Roots and microbes secrete organic compounds into the rhizosphere that influence plant productivity.
Abstract
Microbes and plants have evolved biochemical mechanisms to communicate with each other. The molecules responsible for such communication are secreted during beneficial or harmful interactions. Hundreds of these molecules secreted into the rhizosphere have been identified, and their functions are being studied in order to understand the mechanisms of interaction and communication among the different members of the rhizosphere community. The importance of root and microbe secretion to the underground habitat in improving crop productivity is increasingly recognized, with the discovery and characterization of new secreting compounds found in the rhizosphere. Different omic approaches, such as genomics, transcriptomics, proteomics, and metabolomics, have expanded our understanding of the first signals between microbes and plants. In this review, we highlight the more recent discoveries related to molecules secreted into the rhizosphere and how they affect plant productivity, either negatively or positively. In addition, we include a survey of novel approaches to studying the rhizosphere and emerging opportunities to direct future studies.
Due to the increasing human population, the continued production of food and energy is a basic challenge today (Edgerton, 2009; Ray et al., 2013). With more than seven billion people to feed, the productive yield of crops needs to be higher, more sustainable, and more efficient worldwide. Productivity is not only the plant growth per hectare in the field. It is also defined by the fitness, food production, and healthy development of plants (Boyer, 1982; Edgerton, 2009). Most losses in food production are due to diseases caused by different pathogens and pests, the effect of which is augmented by abiotic stresses such as heat and drought. Although some microorganisms responsible for plant diseases colonize the upper part of the plant, many more are found in the soil and are capable of either destroying a vast number of cultivars (Borneman and Becker, 2007; Berendsen et al., 2012) or building useful symbiotic relationships (Venkateshwaran et al., 2013).
The criticality of the interaction between roots and microorganisms to agricultural output is increasingly recognized. There are many chemicals secreted from microorganisms and roots, such as amino acids, organic acids, flavonols, glucosinolates, indole compounds, fatty acids, polysaccharides, and proteins in the rhizosphere (De-la-Peña et al., 2012a; Nguema-Ona et al., 2013; Weston et al., 2013; Li et al., 2014; Talboys et al., 2014; Zhang et al., 2014), that act as signals; once the recipient organisms recognize them, the process of communication and interaction begins. The type and composition of root secretion can alter the microbial dynamic and diversity of the soil, favoring the growth of microorganisms that can benefit plant health and crop productivity, while, in other cases, root-exuded compounds prevent the growth of harmful microbes (Bais et al., 2006; Chaparro et al., 2012; Dutta et al., 2013; Li et al., 2013). Phytochemicals collected from the root exudates of Arabidopsis (Arabidopsis thaliana) added into the soil have shown how important these compounds are to the modulation of microbe composition (Badri et al., 2013).
Although soil microorganisms are associated with plant disease and productivity losses, numerous bacteria and fungi species are beneficial for plant productivity (van der Heijden et al., 2008; Pérez-García et al., 2011; Venkateshwaran et al., 2013). For instance, the rhizobia bacteria build a social network with legumes in a symbiotic structure called a nodule, where atmospheric nitrogen is fixed and transferred to a plant for its use (Spaink, 2000). To overcome nitrogen deficiency, legumes release specific flavonoids that attract and initiate these symbiotic relationships with rhizobia (Zhang et al., 2009), which secrete exopolysaccharides needed for symbiosis (Cheng and Walker, 1998; Berge et al., 2009). This symbiosis introduces 50 to 70 × 106 tons of nitrogen annually into the soil of agricultural systems (Herridge et al., 2008), which reduces the use of fertilizers that, depending on the dose and type, can have a detrimental environmental impact. On the other hand, arbuscular mycorrhizal fungi (AMF) also benefit plant productivity by spreading their hyphal networks into the soil to acquire nutrients, such as phosphorous, which are then supplied to their hosts (Joner et al., 2000; Klironomos et al., 2000; Jeffries et al., 2003).
Based on this explosion of new knowledge of rhizosphere interactions, the question becomes, how many molecules in the rhizosphere have important roles in plant productivity? And why are some molecules, from microbes and/or plants, harmful enough to destroy hectares of cultivars (Shieh et al., 1997; Weir et al., 2003; Oerke, 2006; Ejeta and Gressel, 2007; Joel et al., 2007)? With the advance of new technologies, such as proteomics and metabolomics, the ability to identify secreted molecules has revealed important clues about their possible functions (De-la-Peña and Loyola-Vargas, 2012; Badri et al., 2013; Martin et al., 2014; Neumann et al., 2014), but the ecological and environmental relevance of many of them remains unresolved. In this review, we will focus on how root exudates are key players involved in the selection of the microbial community during plant-microbe interaction and how this interaction affects the productivity of plants in the field.
BIOTIC INTERACTIONS IN THE RHIZOSPHERE ARE IMPORTANT FOR PLANT PRODUCTIVITY
The rhizosphere is an environment where plants interact with other plants, herbivores, and microorganisms (Lynch and Whipps, 1990; Barea et al., 2005; Bais et al., 2006). The rhizosphere not only represents the trade zone in which pathogens or neighbor roots interact with the plant but is also a preventive microbial buffer zone that protects against infection (Baetz and Martinoia, 2014). In general, the rhizosphere has three main zones: endorhizosphere, rhizoplane, and ectorhizosphere. The first zone, the endorhizosphere, includes the root cortical and endodermal tissue, while the rhizoplane encompasses the root epidermis and associated mucilage. The ectorhizosphere covers the soil near the root (Lynch and Whipps, 1990; Badri and Vivanco, 2009). The rhizosphere can contain up to 1011 microbial cells per gram of root (Egamberdieva et al., 2008) and more than 30,000 prokaryotic species that could influence plant productivity (Mendes et al., 2011, 2013). Although the total number of these microorganisms is likely underestimated due to culturing limitations, it is known that plants are able to shape their rhizosphere microbiome in order to recruit protective bacteria or fungi (Berendsen et al., 2012).
Plant development and growth have benefited from the diversity and complexity of the microbial society. There is not a single organism responsible for a beneficial effect in plants; rather, the multiple interactions between all the actors work together. The interspecies or interkingdom cross talk in the rhizosphere that is produced during plant-microbe or plant-plant interactions is essential for the function, health, stability, and sustainability of ecosystems, including the farming environment (Bais et al., 2004b; Shukla et al., 2008; Broz et al., 2010; Pellegrino and Bedini, 2014). Some of the most studied biotic relationships for plant productivity are those produced by plant growth-promoting rhizobacteria (PGPRs), AMF, and invasive plant species.
PGPR Important in Plant Productivity and Defense
Plant roots are able to recruit beneficial soil bacteria, called PGPRs, from a wide range of genera, including Acinetobacter, Alcaligenes, Azospirillus, Bacillus, Pseudomonas, Rhizobium, and others. These rhizobacteria are important due to their functions as producers of plant growth regulators, solubilizers of phosphorus, biofertilizers, and elicitors of tolerance to abiotic and biotic stresses (Yang et al., 2009; Bhattacharyya and Jha, 2012).
The major classification of the PGPRs is as biofertilizers, phytostimulators, and biopesticides (Bhattacharyya and Jha, 2012; Bhardwaj et al., 2014; Pérez-Montaño et al., 2014). The function of the biofertilizers is to promote plant growth by supplying nutrients to the host; examples of biofertilizers are Allorhizobium spp., Trichoderma spp. (e.g. Trichoderma hamatum and Trichoderma asperellum), Pseudomonas fluorescens, and Rhizobium spp. (Vessey, 2003; Badar and Qureshi, 2012; Bhattacharyya and Jha, 2012; Yadav et al., 2013). The phytostimulators, on the other hand, produce phytohormones such as indole acetic acid (IAA), GA3, and cytokinins, which alter root architecture and promote plant development (Vessey, 2003; Spaepen et al., 2007; Apine and Jadhav, 2011; Duca et al., 2014). In this group are Bacillus, Pseudomonas, Azosporillus, Enterobacter, Azotobacter, Pantoea, Streptomyces, and Rhizobium spp. Finally, examples of biopesticides are Pseudomonas spp. (e.g. P. fluorescens), Streptomyces spp., and Bacillus spp. (e.g. Bacillus subtilis), which function to inhibit pathogen proliferation and help plant growth (Copping and Menn, 2000; Radja Commare et al., 2002; Bhattacharyya and Jha, 2012). Besides the biofertilizers, phytostimulators, and biopesticides, there are other PGPRs that induce tolerance in plants to abiotic stress. For instance, Paenibacillus polymyxa, Achromobacter piechaudii, and Rhizobium tropici confer tolerance to drought stress in Arabidopsis, tomato (Solanum lycopersicum), and common bean (Phaseolus vulgaris), respectively, possibly by abscisic acid accumulation and degradation of reactive oxygen species and 1-aminocyclopropane-1-carboxylate (Timmusk and Wagner, 1999; Mayak et al., 2004b; Figueiredo et al., 2008; Yang et al., 2009). Achromobacter piechaudii and B. subtilis are also involved in salinity tolerance in plants (Mayak et al., 2004a; Zhang et al., 2008; Yang et al., 2009). All these functions are important to improve crop productivity, and in a time of escalated climate change, the need to reduce mineral fertilizers and produce plants tolerant to abiotic stresses is becoming an international priority.
Much discussion has centered on the use of rhizobacteria as commercial biofertilizers. In a recent review, Shen et al. (2013) pointed out that in order to have the highest crop productivity, plants need an optimal nutrient input and resilience under stress. This allows plants to efficiently use soil nutrients through maximizing root/rhizosphere efficiency in nutrient mobilization and acquisition. Such mobilization is done at a high rate by rhizosphere bacteria (Koller et al., 2013; Parmar and Sindhu, 2013). Plant agriculture needs PGPRs to increase plant growth, facilitate nutrient availability, enhance seed emergence, and favor plant health (Suslow et al., 1979; Zehnder et al., 2001; Lugtenberg and Kamilova, 2009; Mia et al., 2010), and the way plants recruit these beneficial bacteria is through chemical root secretion. For instance, legumes release specific flavonoids to attract and initiate symbiotic relationships with rhizobia (Zhang et al., 2009). Other plants, such as maize (Zea mays), secrete a benzoxazinoid called 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one to attract the rhizobacterium Pseudomonas putida KT2440, which helps to repel other pathogenic microbes in the maize rhizosphere (Neal et al., 2012).
The way that PGPRs favor plant health is by calling for beneficial bacteria when a pathogen is attacking. For instance, the infection of Arabidopsis by Pseudomonas syringae pv tomato DC3000 is able to induce the expression of the l-malic acid (MA) transporter ALUMINUM-ACTIVATED MALATE TRANSPORTER1, increasing the secretion of MA by roots (Rudrappa et al., 2008; Lakshmanan et al., 2012). Once the MA is in the rhizosphere, it recruits the beneficial rhizobacterium B. subtilis FB17 in a dose-dependent manner and promotes the biofilm formation of B. subtilis FB17 on Arabidopsis roots (Rudrappa et al., 2008; Lakshmanan et al., 2013), producing a systemic resistance response against the pathogen. Besides MA, some bacteria secrete antimicrobial metabolites (e.g. cyclic lipopeptide surfactin and iturin A) that serve as a protective shield in roots against pathogenic fungi like Rhizoctonia spp. or pathogenic gram-negative bacteria such as P. syringae (Asaka and Shoda, 1996; Bais et al., 2004a). Although P. syringae is one of the most studied pathogenic bacteria, which has been accorded first place on a list of the top 10 plant pathogenic bacteria (Mansfield et al., 2012), this bacterium also has been useful in controlling crop diseases (Ligon et al., 2000). For instance, diverse species of the genus Pseudomonas, including Pseudomonas cepacia, P. fluorescens, Pseudomonas aeruginosa, and Pseudomonas aureofaciens, produce hydrogen cyanide, 2,4-diacetylphloroglucinol, pyrrolnitrin, phenazine, oomocyn A, and other compounds that in some way help protect the plant against diseases (Burkhead et al., 1994; Raaijmakers and Weller, 1998; Haas and Keel, 2003). The production of these compounds depends on different factors; for instance, oomycin A and 2,4-diacetylphloroglucinol are stimulated by Glc (Gutterson, 1990; Duffy and Défago, 1999), hydrogen cyanide is affected by light and temperature (Vickery et al., 1987), and an acid pH seems to enhance the production of pyrrolnitrin (Hwang et al., 2002). Therefore, it is tempting to speculate that changes in the soil environment due to climate changes (Davidson and Janssens, 2006; Frey et al., 2013) also could affect antibiotic production from beneficial bacteria, making plants more susceptible to pathogen attack and damaging plant productivity.
Other important compounds secreted by gram-negative bacteria, such as P. aeruginosa, Erwinia chrysanthemi, and many others, include N-acyl-homoserine lactones (AHLs), which are the principal signal components for quorum sensing (QS), the cell-to-cell communication system in bacteria (Dong and Zhang, 2005). Because the AHL signal is a key factor for virulence gene expression in pathogenic bacteria, this signal provides a way to manipulate the QS systems and interrupt communication between bacteria. There are several chemicals and enzymes that target different components of the bacterial QS system to disrupt QS signaling, a process known as quorum quenching (QQ; Hong et al., 2012). The first QQ enzyme, an AHL-lactonase, was isolated from Bacillus sp. 240B1 (Dong et al., 2000, 2001). These QQ molecules could be developed for agricultural applications through targeting the QS signals by inhibiting AHL biosynthesis or by degrading AHL as a strategy to avoid bacteria pathogenicity; both are likely to be ecologically benign. One of the most efficient quencher strains characterized is Bacillus cereus U92, which has been used as a biocontrol agent in the rhizospheres of tomato and potato (Zamani et al., 2013).
AMF
AMF belong to the ancient phylum Glomeromycota and have established biotic interactions with more than 80% of land plant families, including many agriculturally important crop species, such as maize, rice (Oryza sativa), wheat (Triticum aestivum), and soybean (Glycine max). As such, they contribute to plant nutrition and disease resistance (Gutjahr and Parniske, 2013). The colonization of roots by the AMF is carried out after an exchange of chemical signals among the fungi and the plant. The primary signal is produced and secreted by the roots of the host; it has been identified as various strigolactones (SLs; Akiyama and Hayashi, 2006), which induce germination and hyphal branching in AMF as well as stimulate fungal metabolism (Akiyama et al., 2005; Besserer et al., 2006; Zwanenburg and Pospíšil, 2013). The fungus responds to that signal, secreting tetrameters and pentamers of N-acetylglucosamine and lipochitin-oligosaccharides (Maillet et al., 2011; Czaja et al., 2012; Gutjahr and Parniske, 2013), which activate a signaling pathway in the roots of the host. Once the communication network has been established between the fungus and the plant, the main interface for symbiotic nutrient exchange begins (Bücking et al., 2012). The interaction between plants and AMF is bidirectional; AMF receive carbohydrates from the plant and compensate it with mineral nutrients, abiotic stress resistance (Bücking et al., 2012), and improved water supply (Parniske, 2008). This interaction is so important to the plant that it is able to transfer between 4% and 20% of its photosynthetically fixed carbon to the AMF (Wright et al., 1998).
The formation of AFM depends not only on host root exudates but also on soil phosphorus conditions (Nagahashi et al., 1996; Tamasloukht et al., 2003), normally through phosphorus fertilizer management (Grant et al., 2005). If the phosphorus concentration is too high (e.g. 10 mm), the growth of the fungal hyphae is inhibited and the AMF colonization is reduced (Nagahashi et al., 1996). Among other factors that could affect AFM colonization are the use of monoculture, tillage (Berruti et al., 2014), modern agricultural practices (Toth et al., 1990; Hetrick et al., 1992), and, still controversial, genetically modified crops (e.g. maize; Liu, 2010).
Root Secretion of Invasive Species Increases Crop Losses
The plant-plant relationship is also an important biotic interaction that has significant repercussions on plant productivity. The allelopathy phenomenon, in which “one plant negatively affects another through chemicals exuded or secreted,” may arise during the interaction between invasive and native species and may involve the microflora in the rhizosphere (Bains et al., 2009). Invasive plants, which are responsible for high negative economic effects (Vilà et al., 2009; McLaughlan et al., 2014), have different mechanisms to take possession of the land, frequently through altered pathways or adapted mechanisms that appear in the new environment. All plants secrete compounds; however, some invasive plants have evolved an adaptive mechanism(s) through which they secrete new compounds that adversely affect the native plant populations of their new habitat (enemy-release hypothesis). Allelochemicals can affect metabolite production, respiration, photosynthesis, and membrane transport and can inhibit root and shoot growth in susceptible plants (Einhellig, 1994; Weir et al., 2004).
Many allelochemicals have been identified as components of the root exudation of invasive species (Table I). One of the most studied has been catechin, a flavonoid present in the root exudates of the invasive spotted knapweeds Centaurea maculosa and Centaurea stoebe, which exhibits a strong inhibitory effect on a number of plant species (Bais et al., 2003; Weir et al., 2003; Tharayil and Triebwasser, 2010). It has been found that the concentration and effect of this compound increase in response to light intensity (Tharayil and Triebwasser, 2010). This resolves, in part, the question of why the concentration and the allelopathic effect of catechin change under different conditions (Perry et al., 2007). Perry et al. (2007) found that the concentration of catechin was higher during the months of May to August, when there are more hours of light. Although the molecular mechanism by which this is achieved is unknown, it has been proposed that light regulates flavonoid biosynthesis and, therefore, catechin accumulation (Tharayil and Triebwasser, 2010; Hong et al., 2014).
Table I. Principal compounds found in root exudates with negative and positive effects on plant productivity.
| Species | Compound | Structure | Function | Effect on Productivity | Reference |
|---|---|---|---|---|---|
| Alliaria petiolata | Benzyl isothiocyanate | 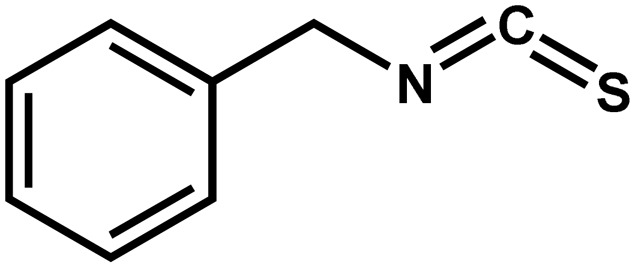 |
Inhibits the growth of ectomycorrhizal fungi | Negative | Stinson et al. (2006); Wolfe et al. (2008) |
| Centaurea maculosa | (−)-Catechin | 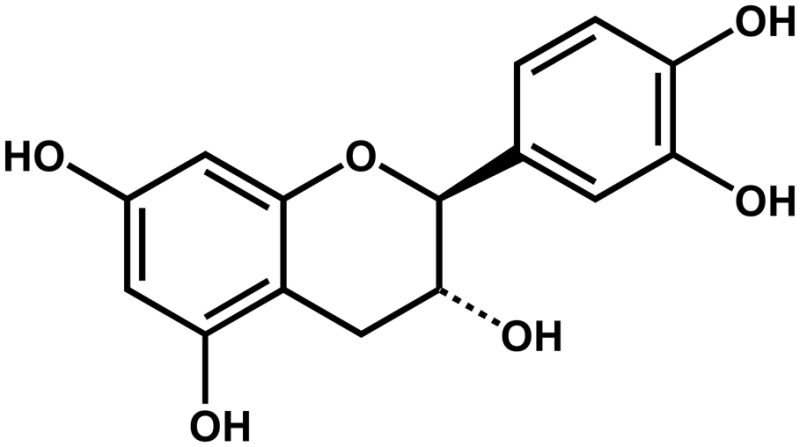 |
Reduces the growth of native plant species | Negative | Bais et al. (2003); Weir et al. (2003) |
| Juglans nigra | Juglone (5-hydroxy-1,4-napthoquinone) | 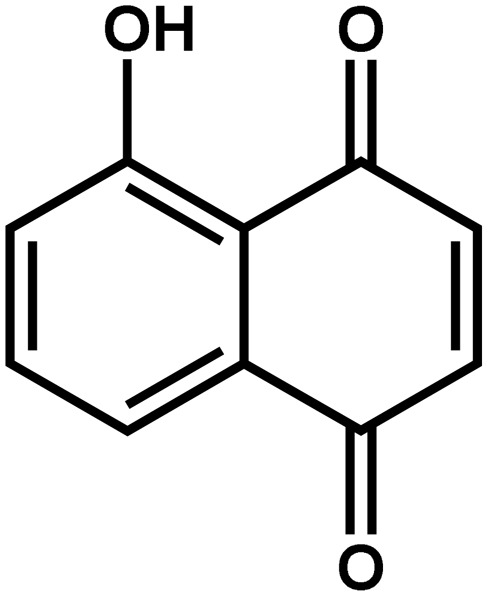 |
Inhibits the growth of crops, mainly shoot elongation | Negative | Rietveld (1983); Jose and Gillespie (1998) |
| Phragmites australis | Gallic acid (3,4,5-trihydroxybenzoic acid) | 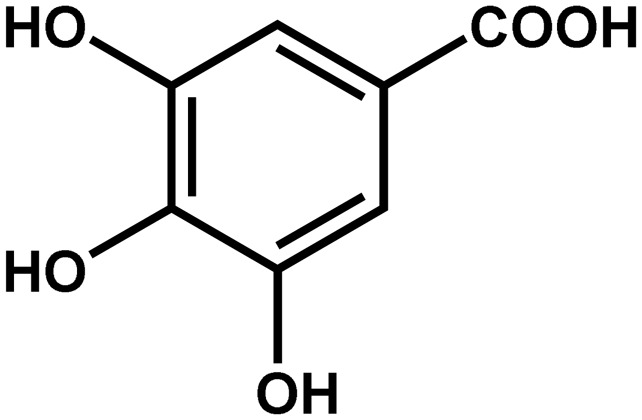 |
Allelopathic agent | Negative | Saltonstall (2002) |
| Arabidopsis | Rhizathalene A | 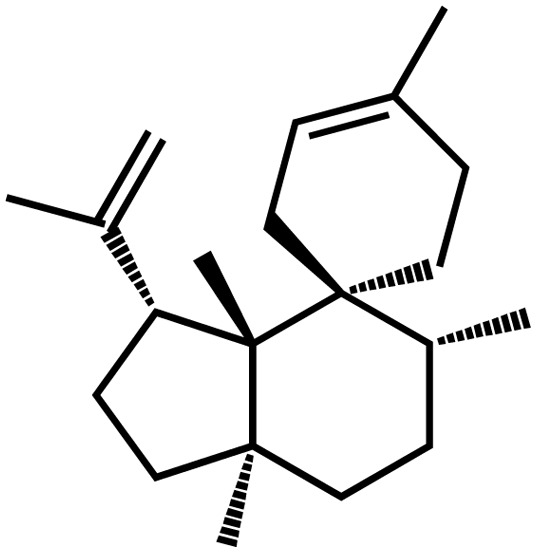 |
Defense against herbivores | Positive | Vaughan et al. (2013) |
| Rice | Momilactone A | 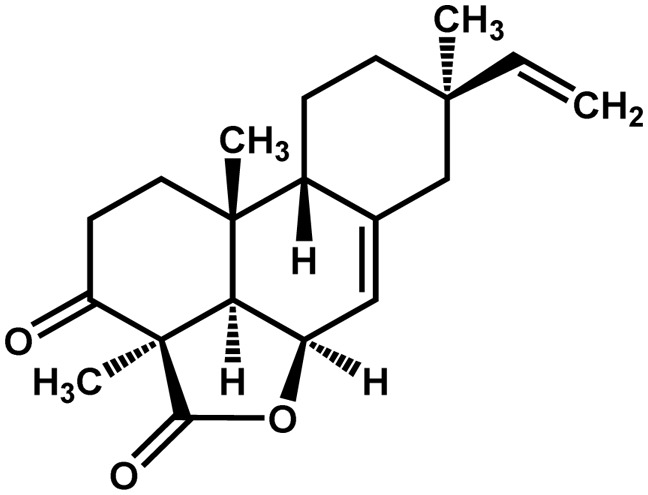 |
Antimicrobial | Positive | Kato-Noguchi et al. (2008) |
| Momilactone B | 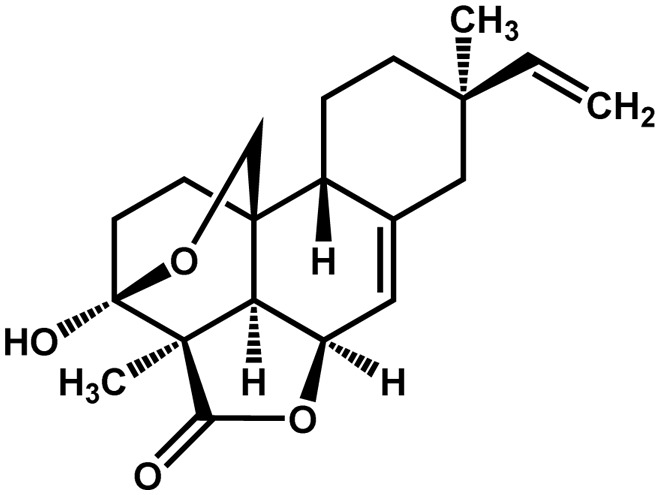 |
Allelopathic agent | Positive | Kato-Noguchi (2004) | |
| Phytocassanes A to Ea | 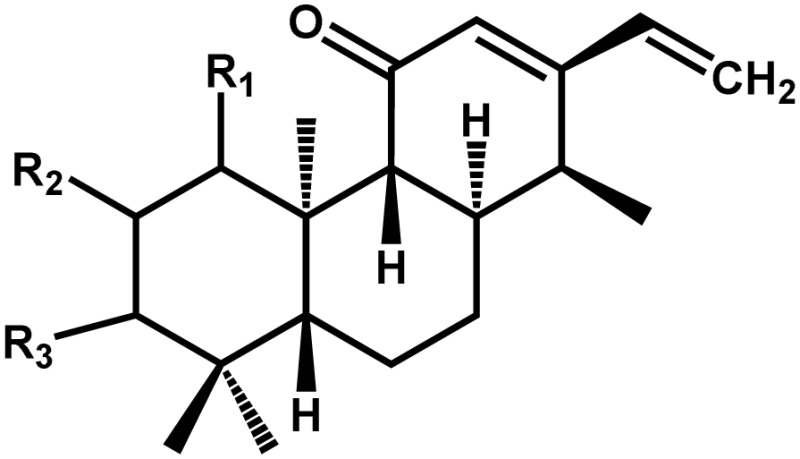 |
Antimicrobial agent | Positive | Toyomasu et al. (2008) | |
| Sorghum | Sorgoleone (2-hydroxy-5-methoxy-3-[(8′Z,11′Z)-8′,11′,14′-pentadecatriene]-p-benzaquinone) | 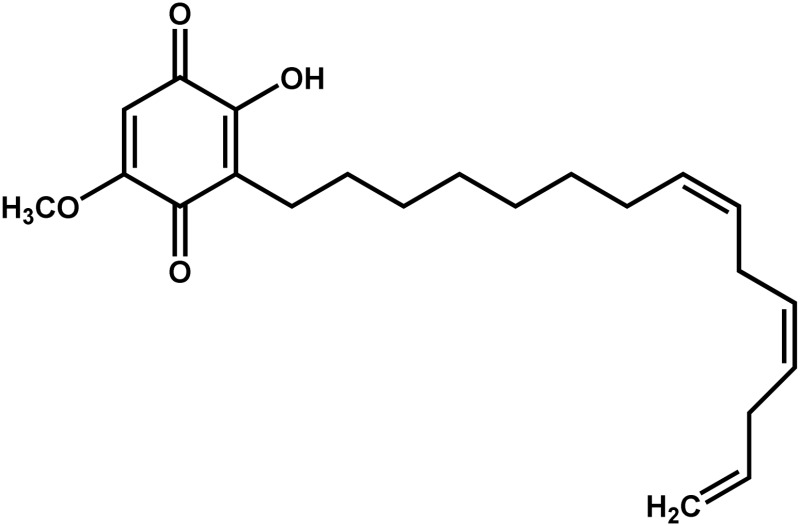 |
Allelopathic agent | Positive | Uddin et al. (2014) |
| Maize | Benzoxazinoids (2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one) | 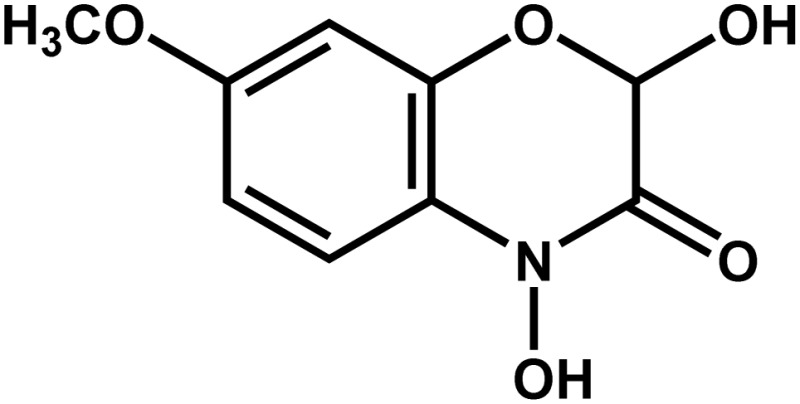 |
Antimicrobial agent | Positive | Neal et al. (2012) |
Phytocassane A, R1, R2, R3 = H, OH, O; phytocassane B, OH, OH, OH; phytocassane C, OH, H, OH; phytocassane D, H, O, OH; phytocassane E, OH, H, O.
Other important allelopathic plants are black walnut (Juglans nigra), which produces juglone (5-hydroxy-1,4-naphthoquinone), and sorghum (Sorghum bicolor), which produces sorgoleone (2-hydroxy-5-methoxy-3-[(8′Z,11′Z)-8′,11′,14′-pentadecatriene]-p-benzaquinone), an oxidized form of a hydrophobic p-benzoquinone (Table I). Both compounds inhibit electron transport reactions of photosynthesis and respiration, killing both maize and soybean and other susceptible plants (Rietveld, 1983; Nimbal et al., 1996; Jose and Gillespie, 1998). Another important allelochemical is the phenolic gallic acid produced by the common reed (Phragmites australis), a large perennial grass found in the United States and Asia. It has been determined that there exist two populations of this plant, one invasive and one noninvasive (Saltonstall, 2002). The invasive one produces more polymeric gallotannin than the noninvasive one. This polymer is transformed to gallic acid by a tannase, an enzyme produced by various native plants and microbial community members of North America, a process that intensifies the aggressiveness of the common reed (Rudrappa et al., 2007; Bains et al., 2009). However, controversial evidence questioning the negative effect of gallic acid on nonnative genotypes of this plant has recently been released (Weidenhamer et al., 2013).
CHEMICAL PLAYERS IN PLANT PRODUCTIVITY
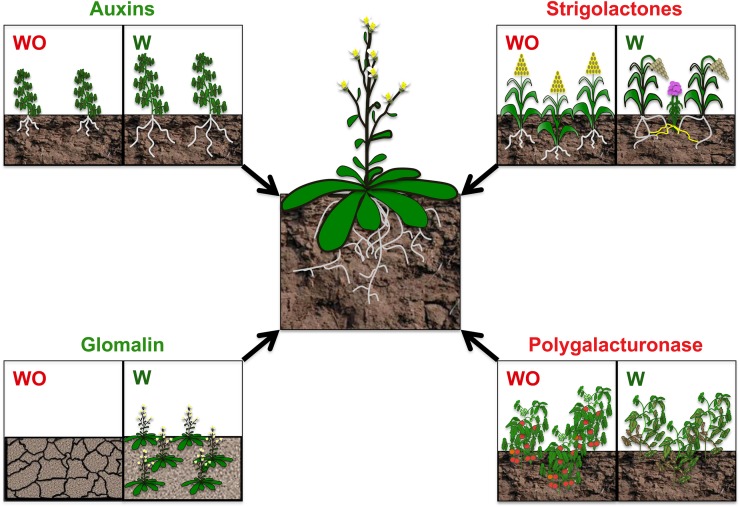
Although we do not pretend to discuss all chemicals secreted by the roots, we highlight below the role of those metabolites and proteins that have been found to be important players in plant productivity, either positively, such as auxins and glomalin, or negatively, such as SLs and polygalacturonase (PG; Fig. 1).
Figure 1.
Effects of auxins, glomalin, SLs, and PGs on plant development and health. The presence (W) or absence (WO) of auxins, glomalin, SLs, and PGs can produce positive (green) or negative (red) effects on plants. The presence of a bacterium that secretes auxins helps the plants to stimulate roots, increasing nutrient acquisition from the soil. On the other hand, the absence of glomalin affects soil structure, facilitating soil erosion. Sorghum secretes SLs that induce the germination of the parasite plant Striga lutea, and PGs can disturb the plant cell wall in tomato, affecting plant productivity. [See online article for color version of this figure.]
Auxins
Auxins are phytohormones generated by plants, some fungi, and PGPRs (Reineke et al., 2008; Simon and Petrášek, 2011; Duca et al., 2014). Some rhizobacteria are able to produce auxins, particularly IAA, in a range from 17 to 719 μmol L−1, thus contributing to plant growth (Ivanova et al., 2001; Ali et al., 2010; Gumiere et al., 2014), although high concentrations of IAA can inhibit root growth and, therefore, plant productivity (Davies, 1995; Xie et al., 1996).
IAA in bacteria is synthesized from l-Trp (Duca et al., 2014), which can be obtained from compost, fertilizers, and root exudates (Jaeger et al., 1999; Kravchenko et al., 2004; Arkhipchenko et al., 2006). It has been reported that the presence of 5 mm Trp can cause a 5-fold increase in the IAA secretion in Bacillus amyloliquefaciens FZB42 (Idris et al., 2007) and up to a 100-fold increase in Azospirillum brasilence (Baca et al., 1994). Field and pot trials have revealed that by adding l-Trp to the soil, the growth and yield of several important crops can be improved, including rice, wheat, soybean, potato, and tomato (Zahir et al., 2000). Therefore, in managed crops, PGPR inoculation can still contribute to IAA production if l-Trp is applied alone to the plants (Zahir et al., 2005) or together with fertilizer (Zahir et al., 2007).
The benefits that auxins from bacteria have had in plant development and productivity have been documented. For instance, an increase in soil IAA is able to enlarge the tree stem diameter and intensify the growth height of 17-year-old Pinus radiata trees (Smaill et al., 2010). In yam (Dioscorea rotundata), it was found that IAA from B. subtilis increases the elongation of shoots 83.3% and the production of roots 63.5% (Swain et al., 2007), both of which favor nutrient uptake and distribution (Singh Gahoonia et al., 1997; Talboys et al., 2014). It is known that several rhizobacteria species increase root biomass by changing the auxin balance and, in consequence, the signaling process, by either producing and secreting the auxin themselves (Dobbelaere et al., 1999; Patten and Glick, 2002; Idris et al., 2007) or changing its homeostasis inside plant cells (Spaepen et al., 2007; Kurepin et al., 2014). The same positive effect of bacterial IAA has been observed in tomato (Pastor et al., 2014), sesame (Sesamum indicum; Kumar et al., 2009), maize (Fallik et al., 1989), wheat (Egamberdieva, 2009), and other crops.
SLs
SLs are sesquiterpene lactones, derived from carotenoids, found in the rhizosphere and secreted in very small amounts (pg plant−1 d−1; Akiyama et al., 2005; Alder et al., 2012). These compounds are able to establish symbiotic and parasitic interactions (Soto et al., 2010; Zwanenburg and Pospíšil, 2013). Although SLs have been related to the stimulation of hyphal branching and nodule formation in alfalfa (Medicago sativa; Akiyama et al., 2005; Besserer et al., 2006; Soto et al., 2010), more attention has been paid to the negative effects that these molecules have on plant productivity (Bouwmeester et al., 2003).
Many important crops, such as sorghum, maize, cotton (Gossypium hirsutum), and cowpea (Vigna unguiculata), secrete SLs through their roots (Cook et al., 1966; Hauck et al., 1992; Müller et al., 1992; Siame et al., 1993). The SLs promote the germination of seeds of parasitic weeds, such as witchweed (Striga lutea), broomrapes (Orobanche and Phelipanche spp.), and Alectra spp. (Cook et al., 1966; Joel et al., 2007; Zwanenburg and Pospíšil, 2013). It has been reported that millions of hectares of crop fields and billions of dollars are lost annually due to Striga and Alectra spp. infestation in sub-Saharan Africa (Ejeta and Gressel, 2007; Joel et al., 2007) and to Orobanche spp. in the Mediterranean and western Asia (Parker, 2009). However, the mechanism by which the SLs induce seed germination is still controversial; it appears that the interaction of SLs with other molecules in the rhizosphere could contribute to their specificity and biological activities (Xie et al., 2010), although the catabolism of abscisic acid, allowing the germination of the parasitic seeds, also has been proposed (Lechat et al., 2012).
There have been scientific advances in understanding the biochemistry and production of SLs in order to avoid crop losses. One example is the use of carotenoid inhibitors (Jamil et al., 2010). Although carotenoid inhibitors such as fluridone, amitrole, clomazone, and norflurazon reduce SL production (Jamil et al., 2010), it is not known how these inhibitors could directly or indirectly affect other processes in the plant, such as abscisic acid biosynthesis (Gamble and Mullet, 1986), and, in consequence, plant productivity.
The secretion and activity of SLs are influenced by several factors, such as temperature, light regime, and even soil moisture (Weerasuriya et al., 1993; Xie et al., 2010; Toh et al., 2012). Therefore, it is tempting to speculate that if we are experiencing severe global climate change, the secretion of these molecules could be affected by means that we do not yet know.
Glomalin
Soil structure impacts the flux of water, gases, and nutrients and is thus an important consideration for agriculture, particularly under current farming practices, which demand more fertilizers, water, and pesticides (Angers and Caron, 1998; Tilman et al., 2002). Soil health is very important not only for crop productivity but also for microbial livelihood. One of the microorganisms responsible for soil physical structure is the AMF, which have positive effects on plant development. These fungi produce an iron-containing glycoprotein named glomalin (Wright et al., 1996; Purin and Rillig, 2007), insoluble in water and resistant to heat degradation (Singh et al., 2013), which moves into the soil and works as a glue among soil particles, contributing to a decrease in soil erosion (Haddad and Sarkar, 2003). It has been found that the glomalin content in the soil is higher in the summer season and in places with high moisture content (Emran et al., 2012; Gispert et al., 2013). This finding could explain why the concentration of this protein is lower in the desert (1 mg g−1; Bai et al., 2009) than in tropical forest (100 mg g−1; Rillig et al., 2001).
Glomalin has been used as an indicator of the effects of land-use change (Rillig et al., 2003) and is considered to contribute to carbon (Rillig et al., 2001) and nitrogen (Nichols and Wright, 2006) storage, which can help reduce the release of nitrous oxide, probably by influencing nitrification and denitrification, into the atmosphere and, therefore, greenhouse gases (Janzen, 2004; Nichols and Wright, 2006; Singh et al., 2013). Studies on sandy loam soils treated for 21 years with continuous fertilization have shown that fertilization affected the community composition of AMF and glomalin-related soil protein content (Dai et al., 2013). Also, it has been suggested that prolonged cultivation can reduce glomalin content in the soil, facilitating the disturbance of the soil and causing a decline in crop production (Preger et al., 2007). Soil glomalin is related to net primary productivity (Treseder and Turner, 2007), and a possible link between coffee (Coffea arabica) productivity and glomalin levels in the soil has been proposed (Banks et al., 2011). Therefore, glomalin, like auxins, also can be considered a positive player in plant productivity.
PGs
PGs are cell wall-degrading enzymes that cleave glycosidic bonds linking d-galacturonic acid residues, which are the main components of pectin. These enzymes are important pathogenicity factors produced by fungi (e.g. Fusarium spp., Botrytis cinerea, Colletotrichum lindemuthianum, and Aspergillus spp.; Lafitte et al., 1984; Di Pietro and Roncero, 1996; ten Have et al., 1998; Lang and Dörnenburg, 2000) and bacteria (e.g. P. syringae, Pseudomonas solanacearum, Erwinia carotovora, and Ralstonia solanacearum; Salmond, 1994; Vasse et al., 1995; Lorenzo et al., 1997; Huang and Allen, 2000).
PGs are one of the factors responsible for causing severe diseases in many plants (Collmer and Keen, 1986; Magro et al., 1994; Shieh et al., 1997; Aysan et al., 2003). For instance, Aspergillus flavus, a saprophytic fungus responsible for important losses in maize, peanut (Arachis hypogaea), and cotton (Lillehoj et al., 1974; Shieh et al., 1997; Asis et al., 2005), produces a PG, P2c, which causes intercarpellary membrane damage due to the loss of pectin integrity, thus helping the spread and invasion of the fungus (Shieh et al., 1997). Furthermore, studies in bacteria have shown that PGs from R. solanacearum and P. solanacearum are necessary to colonize the tomato root cortex, infect vascular parenchyma, and invade xylem elements, causing the devastating disease bacterial wilt (Wallis and Truter, 1978; Vasse et al., 1995; Huang and Allen, 2000). It has been found that PGs can be secreted by P. syringae (De-la-Peña et al., 2008) and P. solanacearum (Schell et al., 1988). Therefore, it could be that PGs secreted into the rhizosphere can reach and degrade the pectin of root cell walls, allowing the bacteria to enter and move throughout the vascular parenchyma.
Although bacteria and fungi have developed a mechanism to use PGs against plants, plants are able to defend themselves using polygalacturonase-inhibiting proteins (PGIPs), which favor the accumulation of oligogalacturonides (elicitors of the defense response) and limit pathogen infection (De Lorenzo et al., 2001; De Lorenzo and Ferrari, 2002; D’Ovidio et al., 2004). Studies in bean and tomato have shown that PGIP protects the plant cell walls from C. lindemuthianum, B. cinerea, and Fusarium oxysporum f. sp. lycopersici (Jones et al., 1972; Lafitte et al., 1984; Powell et al., 2000). These proteins also have been found in the secretome of Arabidopsis roots when the plant is infected with P. syringae (Magro et al., 1994; De-la-Peña et al., 2008), which indicates that PGIPs participate in defense response signaling in the rhizosphere during bacterial attack. Therefore, PGIPs could be exploited as a strategy for protecting crops against PG-producing pathogens.
OMIC APPROACHES TO STUDY RHIZOSPHERE DYNAMICS AND CHEMICAL PLAYERS
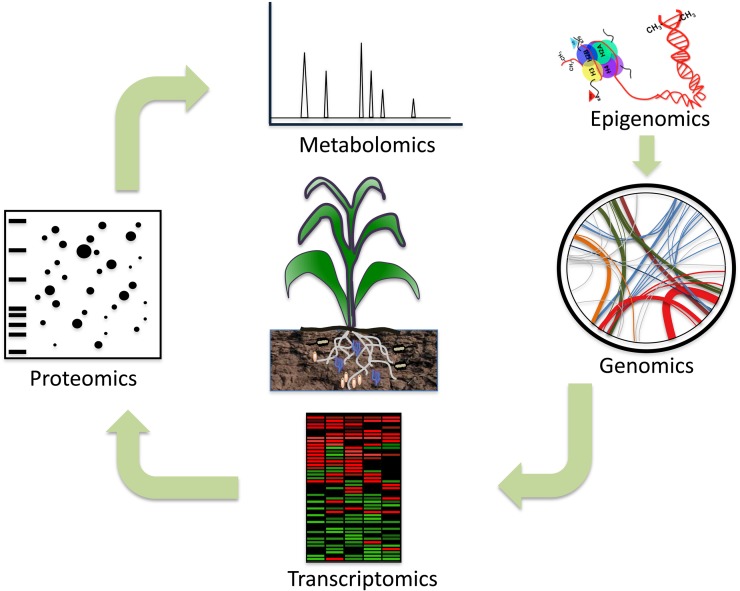
Most of the information about the rhizosphere and the principal compounds, genes, functions, and mechanisms has been achieved using omic approaches, such as genomics, transcriptomics, proteomics, metabolomics, and, very recently, epigenomics (Fig. 2). However, there are still many secrets to be uncovered.
Figure 2.
Omic approaches to the study of rhizosphere dynamics in order to improve plant productivity. Many organisms visit plants; however, some can be used to increase crop productivity, and others just kill the plant. In the case of the rhizosphere, the interactions between bacteria (white and cream), fungi (blue), and roots have been studied extensively from different omic approaches, such as genomics, transcriptomics, proteomics, and metabolomics. Epigenomics, the newest omics in the rhizosphere, is starting to be a focus of attention (De-la-Peña and Loyola-Vargas, 2012), which could have a great impact on what we already know about plant productivity and environmental stress. [See online article for color version of this figure.]
When interest in root secretion first began, the available technologies were based on analytical separation and biological detection (Knudson and Smith, 1919; Flores et al., 1999; Akiyama et al., 2005). One of the pioneering biochemical studies on root secretion ran from 1919 to 1920 and was headed by Dr. Lewis Knudson from Cornell University (Knudson and Smith, 1919; Knudson, 1920). In his articles, he explains step by step how and why root secretion was analyzed. Now the technology is much more specialized, sensitive, and complex (Diggle et al., 2007), and the resulting information is more precise, accurate, and quantitative. In the past, it was enough to use the PCR technique (Mullis and Faloona, 1987; Mullis et al., 1986) to investigate gene expression. Now the use of quantitative reverse transcriptase-coupled PCR is almost mandatory in order to have better and more robust information about a specific gene. This technique, introduced for the first time in 1992 (Higuchi et al., 1992), allows the quantification of RNA transcription levels by monitoring the amplification of a target sequence in real time using fluorescence detection. Now, a new layer of complexity has developed, with information given on chromatin remodeling in order to have more information about regulation in gene transcription. Therefore, advances in technology seem to favor the development of knowledge to improve our understanding of plant development, plant-pathogen interaction, root/microbe secretion, and rhizosphere dynamics (Dennis et al., 2010).
Epigenomics
Epigenomics is one of the newest omics and will likely increase our understanding of the dynamics of the rhizosphere and their function in improving plant productivity. Epigenetics has provided a new layer of complexity to the understanding of gene regulation for agricultural applications (Boyko and Kovalchuk, 2013; Us-Camas et al., 2014). Epigenetics are typically studied by three different mechanisms: DNA methylation, histone modification, and small (sRNA; 50–250 nucleotides) or micro (19–25 nucleotides) noncoding RNAs.
DNA methylation can be analyzed by different methods depending on the type of information needed (Fraga and Esteller, 2002). The most frequently used techniques to analyze DNA methylation are HPLC (Kuo et al., 1980; De-la-Peña et al., 2012b; Chen et al., 2013), methylation-sensitive amplification polymorphism (Sha et al., 2005; Park et al., 2009; Hubbard et al., 2014), and bisulfite sequencing (Frommer et al., 1992; Cokus et al., 2008; Li and Tollefsbol, 2011). In the case of histone modifications (e.g. methylation, acetylation, phosphorylation, etc.), the use of antibodies is necessary to observe global histone modifications or loci-specific modifications. Western blots are normally used to study global histone modifications (Nic-Can and De-la-Peña, 2012; Nallamilli et al., 2014). In the latter case, chromatin immunoprecipitation is useful (He et al., 2003; Saleh et al., 2008; Liu et al., 2014). For sRNA analysis, northern blots or microarrays can be used (Altuvia, 2007), and the microRNAs can be analyzed by quantitative reverse transcriptase-coupled PCR (Monavar Feshani et al., 2012; Verma et al., 2014) or northern blot (Válóczi et al., 2004; Xie et al., 2005).
Plant epigenetics can be modified in response to environmental conditions (Lira-Medeiros et al., 2010; Boyko and Kovalchuk, 2013; Bräutigam et al., 2013), and the progeny can inherit regulatory mechanisms to cope with such stresses. For instance, while in recent decades increasing temperatures have caused major losses in food crops (Peng et al., 2004), it has been found that plants exposed to heat stress can inherit epigenetic changes that result in offspring better adapted to extreme heat, a finding that could improve plant productivity (Lang-Mladek et al., 2010; Pecinka et al., 2010; Tittel-Elmer et al., 2010; Mirouze and Paszkowski, 2011).
In bacteria, epigenetics are also important. For instance, sRNAs have been involved in carbon metabolism and transport, amino acid metabolism, metal sensing, QS, biofilm formation, microbial virulence, and infection (Michaux et al., 2014). It has been found that two sRNAs, RsmY and RsmZ, are regulated by the global activator GacA, which is required for the production of exoenzymes, virulent factors, and secondary metabolites in P. aeruginosa (Reimmann et al., 1997; Kay et al., 2006). These sRNAs sequester the rsmA (for regulator of secondary metabolism) gene, inhibiting secondary metabolism and biofilm formation and activating motility and type III secretion (Sonnleitner and Haas, 2011). In fact, it has been found that when the sRNA rsmZ is overexpressed, biofilm development, which is associated with pathogenicity in P. aeruginosa, is reduced (Petrova and Sauer, 2010).
Although the epigenetics of rhizosphere interactions have not been studied, De-la-Peña and Loyola-Vargas (2012) have suggested that the secretion of certain chemicals into the rhizosphere could be due to epigenetic changes during plant-pathogen interaction. There are data that strongly suggest that changes in protein secretion could be due to epigenetic changes. For instance, De-la-Peña et al. (2008, 2010), working with Arabidopsis defense-impaired mutants nonexpressor of PATHOGENESIS-RELATED GENES1 (npr1-1; a mutant that does not express NPR1), constitutive expressor of PATHOGENESIS-RELATED GENES5 (cpr5-2; a mutant that accumulates large amounts of salicylic acid), and salicylate hydrolase (NahG; a mutant that is unable to accumulate salicylic acid), found that these mutants had a different protein root secretion pattern. Moreover, it was found that the defense-responsive genes in Arabidopsis, like CPR5 and NPR1, could be mediated by epigenetic factors (De-la-Peña et al., 2012c), suggesting that protein root secretion also could be modified due to epigenetic changes.
However, much work remains to be done in this area in order to apply this knowledge to improving plant productivity, as the examples are all complex and highly case specific. Some very new techniques, such as bisulfite sequencing or chromatin immunoprecipitation, hold promise as means to understand the intricate gene regulation during the cross talk of plants with their neighbors.
Genomics
In parallel to the biochemical and classical molecular approaches, genomic contributions, especially metagenomics in relation to rhizosphere interactions, have been enormous (Kakirde et al., 2010; Mendes et al., 2011). Many microbes have been identified using the metagenomic approach, but few have been characterized (Mendes et al., 2011). The microbial biodiversity in the soil seems to have specific biological functions in plants. For instance, recent genomic research on five different Sinorhizobium spp. (Sinorhizobium meliloti, Sinorhizobium medicae, Sinorhizobium freddi, Sinorhizobium terangae, and Sinorhizobium saheli) has found that each bacterium has adopted a slightly different strategy to interact with diverse Medicago spp. host plants and soil environments. This work also shows that the genes involved in the biosynthesis of the Nod factor (lipochitooligosaccharides secreted by rhizobia that are important in the interplay of recognition between roots and microbes) and polysaccharides, denitrification, and secretion systems vary within and between species (Sugawara et al., 2013).
The identification of genes not only in one genus but also across the plant kingdom to improve crop traits is a common goal that scientists need to pursue. Moreover, the techniques of insertion mutagenesis and positional cloning have helped to illuminate the nodulation process (Webb et al., 2000; Stougaard, 2001), which also can give some clues about the species-specific relationship between rhizobacteria and legumes. Takahara et al. (2013), using deleted regions in hypernodulating mutants (tml-1, tml-2, and tml-3), map-based cloning, and next-generation sequencing analysis in Lotus japonicus, characterized and identified a root factor named TOO MUCH LOVE (TML) that acts during the autoregulation of nodulation in this legume. Although these mutants have not been used to study root secretion, it would be interesting to investigate whether the proteins and molecules secreted are affected by these mutations. Probably, the mutations impair or enhance the secretion of an important chemical player, maximizing its potential in plant productivity.
Transcriptomics
The use of transcriptomics has become an important tool to identify genes involved in plant-microbe interactions (Ramachandran et al., 2011; Schenk et al., 2012; Lakshmanan et al., 2013). For instance, transcriptomics has been used to compare R. leguminosarum adaptation in the rhizosphere of a host legume (pea [Pisum sativum]), a nonhost legume (alfalfa), and a nonlegume (sugar beet [Beta vulgaris]; Ramachandran et al., 2011). Although a common core of bacterial genes was identified, 66% were of unknown function and several were plant specific. In the rhizosphere of pea, R. leguminosarum expressed genes related to bacterial metabolism and Nod factor synthesis, while in the presence of the alfalfa rhizosphere, the genes involved in lignin breakdown and metabolite transporters were up-regulated (Ramachandran et al., 2011). Studies on the gram-positive rhizobacterium B. amyloliquefaciens in response to root exudates from maize revealed that 8.2% of the bacterial transcriptome was altered in the presence of these exudates (Fan et al., 2012). The majority of the altered genes were up-regulated, and most of them are involved in nutrient utilization, bacterial chemotaxis and motility, and antimicrobial peptides. However, there were some genes found with unknown functions, opening new questions about the role of root exudates in rhizobacteria behavior.
Transcriptomics is a powerful tool that produces a massive amount of data, which need to be focused in order to find candidate genes useful for plant productivity. Mitra et al. (2004) focused the results of the transcriptome data on one candidate, Ca2+-calmodulin-dependent protein kinase. They show that transcript-based cloning is a valid approach for cloning genes and that this method does not require the construction of a genetic map. This approach also could be used to study plant productivity and rhizosphere dynamics, as was done in some recent reports (Fan et al., 2012; Alavi et al., 2013; Carvalhais et al., 2013). Other important genes discovered with transcriptomics are the nodule Cys-rich antimicrobial peptides (NCR) genes. It has been found that NCRs control the terminal differentiation of intracellular S. meliloti bacteroids by manipulating the bacterial cell cycle (Van de Velde et al., 2010; Penterman et al., 2014). Penterman et al. (2014) provided strong evidence about the molecular mechanism by which NCR peptides control the S. meliloti cell cycle during symbiosis. Working with NCR247, the authors found that these peptides specifically block cell division without affecting DNA replication. Thus, NCR perturbs the expression of genes involved in motility, cell division, and cell cycle regulation.
Proteomics
Proteomics has been frequently used to find proteins secreted into the rhizosphere and involved in plant-microbe interaction (De-la-Peña et al., 2008; De-la-Peña and Vivanco, 2010; Yang et al., 2012; Wang et al., 2013), and it has been considered an important biotechnological tool for crop improvement (Eldakak et al., 2013). The proteomic field has taken advantage of new technologies, and many of them are applicable for studying plant-microbe interaction, such as matrix-assisted laser-desorption ionization-time-of-flight-mass spectrometry (Watt et al., 2005; Noir et al., 2009), liquid chromatography-mass spectrometry (Larrainzar et al., 2007; Koch et al., 2010), isobaric tags for relative and absolute quantification-mass spectrometry (Taylor et al., 2008; Kaffarnik et al., 2009; Li et al., 2014) and multidimensional protein identification technology-tandem mass spectrometry (Wen et al., 2007; Kaschani et al., 2009). Medicago truncatula has been the model legume from a proteomic point of view (Sumner et al., 2007; Colditz and Braun, 2010; Lee et al., 2013), and it has opened an avenue to studying the proteomics of major crops such as wheat, maize, soybean, and rice (Song et al., 2007; Eldakak et al., 2013).
Proteomics also has been useful in the study of bacterial and fungal secretomes. In the case of the bacterial secretome, B. subtilis has been widely used (Hirose et al., 2000; Tjalsma et al., 2000, 2004; Antelmann et al., 2006). It was found that this bacterium secretes around 300 different proteins into the soil. Fungi also secrete many proteins that have been related to virulence factors (Song et al., 2009), adhesion to the plant surface (Newey et al., 2007), host tissue penetration and invasion (van Esse et al., 2008; Panstruga and Dodds, 2009; Dong et al., 2011), and symbiotic formation (Plett et al., 2011). Like transcriptomics, proteomics has a big problem with unknown proteins. In fungi, 208,883 putative secretory proteins have been identified (Choi et al., 2010), but only a small fraction of them have been studied, and so far, nobody knows the biological/ecological role of most of them. On the other hand, there are many reports describing the use of plant mutants, pathogenic bacteria, and symbiotic microbes that highlight the vast diversity of proteins or even the function of a group of proteins during plant-microbe interaction (De-la-Peña et al., 2008; Afroz et al., 2013; Alberton et al., 2013; Dam et al., 2014). These and other proteomic reports can be used to study proteins important to plant productivity. Also, in vitro and controlled systems could be used with other omic analyses to help assign function and/or identification through reducing environmental noise.
Metabolomics
Metabolomics of the rhizosphere is another big omic topic highly studied. The principal technologies used to detect metabolites are NMR spectroscopy, HPLC, liquid chromatography-mass spectrometry, gas chromatography-mass spectrometry, and capillary electrophoresis-time-of-flight-mass spectrometry. Important compounds such as SLs have been identified using liquid chromatography-tandem mass spectrometry (Xie et al., 2007; López-Ráez et al., 2008), and hundreds of metabolites have been found to highlight the importance that chemical cross talk between roots and microbes has in the initial recognition and species-dependent response.
Vauclare et al. (2013), using NMR spectroscopy, analyzed the metabolite profiles of Bradyrhizobium japonicum cells and the bacteroids from soybean nodules. Fan et al. (2001) used this same spectroscopy technology to examine the importance of root exudates from barley (Hordeum vulgare) and wheat in the acquisition of cadmium. Furthermore, Tawaraya et al. (2013) identified by capillary electrophoresis-time-of-flight-mass spectrometry 65 metabolites from root exudates of rice under phosphorous deficiency, and more than 30% of these showed a higher concentration at low phosphorous levels. This novel technique also can be used to determine the metabolites secreted during plant-microbe interaction. Like proteins (De-la-Peña et al., 2010), metabolites secreted from roots change depending on the developmental stage of the plant and have different effects on root microbes (Chaparro et al., 2013; Ziegler et al., 2013). Among the major bioactive metabolites found in the rhizosphere are flavonoids, phenolic compounds, exopolysaccharides, antibiotics, and QS signals. Badri et al. (2013), working with phytochemicals characterized by gas chromatography-mass spectrometry from root exudates, found that some phenolic-related compounds are able to modulate soil microbes. Moreover, the authors propose that the use of natural compounds to establish a positive interaction between plants and soil microbes could improve crop yield and sustainability. Therefore, understanding how microbes are selected in the rhizosphere and how plants affect microbial activity will provide new opportunities to increase crop production (Berendsen et al., 2012). Moreover, this will lead to a better understanding of the ecological relevance of the interaction among all the components of agricultural ecosystems as well as support the development of sustainable management technologies.
CONCLUSION
Although much of what we know about exudates has been done in Arabidopsis, it is necessary expand the research to important crop and nonmodel plants in order to understand more about the dynamics of the rhizosphere. It is known that different types of plants (Haichar et al., 2008) or even different ages of a plant (Chaparro et al., 2013; İnceoğlu et al., 2013) can harbor totally different microbial communities. A recent work argues that Arabidopsis is a limited model for investigating the impact of stress on rhizosphere community composition and function (Blee et al., 2013). Furthermore, expanding rhizosphere investigations to other species will speed the discovery of new molecules or compounds in the rhizosphere, which may enable the expansion of some plants’ abilities in symbiosis, defense, and development to other crops to improve plant fitness, increase tolerance of biotic and abiotic stress, and enhance plant productivity.
It is also important to study the microbiome of different cultivars, under different environmental conditions and in the presence of different neighbors, to analyze the compounds secreted in specific circumstances. Such work will uncover the numerous soil microorganisms, functions, and genes that remain unknown and demonstrate their usefulness for diverse applications. It will also provide the information necessary to engineer plants better adapted to extreme and changing environments, leading to improved agriculture. These improvements might include a better capacity of plants to resist invasive species, efficiently acquire nutrients, detoxify contaminated soils, and attract beneficial PGPRs, improving plant productivity.
Plants as well as microbes have the ability to change in response to other organisms at different omic levels. For instance, mycorrhizal fungi can change not only the transcriptome of a plant by inducing the regulation of many genes (Liu et al., 2003) but also modify the plant metabolome in order to establish a symbiotic contact with roots (Duhamel et al., 2013; Zhao et al., 2013). On the other hand, proteomics and epigenomics studied in Arabidopsis have shown that if the plant is in contact with a pathogen, the secretion of proteins (De-la-Peña et al., 2008) as well as different histone modifications in the plant (De-la-Peña et al., 2012c) are altered. Therefore, all these approaches could be used to improve plant productivity under stressful conditions and enhance the diversity of beneficial microbes in the rhizosphere.
Glossary
- PGPR
plant growth-promoting rhizobacterium
- IAA
indole acetic acid
- MA
l-malic acid
- AHL
N-acyl-homoserine lactone
- QS
quorum sensing
quorum quenching
- AMF
arbuscular mycorrhizal fungi
- SL
strigolactone
- PG
polygalacturonase
- PGIP
polygalacturonase-inhibiting protein
- sRNA
small RNA
Footnotes
This work was supported by the Consejo Nacional de Ciencia y Tecnología (grant no. 178149 to C.D. and grant no. 157014 to V.M.L.-V.).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- Afroz A, Zahur M, Zeeshan N, Komatsu S. (2013) Plant-bacterium interactions analyzed by proteomics. Front Plant Sci 4: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Hayashi H. (2006) Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann Bot (Lond) 97: 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Alavi P, Starcher MR, Zachow C, Müller H, Berg G. (2013) Root-microbe systems: the effect and mode of interaction of stress protecting agent (SPA) Stenotrophomonas rhizophila DSM14405(T.). Front Plant Sci 4: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberton D, Müller-Santos M, Brusamarello-Santos LCC, Valdameri G, Cordeiro FA, Yates MG, de Oliveira Pedrosa F, de Souza EM. (2013) Comparative proteomics analysis of the rice roots colonized by Herbaspirillum seropedicae strain SmR1 reveals induction of the methionine recycling in the plant host. J Proteome Res 12: 4757–4768 [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Ali B, Sabri A, Hasnain S. (2010) Rhizobacterial potential to alter auxin content and growth of Vigna radiata (L.). World J Microb Biot 26: 1379–1384 [Google Scholar]
- Altuvia S. (2007) Identification of bacterial small non-coding RNAs: experimental approaches. Curr Opin Microbiol 10: 257–261 [DOI] [PubMed] [Google Scholar]
- Angers D, Caron J (1998) Plant-induced changes in soil structure: processes and feedbacks. In N Breemen, ed, Plant-Induced Soil Changes: Processes and Feedbacks, Vol 4. Springer, Dordrecht, The Netherlands, pp 55–72 [Google Scholar]
- Antelmann H, van Dijk JM, Bron S, Hecker M (2006) Proteomic survey through secretome of Bacillus subtilis. In I Humphery-Smith, M Hecker, eds, Microbial Proteomics: Functional Biology of Whole Organisms. John Wiley & Sons, New York, pp 179–208 [PubMed] [Google Scholar]
- Apine OA, Jadhav JP. (2011) Optimization of medium for indole-3-acetic acid production using Pantoea agglomerans strain PVM. J Appl Microbiol 110: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Arkhipchenko IA, Shaposhnikov AI, Kravchenko LV. (2006) Tryptophan concentration of animal wastes and organic fertilizers. Appl Soil Ecol 34: 62–64 [Google Scholar]
- Asaka O, Shoda M. (1996) Biocontrol of Rhizoctonia solani damping-off of tomato with Bacillus subtilis RB14. Appl Environ Microbiol 62: 4081–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asis R, Barrionuevo DL, Giorda LM, Nores ML, Aldao MA. (2005) Aflatoxin production in six peanut (Arachis hypogaea L.) genotypes infected with Aspergillus flavus and Aspergillus parasiticus, isolated from peanut production areas of Cordoba, Argentina. J Agric Food Chem 53: 9274–9280 [DOI] [PubMed] [Google Scholar]
- Aysan Y, Karatas A, Cinar O. (2003) Biological control of bacterial stem rot caused by Erwinia chrysanthemi on tomato. Crop Protection 22: 807–811 [Google Scholar]
- Baca BE, Soto-Urzua L, Xochihua-Corona YG, Cuervo-Garcia A. (1994) Characterization of two aromatic amino acid aminotransferases and production of indoleacetic acid in Azospirillum strains. Soil Biol Biochem 26: 57–63 [Google Scholar]
- Badar R, Qureshi SA. (2012) Use of Trichoderma hamatum alone and in combination with rhizobial isolates as bio-fertilizer for improving the growth and strength of sunflower. J Basic Appl Sci Res 2: 6307–6314 [Google Scholar]
- Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM. (2013) Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J Biol Chem 288: 4502–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Vivanco JM. (2009) Regulation and function of root exudates. Plant Cell Environ 32: 666–681 [DOI] [PubMed] [Google Scholar]
- Baetz U, Martinoia E. (2014) Root exudates: the hidden part of plant defense. Trends Plant Sci 19: 90–98 [DOI] [PubMed] [Google Scholar]
- Bai CM, He XL, Tang HT, Shan BQ, Zhao LL. (2009) Spatial distribution of arbuscular mycorrhizal fungi, glomalin and soil enzymes under the canopy of Astragalus adsurgens Pall. in the Mu Us sandland, China. Soil Biol Biochem 41: 941–947 [Google Scholar]
- Bains G, Kumar AS, Rudrappa T, Alff E, Hanson TE, Bais HP. (2009) Native plant and microbial contributions to a negative plant-plant interaction. Plant Physiol 151: 2145–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais HP, Fall R, Vivanco JM. (2004a) Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol 134: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais HP, Park SW, Weir TL, Callaway RM, Vivanco JM. (2004b) How plants communicate using the underground information superhighway. Trends Plant Sci 9: 26–32 [DOI] [PubMed] [Google Scholar]
- Bais HP, Vepachedu R, Gilroy S, Callaway RM, Vivanco JM. (2003) Allelopathy and exotic plant invasion: from molecules and genes to species interactions. Science 301: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266 [DOI] [PubMed] [Google Scholar]
- Banks JE, Cline E, Castro S, Urena N, Nichols K, Hannon L, Singer R, Chandler M. (2011) Effects of synthetic fertilizer on coffee yields and ecosystem services: parasitoids and soil glomalin in a Costa Rican coffee agroecosystem. J Crop Improv 25: 650–663 [Google Scholar]
- Barea JM, Pozo MJ, Azcón R, Azcón-Aguilar C. (2005) Microbial co-operation in the rhizosphere. J Exp Bot 56: 1761–1778 [DOI] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM. (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486 [DOI] [PubMed] [Google Scholar]
- Berge O, Lodhi A, Brandelet G, Santaella C, Roncato MA, Christen R, Heulin T, Achouak W. (2009) Rhizobium alamii sp. nov., an exopolysaccharide-producing species isolated from legume and non-legume rhizospheres. Int J Syst Evol Microbiol 59: 367–372 [DOI] [PubMed] [Google Scholar]
- Berruti A, Borriello R, Orgiazzi A, Barbera AC, Lumini E, Bianciotto V (2014) Arbuscular mycorrhizal fungi and their value for ecosystem management. In O Grillo, ed, Biodiversity: The Dynamic Balance of the Planet. InTech, Rijeta, Croacia pp 159–191 [Google Scholar]
- Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N. (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj D, Ansari MW, Sahoo RK, Tuteja N. (2014) Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb Cell Fact 13: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya PN, Jha DK. (2012) Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28: 1327–1350 [DOI] [PubMed] [Google Scholar]
- Blee K, Hein J, Wolfe GV (2013) Arabidopsis thaliana: a useful but limited model to investigate stress impacts on rhizosphere community composition and function. In FJ de Bruijn, ed, Molecular Microbial Ecology of the Rhizosphere, Vol 1. Wiley, New York, pp 265–270 [Google Scholar]
- Borneman J, Becker JO. (2007) Identifying microorganisms involved in specific pathogen suppression in soil. Annu Rev Phytopathol 45: 153–172 [DOI] [PubMed] [Google Scholar]
- Bouwmeester HJ, Matusova R, Zhongkui S, Beale MH. (2003) Secondary metabolite signalling in host-parasitic plant interactions. Curr Opin Plant Biol 6: 358–364 [DOI] [PubMed] [Google Scholar]
- Boyer JS. (1982) Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Boyko A, Kovalchuk I (2013) Epigenetic modifications in plants under adverse conditions: agricultural applications. In N Tuteja, S Singh Gill, eds, Plant Acclimation to Environmental Stress. Springer, New York, pp 233–267 [Google Scholar]
- Bräutigam K, Vining KJ, Lafon-Placette C, Fossdal CG, Mirouze M, Marcos JG, Fluch S, Fraga MF, Guevara MÁ, Abarca D, et al. (2013) Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol Evol 3: 399–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz AK, Broeckling CD, De-la-Peña C, Lewis MR, Greene E, Callaway RM, Sumner LW, Vivanco JM. (2010) Plant neighbor identity influences plant biochemistry and physiology related to defense. BMC Plant Biol 10: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücking H, Liepold E, Ambilwade P. (2012) The role of the mycorrhizal symbiosis in nutrient uptake of plants and the regulatory mechanisms underlying these transport processes. Plant Sci 4: 108–132 [Google Scholar]
- Burkhead KD, Schisler DA, Slininger PJ. (1994) Pyrrolnitrin production by biological control agent Pseudomonas cepacia B37w in culture and in colonized wounds of potatoes. Appl Environ Microbiol 60: 2031–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhais LC, Dennis PG, Fan B, Fedoseyenko D, Kierul K, Becker A, von Wiren N, Borriss R. (2013) Linking plant nutritional status to plant-microbe interactions. PLoS ONE 8: e68555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro J, Sheflin A, Manter D, Vivanco J. (2012) Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils 48: 489–499 [Google Scholar]
- Chaparro JM, Badri DV, Bakker MG, Sugiyama A, Manter DK, Vivanco JM. (2013) Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 8: e55731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Tao S, Bi X, Xu X, Wang L, Li X. (2013) Research of total levels on DNA methylation in plant based on HPLC analysis. Am J Mol Biol 3: 98–101 [Google Scholar]
- Cheng HP, Walker GC. (1998) Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180: 5183–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Park J, Kim D, Jung K, Kang S, Lee YH. (2010) Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genomics 11: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colditz F, Braun HP. (2010) Medicago truncatula proteomics. J Proteomics 73: 1974–1985 [DOI] [PubMed] [Google Scholar]
- Collmer A, Keen NT. (1986) The role of pectic enzymes in plant pathogenesis. Annu Rev Phytopathol 24: 383–409 [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH. (1966) Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154: 1189–1190 [DOI] [PubMed] [Google Scholar]
- Copping LG, Menn JJ. (2000) Biopesticides: a review of their action, applications and efficacy. Pest Manag Sci 56: 651–676 [Google Scholar]
- Czaja LF, Hogekamp C, Lamm P, Maillet F, Martinez EA, Samain E, Dénarié J, Küster H, Hohnjec N. (2012) Transcriptional responses toward diffusible signals from symbiotic microbes reveal MtNFP- and MtDMI3-dependent reprogramming of host gene expression by arbuscular mycorrhizal fungal lipochitooligosaccharides. Plant Physiol 159: 1671–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Hu J, Lin X, Yang A, Wang R, Zhang J, Wong M. (2013) Arbuscular mycorrhizal fungal diversity, external mycelium length, and glomalin-related soil protein content in response to long-term fertilizer management. J Soils Sed 13: 1–11 [Google Scholar]
- Dam S, Dyrlund TF, Ussatjuk A, Jochimsen B, Nielsen K, Goffard N, Ventosa M, Lorentzen A, Gupta V, Andersen SU, et al. (2014) Proteome reference maps of the Lotus japonicus nodule and root. Proteomics 14: 230–240 [DOI] [PubMed] [Google Scholar]
- Davidson EA, Janssens IA. (2006) Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440: 165–173 [DOI] [PubMed] [Google Scholar]
- Davies PJ (1995) Plant Hormones: Physiology, Biochemistry and Molecular Biology. Kluwer Academic, Dordrecht, The Netherlands [Google Scholar]
- De Lorenzo G, D’Ovidio R, Cervone F. (2001) The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu Rev Phytopathol 39: 313–335 [DOI] [PubMed] [Google Scholar]
- De Lorenzo G, Ferrari S. (2002) Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr Opin Plant Biol 5: 295–299 [DOI] [PubMed] [Google Scholar]
- De-la-Peña C, Badri D, Loyola-Vargas V (2012a) Plant root secretions and their interactions with neighbors. In F Baluska, J Vivanco, eds, Secretions and Exudates in Biological Systems. Springer-Verlag, Berlin, pp 1–26 [Google Scholar]
- De-la-Peña C, Badri DV, Lei Z, Watson BS, Brandão MM, Silva-Filho MC, Sumner LW, Vivanco JM. (2010) Root secretion of defense-related proteins is development-dependent and correlated with flowering time. J Biol Chem 285: 30654–30665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-la-Peña C, Lei Z, Watson BS, Sumner LW, Vivanco JM. (2008) Root-microbe communication through protein secretion. J Biol Chem 283: 25247–25255 [DOI] [PubMed] [Google Scholar]
- De-la-Peña C, Loyola-Vargas VM. (2012) The hidden chemical cross-talk between roots and microbes: a proteomic approach. Curr Proteomics 9: 103–117 [Google Scholar]
- De-la-Peña C, Nic-Can G, Ojeda G, Herrera-Herrera JL, López-Torres A, Wrobel K, Robert-Díaz ML. (2012b) KNOX1 is expressed and epigenetically regulated during in vitro conditions in Agave spp. BMC Plant Biol 12: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-La-Peña C, Rangel-Cano A, Alvarez-Venegas R. (2012c) Regulation of disease-responsive genes mediated by epigenetic factors: interaction of Arabidopsis-Pseudomonas. Mol Plant Pathol 13: 388–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-la-Peña C, Vivanco J. (2010) Root-microbe interactions: the importance of protein secretion. Curr Proteomics 7: 265–274 [Google Scholar]
- Dennis PG, Miller AJ, Hirsch PR. (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72: 313–327 [DOI] [PubMed] [Google Scholar]
- Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Cámara M, Williams P. (2007) The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol 14: 87–96 [DOI] [PubMed] [Google Scholar]
- Di Pietro A, Roncero MIG. (1996) Endopolygalacturonase from Fusarium oxysporum f. sp. lycopersici: purification, characterization, and production during infection of tomato plants. Phytopathology 86: 1324–1330 [Google Scholar]
- Dobbelaere S, Croonenborghs A, Thys A, Vande Broek A, Vanderleyden J. (1999) Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil 212: 153–162 [Google Scholar]
- Dong S, Yin W, Kong G, Yang X, Qutob D, Chen Q, Kale SD, Sui Y, Zhang Z, Dou D, et al. (2011) Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. PLoS Pathog 7: e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Wang LH, Xu JL, Zhang HB, Zhang XF, Zhang LH. (2001) Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411: 813–817 [DOI] [PubMed] [Google Scholar]
- Dong YH, Xu JL, Li XZ, Zhang LH. (2000) AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci USA 97: 3526–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Zhang LH. (2005) Quorum sensing and quorum-quenching enzymes. J Microbiol 43: 101–109 [PubMed] [Google Scholar]
- D’Ovidio R, Mattei B, Roberti S, Bellincampi D. (2004) Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant–pathogen interactions. Biochim Biophys Acta 1696: 237-244 [DOI] [PubMed] [Google Scholar]
- Duca D, Lorv J, Patten CL, Rose D, Glick BR. (2014) Indole-3-acetic acid in plant-microbe interactions. Antonie van Leeuwenhoek 106: 85–125 [DOI] [PubMed] [Google Scholar]
- Duffy BK, Défago G. (1999) Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl Environ Microbiol 65: 2429–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhamel M, Pel R, Ooms A, Bücking H, Jansa J, Ellers J, van Straalen NM, Wouda T, Vandenkoornhuyse P, Kiers ET. (2013) Do fungivores trigger the transfer of protective metabolites from host plants to arbuscular mycorrhizal hyphae? Ecology 94: 2019–2029 [DOI] [PubMed] [Google Scholar]
- Dutta S, Rani TS, Podile AR. (2013) Root exudate-induced alterations in Bacillus cereus cell wall contribute to root colonization and plant growth promotion. PLoS ONE 8: e78369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgerton MD. (2009) Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiol 149: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egamberdieva D. (2009) Alleviation of salt stress by plant growth regulators and IAA producing bacteria in wheat. Acta Physiol Plant 31: 861–864 [Google Scholar]
- Egamberdieva D, Kamilova F, Validov S, Gafurova L, Kucharova Z, Lugtenberg B. (2008) High incidence of plant growth-stimulating bacteria associated with the rhizosphere of wheat grown on salinated soil in Uzbekistan. Environ Microbiol 10: 1–9 [DOI] [PubMed] [Google Scholar]
- Einhellig FA (1994) Mechanism of action of allelochemicals in allelopathy. In KMM Dakshini, FA Einhellig, eds, Allelopathy, Vol 582. American Chemical Society, Washington, DC, pp 96–116 [Google Scholar]
- Ejeta G, Gressel J (2007) Integrating New Technologies for Striga Control: Towards Ending the Witch-Hunt. World Scientific, Singapore [Google Scholar]
- Eldakak M, Milad SI, Nawar AI, Rohila JS. (2013) Proteomics: a biotechnology tool for crop improvement. Front Plant Sci 4: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emran M, Gispert M, Pardini G. (2012) Patterns of soil organic carbon, glomalin and structural stability in abandoned Mediterranean terraced lands. Eur J Soil Sci 63: 637–649 [Google Scholar]
- Fallik E, Okon Y, Epstein E, Goldman A, Fischer M. (1989) Identification and quantification of IAA and IBA in Azospirillum brasilense-inoculated maize roots. Soil Biol Biochem 21: 147–153 [Google Scholar]
- Fan B, Carvalhais LC, Becker A, Fedoseyenko D, von Wirén N, Borriss R. (2012) Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol 12: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TWM, Lane AN, Shenker M, Bartley JP, Crowley D, Higashi RM. (2001) Comprehensive chemical profiling of gramineous plant root exudates using high-resolution NMR and MS. Phytochemistry 57: 209–221 [DOI] [PubMed] [Google Scholar]
- Figueiredo MVB, Burity HA, Martínez CR, Chanway CP. (2008) Alleviation of drought stress in the common bean (Phaseolus vulgaris L.) by co-inoculation with Paenibacillus polymyxa and Rhizobium tropici. Appl Soil Ecol 40: 182–188 [Google Scholar]
- Flores HE, Vivanco JM, Loyola-Vargas VM. (1999) ‘Radicle’ biochemistry: the biology of root-specific metabolism. Trends Plant Sci 4: 220–226 [DOI] [PubMed] [Google Scholar]
- Fraga MF, Esteller M. (2002) DNA methylation: a profile of methods and applications. Biotechniques 33: 632–649 [DOI] [PubMed] [Google Scholar]
- Frey SD, Lee J, Melillo JM, Six J. (2013) The temperature response of soil microbial efficiency and its feedback to climate. Nature Climate Change 3: 395–398 [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA 89: 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble PE, Mullet JE. (1986) Inhibition of carotenoid accumulation and abscisic acid biosynthesis in fluridone-treated dark-grown barley. Eur J Biochem 160: 117–121 [DOI] [PubMed] [Google Scholar]
- Gispert M, Emran M, Pardini G, Doni S, Ceccanti B. (2013) The impact of land management and abandonment on soil enzymatic activity, glomalin content and aggregate stability. Geoderma 202–203: 51–61 [Google Scholar]
- Grant C, Bittman S, Montreal M, Plenchette C, Morel C. (2005) Soil and fertilizer phosphorus: effects on plant P supply and mycorrhizal development. Can J Plant Sci 85: 3–14 [Google Scholar]
- Gumiere T, Ribeiro CM, Vasconcellos RL, Cardoso EJ. (2014) Indole-3-acetic acid producing root-associated bacteria on growth of Brazil pine (Araucaria angustifolia) and slash pine (Pinus elliottii). Antonie van Leeuwenhoek 105: 663–669 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Parniske M. (2013) Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol 29: 593–617 [DOI] [PubMed] [Google Scholar]
- Gutterson N. (1990) Microbial fungicides: recent approaches to elucidating mechanisms. Crit Rev Biotechnol 10: 69–91 [Google Scholar]
- Haas D, Keel C. (2003) Regulation of antibiotic production in root-colonizing Peudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol 41: 117–153 [DOI] [PubMed] [Google Scholar]
- Haddad MJ, Sarkar D. (2003) Glomalin, a newly discovered component of soil organic matter: environmental significance. Environ Geosci 10: 91–98 [Google Scholar]
- Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W. (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Hauck C, Müller S, Schildknecht H. (1992) A germination stimulant for parasitic flowering plants from Sorghum bicolor, a genuine host plant. J Plant Physiol 139: 474–478 [Google Scholar]
- He Y, Michaels SD, Amasino RM. (2003) Regulation of flowering time by histone acetylation in Arabidopsis. Science 302: 1751–1754 [DOI] [PubMed] [Google Scholar]
- Herridge D, Peoples M, Boddey R. (2008) Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311: 1–18 [Google Scholar]
- Hetrick BAD, Wilson GWT, Cox TS. (1992) Mycorrhizal dependence of modern wheat varieties, landraces, and ancestors. Can J Bot 70: 2032–2040 [Google Scholar]
- Higuchi R, Dollinger G, Walsh PS, Griffith R. (1992) Simultaneous amplification and detection of specific DNA sequences. Biotechnology (N Y) 10: 413–417 [DOI] [PubMed] [Google Scholar]
- Hirose I, Sano K, Shioda I, Kumano M, Nakamura K, Yamane K. (2000) Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 146: 65–75 [DOI] [PubMed] [Google Scholar]
- Hong G, Wang J, Zhang Y, Hochstetter D, Zhang S, Pan Y, Shi Y, Xu P, Wang Y. (2014) Biosynthesis of catechin components is differentially regulated in dark-treated tea (Camellia sinensis L.). Plant Physiol Biochem 78: 49–52 [DOI] [PubMed] [Google Scholar]
- Hong KW, Koh CL, Sam CK, Yin WF, Chan KG. (2012) Quorum quenching revisited: from signal decays to signalling confusion. Sensors (Basel) 12: 4661–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Allen C. (2000) Polygalacturonases are required for rapid colonization and full virulence of Ralstonia solanacearum on tomato plants. Physiol Mol Plant Pathol 57: 77–83 [Google Scholar]
- Hubbard M, Germida JJ, Vujanovic V. (2014) Fungal endophyte colonization coincides with altered DNA methylation in drought-stressed wheat seedlings. Can J Plant Sci 94: 223–234 [Google Scholar]
- Hwang J, Chilton WS, Benson DM. (2002) Pyrrolnitrin production by Burkholderia cepacia and biocontrol of Rhizoctonia stem rot of poinsettia. Biol Control 25: 56–63 [Google Scholar]
- Idris EE, Iglesias DJ, Talon M, Borriss R. (2007) Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol Plant Microbe Interact 20: 619–626 [DOI] [PubMed] [Google Scholar]
- İnceoğlu Ö, van Overbeek LS, Falcão Salles J, van Elsas JD. (2013) Normal operating range of bacterial communities in soil used for potato cropping. Appl Environ Microbiol 79: 1160–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova EG, Doronina NV, Trotsenko IUA. (2001) [Aerobic methylobacteria are capable of synthesizing auxins] (in Russian). Mikrobiologiia 70: 452–458 [PubMed] [Google Scholar]
- Jaeger CH, III, Lindow SE, Miller W, Clark E, Firestone MK. (1999) Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol 65: 2685–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil M, Charnikhova T, Verstappen F, Bouwmeester H. (2010) Carotenoid inhibitors reduce strigolactone production and Striga hermonthica infection in rice. Arch Biochem Biophys 504: 123–131 [DOI] [PubMed] [Google Scholar]
- Janzen HH. (2004) Carbon cycling in earth systems: a soil science perspective. Agric Ecosyst Environ 104: 399–417 [Google Scholar]
- Jeffries P, Gianinazzi S, Perotto S, Turnau K, Barea JM. (2003) The contribution of arbuscular mycorrhizal fungi in sustainable maintenance of plant health and soil fertility. Biol Fertil Soils 37: 1–16 [Google Scholar]
- Joel D, Hershenhorn J, Eizenberg H, Aly R, Ejeta G, Rich P, Ransom J, Sauerborn J, Rubiales D. (2007) Biology and management of weedy root parasites. Hortic Rev (Am Soc Hortic Sci) 33: 267–349 [Google Scholar]
- Joner E, van Aarle I, Vosatka M. (2000) Phosphatase activity of extra-radical arbuscular mycorrhizal hyphae: a review. Plant Soil 226: 199–210 [Google Scholar]
- Jones TM, Anderson AJ, Albersheim P. (1972) Host-pathogen interactions. IV. Studies on the polysaccharide-degrading enzymes secreted by Fusarium oxysporum f. sp. lycopersici. Physiol Plant Pathol 2: 153–166 [Google Scholar]
- Jose S, Gillespie A. (1998) Allelopathy in black walnut (Juglans nigra L.) alley cropping. II. Effects of juglone on hydroponically grown corn (Zea mays L.) and soybean (Glycine max L. Merr.) growth and physiology. Plant Soil 203: 199–206 [Google Scholar]
- Kaffarnik FAR, Jones AME, Rathjen JP, Peck SC. (2009) Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol Cell Proteomics 8: 145–156 [DOI] [PubMed] [Google Scholar]
- Kakirde KS, Parsley LC, Liles MR. (2010) Size does matter: application-driven approaches for soil metagenomics. Soil Biol Biochem 42: 1911–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F, Gu C, Niessen S, Hoover H, Cravatt BF, van der Hoorn RAL. (2009) Diversity of serine hydrolase activities of unchallenged and Botrytis-infected Arabidopsis thaliana. Mol Cell Proteomics 8: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Noguchi H. (2004) Allelopathic substance in rice root exudates: rediscovery of momilactone B as an allelochemical. J Plant Physiol 161: 271–276 [DOI] [PubMed] [Google Scholar]
- Kato-Noguchi H, Ino T, Ota K. (2008) Secretion of momilactone A from rice roots to the rhizosphere. J Plant Physiol 165: 691–696 [DOI] [PubMed] [Google Scholar]
- Kay E, Humair B, Dénervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. (2006) Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol 188: 6026–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klironomos JN, McCune J, Hart M, Neville J. (2000) The influence of arbuscular mycorrhizae on the relationship between plant diversity and productivity. Ecol Lett 3: 137–141 [Google Scholar]
- Knudson L. (1920) The secretion of invertase by plant roots. Am J Bot 7: 371–379 [Google Scholar]
- Knudson L, Smith RS. (1919) Secretion of amylase by plant roots. Bot Gaz 68: 460–466 [Google Scholar]
- Koch M, Delmotte N, Rehrauer H, Vorholt JA, Pessi G, Hennecke H. (2010) Rhizobial adaptation to hosts, a new facet in the legume root-nodule symbiosis. Mol Plant Microbe Interact 23: 784–790 [DOI] [PubMed] [Google Scholar]
- Koller R, Robin C, Bonkowski M, Ruess L, Scheu S. (2013) Litter quality as driving factor for plant nutrition via grazing of protozoa on soil microorganisms. FEMS Microbiol Ecol 85: 241–250 [DOI] [PubMed] [Google Scholar]
- Kravchenko LV, Azarova TS, Makarova NM, Tikhonovich IA. (2004) The effect of tryptophan present in plant root exudates on the phytostimulating activity of rhizobacteria. Microbiology 73: 156–158 [PubMed] [Google Scholar]
- Kumar S, Pandey P, Maheshwari DK. (2009) Reduction in dose of chemical fertilizers and growth enhancement of sesame (Sesamum indicum L.) with application of rhizospheric competent Pseudomonas aeruginosa LES4. Eur J Soil Biol 45: 334–340 [Google Scholar]
- Kuo KC, McCune RA, Gehrke CW, Midgett R, Ehrlich M. (1980) Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res 8: 4763–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepin LV, Zaman M, Pharis RP. (2014) Phytohormonal basis for the plant growth promoting action of naturally occurring biostimulators. J Sci Food Agric 94: 1715–1722 [DOI] [PubMed] [Google Scholar]
- Lafitte C, Barthe JP, Montillet JL, Tou A. (1984) Glycoprotein inhibitors of Colletotrichum lindemuthianum endopolygalacturonase in near isogenic lines of Phaseolus vulgaris resistant and susceptible to anthracnose. Physiol Plant Pathol 25: 39–53 [Google Scholar]
- Lakshmanan V, Castaneda R, Rudrappa T, Bais HP. (2013) Root transcriptome analysis of Arabidopsis thaliana exposed to beneficial Bacillus subtilis FB17 rhizobacteria revealed genes for bacterial recruitment and plant defense independent of malate efflux. Planta 238: 657–668 [DOI] [PubMed] [Google Scholar]
- Lakshmanan V, Kitto SL, Caplan JL, Hsueh YH, Kearns DB, Wu YS, Bais HP. (2012) Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol 160: 1642–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C, Dörnenburg H. (2000) Perspectives in the biological function and the technological application of polygalacturonases. Appl Microbiol Biotechnol 53: 366–375 [DOI] [PubMed] [Google Scholar]
- Lang-Mladek C, Popova O, Kiok K, Berlinger M, Rakic B, Aufsatz W, Jonak C, Hauser MT, Luschnig C. (2010) Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol Plant 3: 594–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar E, Wienkoop S, Weckwerth W, Ladrera R, Arrese-Igor C, González EM. (2007) Medicago truncatula root nodule proteome analysis reveals differential plant and bacteroid responses to drought stress. Plant Physiol 144: 1495–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechat MM, Pouvreau JB, Péron T, Gauthier M, Montiel G, Véronési C, Todoroki Y, Le Bizec B, Monteau F, Macherel D, et al. (2012) PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J Exp Bot 63: 5311–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lei Z, Watson BS, Sumner LW. (2013) Sub-cellular proteomics of Medicago truncatula. Front Plant Sci 4: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XG, Zhang TL, Wang XX, Hua K, Zhao L, Han ZM. (2013) The composition of root exudates from two different resistant peanut cultivars and their effects on the growth of soil-borne pathogen. Int J Biol Sci 9: 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tollefsbol TO. (2011) DNA methylation detection: bisulfite genomic sequencing analysis. Methods Mol Biol 791: 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Czarnecki O, Chourey K, Yang J, Tuskan GA, Hurst GB, Pan C, Chen JG. (2014) Strigolactone-regulated proteins revealed by iTRAQ-based quantitative proteomics in Arabidopsis. J Proteome Res 13: 1359–1372 [DOI] [PubMed] [Google Scholar]
- Ligon JM, Hill DS, Hammer PE, Torkewitz NR, Hofmann D, Kempf HJ, van Pée KH. (2000) Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag Sci 56: 688–695 [Google Scholar]
- Lillehoj EB, Garcia WJ, Lambrow M. (1974) Aspergillus flavus infection and aflatoxin production in corn: influence of trace elements. Appl Microbiol 28: 763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira-Medeiros CF, Parisod C, Fernandes RA, Mata CS, Cardoso MA, Ferreira PC. (2010) Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE 5: e10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ. (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Fromm M, Avramova Z. (2014) H3K27me3 and H3K4me3 chromatin environment at super-induced dehydration stress memory genes of Arabidopsis thaliana. Mol Plant 7: 502–513 [DOI] [PubMed] [Google Scholar]
- Liu W. (2010) Do genetically modified plants impact arbuscular mycorrhizal fungi? Ecotoxicology 19: 229–238 [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, Matusova R, Kohlen W, De Vos R, Verstappen F, Puech-Pages V, Bécard G, Mulder P, et al. (2008) Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol 178: 863–874 [DOI] [PubMed] [Google Scholar]
- Lorenzo G, Castoria R, Bellincampi D, Cervone F (1997) Fungal invasion enzymes and their inhibition. In G Carroll, P Tudzynski, eds, Plant Relationships, Vol 5. Springer, Berlin, pp 61–83 [Google Scholar]
- Lugtenberg B, Kamilova F. (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63: 541–556 [DOI] [PubMed] [Google Scholar]
- Lynch JM, Whipps JM. (1990) Substrate flow in the rhizosphere. Plant Soil 129: 1–10 [Google Scholar]
- Magro P, Varvaro L, Chilosi G, Avanzo C, Balestra GM. (1994) Pectolytic enzymes produced by Pseudomonas syringae pv. glycinea. FEMS Microbiol Lett 117: 1–5 [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, et al. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13: 614–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BC, George SJ, Price CA, Ryan MH, Tibbett M. (2014) The role of root exuded low molecular weight organic anions in facilitating petroleum hydrocarbon degradation: current knowledge and future directions. Sci Total Environ 472: 642–653 [DOI] [PubMed] [Google Scholar]
- Mayak S, Tirosh T, Glick BR. (2004a) Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol Biochem 42: 565–572 [DOI] [PubMed] [Google Scholar]
- Mayak S, Tirosh T, Glick BR. (2004b) Plant growth-promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Sci 166: 525–530 [Google Scholar]
- McLaughlan C, Gallardo B, Aldridge DC. (2014) How complete is our knowledge of the ecosystem services impacts of Europe's top 10 invasive species? Acta Oecol 54: 119–130 [Google Scholar]
- Mendes R, Garbeva P, Raaijmakers JM. (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37: 634–663 [DOI] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, et al. (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332: 1097–1100 [DOI] [PubMed] [Google Scholar]
- Mia MB, Shamsuddin Z, Wahab Z, Marziah M. (2010) Effect of plant growth promoting rhizobacterial (PGPR) inoculation on growth and nitrogen incorporation of tissue cultured Musa plantlets under nitrogen-free hydroponics condition. Aust J Crop Sci 4: 85–90 [Google Scholar]
- Michaux C, Verneuil N, Hartke A, Giard JC. (2014) Physiological roles of small RNA molecules. Microbiology 160: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Mirouze M, Paszkowski J. (2011) Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol 14: 267–274 [DOI] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GED, Long SR. (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monavar Feshani A, Mohammadi S, Frazier TP, Abbasi A, Abedini R, Karimi Farsad L, Ehya F, Salekdeh GH, Mardi M. (2012) Identification and validation of Asteraceae miRNAs by the expressed sequence tag analysis. Gene 493: 253–259 [DOI] [PubMed] [Google Scholar]
- Müller S, Hauck C, Schildknecht H. (1992) Germination stimulants produced by Vigna unguiculata Walp cv Saunders Upright. J Plant Growth Regul 11: 77–84 [Google Scholar]
- Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. (1986) Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol 51: 263–273 [DOI] [PubMed] [Google Scholar]
- Mullis KB, Faloona FA. (1987) Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol 155: 335–350 [DOI] [PubMed] [Google Scholar]
- Nagahashi G, Douds DD, Jr, Abney GD. (1996) Phosphorus amendment inhibits hyphal branching of the VAM fungus Gigaspora margarita directly and indirectly through its effect on root exudation. Mycorrhiza 6: 403–408 [Google Scholar]
- Nallamilli BRR, Edelmann MJ, Zhong X, Tan F, Mujahid H, Zhang J, Nanduri B, Peng Z. (2014) Global analysis of lysine acetylation suggests the involvement of protein acetylation in diverse biological processes in rice (Oryza sativa). PLoS ONE 9: e89283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal AL, Ahmad S, Gordon-Weeks R, Ton J. (2012) Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 7: e35498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Bott S, Ohler MA, Mock HP, Lippmann R, Grosch R, Smalla K. (2014) Root exudation and root development of lettuce (Lactuca sativa L. cv. Tizian) as affected by different soils. Front Microbiol 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newey LJ, Caten CE, Green JR. (2007) Rapid adhesion of Stagonospora nodorum spores to a hydrophobic surface requires pre-formed cell surface glycoproteins. Mycol Res 111: 1255–1267 [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E, Vicré-Gibouin M, Cannesan MA, Driouich A. (2013) Arabinogalactan proteins in root-microbe interactions. Trends Plant Sci 18: 440–449 [DOI] [PubMed] [Google Scholar]
- Nic-Can GI, De-la-Peña C (2012) Determination of histone methylation in mono- and dicotyledonous plants. In VM Loyola-Vargas, N Ochoa-Alejo, eds, Plant Cell Culture Protocols, Vol 877. Humana Press, London, pp 313–324 [DOI] [PubMed] [Google Scholar]
- Nichols KA, Wright SF. (2006) Carbon and nitrogen in operationally defined soil organic matter pools. Biol Fertil Soils 43: 215–220 [Google Scholar]
- Nimbal CI, Yerkes CN, Weston LA, Weller SC. (1996) Herbicidal activity and site of action of the natural product sorgoleone. Pestic Biochem Physiol 54: 73–83 [Google Scholar]
- Noir S, Colby T, Harzen A, Schmidt J, Panstruga R. (2009) A proteomic analysis of powdery mildew (Blumeria graminis f.sp. hordei) conidiospores. Mol Plant Pathol 10: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerke EC. (2006) Crop losses to pests. J Agric Sci 144: 31–43 [Google Scholar]
- Panstruga R, Dodds PN. (2009) Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science 324: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Murthy H, Chakrabarthy D, Paek K. (2009) Detection of epigenetic variation in tissue-culture-derived plants of Doritaenopsis by methylation-sensitive amplification polymorphism (MSAP) analysis. In Vitro Cell Dev Biol Plant 45: 104–108 [Google Scholar]
- Parker C. (2009) Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag Sci 65: 453–459 [DOI] [PubMed] [Google Scholar]
- Parmar P, Sindhu S. (2013) Potassium solubilization by rhizosphere bacteria: influence of nutritional and environmental conditions. J Microbiol Res 3: 25–31 [Google Scholar]
- Parniske M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Pastor N, Rosas S, Luna V, Rovera M. (2014) Inoculation with Pseudomonas putida PCI2, a phosphate solubilizing rhizobacterium, stimulates the growth of tomato plants. Symbiosis 62: 157–167 [Google Scholar]
- Patten CL, Glick BR. (2002) Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl Environ Microbiol 68: 3795–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O. (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino E, Bedini S. (2014) Enhancing ecosystem services in sustainable agriculture: biofertilization and biofortification of chickpea (Cicer arietinum L.) by arbuscular mycorrhizal fungi. Soil Biol Biochem 68: 429–439 [Google Scholar]
- Peng S, Huang J, Sheehy JE, Laza RC, Visperas RM, Zhong X, Centeno GS, Khush GS, Cassman KG. (2004) Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci USA 101: 9971–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J, Abo RP, De Nisco NJ, Arnold MFF, Longhi R, Zanda M, Walker GC. (2014) Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc Natl Acad Sci USA 111: 3561–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-García A, Romero D, de Vicente A. (2011) Plant protection and growth stimulation by microorganisms: biotechnological applications of bacilli in agriculture. Curr Opin Biotechnol 22: 187–193 [DOI] [PubMed] [Google Scholar]
- Pérez-Montaño F, Alías-Villegas C, Bellogín RA, del Cerro P, Espuny MR, Jiménez-Guerrero I, López-Baena FJ, Ollero FJ, Cubo T. (2014) Plant growth promotion in cereal and leguminous agricultural important plants: from microorganism capacities to crop production. Microbiol Res 169: 325–336 [DOI] [PubMed] [Google Scholar]
- Perry LG, Thelen GC, Ridenour WM, Callaway RM, Paschke MW, Vivanco JM. (2007) Concentrations of the allelochemical (+/−)-catechin in Centaurea maculosa soils. J Chem Ecol 33: 2337–2344 [DOI] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. (2010) The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J Bacteriol 192: 5275–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plett JM, Kemppainen M, Kale SD, Kohler A, Legué V, Brun A, Tyler BM, Pardo AG, Martin F. (2011) A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol 21: 1197–1203 [DOI] [PubMed] [Google Scholar]
- Powell ALT, van Kan J, ten Have A, Visser J, Greve LC, Bennett AB, Labavitch JM. (2000) Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol Plant Microbe Interact 13: 942–950 [DOI] [PubMed] [Google Scholar]
- Preger AC, Rillig MC, Johns AR, Du Preez CC, Lobe I, Amelung W. (2007) Losses of glomalin-related soil protein under prolonged arable cropping: a chronosequence study in sandy soils of the South African highveld. Soil Biol Biochem 39: 445–453 [Google Scholar]
- Purin S, Rillig MC. (2007) The arbuscular mycorrhizal fungal protein glomalin: limitations, progress, and a new hypothesis for its function. Pedobiologia (Jena) 51: 123–130 [Google Scholar]
- Raaijmakers JM, Weller DM. (1998) Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant Microbe Interact 11: 144–152 [Google Scholar]
- Radja Commare R, Nandakumar R, Kandan A, Suresh S, Bharathi M, Raguchander T, Samiyappan R. (2002) Pseudomonas fluorescens based bio-formulation for the management of sheath blight disease and leaffolder insect in rice. Crop Protection 21: 671–677 [Google Scholar]
- Ramachandran VK, East AK, Karunakaran R, Downie JA, Poole PS. (2011) Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol 12: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DK, Mueller ND, West PC, Foley JA. (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8: e66428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. (1997) The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol 24: 309–319 [DOI] [PubMed] [Google Scholar]
- Reineke G, Heinze B, Schirawski J, Buettner H, Kahmann R, Basse CW. (2008) Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol Plant Pathol 9: 339–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld WJ. (1983) Allelopathic effects of juglone on germination and growth of several herbaceous and woody species. J Chem Ecol 9: 295–308 [DOI] [PubMed] [Google Scholar]
- Rillig M, Ramsey P, Morris S, Paul E. (2003) Glomalin, an arbuscular-mycorrhizal fungal soil protein, responds to land-use change. Plant Soil 253: 293–299 [Google Scholar]
- Rillig M, Wright S, Nichols K, Schmidt W, Torn M. (2001) Large contribution of arbuscular mycorrhizal fungi to soil carbon pools in tropical forest soils. Plant Soil 233: 167–177 [Google Scholar]
- Rudrappa T, Bonsall J, Gallagher JL, Seliskar DM, Bais HP. (2007) Root-secreted allelochemical in the noxious weed Phragmites australis deploys a reactive oxygen species response and microtubule assembly disruption to execute rhizotoxicity. J Chem Ecol 33: 1898–1918 [DOI] [PubMed] [Google Scholar]
- Rudrappa T, Czymmek KJ, Paré PW, Bais HP. (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Salmond GP. (1994) Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu Rev Phytopathol 32: 181–200 [Google Scholar]
- Saltonstall K. (2002) Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc Natl Acad Sci USA 99: 2445–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA, Roberts DP, Denny TP. (1988) Analysis of the Pseudomonas solanacearum polygalacturonase encoded by pglA and its involvement in phytopathogenicity. J Bacteriol 170: 4501–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Carvalhais LC, Kazan K. (2012) Unraveling plant-microbe interactions: can multi-species transcriptomics help? Trends Biotechnol 30: 177–184 [DOI] [PubMed] [Google Scholar]
- Sha AH, Lin XH, Huang JB, Zhang DP. (2005) Analysis of DNA methylation related to rice adult plant resistance to bacterial blight based on methylation-sensitive AFLP (MSAP) analysis. Mol Genet Genomics 273: 484–490 [DOI] [PubMed] [Google Scholar]
- Shen J, Li C, Mi G, Li L, Yuan L, Jiang R, Zhang F. (2013) Maximizing root/rhizosphere efficiency to improve crop productivity and nutrient use efficiency in intensive agriculture of China. J Exp Bot 64: 1181–1192 [DOI] [PubMed] [Google Scholar]
- Shieh MT, Brown RL, Whitehead MP, Cary JW, Cotty PJ, Cleveland TE, Dean RA. (1997) Molecular genetic evidence for the involvement of a specific polygalacturonase, P2c, in the invasion and spread of Aspergillus flavus in cotton bolls. Appl Environ Microbiol 63: 3548–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SK, Yadav RL, Suman A, Singh PN. (2008) Improving rhizospheric environment and sugarcane ratoon yield through bioagents amended farm yard manure in Udic Ustochrept soil. Soil Tillage Res 99: 158–168 [Google Scholar]
- Siame BA, Weerasuriya Y, Wood K, Ejeta G, Butler LG. (1993) Isolation of strigol, a germination stimulant for Striga asiatica, from host plants. J Agric Food Chem 41: 1486–1491 [Google Scholar]
- Simon S, Petrášek J. (2011) Why plants need more than one type of auxin. Plant Sci 180: 454–460 [DOI] [PubMed] [Google Scholar]
- Singh PK, Singh M, Tripathi BN. (2013) Glomalin: an arbuscular mycorrhizal fungal soil protein. Protoplasma 250: 663–669 [DOI] [PubMed] [Google Scholar]
- Singh Gahoonia T, Care D, Nielsen N. (1997) Root hairs and phosphorus acquisition of wheat and barley cultivars. Plant Soil 191: 181–188 [Google Scholar]
- Smaill SJ, Leckie AC, Clinton PW, Hickson AC. (2010) Plantation management induces long-term alterations to bacterial phytohormone production and activity in bulk soil. Appl Soil Ecol 45: 310–314 [Google Scholar]
- Song J, Win J, Tian M, Schornack S, Kaschani F, Ilyas M, van der Hoorn RAL, Kamoun S. (2009) Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc Natl Acad Sci USA 106: 1654–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Ni Z, Yao Y, Xie C, Li Z, Wu H, Zhang Y, Sun Q. (2007) Wheat (Triticum aestivum L.) root proteome and differentially expressed root proteins between hybrid and parents. Proteomics 7: 3538–3557 [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Haas D. (2011) Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl Microbiol Biotechnol 91: 63–79 [DOI] [PubMed] [Google Scholar]
- Soto MJ, Fernández-Aparicio M, Castellanos-Morales V, García-Garrido JM, Ocampo JA, Delgado MJ, Vierheilig H. (2010) First indications for the involvement of strigolactones on nodule formation in alfalfa (Medicago sativa). Soil Biol Biochem 42: 383–385 [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31: 425–448 [DOI] [PubMed] [Google Scholar]
- Spaink HP. (2000) Root nodulation and infection factors produced by rhizobial bacteria. Annu Rev Microbiol 54: 257–288 [DOI] [PubMed] [Google Scholar]
- Stinson KA, Campbell SA, Powell JR, Wolfe BE, Callaway RM, Thelen GC, Hallett SG, Prati D, Klironomos JN. (2006) Invasive plant suppresses the growth of native tree seedlings by disrupting belowground mutualisms. PLoS Biol 4: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard J. (2001) Genetics and genomics of root symbiosis. Curr Opin Plant Biol 4: 328–335 [DOI] [PubMed] [Google Scholar]
- Sugawara M, Epstein B, Badgley BD, Unno T, Xu L, Reese J, Gyaneshwar P, Denny R, Mudge J, Bharti AK, et al. (2013) Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol 14: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner L, Watson B, Lei Z, Nagaraj S (2007) Proteomics of Medicago truncatula. In J Samaj, JJ Thelen, eds, Plant Proteomics. Springer, Berlin, pp 121–136 [Google Scholar]
- Suslow T, Kloepper J, Schroth M, Burr T. (1979) Beneficial bacteria enhance plant growth. Calif Agric 33: 15–17 [Google Scholar]
- Swain MR, Naskar SK, Ray RC. (2007) Indole-3-acetic acid production and effect on sprouting of yam (Dioscorea rotundata L.) minisetts by Bacillus subtilis isolated from culturable cowdung microflora. Pol J Microbiol 56: 103–110 [PubMed] [Google Scholar]
- Takahara M, Magori S, Soyano T, Okamoto S, Yoshida C, Yano K, Sato S, Tabata S, Yamaguchi K, Shigenobu S, et al. (2013) Too much love, a novel Kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume-Rhizobium symbiosis. Plant Cell Physiol 54: 433–447 [DOI] [PubMed] [Google Scholar]
- Talboys PJ, Owen DW, Healey JR, Withers PJ, Jones DL. (2014) Auxin secretion by Bacillus amyloliquefaciens FZB42 both stimulates root exudation and limits phosphorus uptake in Triticum aestivum. BMC Plant Biol 14: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamasloukht M, Séjalon-Delmas N, Kluever A, Jauneau A, Roux C, Bécard G, Franken P. (2003) Root factors induce mitochondrial-related gene expression and fungal respiration during the developmental switch from asymbiosis to presymbiosis in the arbuscular mycorrhizal fungus Gigaspora rosea. Plant Physiol 131: 1468–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawaraya K, Horie R, Saito A, Shinano T, Wagatsuma T, Saito K, Oikawa A. (2013) Metabolite profiling of shoot extracts, root extracts, and root exudates of rice plant under phosphorus deficiency. J Plant Nutr 36: 1138–1159 [Google Scholar]
- Taylor RD, Saparno A, Blackwell B, Anoop V, Gleddie S, Tinker NA, Harris LJ. (2008) Proteomic analyses of Fusarium graminearum grown under mycotoxin-inducing conditions. Proteomics 8: 2256–2265 [DOI] [PubMed] [Google Scholar]
- ten Have A, Mulder W, Visser J, van Kan JA. (1998) The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol Plant Microbe Interact 11: 1009–1016 [DOI] [PubMed] [Google Scholar]
- Tharayil N, Triebwasser DJ. (2010) Elucidation of a diurnal pattern of catechin exudation by Centaurea stoebe. J Chem Ecol 36: 200–204 [DOI] [PubMed] [Google Scholar]
- Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S. (2002) Agricultural sustainability and intensive production practices. Nature 418: 671–677 [DOI] [PubMed] [Google Scholar]
- Timmusk S, Wagner EGH. (1999) The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: a possible connection between biotic and abiotic stress responses. Mol Plant Microbe Interact 12: 951–959 [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I. (2010) Stress-induced activation of heterochromatic transcription. PLoS Genet 6: e1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H, Antelmann H, Jongbloed JDH, Braun PG, Darmon E, Dorenbos R, Dubois JY, Westers H, Zanen G, Quax WJ, et al. (2004) Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol Mol Biol Rev 68: 207–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H, Bolhuis A, Jongbloed JDH, Bron S, van Dijl JM. (2000) Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol Mol Biol Rev 64: 515–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S, Kamiya Y, Kawakami N, Nambara E, McCourt P, Tsuchiya Y. (2012) Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant Cell Physiol 53: 107–117 [DOI] [PubMed] [Google Scholar]
- Toth R, Toth D, Starke D, Smith DR. (1990) Vesicular-arbuscular mycorrhizal colonization in Zea mays affected by breeding for resistance to fungal pathogens. Can J Bot 68: 1039–1044 [Google Scholar]
- Toyomasu T, Kagahara T, Okada K, Koga J, Hasegawa M, Mitsuhashi W, Sassa T, Yamane H. (2008) Diterpene phytoalexins are biosynthesized in and exuded from the roots of rice seedlings. Biosci Biotechnol Biochem 72: 562–567 [DOI] [PubMed] [Google Scholar]
- Treseder KK, Turner KM. (2007) Glomalin in ecosystems. Soil Sci Soc Am J 71: 1257–1266 [Google Scholar]
- Uddin MR, Park SU, Dayan FE, Pyon JY. (2014) Herbicidal activity of formulated sorgoleone, a natural product of sorghum root exudate. Pest Manag Sci 70: 252–257 [DOI] [PubMed] [Google Scholar]
- Us-Camas R, Rivera-Solís G, Duarte-Aké F, De-la-Peña C. (2014) In vitro culture: an epigenetic challenge for plants. Plant Cell Tissue Organ Cult 118: 187–201 [Google Scholar]
- Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z. (2004) Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res 32: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden MGA, Bardgett RD, van Straalen NM. (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11: 296–310 [DOI] [PubMed] [Google Scholar]
- Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z, Farkas A, Mikulass K, Nagy A, Tiricz H, et al. (2010) Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122–1126 [DOI] [PubMed] [Google Scholar]
- van Esse HP, Van’t Klooster JW, Bolton MD, Yadeta KA, van Baarlen P, Boeren S, Vervoort J, de Wit PJGM, Thomma BPHJ. (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20: 1948–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse J, Frey P, Trigalet A. (1995) Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol Plant Microbe Interact 8: 241–251 [Google Scholar]
- Vauclare P, Bligny R, Gout E, Widmer F. (2013) An overview of the metabolic differences between Bradyrhizobium japonicum 110 bacteria and differentiated bacteroids from soybean (Glycine max) root nodules: an in vitro 13C- and 31P-nuclear magnetic resonance spectroscopy study. FEMS Microbiol Lett 343: 49–56 [DOI] [PubMed] [Google Scholar]
- Vaughan MM, Wang Q, Webster FX, Kiemle D, Hong YJ, Tantillo DJ, Coates RM, Wray AT, Askew W, O’Donnell C, et al. (2013) Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 25: 1108–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateshwaran M, Volkening JD, Sussman MR, Ané JM. (2013) Symbiosis and the social network of higher plants. Curr Opin Plant Biol 16: 118–127 [DOI] [PubMed] [Google Scholar]
- Verma SS, Rahman MH, Deyholos MK, Basu U, Kav NNV. (2014) Differential expression of miRNAs in Brassica napus root following infection with Plasmodiophora brassicae. PLoS ONE 9: e86648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JK. (2003) Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255: 571–586 [Google Scholar]
- Vickery P, Wheeler J, Mulcahy C. (1987) Factors affecting the hydrogen cyanide potential of white clover (Trifolium repens L.). Aust J Agric Res 38: 1053–1059 [Google Scholar]
- Vilà M, Basnou C, Pyšek P, Josefsson M, Genovesi P, Gollasch S, Nentwig W, Olenin S, Roques A, Roy D, et al. (2009) How well do we understand the impacts of alien species on ecosystem services? A pan-European, cross-taxa assessment. Front Ecol Environ 8: 135–144 [Google Scholar]
- Wallis FM, Truter SJ. (1978) Histopathology of tomato plants infected with Pseudomonas solanacearum, with emphasis on ultrastructure. Physiol Plant Pathol 13: 307–317 [Google Scholar]
- Wang Y, Kim SG, Wu J, Huh HH, Lee SJ, Rakwal R, Agrawal GK, Park ZY, Young Kang K, Kim ST. (2013) Secretome analysis of the rice bacterium Xanthomonas oryzae (Xoo) using in vitro and in planta systems. Proteomics 13: 1901–1912 [DOI] [PubMed] [Google Scholar]
- Watt SA, Wilke A, Patschkowski T, Niehaus K. (2005) Comprehensive analysis of the extracellular proteins from Xanthomonas campestris pv. campestris B100. Proteomics 5: 153–167 [DOI] [PubMed] [Google Scholar]
- Webb KJ, Skøt L, Nicholson MN, Jørgensen B, Mizen S. (2000) Mesorhizobium loti increases root-specific expression of a calcium-binding protein homologue identified by promoter tagging in Lotus japonicus. Mol Plant Microbe Interact 13: 606–616 [DOI] [PubMed] [Google Scholar]
- Weerasuriya Y, Siame BA, Hess D, Ejeta G, Butler LG. (1993) Influence of conditions and genotype on the amount of Striga germination stimulants exuded by roots of several host crops. J Agric Food Chem 41: 1492–1496 [Google Scholar]
- Weidenhamer JD, Li M, Allman J, Bergosh RG, Posner M. (2013) Evidence does not support a role for gallic acid in Phragmites australis invasion success. J Chem Ecol 39: 323–332 [DOI] [PubMed] [Google Scholar]
- Weir TL, Bais HP, Vivanco JM. (2003) Intraspecific and interspecific interactions mediated by a phytotoxin, (−)-catechin, secreted by the roots of Centaurea maculosa (spotted knapweed). J Chem Ecol 29: 2397–2412 [DOI] [PubMed] [Google Scholar]
- Weir TL, Park SW, Vivanco JM. (2004) Biochemical and physiological mechanisms mediated by allelochemicals. Curr Opin Plant Biol 7: 472–479 [DOI] [PubMed] [Google Scholar]
- Wen F, VanEtten HD, Tsaprailis G, Hawes MC. (2007) Extracellular proteins in pea root tip and border cell exudates. Plant Physiol 143: 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston LA, Alsaadawi IS, Baerson SR. (2013) Sorghum allelopathy: from ecosystem to molecule. J Chem Ecol 39: 142–153 [DOI] [PubMed] [Google Scholar]
- Wolfe BE, Rodgers VL, Stinson KA, Pringle A. (2008) The invasive plant Alliaria petiolata (garlic mustard) inhibits ectomycorrhizal fungi in its introduced range. J Ecol 96: 777–783 [Google Scholar]
- Wright DP, Read DJ, Scholes JD. (1998) Mycorrhizal sink strength influences whole plant carbon balance of Trifolium repens L. Plant Cell Environ 21: 881–891 [Google Scholar]
- Wright SF, Franke-Snyder M, Morton JB, Upadhyaya A. (1996) Time-course study and partial characterization of a protein on hyphae of arbuscular mycorrhizal fungi during active colonization of roots. Plant Soil 181: 193–203 [Google Scholar]
- Xie H, Pasternak JJ, Glick BR. (1996) Isolation and characterization of mutants of the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2 that overproduce indoleacetic acid. Curr Microbiol 32: 67–71 [Google Scholar]
- Xie X, Kusumoto D, Takeuchi Y, Yoneyama K, Yamada Y, Yoneyama K. (2007) 2′-Epi-orobanchol and solanacol, two unique strigolactones, germination stimulants for root parasitic weeds, produced by tobacco. J Agric Food Chem 55: 8067–8072 [DOI] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Yoneyama K. (2010) The strigolactone story. Annu Rev Phytopathol 48: 93–117 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SK, Dave A, Sarkar A, Singh HB, Sarma BK. (2013) Co-inoculated biopriming with Trichoderma, Pseudomonas and Rhizobium improves crop growth in Cicer arietinum and Phaseolus vulgaris. Int J Agric Environ Biotech 6: 255–259 [Google Scholar]
- Yang F, Jensen JD, Svensson B, Jørgensen HJL, Collinge DB, Finnie C. (2012) Secretomics identifies Fusarium graminearum proteins involved in the interaction with barley and wheat. Mol Plant Pathol 13: 445–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kloepper JW, Ryu CM. (2009) Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci 14: 1–4 [DOI] [PubMed] [Google Scholar]
- Zahir Z, Malik M, Arshad M. (2000) Improving crop yields by the application of an auxin precursor L-tryptophan. Pak J Biol Sci 3: 133–135 [Google Scholar]
- Zahir ZA, Asghar HN, Akhtar MJ, Arshad M. (2005) Precursor (L-tryptophan)-inoculum (Azotobacter) interaction for improving yields and nitrogen uptake of maize. J Plant Nutr 28: 805–817 [Google Scholar]
- Zahir ZA, Iqbal M, Arshad M, Naveed M, Khalid M. (2007) Effectiveness of IAA, GA3 and kinetin blended with recycled organic waste for improving growth and yield of wheat (Triticum aestivum L.). Pak J Bot 39: 761–768 [Google Scholar]
- Zamani M, Behboudi K, Ahmadzadeh M. (2013) Quorum quenching by Bacillus cereus U92: a double-edged sword in biological control of plant diseases. Biocontrol Sci Technol 23: 555–573 [Google Scholar]
- Zehnder GW, Murphy JF, Sikora EJ, Kloepper JW. (2001) Application of rhizobacteria for induced resistance. Eur J Plant Pathol 107: 39–50 [Google Scholar]
- Zhang H, Kim MS, Sun Y, Dowd SE, Shi H, Paré PW. (2008) Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol Plant Microbe Interact 21: 737–744 [DOI] [PubMed] [Google Scholar]
- Zhang J, Subramanian S, Stacey G, Yu O. (2009) Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J 57: 171–183 [DOI] [PubMed] [Google Scholar]
- Zhang N, Wang D, Liu Y, Li S, Shen Q, Zhang R. (2014) Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil 374: 689–700 [Google Scholar]
- Zhao N, Wang G, Norris A, Chen X, Chen F. (2013) Studying plant secondary metabolism in the age of genomics. Crit Rev Plant Sci 32: 369–382 [Google Scholar]
- Ziegler M, Engel M, Welzl G, Schloter M. (2013) Development of a simple root model to study the effects of single exudates on the development of bacterial community structure. J Microbiol Methods 94: 30–36 [DOI] [PubMed] [Google Scholar]
- Zwanenburg B, Pospíšil T. (2013) Structure and activity of strigolactones: new plant hormones with a rich future. Mol Plant 6: 38–62 [DOI] [PubMed] [Google Scholar]