; Philippot et al., 2013; Spence and Bais, 2013; Turner et al., 2013a). However, the extent to which both abiotic and biotic factors contribute to microbial communities is not fully understood.
DETERMINATION OF MICROBIOME BY HOST
Host Genotype
Plant host specificity is well understood in the case of phytopathogenic interaction with fungi or bacteria (Raaijmakers et al., 2009). The classical case of rhizobium-legume symbiotic interactions is well studied and shows highly host-specific interactions (Long, 1989). Earlier studies that determined microbial communities using automated ribosomal intergenic spacer showed that differential plant developmental stages influence the rhizospheric microbial communities in maize roots (Baudoin et al., 2002), pea (Pisum sativum), wheat, and sugar beet (Beta vulgaris; Houlden et al., 2008), M. truncatula (Mougel et al., 2006), and Arabidopsis (Arabidopsis thaliana; Micallef et al., 2009; Chaparro et al., 2014). Recently initiated high-throughput analysis characterized the core microbiome in the model plant Arabidopsis and indicated that the host genotype has a small but measurable effect on the microbes inhabiting the endophyte compartment of the root (Bulgarelli et al., 2012; Lundberg et al., 2012). In another study involving wheat, pea, and oat (Avena sativa), plants were grown for 4 weeks in similar bulk soil and the microbiomes were evaluated. Interestingly, the microbiomes were found to be different from each other, with a profound change in the balance of prokaryotes and eukaryotes between different plant species. Oat and pea exerted strong selection on eukaryotes, whereas selection by wheat was much weaker (Turner et al., 2013b). In a similar way, modern maize inbreds planted in five environmental locations and estimated for the microbiome at flowering time were found to have heritable variation in the rhizosphere microbial community composition (Peiffer et al., 2013). However, the conclusion that the genetic component of the maize allele may play a role in variation in the microbiome is still hypothetical (Peiffer et al., 2013; Peiffer and Ley, 2013). A recent experiment with the establishment of rhizosphere communities in three cultivars of potato (Solanum tuberosum) grown in two distinct field sites revealed that only 4% of operational taxonomic units were dependent on the host genotype by 40% soil-specific abundance (Weinert et al., 2011). Interestingly, potato cultivars showed differences in microbes belonging to the families of bacteria that have been studied extensively for their ability to control plant pathogens (Weinert et al., 2011), and in another study, it was shown that plant age and genotype of sweet potato (Ipomoea batatas) also influenced the root microbiome (Marques et al., 2014). Similarly, the structure and function of the rhizospheric bacterial community associated with Arabidopsis at four different plant development stages (seedling, vegetative, bolting, and flowering) were analyzed and showed that there were no significant differences in bacterial community structure (Chaparro et al., 2014). Interestingly, the microbial community at the seedling stage was found to be distinct from the other developmental time points (Chaparro et al., 2014). Intriguingly, rice (Oryza sativa) mutant lines of a common symbiosis pathway gene, calcium/calmodulin-dependent protein kinase (CCaMK), had significant impact on the rhizospheric microbiome while testing under both paddy and upland field conditions (Ikeda et al., 2011). That study showed a significant decrease in the population of class A proteobacteria (an environmentally ubiquitous group) in a recessive homozygous (R) CCaMK mutant under both tested conditions due to a crucial shift in the population of its component orders: Sphingomonadales and Rhizobiales. Likewise, there was an increased abundance of Anaerolineae (Chloroflexi), Clostridia (Firmicutes), and a subpopulation of Actinobacteria (Saccharothrix spp., unclassified Actinosynnemataceae) in R plants under paddy and upland conditions.
In another study, Shakya et al. (2013) showed that a high percentage (more than 90%) of operational taxonomic units (OTUs) specific to sampled mature Populus deltoides trees from two watersheds of North Carolina and Tennessee in two seasons (spring and fall) had the dominant phyla Proteobacteria (56.1%), Actinobacteria (17.5%), and Acidobacteria (10%). However, dominance of Proteobacteria was replaced by Actinobacteria from spring to fall in the Tennessee samples. The core rhizosphere OTUs were within the orders Burkholderiales and Rhizobiales. This study further attempted to investigate the impact of host genotype and phenotype on community structure, where more than 40% variation of factors was statistically unexplained and only approximately 20% was significantly contributed in two habitats of the tree belowground: rhizosphere and endosphere. This study did not directly explain any host-specific variance in rhizospheric bacterial community structure, instead postulating an indirect influence of changing soil properties (through rhizodeposits and exudates) that showed statistically significant influence on the rhizospheric microbiome. These studies (Ikeda et al., 2011; Shakya et al., 2013) still pose open-ended questions. What are the specific genes/alleles that control microbial communities? And what are the specific host factors that are involved in the orchestration of the microbial populations? Therefore, the hypothesis that microbial community composition could be related directly to host genotype requires further appraisal based on a wider range of plant genotypes.
Alteration in Host Signaling Pathways
In response to biotic and abiotic stress, plants activate complex jasmonic acid (JA) and salicylic acid signaling pathways that lead to localized and systemic defenses (Dangl and Jones, 2001; Glazebrook, 2005; De Vos et al., 2006; van Loon et al., 2006; Pozo et al., 2008; Pieterse et al., 2012). The variation in the salicylic acid and JA signaling defense pathways affects the abundance, diversity, and composition of the natural bacterial microbiome of Arabidopsis, and susceptible genotypes have a higher abundance of microbial communities (Kniskern et al., 2007). It was shown that activation of a plant’s JA defense pathway significantly altered the rhizosphere microbial community (Carvalhais et al., 2013; for review, see Ramamoorthy et al., 2001; Pieterse et al., 2003; Lugtenberg and Kamilova, 2009). On the other hand, production of a single exogenous glucosinolate significantly altered the microbial community on the roots of transgenic Arabidopsis (Bressan et al., 2009). Recently, the first evidence of the recruitment of beneficial root microbes after aboveground herbivory and pathogenic bacterial attack was shown: aerial aphid feeding and pathogenic microbial attack increased the population of the nonpathogenic rhizobacterium Bacillus subtilis in the rhizosphere of sweet pepper (Capsicum annuum) and Arabidopsis plants (Rudrappa et al., 2008; Yang et al., 2011; Lakshmanan et al., 2012; Lee et al., 2012). These studies reveal a new type of interaction and raise the question of how multiple herbivory/pathogen attacks would affect the colonization of root-associated microbes. These studies showed that even a minor modification in plant roots could have important repercussions for soil microbial communities. However, the chemical cue that triggers the increased colonization under aerial herbivory has not been discovered yet.
Alteration in Root Secretions
Rhizodeposition represents approximately 11% of net fixed carbon and 27% of carbon allocated to roots (Jones et al., 2009). This rhizodeposition contains both low- and high-Mr compounds. The low-Mr compounds are more abundant in exudates and include amino acids, organic acids, phenolic compounds, simple sugars, and other small secondary metabolites (Walker et al., 2003; Bais et al., 2006). Root exudates generally vary substantially between different plant species and genotypes and can depend on a variety of factors, such as nutrient levels, disease, stress, and even the microbial community itself (Lankau, 2011). This variation in chemicals, such as chemotactic or signaling molecules to orchestrate changes in microbial composition, may influence the composition and dynamics of microbial communities (Shaw, 1991; de Weert et al., 2002; Jain and Nainawatee, 2002; Horiuchi et al., 2005; Bais et al., 2006; Badri and Vivanco, 2009; Neal et al., 2012; Badri et al., 2013a). The secretion of sugars and sugar alcohols is regulated by plant developmental stages, which may help to orchestrate the assemblage of the rhizospheric microbiome on roots (Chaparro et al., 2013, 2014). In addition, it was concluded that specific developmental stages in plants may secrete different phytochemicals (Chaparro et al., 2014). The specific role of root exudates in the shaping of the rhizosphere is further confirmed by showing different groups of natural compounds derived from plant root exudates synergistically modifying the root microbiome (Badri et al., 2013b). The known components of cucumber (Cucumis sativus) root exudates p-coumaric acid and vanillic acid showed differential effects on the soil microbiome: p-coumaric acid increased the pathogenic fungal taxa that degrades the p-coumaric acid (Zhou and Wu, 2012), while vanillic acid promoted the plant growth-promoting rhizobacteria Bacillus spp. (Zhou and Wu, 2013). Microbiome composition was also affected by altering the composition of root exudates. Specifically, this was done by increasing the phenolic compounds as compared with sugars by creating an ATP-binding cassette transporter mutant in Arabidopsis (Badri et al., 2009a). It was further supported by the reduction of phenolic exudates in transgenic rice (inhibition of Phe ammonia lyase gene expression) that resulted in decreased microbial communities (Fang et al., 2013). The above-mentioned studies supported the concept that plant root secretion may play a strong role in shaping the rhizospheric community structure and function (for review, see Berendsen et al., 2012; Huang et al., 2014).
In recent years, plant-microbe interaction studies were carried out with specific plants and microbes, and low-Mr organic acids in the root exudates, such as l-malic acid, citric acid, and fumaric acid, were shown to act as chemoattractants to establish root colonization (Rudrappa et al., 2008; Lakshmanan et al., 2012; Lakshmanan and Bais, 2013; Tan et al., 2013; Liu et al., 2014). These studies suggest that biotic or abiotic stress regimes may modify the secretion of organic acids in the root exudates and attract specific microbes and alter the structure and composition of the microbiome (Broeckling et al., 2008; Houlden et al., 2008; Rudrappa et al., 2008; Lakshmanan et al., 2012; Lakshmanan and Bais, 2013; Tan et al., 2013; Liu et al., 2014). Tricarboxylic acids, such as malic acid and citrate, are suitable carbon sources for many microorganisms (López-Bucio et al., 2000; for review, see Pineda et al., 2010). Carbon enrichment of the rhizosphere, especially carboxylate excretion and acidification at the root surface, might have a strong impact on structuring rhizospheric microbial communities (Marschner et al., 2002).
POTENTIAL IMPACT OF THE SOIL MICROBIOME ON THE HOST: FROM GENOME TO PHENOME
The rhizospheric microbiome can impact plant growth and development, as their interactions are coevolved and coadapted over time and space (Bakker et al., 2012). Microbiomes vary in composition, diversity, and abundance according to many factors. Consequently, the impact on the host also inflicts changes in the microbiome. Interdependence and interplay between the soil microbiome, edaphic factors, and the host result in the overall quality of plant productivity. Apart from being a predictor of soil quality (Sharma et al., 2010), the soil microbiome exerts increased disease suppressiveness against pathogens with respect to elevated microbial richness and diversity (Nannipieri et al., 2003; Garbeva et al., 2004; Mendes et al., 2011). Mendes et al. (2011) analyzed and compared microbiomes of disease-suppressive and -conducive soils (against Rhizoctonia solani) of sugar beet. The soils were subsequently treated to remove suppressiveness or mixed to obtain six different soil types based on a PhyloChip-based metagenomic approach. Although there was no significant difference found in the number of OTUs (more than 30,000 in all), the abundance of particular classes, which correlates to the soil suppressiveness, showed significant differences between the different soil types. The major microbial components of soil suppression found in this study include the γ- and β-proteobacteria (Pseudomonadaceae, Burkholderiaceae, and Xanthomonadales) and the Firmicutes (Lactobacillaceae), especially more abundant during R. solani infection, implying the possibility of a host-induced microbiome to combat pathogenic attack. This study was further extended by culture-dependent methods, where one of the major Pseudomonadaceae group members was shown to be antagonistic against R. solani infection and tracked the key genetic element as a nonribosomal peptide synthetase gene.
This study (Mendes et al., 2011) is particularly thorough, as it applied concurrent analysis using both culture-dependent and -independent approaches and demonstrated the ability of a sympatric soil microbiome to increase Arabidopsis growth under drought conditions (Zolla et al., 2013). Against the convention of a single bacterial application to combat drought, this study unraveled the importance of the soil microbiome as a whole in alleviating drought stress. The study considered analyzing both the drought-response genes in the host and the molecular profile of the soil microbiome involved in the process. In the microbiome analysis, there were 33 genera in the core microbiome of Arabidopsis soil that were already reported to be part of the core microbiome of this species. Among them, the 14 OTUs were more highly abundant in the Arabidopsis microbiome compared with other nonsympatric (pine [Pinus spp.] and maize) soil microbiomes. These 14 OTUs cover species including Micromonospora, Streptomyces, Bacillus, Hyphomicrobium, Rhizobium, Burkholderia, and Azohydromonas spp. Furthermore, it was concluded that various soil microbes play a role in improving the host’s ability to sense and respond to drought. Another study, by Badri et al. (2013a), has demonstrated the significant effect of various soil microbiomes (from different hosts) on the metabolome of Arabidopsis in response to herbivory. The production of phenolics, amino acids, sugars, and sugar alcohols (components of the leaf metabolome tested) is significantly altered by the application of various soil microbes, in turn influencing the feeding of herbivores. In particular, there was a positive correlation observed between the production of amino acids and the reduction in herbivory. This allows us to speculate that such a connection and impact from belowground and aboveground plant organs may not only be involved in herbivory but also in manipulating other multitrophic interactions with microbes, animals, and neighboring plant species.
There are some interesting pieces of evidence showing the significant contribution of the soil microbiome to plant community dynamics through a negative feedback mechanism (Janzen-Connell effect), leading to the coexistence of strong and diverse competitors in close proximity sharing an ecological niche (Bever, 2003; Fitzsimons and Miller, 2010). This effect comes from the negative impact of soil-borne pathogens and predators, which limits the establishment of a diverse plant community, whereas the positive feedback is from host mutualists, thus delineating both effects originating at the multidimensional cost of virulence and mutualism (Bever et al., 2012). There are both positive and negative soil community feedback activities playing crucial roles in the establishment of plant population structure. Some studies have demonstrated the inevitable role of certain endosymbionts (either arbuscular mycorrhizal fungi or diazotrophs) in the initial establishment of species in a new environment or community conversion due to positive feedback, but likely leading to exotic species dominance instead of establishing a diverse plant community (Rejmanek and Richardson, 1996; Larson and Siemann, 1998; Klironomos, 2002; Fitzsimons and Miller, 2010). However, this positive feedback will have a potential role to play in an agricultural system, where single monoculture crops are used instead of a diverse species population. In contrast, through negative feedback, the remnant whole-soil microbial communities from native ecosystems can help achieve the restoration of native plant communities. The plant diversity was well restored in a tallgrass prairie by microbia-mediated negative feedback from native plant soil (Fitzsimons and Miller, 2010). However, a separate study explored the microbial community structure and composition (Rosenzweig et al., 2013), and there were no connective studies between the soil microbiome and ecological restoration projects. Hence, these feedback mechanisms should be analyzed along with microbiome structure and function by coupling metagenomics, which will enhance our knowledge of the signature microbiome involved in such mechanisms. In addition, the negative feedback is almost essential to ecosystem restoration and engineering, which is a serious global concern due to the pressure of global warming and other growing anthropogenic activities disturbing the integrity of many ecosystems. Sometimes the positive soil community feedback is also believed to confer host-specific symbionts to certain species that are at risk of extinction. On realizing the significance of microbiomes as a whole, it is necessary to find and design new preservation techniques for those microbiomes. Since techniques for preserving individual microbes are advancing, these improvements can be adopted and modified accordingly to be used for the preservation of the microbiome for both future applications and scientific experiments.
The endosymbiotic microbiome of a plant host is another important resource of many functional genes and metabolites, with specific roles established in host stress tolerance, defense against pathogens, and nutrition. The endosymbiotic microbiome, for its contribution toward stress tolerance, defense, and nutrient acquisition, is often referred to as stress tolerance endosymbiotic systems, defensive endosymbiotic systems, and nutritional endosymbiotic systems, respectively (White et al., 2014). Interestingly, plants have evolved to extract nutrients from endosymbiotic bacteria by either oxidative nitrogen scavenging (especially from diazotrophs) or other phagocytic digestive systems (Paungfoo-Lonhienne et al., 2010; White et al., 2012). Many of the endosymbiosis-based activities occur at plant roots, due to their entangled communications with the abundant soil microbiome, which acts as a source to provide host-specific microbial partners. These observations ascertain that our understanding of the evolutionary and functional significance of the root endophytic microbiome is scant and demands a lot more technological advancement in the future. Both the epiphytic and endophytic microbiomes may have significant impact on plant growth and defense, but studies in this area are scarce. This necessitates a timely surge to expand our knowledge from metagenomics to metaphenomics, which comprises the functional (phenotypic) components of the soil/rhizospheric microbiome with regard to a plant host’s phenome. Furthermore, there is increasing evidence demonstrating the effect of the microbiome in metaorganisms for its role in shaping fundamental physiological phenotypes such as aging (Heintz and Mair, 2014). The basic search in this direction will start with phenome-based sampling of the microbiome and subsequent scrutiny to address how the functional microbiome is shaped and how it connects to the plant host phenome (Fig. 1). It was understood from earlier isolated experiments and applications of individual microbes (by their plant growth-modulating phenotypes like nitrogen fixation, phosphate solubilization, etc.) that microbes play a role in shaping a host’s phenotype (Fig. 2). Now, it is time to move forward from this dissected approach to a holistic one, as it exists in nature.
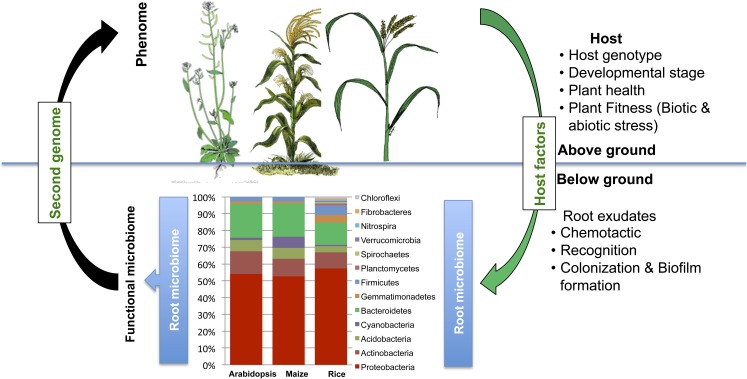
Figure 1.
Rhizospheric microbiome composition based on a high-throughput analysis of three plant species, Arabidopsis, maize, and rice (adapted from Ikeda et al. [2011], Lundberg et al. [2012], and Peiffer et al. [2013]). The possible involvement of plant-derived factors involved in shaping the rhizospheric microbiome composition is shown. The usefulness and potential applications of the microbiome to host phenome establishment are discussed. [See online article for color version of this figure.]
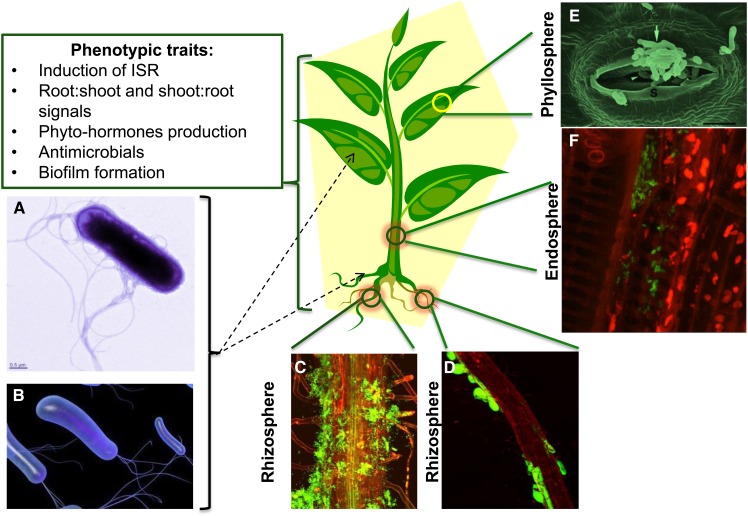
Figure 2.
Impact of single-isolate studies on model systems. To date, the majority of single-isolate studies have shown the impact on plants of both biotic and abiotic stress regimes. The phenotypes induced by single-isolate treatments are encouraging, yet several fundamental questions remain unanswered and are discussed. A and B, Electron microscopic images of B. subtilis FB17 (A) and a single isolate of Pseudomonas fluorescens (B). C and D, Visualization on biofilm of B. subtilis on Arabidopsis roots (C) and P. fluorescens on potato roots (D). E and F, Phyllosphere-associated Methylobacterium spp. (E) and endophytic Burkholderia phytofirmans strain PsJN-inside grapevine (Vitis vinifera; F). The dotted arrows show the application of single isolates through roots or leaves. Image sources are as follows: B is from http://www.buzzle.com/articles/pseudomonas-fluorescens.html; D is reproduced, with permission, from Krzyzanowska et al. (2012); E is reproduced, with permission, from Kutschera (2007); and F is adapted from http://endophytes.eu. [See online article for color version of this figure.]
IMPLICATION OF THE SOIL MICROBIOME ON SUSTAINABLE AGRICULTURE AND FOOD SECURITY
In order to feed a present population of 6.9 billion, the world will need a new vision for agriculture. Delivering food security, the process of increasing food production, and improving food quality to sustain population growth without compromising environmental safety has been called a global green revolution (Gupta, 2012). Sustainable agriculture development is needed to mitigate these issues. The ultimate goal of sustainable agriculture, according to the U.S. National Research Council, is to develop farming systems that are productive, profitable, energy conserving, environmentally sound, conserving of natural resources, and that ensure food safety and quality. It is our view that the most promising strategy to reach this goal is to substitute hazardous agrochemicals (chemical fertilizers and pesticides) with environmentally friendly preparations of beneficial microbes, which could improve the nutrition of crops and livestock and also confer protection from biotic (pathogens and pests) and abiotic (including pollution and climatic change) stresses. There is a vast literature available for the identification, isolation, and utilization of microbes as a major substitute for chemical input for crop protection (for review, see Bhattacharyya and Jha, 2012; Doornbos et al., 2012). Increasing the soil microbial species richness was shown to be a predictor of plant health and productivity (van der Heijden et al., 2008; Lau and Lennon, 2011; Schnitzer et al., 2011; Wagg et al., 2011). The beneficial effects and mechanisms of microbes on plant health and fitness and their utilization in agriculture are widely studied and documented (Higa and Parr, 1994; Horrigan et al., 2002; Johansson et al., 2004; Bhattacharyya and Jha, 2012; Chaparro et al., 2012; Wu et al., 2013). The potential microbial isolates are formulated using different organic and inorganic carriers either through solid or liquid fermentation technologies (summarized in Table I). They are delivered as individual strains or mixtures of strains through seed treatment, biopriming, seedling dip, or soil application. Further optimization of microbial isolates and the formulation process is needed through extensive research to introduce them in sustainable agricultural practices. Apart from the application of individual microbes, identifying healthy and functionally diverse microbiomes and their application for enhancing crop yield represent another big and necessary challenge to venture, after finding that the whole microbiome is an essential and indispensable portion, as being a second genome of the plant host, the metaorganism.
Table I.
Commercial products of plant growth-promoting rhizobacteria in plant health and disease management
| Bioagent | Trade Name/Formulation |
|---|---|
| Agrobacterium radiobacter strain K1026 | Nogall, |
| A. radiobacter strain K84 | Galltrol, Diegall |
| Azospirillum brasilense/Azotobacter chroococcum | Gmax Nitromax |
| A. brasilense | Azo-Green |
| B. subtilis MB1600 | BaciGold, HiStick N/T, Subtilex |
| B. subtilis strain FZB24 | Rhizo-Plus, Serenade, Rhapsody, Taegro, Tae-Technical |
| Bacillus chlororaphis 63-28 | AtEze |
| Bacillus cereus BPO1 | Pix plus |
| Bacillus pumilus GB 34 | Concentrate; YieldShield |
| B. pumilus QST2808 | Sonata ASO, Ballard |
| B. subtilis GB03 | Companion, System 3, Kodiak, Kodiak HB, Epic |
| Bacillus amyloliquefaciens GB99 | Quantum 4000 |
| Bacillus licheniformis SB3086 | EcoGuard, Green Releaf |
| Burkholderia cepacia | Blue Circle, Deny, Intercept |
| P. fluorescens A506 | BlightBan A506, Conquer, Victus |
| Pseudomonas syringae ESC-100 | Bio-Save 10, 11, 100, 110,1000, and 10 LP |
| Pseudomonas chlororaphis | Cedomon |
| Pseudomonas cepacia | Intercept |
| Streptomyces griseovirdis K61 | Mycostop |
| B. subtilis + B. amyloliquefaciens | Bio Yield |
| Pseudomonas spp. + Azospirillum spp. | BioJet |
CONCLUSION AND FUTURE PERSPECTIVES
The plant root microbiome is a complex community formed by the organism that may be detrimental or beneficial to the host plant. Unfortunately, studies on the interactions of host-beneficial and host-pathogenic organisms have been carried out in isolation. Experimental evidence is needed to understand the root microbiome in plant health and how each plant is able to control the composition of its belowground associates. However, recent studies have focused on evaluating belowground microbial community diversity. Its influence on host structure has advanced our understanding of this exciting interaction (Bulgarelli et al., 2012; Lundberg et al., 2012; Peiffer et al., 2013; Chaparro et al., 2014; for review, see Chaparro et al., 2012; Turner et al., 2013a; Huang et al., 2014; Spence et al., 2014). Furthermore, scientific attempts are required to expand our molecular understanding of the plant-microbiome interaction and its impact on plant health and productivity. Still, the key player(s) in terms of microbiome structure have not been identified. Accordingly, there is a big gap in the identification of the molecular components involved in the interaction between the host plant and the microbial population. Moreover, these recent microbiome analyses tried merely to identify its structure and complexity rather than to determine how these microbial assemblages are altering the plant phenome, which is essential to explore toward its utilization. In addition, there would be cross talk via signal transduction between aboveground and belowground plant tissues that can be altered by an external biotic or abiotic stress influencing the rhizospheric microbiome (Rudrappa et al., 2008; Yang et al., 2011; Lakshmanan et al., 2012; Lee et al., 2012). Plant health and fitness are greatly impacted by the microbiota, and this will continue to be an important research area, considering that plant fitness and crop productivity need to be carefully monitored for food security. A comprehensive understanding of the effects of the microbes on staple crops will allow the development of technologies that can exploit the natural alliances among microbes and plants and provide new avenues to increase yields beyond conventional plant genetics and breeding. Conclusively, further scientific endeavors in the direction of preserving, applying, and analyzing the whole microbiome for improving plant health and stress tolerance will ensure actualizing the human resources and financial and intellectual investments that we made in studying microbiomes.
Acknowledgments
We thank Shannon Modla (Delaware Biotechnology Institute) for capturing the electron micrograph of B. subtilis used in Figure 2 and Vidhyavathi Raman and Carla Spence (University of Delaware) for helping to draw a comparative microbiome in Figure 1 and for a critical reading of the article, respectively.
Glossary
- rRNA
ribosomal RNA
- DGGE
denaturing gradient gel electrophoresis
- SIP
stable isotope probing
- HTS
high-throughput sequencing
- OTUs
operational taxonomic units
- JA
jasmonic acid
Footnotes
This work was supported by the National Science Foundation (grant no. PGPR–0923806 to H.P.B.), the United States-India Educational Foundation (to G.S.), and a Fulbright-Nehru postdoctoral fellowship (grant no. 1847/FNPDR/2013 to G.S.).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- Ali B, Sabri AN, Ljung K, Hasnain S. (2009) Auxin production by plant associated bacteria: impact on endogenous IAA content and growth of Triticum aestivum L. Lett Appl Microbiol 48: 542–547 [DOI] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer K-H. (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM. (2013a) Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J Biol Chem 288: 4502–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Quintana N, El Kassis EG, Kim HK, Choi YH, Sugiyama A, Verpoorte R, Martinoia E, Manter DK, Vivanco JM. (2009a) An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol 151: 2006–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Vivanco JM. (2009) Regulation and function of root exudates. Plant Cell Environ 32: 666–681 [DOI] [PubMed] [Google Scholar]
- Badri DV, Weir TL, van der Lelie D, Vivanco JM. (2009b) Rhizosphere chemical dialogues: plant-microbe interactions. Curr Opin Biotechnol 20: 642–650 [DOI] [PubMed] [Google Scholar]
- Badri DV, Zolla G, Bakker MG, Manter DK, Vivanco JM. (2013b) Potential impact of soil microbiomes on the leaf metabolome and on herbivore feeding behavior. New Phytol 198: 264–273 [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266 [DOI] [PubMed] [Google Scholar]
- Bakker MG, Manter DK, Sheflin AM, Weir TL, Vivanco JM. (2012) Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 360: 1–13 [Google Scholar]
- Barret M, Morrissey JP, O’Gara F. (2011) Functional genomics analysis of plant growth-promoting rhizobacterial traits involved in rhizosphere competence. Biol Fertil Soils 47: 729–743 [Google Scholar]
- Baudoin E, Benizri E, Guckert A. (2002) Impact of growth stage on the bacterial community structure along maize roots, as determined by metabolic and genetic fingerprinting. Appl Soil Ecol 19: 135–145 [Google Scholar]
- Beattie GA, Lindow SE. (1995) The secret life of foliar bacterial pathogens on leaves. Annu Rev Phytopathol 33: 145–172 [DOI] [PubMed] [Google Scholar]
- Beneduzi A, Peres D, Vargas LK, Bodanese-Zanettini MH, Passaglia LMP. (2008) Evaluation of genetic diversity and plant growth promoting activities of nitrogen-fixing bacilli isolated from rice fields in South Brazil. Appl Soil Ecol 39: 311–320 [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM. (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486 [DOI] [PubMed] [Google Scholar]
- Berg G, Smalla K. (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68: 1–13 [DOI] [PubMed] [Google Scholar]
- Berlec A. (2012) Novel techniques and findings in the study of plant microbiota: search for plant probiotics. Plant Sci 193-194: 96–102 [DOI] [PubMed] [Google Scholar]
- Bever JD. (2003) Soil community feedback and the coexistence of competitors: conceptual frameworks and empirical tests. New Phytol 157: 465–473 [DOI] [PubMed] [Google Scholar]
- Bever JD, Platt TG, Morton ER. (2012) Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu Rev Microbiol 66: 265–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj D, Ansari MW, Sahoo RK, Tuteja N. (2014) Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb Cell Fact 13: 66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya PN, Jha DK. (2012) Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28: 1327–1350 [DOI] [PubMed] [Google Scholar]
- Bressan M, Roncato MA, Bellvert F, Comte G, Haichar FZ, Achouak W, Berge O. (2009) Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J 3: 1243–1257 [DOI] [PubMed] [Google Scholar]
- Broeckling CD, Broz AK, Bergelson J, Manter DK, Vivanco JM. (2008) Root exudates regulate soil fungal community composition and diversity. Appl Environ Microbiol 74: 738–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, et al. (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91–95 [DOI] [PubMed] [Google Scholar]
- Carvalhais LC, Dennis PG, Badri DV, Tyson GW, Vivanco JM, Schenk PM. (2013) Activation of the jasmonic acid plant defence pathway alters the composition of rhizosphere bacterial communities. PLoS ONE 8: e56457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro JM, Badri DV, Bakker MG, Sugiyama A, Manter DK, Vivanco JM. (2013) Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 8: e55731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro JM, Badri DV, Vivanco JM. (2014) Rhizosphere microbiome assemblage is affected by plant development. ISME J 8: 790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro JM, Sheflin AM, Manter DK, Vivanco JM. (2012) Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils 48: 489–499 [Google Scholar]
- Dangl JL, Jones JD. (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Dennis PG, Miller AJ, Hirsch PR. (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72: 313–327 [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Zaanen W, Koornneef A, Korzelius JP, Dicke M, Van Loon LC, Pieterse CMJ. (2006) Herbivore-induced resistance against microbial pathogens in Arabidopsis. Plant Physiol 142: 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weert S, Vermeiren H, Mulders IHM, Kuiper I, Hendrickx N, Bloemberg GV, Vanderleyden J, De Mot R, Lugtenberg BJJ. (2002) Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant Microbe Interact 15: 1173–1180 [DOI] [PubMed] [Google Scholar]
- Dini-Andreote F, van Elsas JD. (2013) Back to the basics: the need for ecophysiological insights to enhance our understanding of microbial behavior in the rhizosphere. Plant Soil 373: 1–15 [Google Scholar]
- Doornbos RF, van Loon LC, Bakker PAHM. (2012) Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere: a review. Agron Sustain Dev 32: 227–243 [Google Scholar]
- East R. (2013) Microbiome: soil science comes to life. Nature 501: S18–S19 [DOI] [PubMed] [Google Scholar]
- Fang C, Zhuang Y, Xu T, Li Y, Li Y, Lin W. (2013) Changes in rice allelopathy and rhizosphere microflora by inhibiting rice phenylalanine ammonia-lyase gene expression. J Chem Ecol 39: 204–212 [DOI] [PubMed] [Google Scholar]
- Fitzsimons MS, Miller RM. (2010) The importance of soil microorganisms for maintaining diverse plant communities in tallgrass prairie. Am J Bot 97: 1937–1943 [DOI] [PubMed] [Google Scholar]
- Forchetti G, Masciarelli O, Alemano S, Alvarez D, Abdala G. (2007) Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl Microbiol Biotechnol 76: 1145–1152 [DOI] [PubMed] [Google Scholar]
- Friedrich MW. (2006) Stable-isotope probing of DNA: insights into the function of uncultivated microorganisms from isotopically labeled metagenomes. Curr Opin Biotechnol 17: 59–66 [DOI] [PubMed] [Google Scholar]
- Gamalero E, Berta G, Glick BR (2009) The use of microorganisms to facilitate the growth of plants in saline soils. In MS Khan, A Zaidi, J Musarrat, eds, Microbial Strategies for Crop Improvement. Springer, Berlin, pp 1–22 [Google Scholar]
- Garbeva P, van Veen JA, van Elsas JD. (2004) Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42: 243–270 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glick BR. (1995) The enhancement of plant growth by free-living bacteria. Can J Microbiol 41: 109–117 [Google Scholar]
- Gupta VV. (2012) Beneficial microorganisms for sustainable agriculture. Microbiol Australia 3: 113–115 [Google Scholar]
- Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W. (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Heintz C, Mair W. (2014) You are what you host: microbiome modulation of the aging process. Cell 156: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa T, Parr JF (1994) Beneficial and Effective Microorganisms for a Sustainable Agriculture and Environment. International Nature Farming Research Center, Atami, Japan [Google Scholar]
- Hirano SS, Upper CD. (2000) Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae: a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev 64: 624–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi J, Prithiviraj B, Bais HP, Kimball BA, Vivanco JM. (2005) Soil nematodes mediate positive interactions between legume plants and rhizobium bacteria. Planta 222: 848–857 [DOI] [PubMed] [Google Scholar]
- Horrigan L, Lawrence RS, Walker P. (2002) How sustainable agriculture can address the environmental and human health harms of industrial agriculture. Environ Health Perspect 110: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden A, Timms-Wilson TM, Day MJ, Bailey MJ. (2008) Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiol Ecol 65: 193–201 [DOI] [PubMed] [Google Scholar]
- Huang XF, Chaparro JM, Reardon KF, Zhang R, Shen Q, Vivanco JM. (2014) Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92: 267–275 [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. (1998) Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180: 4765–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Okubo T, Takeda N, Banba M, Sasaki K, Imaizumi-Anraku H, Fujihara S, Ohwaki Y, Ohshima K, Fukuta Y, et al. (2011) The genotype of the calcium/calmodulin-dependent protein kinase gene (CCaMK) determines bacterial community diversity in rice roots under paddy and upland field conditions. Appl Environ Microbiol 77: 4399–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Nainawatee HS. (2002) Plant flavonoids: signals to legume nodulation and soil microorganisms. J Plant Biochem Biotechnol 11: 1–10 [Google Scholar]
- Jetiyanon K, Kloepper JW. (2002) Mixtures of plant growth-promoting rhizobacteria for induction of systemic resistance against multiple plant diseases. Biol Control 24: 285–291 [Google Scholar]
- Johansson JF, Paul LR, Finlay RD. (2004) Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol Ecol 48: 1–13 [DOI] [PubMed] [Google Scholar]
- Jones DL, Nguyen C, Finlay RD. (2009) Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant Soil 321: 5–33 [Google Scholar]
- Keiblinger KM, Wilhartitz IC, Schneider T, Roschitzki B, Schmid E, Eberl L, Riedel K, Zechmeister-Boltenstern S. (2012) Soil metaproteomics: comparative evaluation of protein extraction protocols. Soil Biol Biochem 54: 14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klironomos JN. (2002) Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417: 67–70 [DOI] [PubMed] [Google Scholar]
- Kloepper JW, Ryu CM, Zhang S. (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94: 1259–1266 [DOI] [PubMed] [Google Scholar]
- Kloepper JW, Schroth MN (1978) Plant growth-promoting rhizobacteria in radish. In Station de pathologie vegetale et phyto-bacteriologie (ed.), Proceedings of the 4th International Conference on Plant Pathogenic Bacteria, vol II. Gilbert-Clarey, Tours, France, pp 879–882 [Google Scholar]
- Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, von Mering C, Vorholt JA. (2012) Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6: 1378–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniskern JM, Traw MB, Bergelson J. (2007) Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol Plant Microbe Interact 20: 1512–1522 [DOI] [PubMed] [Google Scholar]
- Kolmeder CA, de Vos WM. (2014) Metaproteomics of our microbiome: developing insight in function and activity in man and model systems. J Proteomics 97: 3–16 [DOI] [PubMed] [Google Scholar]
- Krzyzanowska D, Obuchowski M, Bikowski M, Rychlowski M, Jafra S. (2012) Colonization of potato rhizosphere by GFP-tagged Bacillus subtilis MB73/2, Pseudomonas sp. P482 and Ochrobactrum sp. A44 shown on large sections of roots using enrichment sample preparation and confocal laser scanning microscopy. Sensors (Basel) 12: 17608–17619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U. (2007) Plant-associated methylobacteria as co-evolved phytosymbionts: a hypothesis. Plant Signal Behav 2: 74–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan V, Bais HP. (2013) Factors other than root secreted malic acid that contributes toward Bacillus subtilis FB17 colonization on Arabidopsis roots. Plant Signal Behav 8: e27277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan V, Kitto SL, Caplan JL, Hsueh YH, Kearns DB, Wu YS, Bais HP. (2012) Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol 160: 1642–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankau RA. (2011) Conflicts in maintaining biodiversity at multiple scales. Mol Ecol 20: 2035–2037 [DOI] [PubMed] [Google Scholar]
- Larson JL, Siemann E. (1998) Legumes may be symbiont-limited during old-field succession. Am Midl Nat 140: 90–95 [Google Scholar]
- Lau JA, Lennon JT. (2011) Evolutionary ecology of plant-microbe interactions: soil microbial structure alters selection on plant traits. New Phytol 192: 215–224 [DOI] [PubMed] [Google Scholar]
- Lavelle P, Spain AV (2001) Soil Ecology. Kluwer, Dordrecht, The Netherlands [Google Scholar]
- Lee B, Lee S, Ryu CM. (2012) Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non-pathogenic bacteria in pepper. Ann Bot (Lond) 110: 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809 [DOI] [PubMed] [Google Scholar]
- Lindow SE, Brandl MT. (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69: 1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang N, Qiu M, Feng H, Vivanco JM, Shen Q, Zhang R. (2014) Enhanced rhizosphere colonization of beneficial Bacillus amyloliquefaciens SQR9 by pathogen infection. FEMS Microbiol Lett 353: 49–56 [DOI] [PubMed] [Google Scholar]
- Long SR. (1989) Rhizobium-legume nodulation: life together in the underground. Cell 56: 203–214 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, de La Vega OM, Guevara-García A, Herrera-Estrella L. (2000) Enhanced phosphorus uptake in transgenic tobacco plants that overproduce citrate. Nat Biotechnol 18: 450–453 [DOI] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F. (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63: 541–556 [DOI] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, et al. (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JM, da Silva TF, Vollu RE, Blank AF, Ding GC, Seldin L, Smalla K. (2014) Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol Ecol 88: 424–435 [DOI] [PubMed] [Google Scholar]
- Marschner P, Neumann G, Kania A, Weiskopf L, Lieberei R. (2002) Spatial and temporal dynamics of the microbial community structure in the rhizosphere of cluster roots of white lupin (Lupinus albus L.). Plant Soil 246: 167–174 [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, et al. (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332: 1097–1100 [DOI] [PubMed] [Google Scholar]
- Micallef SA, Channer S, Shiaris MP, Colón-Carmona A. (2009) Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal Behav 4: 777–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minz D, Ofek M, Hadar Y (2013) Plant rhizosphere microbial communities. In EF DeLong, S Lory, E Stackebrandt, F Thompson, eds, The Prokaryotes: Prokaryotic Communities and Ecophysiology. Springer-Verlag, Berlin, pp 57–84 [Google Scholar]
- Mishra PK, Mishra S, Selvakumar G, Kundub S, Gupta HS. (2009) Enhanced soybean (Glycine max L.) plant growth and nodulation by Bradyrhizobium japonicum-SB1 in presence of Bacillus thuringiensis-KR1. Acta Agric Scand B Soil Plant Sci 59: 189–196 [Google Scholar]
- Moe LA. (2013) Amino acids in the rhizosphere: from plants to microbes. Am J Bot 100: 1692–1705 [DOI] [PubMed] [Google Scholar]
- Mougel C, Offre P, Ranjard L, Corberand T, Gamalero E, Robin C, Lemanceau P. (2006) Dynamic of the genetic structure of bacterial and fungal communities at different developmental stages of Medicago truncatula Gaertn. cv. Jemalong line J5. New Phytol 170: 165–175 [DOI] [PubMed] [Google Scholar]
- Mullis KB, Erlich HA, Arnheim N, Horn GT, Saiki RK, Scharf SJ July 28, 1987. Process for amplifying, detecting, and/or-cloning nucleic acid sequences. US Patent Application No. US4683195 A
- Muyzer G. (1999) DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol 2: 317–322 [DOI] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G. (2003) Microbial diversity and soil functions. Eur J Soil Sci 54: 655–670 [Google Scholar]
- Narula N, Deubel A, Gans W, Behl RK, Merbach W. (2006) Paranodules and colonization of wheat roots by phytohormone producing bacteria in soil. Plant Soil Environ 52: 119–129 [Google Scholar]
- Neal AL, Ahmad S, Gordon-Weeks R, Ton J. (2012) Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 7: e35498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortíz-Castro R, Martínez-Trujillo M, López-Bucio J. (2008) N-Acyl-L-homoserine lactones: a class of bacterial quorum-sensing signals alter post-embryonic root development in Arabidopsis thaliana. Plant Cell Environ 31: 1497–1509 [DOI] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C, Rentsch D, Robatzek S, Webb RI, Sagulenko E, Näsholm T, Schmidt S, Lonhienne TGA. (2010) Turning the table: plants consume microbes as a source of nutrients. PLoS ONE 5: e11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer JA, Ley RE. (2013) Exploring the maize rhizosphere microbiome in the field: a glimpse into a highly complex system. Commun Integr Biol 6: e25177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 110: 6548–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11: 789–799 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van Pelt JA, Van Wees SCM, Ton J, Léon-Kloosterziel KM, Keurentjes JJB, Verhagen BWM, Knoester M, Van der Sluis I, Bakker PAHM, et al. (2001) Rhizobacteria-mediated induced systemic resistance: triggering, signalling and expression. Eur J Plant Pathol 107: 51–61 [Google Scholar]
- Pieterse CMJ, van Pelt JA, Verhagen BWM, Ton J, van Wees SCM, Leon-Kloosterziel KM, van Loon LC. (2003) Induced systemic resistance by plant growth-promoting rhizobacteria. Symbiosis 35: 39–54 [Google Scholar]
- Pineda A, Zheng SJ, van Loon JJA, Pieterse CMJ, Dicke M. (2010) Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci 15: 507–514 [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Van Der Ent S, Van Loon LC, Pieterse CMJ. (2008) Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol 180: 511–523 [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y. (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321: 341–361 [Google Scholar]
- Ramamoorthy V, Viswanathan R, Raguchander T, Prakasam V, Samiyappan R. (2001) Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop Prot 20: 1–11 [Google Scholar]
- Rejmanek M, Richardson DM. (1996) What attributes make some plant species more invasive? Ecology 77: 1655–1661 [Google Scholar]
- Rincon-Florez VA, Carvalhais LC, Schenk PM. (2013) Culture-independent molecular tools for soil and rhizosphere microbiology. Diversity 5: 581–612 [Google Scholar]
- Rosenblueth M, Martínez-Romero E. (2006) Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact 19: 827–837 [DOI] [PubMed] [Google Scholar]
- Rosenzweig N, Bradeen JM, Tu ZJ, McKay SJ, Kinkel LL. (2013) Rhizosphere bacterial communities associated with long-lived perennial prairie plants vary in diversity, composition, and structure. Can J Microbiol 59: 494–502 [DOI] [PubMed] [Google Scholar]
- Rout ME, Southworth D. (2013) The root microbiome influences scales from molecules to ecosystems: the unseen majority. Am J Bot 100: 1689–1691 [DOI] [PubMed] [Google Scholar]
- Rudrappa T, Biedrzycki ML, Kunjeti SG, Donofrio NM, Czymmek KJ, Paré PW, Bais HP. (2010) The rhizobacterial elicitor acetoin induces systemic resistance in Arabidopsis thaliana. Commun Integr Biol 3: 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa T, Czymmek KJ, Paré PW, Bais HP. (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. (2014) Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci USA 111: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer SA, Klironomos JN, Hillerislambers J, Kinkel LL, Reich PB, Xiao K, Rillig MC, Sikes BA, Callaway RM, Mangan SA, et al. (2011) Soil microbes drive the classic plant diversity-productivity pattern. Ecology 92: 296–303 [DOI] [PubMed] [Google Scholar]
- Shakya M, Gottel N, Castro H, Yang ZK, Gunter L, Labbé J, Muchero W, Bonito G, Vilgalys R, Tuskan G, et al. (2013) A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS ONE 8: e76382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, Ramesh A, Sharma MP, Joshi OP, Govaerts B, Steenwerth KL, Karlen DL (2010) Microbial community structure and diversity as indicators for evaluating soil quality. In E Lichtfouse, ed, Biodiversity, Biofuels, Agroforestry and Conservation Agriculture, Vol 5. Springer, Dordrecht, The Netherlands, pp 317–358 [Google Scholar]
- Shaw CH. (1991) Swimming against the tide: chemotaxis in Agrobacterium. BioEssays 13: 25–29 [DOI] [PubMed] [Google Scholar]
- Shi S, Richardson AE, O’Callaghan M, DeAngelis KM, Jones EE, Stewart A, Firestone MK, Condron LM. (2011) Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol Ecol 77: 600–610 [DOI] [PubMed] [Google Scholar]
- Spence C, Alff E, Johnson C, Ramos C, Donofrio N, Sundaresan V, Bais H. (2014) Natural rice rhizospheric microbes suppress rice blast infections. BMC Plant Biol 14: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence C, Bais H (2013) Probiotics for plants: rhizospheric microbiome and plant fitness. In FJ de Bruijn, ed, Molecular Microbial Ecology of the Rhizosphere Vols 1 and 2. John Wiley & Sons, Hoboken, NJ, pp 713–721 10.1002/9781118297674.ch67 [Google Scholar]
- Tan S, Yang C, Mei X, Shen S, Raza W, Shen O, Xu Y. (2013) The effect of organic acids from tomato root exudates on rhizosphere colonization of Bacillus amyloliquefaciens T5. Appl Soil Ecol 64: 15–22 [Google Scholar]
- Taulé C, Mareque C, Barlocco C, Hackembruch F, Reis VM, Sicardi M, Battistoni F. (2012) The contribution of nitrogen fixation to sugarcane (Saccharum officinarum L.), and the identification and characterization of part of the associated diazotrophic bacterial community. Plant Soil 356: 35–49 [Google Scholar]
- Turner TR, James EK, Poole PS. (2013a) The plant microbiome. Genome Biol 14: 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, Swarbreck D, Osbourn A, Grant A, Poole PS. (2013b) Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J 7: 2248–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlik O, Leewis MC, Strejcek M, Musilova L, Mackova M, Leigh MB, Macek T. (2013) Stable isotope probing in the metagenomics era: a bridge towards improved bioremediation. Biotechnol Adv 31: 154–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren NC (2001) Types, amounts and possible functions of compounds released into the rhizosphere by soil-grown plants. In R Pinton, Z Varanini, P Nannipieri, eds, The Rhizosphere: Biochemistry and Organic Substances at the Soil-Plant Interface. Marcel Dekker, New York, pp 19–40 [Google Scholar]
- van der Heijden MGA, Bardgett RD, van Straalen NM. (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11: 296–310 [DOI] [PubMed] [Google Scholar]
- van Loon LC, Bakker PAHM, Pieterse CMJ. (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36: 453–483 [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ. (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Van Oosten VR, Bodenhausen N, Reymond P, Van Pelt JA, Van Loon LC, Dicke M, Pieterse CMJ. (2008) Differential effectiveness of microbially induced resistance against herbivorous insects in Arabidopsis. Mol Plant Microbe Interact 21: 919–930 [DOI] [PubMed] [Google Scholar]
- Vessey JK. (2003) Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255: 571–586 [Google Scholar]
- Wagg C, Jansa J, Schmid B, van der Heijden MG. (2011) Belowground biodiversity effects of plant symbionts support aboveground productivity. Ecol Lett 14: 1001–1009 [DOI] [PubMed] [Google Scholar]
- Walker TS, Bais HP, Grotewold E, Vivanco JM. (2003) Root exudation and rhizosphere biology. Plant Physiol 132: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert N, Piceno Y, Ding GC, Meincke R, Heuer H, Berg G, Schloter M, Andersen G, Smalla K. (2011) PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: many common and few cultivar-dependent taxa. FEMS Microbiol Ecol 75: 497–506 [DOI] [PubMed] [Google Scholar]
- White JF, Jr, Crawford H, Torres MS, Mattera R, Irizarry I, Bergen M. (2012) A proposed mechanism for nitrogen acquisition by grass seedlings through oxidation of symbiotic bacteria. Symbiosis 57: 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JF, Torres MS, Johnson H, Irizarry I, Tadych M (2014) A functional view of plant microbiomes: endosymbiotic systems that enhance plant growth and survival. In VC Verma, AC Gange, eds, Advances in Endophytic Research. Springer, Bangalore, India, pp 425–439 [Google Scholar]
- Whiteley AS, Manefield M, Lueders T. (2006) Unlocking the ‘microbial black box’ using RNA-based stable isotope probing technologies. Curr Opin Biotechnol 17: 67–71 [DOI] [PubMed] [Google Scholar]
- Woese CR, Fox GE. (1977) Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA 74: 5088–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Wang W, Ma Y, Liu Y, Ma X, An L, Feng H. (2013) Prospect of beneficial microorganisms applied in potato cultivation for sustainable agriculture. Afr J Microbiol Res 7: 2150–2158 [Google Scholar]
- Yang JW, Yi HS, Lee B, Lee S, Ghim SY, Ryu CM. (2011) Whitefly infestation of pepper plants elicits defence responses against bacterial pathogens in leaves and roots and changes the below-ground microflora. J Ecol 99: 46–56 [Google Scholar]
- Yergeau E, Sanschagrin S, Maynard C, St-Arnaud M, Greer CW. (2014) Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J 8: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wu F. (2012) p-Coumaric acid influenced cucumber rhizosphere soil microbial communities and the growth of Fusarium oxysporum f.sp. cucumerinum Owen. PLoS ONE 7: e48288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wu F. (2013) Artificially applied vanillic acid changed soil microbial communities in the rhizosphere of cucumber (Cucumis sativus L.). Can J Soil Sci 93: 13–21 [Google Scholar]
- Zolla G, Badria DV, Bakker MG, Manter DK, Vivanco JM. (2013) Soil microbiomes vary in their ability to confer drought tolerance to Arabidopsis. Appl Soil Ecol 68: 1–9 [Google Scholar]