An enzyme of cytosolic hexose phosphate metabolism is essential for plant reproduction and important for photosynthesis.
Abstract
Carbohydrate metabolism in plants is tightly linked to photosynthesis and is essential for energy and carbon skeleton supply of the entire organism. Thus, the hexose phosphate pools of the cytosol and the chloroplast represent important metabolic resources that are maintained through action of phosphoglucose isomerase (PGI) and phosphoglucose mutase interconverting glucose 6-phosphate, fructose 6-phosphate, and glucose 1-phosphate. Here, we investigated the impact of disrupted cytosolic PGI (cPGI) function on plant viability and metabolism. Overexpressing an artificial microRNA targeted against cPGI (amiR-cpgi) resulted in adult plants with vegetative tissue essentially free of cPGI activity. These plants displayed diminished growth compared with the wild type and accumulated excess starch in chloroplasts but maintained low sucrose content in leaves at the end of the night. Moreover, amiR-cpgi plants exhibited increased nonphotochemical chlorophyll a quenching during photosynthesis. In contrast to amiR-cpgi plants, viable transfer DNA insertion mutants disrupted in cPGI function could only be identified as heterozygous individuals. However, homozygous transfer DNA insertion mutants could be isolated among plants ectopically expressing cPGI. Intriguingly, these plants were only fertile when expression was driven by the ubiquitin10 promoter but sterile when the seed-specific unknown seed protein promoter or the Cauliflower mosaic virus 35S promoter were employed. These data show that metabolism is apparently able to compensate for missing cPGI activity in adult amiR-cpgi plants and indicate an essential function for cPGI in plant reproduction. Moreover, our data suggest a feedback regulation in amiR-cpgi plants that fine-tunes cytosolic sucrose metabolism with plastidic starch turnover.
Starch and Suc turnover are major pathways of primary metabolism in all higher plants. As such, they are essential for carbohydrate storage and the energy supply of sink tissues and as building blocks for amino acid, fatty acid, or cell wall biosynthesis (Stitt and Zeeman, 2012).
A core reaction in both starch and Suc biosynthesis is the reversible interconversion of the hexose phosphate pool metabolites Fru 6-phosphate (Fru6P) and Glc 6-phosphate (Glc6P), which is mediated by phosphoglucose isomerase (PGI). Arabidopsis (Arabidopsis thaliana) contains two isoforms of PGI, one in the plastids and one in the cytosol (Caspar et al., 1985).
During the light period, the plastid isoform of PGI (PGI1) is involved in starch biosynthesis by generating Glc6P from the primary photosynthetic product Fru6P. Glc6P is further converted to Glc 1-phosphate (Glc1P) and ADP-glucose via action of phosphoglucomutase (PGM) and ADP-glucose pyrophosphorylase (AGPase), respectively (Stitt and Zeeman, 2012). Finally, transfer of the glucosyl moiety of ADP-glucose to the growing carbohydrate chain of starch is mediated by starch synthases. Any of the enzymatic reactions of this linear pathway is essential for starch synthesis, as illustrated by the virtual absence of transitory starch in chloroplasts of mutant plant lines with impaired function of PGI1 (Yu et al., 2000; Kunz et al., 2010), PGM (Caspar et al., 1985; Kofler et al., 2000), or AGPase (Lin et al., 1988). Interestingly, in a few specific cell types, e.g. leaf guard cells and root columella cells, loss of PGI1 activity can be bypassed by the presence of the plastid Glc6P/phosphate translocator GPT1 (Niewiadomski et al., 2005; Kunz et al., 2010).
The cytosolic isoform of PGI (cPGI) is involved in anabolism and catabolism of Suc, the major transport form of carbohydrates in plants. Glc6P and Fru6P interconversion is necessary for both Suc synthesis during the day and during the night. During the day, Suc synthesis in source leaves is fueled mainly by triose phosphates exported from chloroplasts that are eventually converted to Fru6P in the cytosol. However, Fru6P is only one substrate for the Suc-generating enzyme Suc phosphate synthase. The second substrate, UDP-glucose, is synthesized from Fru6P via Glc6P and Glc1P by the cytosolic isoenzymes of PGI1 and PGM as well as UDP-glucose pyrophosphorylase.
Because Suc is the major long-distance carbon transport form, its synthesis has to continue throughout the night to supply energy and carbohydrates to all tissues. The nocturnal synthesis of Suc is dependent on breakdown and mobilization of transitory starch from chloroplasts (Zeeman et al., 2007) via export of maltose and Glc (Weber et al., 2000; Niittylä et al., 2004; Weise et al., 2004; Cho et al., 2011). Exported maltose is temporarily integrated into cytosolic heteroglycans (Fettke et al., 2005) mediated by disproportionating enzyme2 (DPE2; Chia et al., 2004; Lu and Sharkey, 2004) yielding Glc and a heteroglycan molecule elongated by an α1-4-bound glucosyl residue. Cytosolic Glc can directly be phosphorylated to Glc6P by the action of hexokinase, while temporarily stored Glc in heteroglycans is released as Glc1P mediated by cytosolic glucan phosphorylase2 (PHS2; Fettke et al., 2004; Lu et al., 2006). Both Glc6P and Glc1P can then be converted to UDP-glucose as during the day.
Generation of Fru6P, the second substrate for Suc synthesis, can proceed only to a limited extent from triose phosphates during the night. This limitation is caused mainly by the nocturnal inactivation of Fru 1,6-bisphosphatase (Cséke et al., 1982; Stitt, 1990), a key enzyme in Suc biosynthesis during the day. Hence, in contrast to the situation in the light, cPGI activity is now crucial for providing Fru6P from Glc6P.
On the catabolic side, degradation of Suc into its monosaccharides in sink tissues yields both Glc6P and Fru6P, of which only Fru6P can be utilized in glycolytic degradation. Therefore, cPGI is also required for Glc6P conversion to Fru6P in glycolysis, which, in combination with respiration, is the major path of energy production in heterotrophic tissues.
Impairment or loss of function of enzymes contributing to the cytosolic hexose phosphate pool has recently been investigated for the Glc1P-forming enzyme PGM (Egli et al., 2010). The Arabidopsis genome encodes three PGM isoforms, with PGM1 localized to plastids and PGM2 and PGM3 localized to the cytosol (Caspar et al., 1985; Egli et al., 2010). Analyses of transfer DNA (T-DNA) mutants showed that homozygous pgm2/pgm3 double mutants were nonviable because of impaired gametophyte development. However, pgm2 and pgm3 single mutants grew like ecotype Columbia (Col-0) wild-type plants, indicating overlapping functions of PGM2 and PGM3 (Egli et al., 2010).
By contrast, cPGI is encoded only by a single locus in Arabidopsis (Kawabe et al., 2000). Higher plant mutants reduced in cPGI activity have so far been characterized only in ethyl methanesulfonate-mutagenized Clarkia xantiana (Jones et al., 1986a; Kruckeberg et al., 1989; Neuhaus et al., 1989). The C. xantiana genome encodes for two isoenzymes of cPGI, and homozygous point mutations in each individual cPGI led to significant decrease in cPGI enzyme activity, which was further reduced to a residual activity of 18% in cpgi2/cpgi3 double mutants, where the cPGI3 locus was heterozygous for the mutation (Jones et al., 1986a; Kruckeberg et al., 1989). Detailed physiological analyses of these mutants indicated a negative impact on Suc biosynthesis and elevated starch levels when cPGI activity was decreased at least 3- to 5-fold (Kruckeberg et al., 1989).
The physiological impact of decreased or even absent cPGI activity has not been characterized in the genetic model organism Arabidopsis. Here, we show that homozygous T-DNA insertion mutants in the cPGI locus are nonviable and present data from analyses of mature Arabidopsis plants constitutively expressing artificial microRNAs (amiRNAs) targeted against cPGI. These mutants reveal altered photosynthesis, a strong impact on nocturnal leaf starch degradation, and impaired Suc metabolism.
RESULTS
Tissue-Specific Expression and Subcellular Localization of PGI Isoenzymes
According to publicly available expression data (eFP-Browser, http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007), both PGI homologs are expressed at a similarly high level throughout somatic leaf tissue. Intriguingly, while cPGI (At5g42740) transcript is also highly abundant in developing embryos and seeds, plastidic PGI1 (At4g24620) expression is much lower in these tissues. In roots, highest transcript accumulation for the plastidic isoform is found in vascular tissues and columella cells, which build starch statoliths to perceive gravitropism. Expression of the cytosolic isoform in roots is strongest in the meristematic, elongation, and maturation zones.
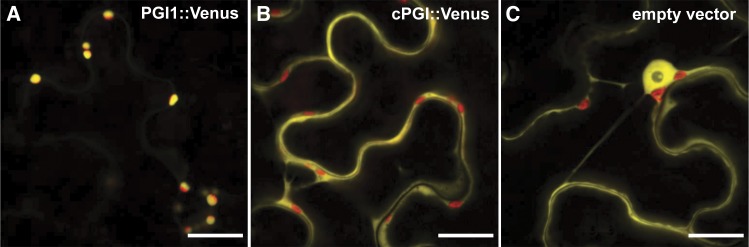
To study the subcellular localization of both PGI isoenzymes in vivo, the Venus reporter protein was fused to the C terminus of either PGI protein. According to activity gel assays, PGI1 localizes to the chloroplast (Caspar et al., 1985; Yu et al., 2000). Yellow fluorescence signal for the PGI1::Venus fusion protein was detected specifically in chloroplasts of transiently expressing Nicotiana benthamiana leaves as demonstrated by the colocalization with chlorophyll autofluorescence (Fig. 1A). For the cPGI::Venus fusion protein, on the other hand, fluorescence could be detected in the cytosol, clearly not colocalizing with the red autofluorescence from chlorophyll in the chloroplasts (Fig. 1B). Cytosolic localization of cPGI is in agreement with in silico predictions (Schwacke et al., 2003) and proteome studies of the Arabidopsis cytosol (Ito et al., 2011). By contrast, free Venus protein was detected in the cytosol and also strongly in the nucleus (Fig. 1C).
Figure 1.
Subcellular localization of PGI1 and cPGI. PGI1 (A) and cPGI (B) Venus fusion proteins (yellow) localize to chloroplasts (red) and the cytosol, respectively. C, An unfused Venus protein expressed from the empty vector was used as cytosolic control. Constructs were transiently expressed in N. benthamiana leaves. All images are merged with the corresponding chlorophyll autofluorescence image (red). Bar = 20 µm.
Isolation of T-DNA Insertion Lines and Generation of cPGI amiRNA Plants
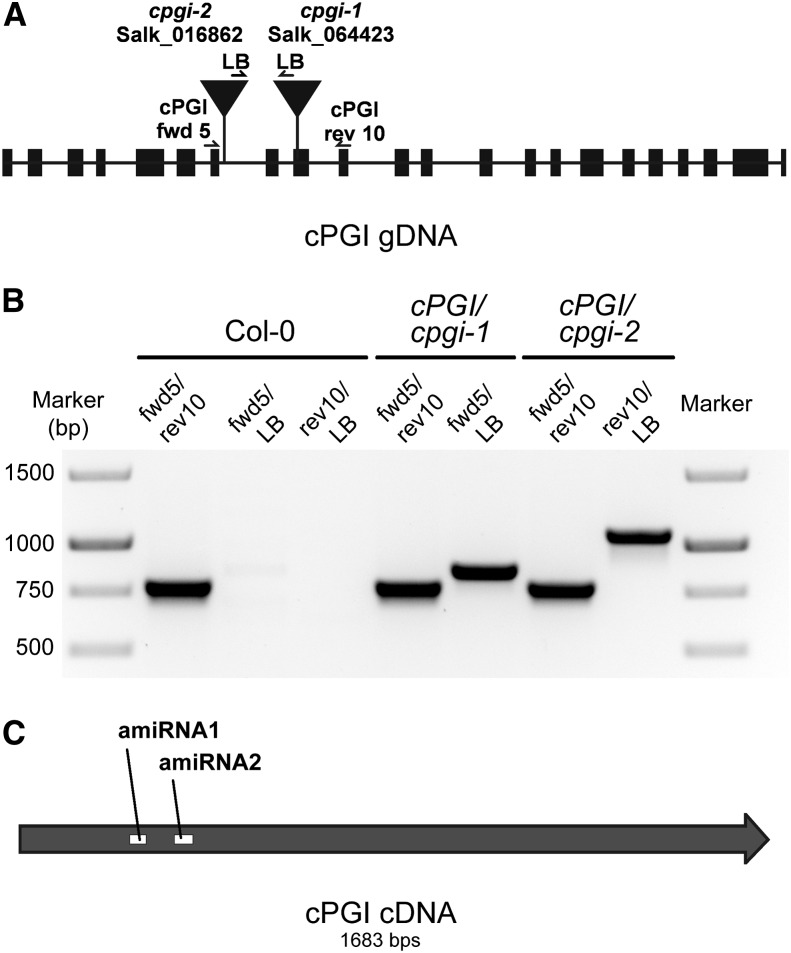
Two independent T-DNA insertion lines from the SALK collection (Alonso et al., 2003) were obtained from the Arabidopsis stock center. T-DNA insertion sites were located in the ninth exon (cpgi-1) and the seventh intron (cpgi-2), respectively (Fig. 2A). Heterozygous cpgi plants could be identified from the segregating progeny of self-pollinated heterozygous parent plants at a ratio of 148:156 and 159:146 (heterozygotes:wild type) for cpgi-1 and cpgi-2, respectively. However, no homozygous individuals could be recovered from the same segregating seed population (Fig. 2B; Supplemental Table S1), suggesting that a homozygous mutation in cPGI is lethal. To exclude that homozygous individuals are missed due to the inability of homozygous seeds to germinate, we compared the germination rates of seeds from heterozygous cpgi mutants and Col-0 wild-type plants. Results show that germination rates of segregating cpgi seeds were not different from the wild type (Supplemental Table S2), indicating that no homozygous seeds are among the segregating cpgi seed population.
Figure 2.
Localization and identification of T-DNA insertion and amiRNA target sites for cPGI. A, Two independent cPGI insertion lines (cpgi-1 and cpgi-2) were isolated, and the insertion site was confirmed at the positions indicated. B, Confirmation of T-DNA insertions in cPGI/cpgi-1 and cPGI/cpgi-2 by genomic DNA PCR using primers indicated in A. No homozygous plants could be identified. C, Position of target sites of two independent amiRNAs designed against cPGI mRNA. Two independent lines of amiRNA1 (amiR-cpgi6 and amiR-cpgi10) and three lines of amiRNA2 (amiR2-cpgi8, amiR2-cpgi9, and amiR2-cpgi10) were analyzed.
To overcome lethality in homozygous T-DNA insertion mutants, two independent amiRNAs were designed targeting different regions of the cPGI coding region (Fig. 2C). In contrast to loss-of-function T-DNA mutants, amiRNA mutants allow analysis of only partial or regulated functional loss of the gene of interest when full knockout mutants are lethal (Schwab et al., 2010). Plants expressing these amiRNAs under the control of the Cauliflower mosaic virus (CaMV) 35S promoter were isolated on selective medium and the progeny of individual plants analyzed. In total, five independent lines of the two amiRNA target sites (two lines of amiRNA1 and three lines of amiRNA2) were analyzed for core metabolic data. Two independent lines carrying the amiRNA1 transgene, designated as amiR-cpgi6 and amiR-cpgi10, were studied in detail further.
Mature amiR-cpgi Plants Display Reduced Growth and Increased Nonphotochemical Chlorophyll a Quenching in the Virtual Absence of cPGI Activity
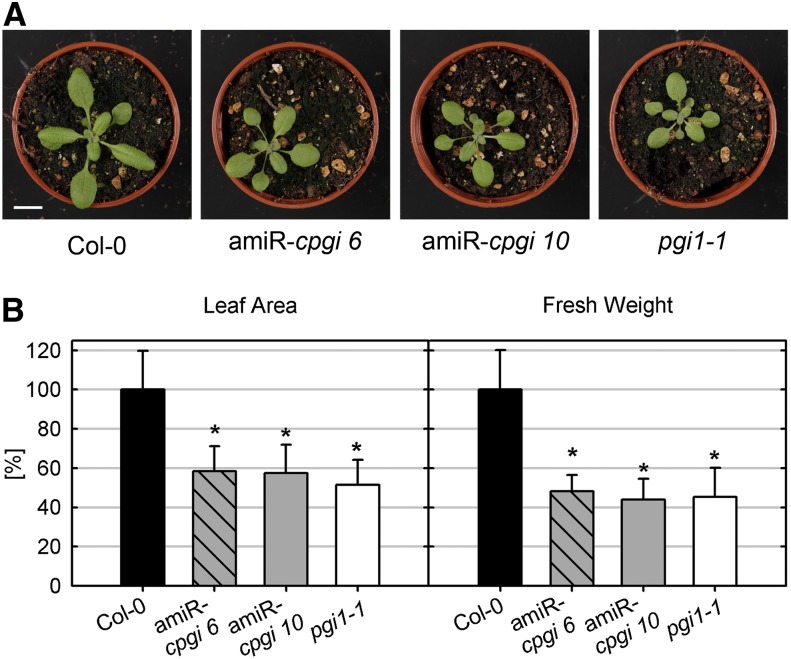
At first glance, no growth abnormalities were found in amiR-cpgi mutants (Fig. 3A). However, when measuring rosette leaf area or rosette fresh weight, it was discovered that amiR-cpgi mutants displayed slight but significant growth retardation compared with wild-type plants that was comparable to that of starch-free pgi1-1 plants (Fig. 3B; Yu et al., 2000). In addition, growth was even further diminished under short-day conditions compared with long-day conditions (Supplemental Fig. S1, B and C). This is characteristic for mutants impaired in carbohydrate metabolism such as pgi1-1 or pgm (Gibon et al., 2009; Izumi et al., 2013).
Figure 3.
Growth phenotype of amiR-cpgi and pgi1-1 mutants. A, Col-0, amiR-cpgi6, amiR-cpgi10, and pgi1-1 grown on soil in long-day conditions for 21 d. Bar = 1 cm. B, Relative leaf area and rosette leaf fresh weight of wild-type and mutant plants grown on soil in long-day conditions for 21 d. Bars represent average ± sd from three independent experiments (n = 15–30). Asterisks indicate significant difference to the wild type (Student’s t test, α ≤ 0.01). [See online article for color version of this figure.]
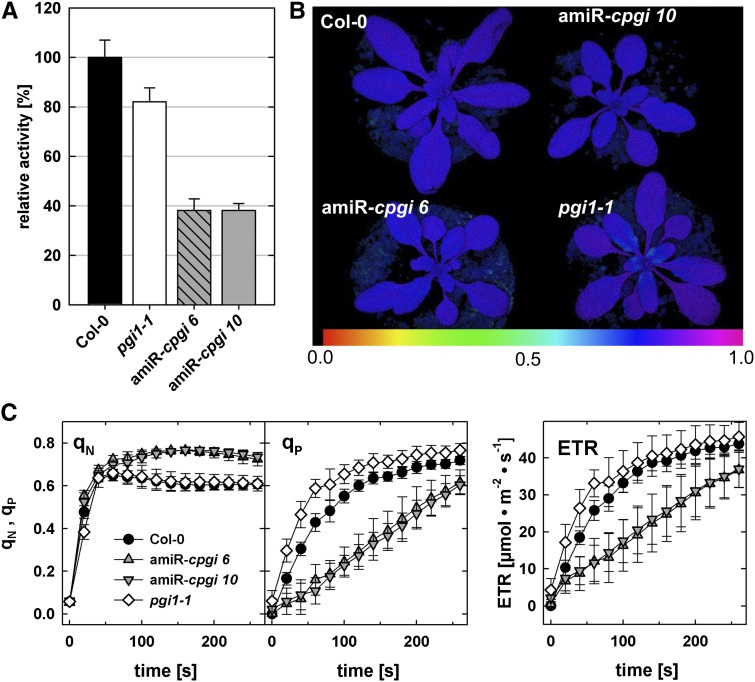
Analyses of total PGI activity (i.e. activity of the plastidic and cytosolic isoform) in leaf extracts of wild-type, amiRNA-cpgi, and pgi1-1 plants revealed that total PGI activity is strongly decreased in amiR-cpgi6 and amiR-cpgi10 lines to less than 40% of wild-type activity (Fig. 4A; Supplemental Fig. S2A). These measurements also showed that total PGI activity in pgi1-1 plants is decreased to approximately 80% of the wild-type level. The plastidic PGI can be heat inactivated by incubating the protein extract for 10 min at 50°C prior to the enzyme assay (Jones et al., 1986b; Yu et al., 2000). Leaf extracts pretreated accordingly showed no detectable residual PGI activity in amiR-cpgi plants (Supplemental Fig. S2B), suggesting that leaves were essentially free of cPGI activity. The residual total PGI activity found in those plants (Fig. 4A) may be attributed to plastidic PGI activity.
Figure 4.
amiR-cpgi mutants display strongly reduced PGI activity in leaves and increased nonphotochemical quenching. A, Total enzymatic PGI (cPGI and PGI) activity in leaf extracts of pgi1-1, amiR-cpgi6, and amiR-cpgi10 plants relative to Col-0 wild-type leaves. Average ± se of the mean (n = 4). B, PAM fluorescence imaging displaying the variable over maximal fluorescence ratio (Fv/Fm). Blue-purple false colors in leaves of all plants indicate an Fv/Fm of about 0.8, which is typical for plants with intact PSII. C, qN, qP, and photosynthetic ETRs during light induction curves determined by PAM fluorescence imaging in rosette leaves of wild-type, amiR-cpgi6, amiR-cpgi10, and pgi1-1 plants. Average ± sd (n = 15).
To investigate whether reduced leaf area and rosette fresh weight might be the result of changes in photosynthetic performance, PSII properties were investigated by pulse amplitude modulation (PAM) fluorometry. Determination of the maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm) showed no difference between the wild type and mutants (Fig. 4B). Thus, the Fv/Fm ratio of approximately 0.8 indicated full functional integrity of PSII for all genotypes. Intriguingly, PAM measurements of photosynthetic induction curves revealed increased nonphotochemical quenching (qN) for all amiRNA lines (Fig. 4C; Supplemental Fig. S3A). Concomitantly, the photochemical quench coefficient (qP) and the electron transport rate (ETR) were decreased in all amiR-cpgi lines compared with wild-type and pgi1-1 plants (Fig. 4C; Supplemental Fig. S3A). These data indicate that decreased cytosolic PGI activity exhibits an effect on the photosynthetic light reaction in the thylakoids of chloroplasts.
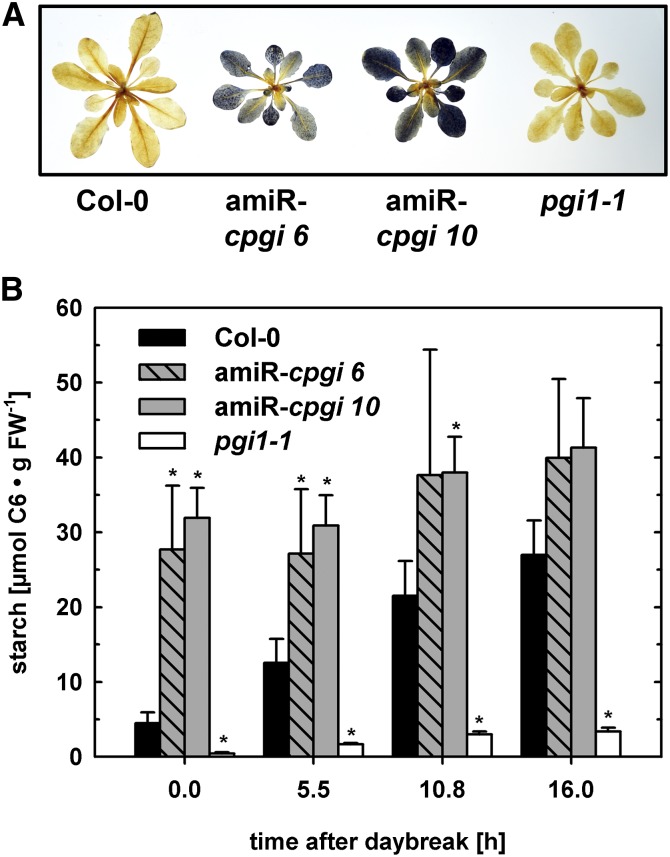
amiRNA-cpgi Plants Display a Starch-Excess Phenotype in Mature Leaves
In addition to the macroscopic growth retardation and photochemical phenotypes of amiR-cpgi plants, whole rosette starch staining revealed strongly increased starch levels at the end of the night in amiR-cpgi plants (Fig. 5A; Supplemental Fig. S3B). Usually transitory leaf starch is depleted at the end of the night period as it was found in wild-type plants (Fig. 5A; Supplemental Fig. S3B). Measurement of starch content in leaves of mutants and the wild type over the course of a day confirmed significant starch excess in amiR-cpgi lines of both amiRNA targets at the end of the night (Fig. 5B; Supplemental Fig. S3C). Moreover, starch levels were generally elevated in leaves of amiR-cpgi plants compared with those in the wild type even at the end of the day, when starch levels are usually highest in wild-type leaves (Fig. 5B; Supplemental Fig. S3C). By contrast, pgi1-1 plants, used as an almost starch-free reference, displayed only very low starch levels in leaves (Fig. 5B; Supplemental Fig. S3C).
Figure 5.
Starch excess in leaves of amiR-cpgi plants. A, Starch staining of whole leaf rosettes harvested at the end of the night period. B, Starch content of wild-type, amiR-cpgi6, amiR-cpgi10, and pgi1-1 leaves at different time points during the light period. Average ± se of the mean (n = 3). Asterisks indicate significant difference to the wild type of the same time point (Student’s t test, α ≤ 0.05). [See online article for color version of this figure.]
To investigate if leaf starch excess (Fig. 5A) and the increased nonphotochemical quenching phenotype (Fig. 4C) are functionally connected, mutants with impaired activity of other cytosolic enzymes known to display high-starch phenotypes, such as fum2 mutants (Pracharoenwattana et al., 2010), defective in cytosolic fumarase activity, or dpe2 mutants, defective in cytosolic heteroglycan metabolism (Lu and Sharkey, 2004), were analyzed. Although all different plant lines were grown side-by-side under the same conditions, we did not detect elevated starch in rosettes of three independent fum2 mutant lines and hence focused on dpe2 mutants that showed a starch excess phenotype as reported before (Supplemental Fig. S1A). dpe2 mutants, just as amiR-cpgi plants, displayed reduced growth in long-day and short-day conditions compared with their respective wild type. However, contrary to amiR-cpgi plants, dpe2 mutants displayed no or only mildly increased nonphotochemical quenching compared with the wild type (Supplemental Fig. S4). In contrast to the starch-free and starch-excess mutants investigated, fum2 mutants appeared to produce more biomass and rosette leaf area compared with the wild type (Supplemental Fig. S1, B and C).
Leaf Suc Concentration and Glc Content of Cytosolic Heteroglycans Is Altered in amiR-cpgi Plants
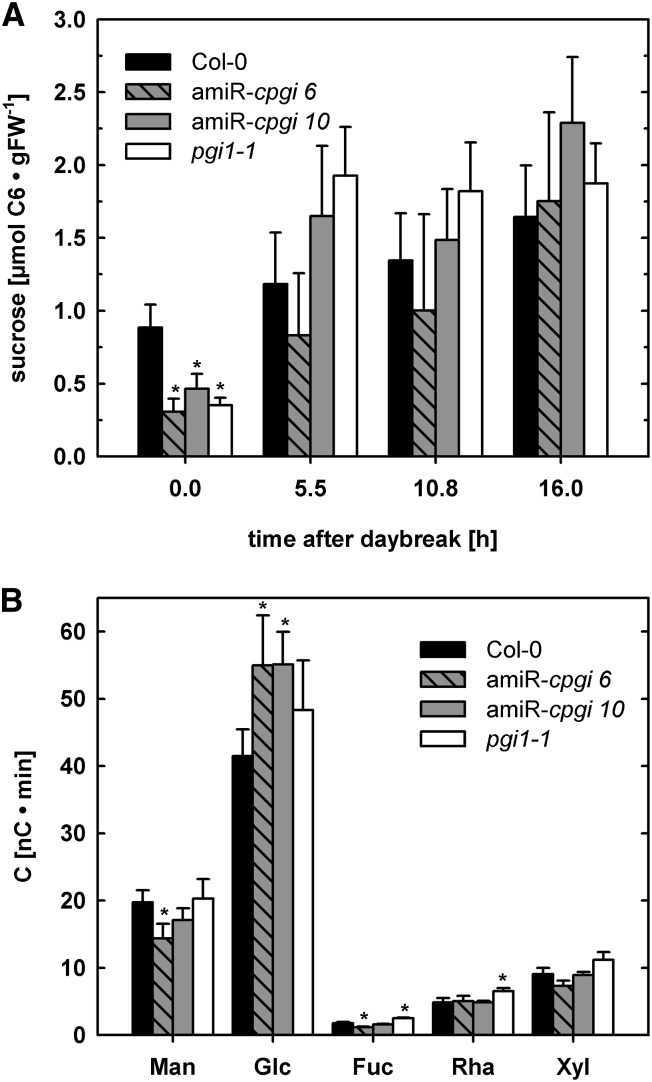
Because cPGI is an important enzyme in Suc biosynthesis and the starch-excess phenotype of amiR-cpgi plants indicated impaired starch turnover, we analyzed leaf Suc at different time points during the day/night cycle and the monosaccharide composition of cytosolic heteroglycans. At the end of the night, the Suc concentration in leaves was significantly reduced compared with the wild type in all amiRNA lines and pgi1-1 (Fig. 6A; Supplemental Fig. S3D). No significant difference in Suc content could be observed at any other time point during the light period (Fig. 6A).
Figure 6.
Suc concentration and relative monosaccharide composition of cytosolic heteroglycans in leaves. A, Suc concentration in wild-type, amiR-cpgi6, amiR-cpgi10, and pgi1-1 leaves at different time points during the light period. Average ± se of the mean (n = 3). B, Monomer composition of low Mr (<10 kD) cytosolic heteroglycans (SHGs) in leaves of wild-type, amiR-cpgi6, amiR-cpgi10, and pgi1-1 plants at the end of the night. Data were normalized to the major heteroglycan-monomer Gal. Average ± se of the mean (n = 6).
Cytosolic heteroglycans are involved in turnover of the starch degradation product maltose and serve as a temporary storage for glucosyl moieties. Analyses of the cytosolic fraction of low (<10 kD, soluble heteroglycans [SHGs])- and high (>10 kD, SHGL)-Mr soluble heteroglycans revealed that both amiR-cpgi6 and amiR-cpgi10 harbored significantly increased Glc concentrations in their SHGs at the end of the night (Fig. 6B). The content of the major heteroglycan monomer Gal was constant between ecotypes and time points and used for normalization. Sugar monomers other than Glc were not consistently changed but showed considerable variation. The observed elevated Glc content in SHGs may result from diminished cPGI activity leading to decreased carbon flux through Suc synthesis at night and hence to maltose-derived Glc being accumulated in heteroglycans.
Simultaneous Loss of Transitory Starch Formation and cPGI Activity Severely Impairs Viability of amiR-cpgi Plants on Soil
Because loss of cPGI activity apparently resulted in starch overaccumulation, we investigated whether the absence of transitory starch had an impact on the severity of the amiR-cpgi plant phenotype. Homozygous plants defective in either plastidic PGI1 (pgi1-1) or AGPase (adg1-1) were transformed with cPGI amiRNA1 and transformed individuals identified on selective one-half-strength Murashige and Skoog (MS) medium (supplemented with 2% [w/v] Suc). In initial experiments, plants transferred from selective media to soil hardly developed into adult plants. Rosette leaves remained very small, and the plants eventually died before producing seeds (Fig. 7). To investigate if exogenous carbohydrate supply could overcome this lethality, plants were transferred from selective plates either to soil or to one-half-strength MS medium supplemented with 2% (w/v) Suc in subsequent experiments. Three weeks after sowing, antibiotic-resistant plants transferred to MS medium containing Suc without antibiotics had developed rosette leaves and had considerably increased in size since transfer (Fig. 7). Starch-free amiR-cpgi × adg1-1 or amiR-cpgi × pgi1-1 plants transferred to Suc-containing medium eventually flowered and developed a small number of seeds. By contrast, plants transferred to soil instead had developed only very tiny leaves that displayed almost no increase in size and died before producing seeds (Fig. 7).
Figure 7.
Phenotype of plants expressing cpgi amiRNA1 in the virtually starch-free mutant background of pgi1-1 or adg1-1 after 21 d of growth in long-day conditions on soil or Suc-supplemented (2% [w/v]), solidified, one-half-strength MS medium. [See online article for color version of this figure.]
Analyses of cpgi T-DNA Mutants Reveal Reduced Transmission Efficiency for Mutant Male and Female Gametophytes
Loss of cPGI activity in mature leaves of amiR-cpgi plants led to viable though size-reduced plants. However, no homozygous cpgi T-DNA insertion mutants could be isolated, and genotyping of the progeny of self-pollinated heterozygous mutants revealed a segregation ratio of approximately 1:1 (cpgi/cPGI:cPGI/cPGI), clearly diverting from the expected 2:1 ratio (Supplemental Table S1). Thus, we investigated the mutant allele transmission efficiency, performing reciprocal crosses either using pollen from heterozygous cPGI/cpgi-1 or cPGI/cpgi-2 to pollinate wild-type pistils or pollinating pistils of heterozygous cPGI/cpgi-1 or cPGI/cpgi-2 plants with wild-type pollen. Genotyping of the progeny of these reciprocal crosses revealed that the transmission efficiency of the mutant allele (ratio of number of heterozygous over wild-type plants; Howden et al., 1998) was strongly decreased through both the male and female gametophyte. While an evenly split genotype distribution in the progeny of crosses would be expected at a transmission efficiency of 100%, transmission efficiency was in fact reduced to at least 32% for male and 49% for female cpgi gametophytes (Table I). However, when testing the vitality of pollen from heterozygous cPGI/cpgi-1 and cPGI/cpgi-2 plants, we did not detect an increased number of nonvital pollen grains by Alexander staining (Fig. 8A).
Table I. Reciprocal crosses between the wild type and heterozygous cpgi T-DNA mutants.
The F1 progeny of crosses between the wild type and heterozygous cpgi T-DNA mutants was genotyped by PCR and transmission efficiency (TE) calculated as the ratio of number of heterozygous over number of wild-type plants. Transmission efficiency of the cpgi allele is strongly reduced through both male and female gametes.
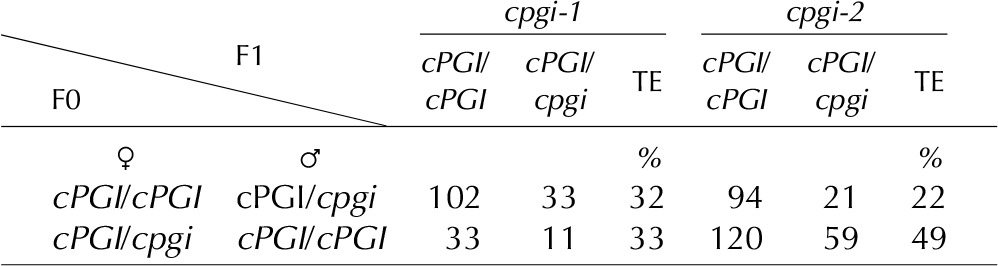
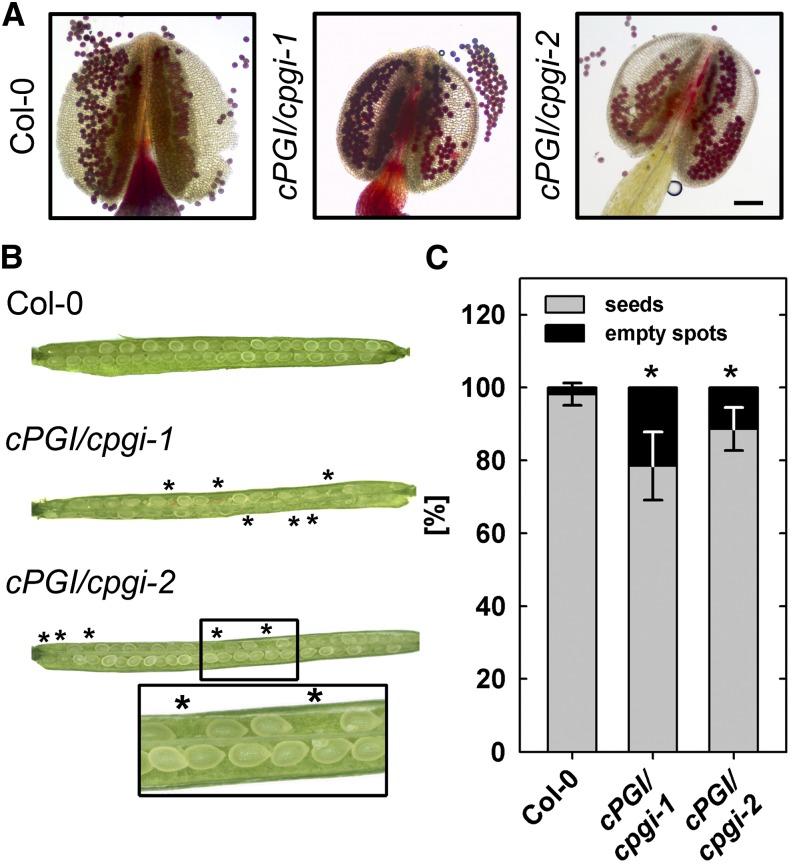
Figure 8.
Effect of T-DNA insertion in cPGI on pollen viability and ovule/seed development. A, Bright-field image of Alexander-stained pollen grains from whole-mount filaments. Viable pollen accumulates red stain. Bar = 100 µm. B, Representative siliques from wild-type, cPGI/cpgi-1, and cPGI/cpgi-2 plants. Asterisks mark empty spots in opened siliques. C, Rate of seed abortion/development in cPGI/cpgi T-DNA mutants and the wild type. Average ± sd (n = 30). Asterisks indicate significant difference to the wild type (Student’s t test, α ≤ 0.01).
Expression of cPGI is higher in developing seeds/embryos compared with other tissues, according to publicly available databases (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi: Winter et al., 2007). Therefore, to assess the viability of the early embryo, we scored the occurrence of empty spots in self-pollinated siliques from heterozygous cPGI/cpgi-1 and cPGI/cpgi-2 plants. With an observed percentage of 21% (cpgi-1) and 11% (cpgi-2) aborted seeds per cPGI/cpgi silique, a significantly higher percentage of empty spots occurred in siliques of both cPGI/cpgi T-DNA insertion lines compared with wild-type siliques (Fig. 8, B and C). To gain insight into the stage of gametophyte or embryo development depending on cPGI function, we transformed the cPGI complementary DNA (cDNA) into heterozygous cPGI/cpgi plants and screened the progeny for complementation, i.e. homozygous cpgi T-DNA plants. Expression of the cPGI cDNA was driven by three different promoters, the ubiquitously active ubiquitin 10 promoter (UBQ10), the CaMV 35S promoter active throughout most developmental stages, and the unknown seed protein promoter (USP; Bäumlein et al., 1991) mainly active during early embryo development in seeds. We could isolate primary transformants (T1) for all three promoter:cPGI complementation constructs that were homozygous for the cpgi T-DNA insertion. However, only UBQ10:cPGI plants were able to set seed. By contrast, siliques of CaMV35S:cPGI and pUSP:cPGI plants remained short and dried back without producing seeds (Supplemental Fig. S5A). Closer inspection of flowers of these plants revealed that pollen grains were absent from their anthers (Supplemental Fig. S5B), explaining the inability to produce seeds by self-fertilization. Notably, pollination of stigmas of homozygous cpgi-1 mutants expressing the CaMV35:cPGI construct with wild-type pollen led to elongating siliques (Supplemental Fig. S5) that harbored a uniformly heterozygous progeny.
DISCUSSION
A balanced carbohydrate turnover in the cytosol that is well integrated into overall metabolism is of major importance for plant fitness and viability (Gibon et al., 2006; Usadel et al., 2008). Thus, maintenance and proper adjustment of the cytosolic hexose phosphate pool is an essential process ensured by cPGI and cytosolic PGM (cPGM) activities interconverting Fru6P, Glc6P, and Glc1P. In this study, we have investigated cPGI knockdown mutants because viable homozygous cpgi T-DNA mutants could not be identified. We provide evidence that cPGI activity impacts photosynthesis and starch and Suc metabolism, but its absence can be compensated in adult plants, at least during the light period.
Analyses of cpgi T-DNA Mutants Indicate an Essential Role of cPGI Activity for Reproduction and Embryo Development
Despite screening more than 300 individual plants from the progeny of two independent heterozygous cPGI/cpgi T-DNA insertion lines, we could not identify plants homozygous for the T-DNA insertion (Fig. 2B; Supplemental Table S1). Attempts to complement T-DNA lines by cPGI overexpression led to plants homozygous for the T-DNA insertion. These complemented lines were unable to produce pollen and self-fertilize when cPGI expression was driven by either the CaMV35S or the USP promoter. However, when cPGI expression was driven by the constitutive UBQ10 promoter (Krebs et al., 2012), complemented homozygous lines were able to set seed and produce a homozygous progeny.
The initial study describing the USP promoter reported its activity mainly in developing embryos of Arabidopsis and seeds of tobacco (Nicotiana tabacum; Bäumlein et al., 1991). In addition, Zakharov et al. (2004) detected USP promoter activity in mature pollen of tobacco and also weakly in developing flowers by testing for transcript of a USP promoter-driven reporter gene. Congruent with the initial report by Bäumlein et al. (1991), we could detect USP promoter-driven GUS activity in Arabidopsis only in developing embryos but not in pollen or flowers (Supplemental Figure S6), indicating that the USP promoter induces only weak or no expression during pollen development in Arabidopsis.
As with the USP promoter, varying results regarding the activity of the CaMV 35S promoter during male gametophyte development have been observed. Early studies reported the absence of CaMV 35S promoter activity in mature Arabidopsis pollen or during pollen development (Wilkinson et al., 1997; Custers et al., 1999) that were later corroborated by studies unsuccessfully using the CaMV 35S promoter to complement Arabidopsis mutants defective in pollen development (Harrison-Lowe and Olsen, 2008), induce a pollen-lethal phenotype (Zhang et al., 2009), or label pollen peroxisomes with a GFP reporter gene fusion (Footitt et al., 2007). However, other studies suggest some activity of the CaMV 35S promoter during pollen maturation in Arabidopsis because pollen callose deposition can be altered by overexpressing callose synthase5 (Xie et al., 2010) or male sterility can be induced by expressing an RNA interference construct hampering meiosis (Wang et al., 2012). Low activity or even inactivity of the CaMV 35S promoter may also be the reason for the fact that we could isolate viable and fully fertile amiR-cpgi plants because the amiRNA would not be sufficiently expressed during cPGI-critical stages of plant development.
Thus, available data on CaMV 35S and USP promoter activity in Arabidopsis and our own data essentially suggest that cPGI expression from both promoters is insufficient to complement cPGI loss during pollen development and indicate that cPGI activity is essential during this phase of male gametophyte development. Activity of cPGI in mature pollen appears to be of minor importance because vitality of pollen from heterozygous cpgi mutants was indistinguishable from wild-type pollen (Fig. 8A) and homozygous cpgi T-DNA mutants could be identified in the T1 generation of complementation lines, which requires viable mutant cpgi pollen.
Despite unaltered pollen vitality, we detected strongly decreased transmission efficiency of the mutant allele through both male and female gametophytes (Fig. 8; Table I). Calculating the expected genotype distribution by taking the decreased transmission efficiency into account (Howden et al., 1998) revealed that the found distribution differs from the expected distribution (Supplemental Table S1; Supplemental Fig. S7). Homozygous cpgi plants that would be expected according to these data but that were not identified (Supplemental Table S1; Supplemental Fig. S7) are likely among the aborted seeds observed at a higher percentage in cPGI/cpgi compared with wild-type siliques (Fig. 8). Thus, aborted seeds/ovules observed in siliques of cPGI/cpgi plants most likely result from both embryo lethality and reduced transmission through the female gametophyte. Because both effects overlay, the observed percentage of aborted seeds/ovules diverts from expected numbers (e.g. 25% if only embryo lethality would be considered). Notably, the impact on cpgi mutant fertility observed in this study is in line with earlier results from cpgm mutants, where homozygous cpgm2/cpgm3 double mutants could not be identified due to impaired gametophyte fertilization ability (Egli et al., 2010). In essence, analyses of cpgi T-DNA mutants support the conclusion that cPGI activity is important for mutant allele transmission through male and female gametophytes and essential for pollen and embryo development. However, elucidation of the exact processes for which cPGI activity is necessary during these developmental stages requires further investigation.
Because homozygous cpgi T-DNA insertion mutants were not available to investigate loss of cPGI function in adult plants, cPGI activity was decreased by employing an amiRNA-based approach. Analyses of cpgi mutants had indicated that cPGI activity might be important for proper pollen development, and the CaMV 35S promoter does not induce expression throughout the entire microspore and pollen development (Custers et al., 1999; Muñoz-Bertomeu et al., 2010; Krebs et al., 2012). This feature was used to generate plants that expressed amiRNAs targeted against cPGI and did not display any detectable cPGI activity in protein extracts of mature leaves (Fig. 4A; Supplemental Fig. S2B). Hence, these plants allowed us to investigate the impact of strongly decreased cPGI activity in somatic tissue even though homozygous T-DNA mutants were nonviable.
Lack of cPGI Activity in Leaves Results in Starch Excess and Increased Nonphotochemical Quenching in Photosynthesis
Expression of amiRNAs targeted against cPGI led to plants without detectable cPGI activity (Fig. 4A; Supplemental Fig. S2) that displayed growth reduction (Fig. 3) but apart from that appeared healthy. This seems to be a common feature of mutants impaired in primary carbohydrate metabolism, such as pgi1, pgm1, and adg1 (Caspar et al., 1985; Lin et al., 1988; Yu et al., 2000), or sugar transport, such as suc2 and sweet1 sweet2 (Gottwald et al., 2000; Chen et al., 2012), which results from the inability to properly allocate and distribute fixed carbon as energy or carbon skeletons to growing tissues (Ludewig and Flügge, 2013).
Additionally, amiR-cpgi plants overaccumulated starch, most significantly at the end of the night but also throughout the entire day/night cycle (Fig. 5; Supplemental Fig. S3, B and C). Starch excess in mutants with an impaired cytosolic enzyme activity that is not directly involved in starch biosynthesis or degradation may seem unexpected at first sight. However, this has also been described for other processes such as Suc export from source tissue mesophyll cells (Gottwald et al., 2000; Srivastava et al., 2008) or transient integration of maltose-derived Glc into heteroglycans mediated by DPE2 (Chia et al., 2004; Lu and Sharkey, 2004). In the process of nocturnal starch mobilization, cPGI acts downstream of DPE2, hence loss of cPGI activity apparently phenocopies the starch-excess phenotype of dpe2 plants (Supplemental Fig. S1A). DPE2-defective plants are also characterized by a strongly decreased or even absent turnover of glucosyl residues in their cytosolic heteroglycans (Fettke et al., 2006; Malinova et al., 2013). Thus, we analyzed the monomer composition of cytosolic heteroglycans and found that their Glc content was significantly increased in amiR-cpgi plants compared with wild-type plants at the end of the night (Fig. 6B). Accumulation of Glc moieties in cytosolic heteroglycans of amiR-cpgi plants indicates decreased Glc turnover and, as a consequence, leads to decreased export of starch degradation products from chloroplasts. In turn, this argues for a feedback mechanism originating in the cytosol and inhibiting cytosolic metabolism of starch degradation products but also the chloroplast-localized starch degradation itself.
However, impairment of cPGI activity affected not only carbohydrate metabolism in chloroplasts and the cytosol, but also photosynthesis performance. After dark incubation, light induction curve measurements revealed a significant increase in qN concomitant with a decrease in qP in both amiR-cpgi lines (Fig. 4C; Supplemental Fig. S3A). By contrast, comparison between dpe2 mutants and corresponding wild-type plants (ecotype Wassilewskija) did not reveal any increase in qN (Supplemental Fig. S4).
Chloroplasts have to protect themselves from energy overload and accompanied damage. This is achieved by dissipation of excess energy in the form of heat as a way to cope with such unfavorable situations. In the light, the diminished formation of Suc from exported triose phosphates feeds back directly on photophosphorylation in the chloroplasts (i.e. by stromal Pi depletion, which occurs at least temporarily until starch biosynthesis is fully induced) and thereby inhibits photosynthetic electron transport (Fig. 4C; Supplemental Fig. S3A). Hence, our data indicate a role for Suc synthesis as a short-term energy buffer, which underpins the complex, fast, and well-coordinated reactions of two compartments involved in proper photosynthesis.
The decrease in qP, which reflects an increased reduction state of QA (the primary quinone electron acceptor of PSII), points at a limitation of ETR at the site of PSII (Fig. 4C; Supplemental Fig. S3A). In addition, the increase in qN would be consistent with a steeper proton gradient across the thylakoid membrane, suggesting a diminished ATP synthesis. By contrast, in the dpe2 mutant, the induction of photosynthesis is not severely impaired because the DPE2 protein fulfills its major function in the dark, when maltose deriving from starch degradation is further processed after export in the cytosol. In the light, maltose production from starch turnover ceases.
Suc Synthesis Is Impaired But Not Abolished in amiR-cpgi Plants
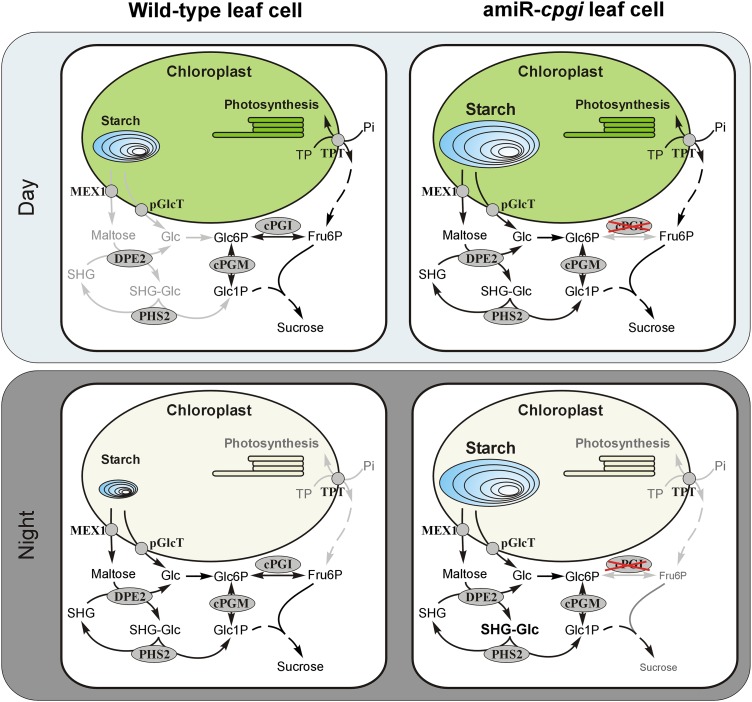
Although cytosolic PGI activity was virtually absent from amiR-cpgi leaf extracts (Supplemental Fig. S2), Suc could still be detected in leaves (Fig. 6A). While at most time points during the day leaf Suc contents in amiR-cpgi plants were comparable to those in wild-type plants, Suc content was significantly reduced at the end of the night in amiR-cpgi plants (Fig. 6A; Supplemental Fig. S3D). These data led to the conclusion that Suc synthesis during the day is apparently operating without cPGI-mediated conversion of Fru6P to Glc6P, which serves as substrate for UDP-glucose synthesis via cytosolic PGM and UDP-glucose pyrophosphorylase. Fru6P is easily generated during the day from chloroplast-exported triose phosphates independent from cPGI activity. Flexibility of plant carbohydrate metabolism could enable amiR-cpgi plants to generate the Suc phosphate synthase substrate UDP-glucose from Glc1P originating from transitory starch degradation during the day (Fig. 9). amiR-cpgi plants may be able to use the night path of photoassimilate export from chloroplasts already during the day. This includes the export of the starch breakdown products Glc (via the chloroplastic Glc transporter pGlcT; Weber et al., 2000) and maltose (via the maltose exporter MEX1; Niittylä et al., 2004) from chloroplasts. Glc can subsequently be converted to Glc1P via hexokinase and cPGM. Alternatively, cytosolic Glc may originate from DPE2-catalyzed maltose turnover (Chia et al., 2004; Lu and Sharkey, 2004) involving SHGs (Fettke et al., 2006) and the cytosolic glucan phosphorylase PHS2 (Fig. 9). Recently, the possibility of direct export of Glc1P from chloroplasts has been discussed (Stitt and Zeeman, 2012) in face of apparent Glc1P import into isolated Arabidopsis protoplasts and chloroplasts (Fettke et al., 2011). This could avoid the necessity for partial starch degradation during the day because Glc1P could be channeled directly from the chloroplast to the cytosol and into Suc biosynthesis instead of plastidic starch synthesis. However, the molecular entity mediating Glc1P transport has not been identified yet.
Figure 9.
Model of cPGI involvement in Suc and starch metabolism in wild-type and amiR-cpgi plants. Light-gray arrows and labels indicate inactive steps and pathways during the respective period.
Starch breakdown during the day as compensatory mechanism has clearly been demonstrated in mutant plants defective in TPT, the chloroplast envelope triose phosphate/phosphate translocator, which mediates the day path of photoassimilate export from chloroplasts (Schneider et al., 2002; Walters et al., 2004). The tpt-1 mutant only showed severe growth retardation with simultaneously defective starch synthesis and less pronounced with starch degradation (Schmitz et al., 2012). In agreement with this, amiR-cpgi mutants in the starch-free genetic background of the pgi1-1 or adg1-1 mutation displayed massive growth retardation, which was rescued to some extent by exogenous Suc supply (Fig. 7).
In contrast to the situation in the light, the lack of cPGI activity during the dark period appears to be less well compensated because Suc was strongly decreased at the end of the night (Fig. 6A). During darkness, the cytosolic pathway, which uses exported triose phosphates from the chloroplast to yield Fru6P throughout the day, is mainly blocked because of the nocturnal inactivation of the key enzyme Fru 1,6-bisphosphatase (Cséke et al., 1982; Stitt, 1990). Fru 1,6-bisphosphtase activity is tightly regulated by Fru 2,6-bisphosphate and strongly repressed in presence of the low triose phosphate and high Fru 2,6-bisphosphate concentrations found in leaf tissue during the night (Stitt et al., 1985). Hence, the growth depression observed in amiR-cpgi plants (Fig. 3) may predominantly be explained by the inability to provide sufficient energy in the form of Suc at night (Fig. 9), as has been demonstrated for starch metabolism mutants such as starch excess1 through metabolic modeling (Rasse and Tocquin, 2006; Streb et al., 2012). This energy shortage of metabolism in amiR-cpgi plants may further be aggravated by the elevated dissipation of incident light energy in the form of heat as shown in increased qN (Fig. 4, B and C). This argument is in line with the more dramatic growth retardation observed in plants grown in longer nights, i.e. in short-day growth conditions (Supplemental Fig. S1, B and C).
In summary, our data provide evidence for an essential role of cPGI that cannot be compensated by gametophyte and early embryo metabolism. However, in mature somatic tissue, cytosolic hexose phosphate metabolism revealed a much higher adaptive ability. Here, cPGI activity-free amiR-cpgi plants performed well, although plant growth was generally reduced, mainly due to the inability to generate Suc during the night. Moreover, the starch excess phenotype of amiR-cpgi plants observed in this study and also previously identified for other cytosolic enzymes such as DPE2 (Chia et al., 2004; Lu and Sharkey, 2004) supports the existence of a yet unidentified feedback mechanism, which originates in the cytosol and affects transitory starch turnover.
MATERIALS AND METHODS
Plant Material and Growth Conditions
For all investigations, Arabidopsis (Arabidopsis thaliana) Col-0 or ecotype Wassilewskija were used as wild-type control and mutant alleles of pgi1-1 (Yu et al., 2000) and adg1-1 (Lin et al., 1988). T-DNA insertion lines cpgi-1 (Salk_064423) and cpgi-2 (Salk_016862) of the SALK collection were ordered from the Nottingham Arabidopsis Stock Center (Alonso et al., 2003). Both plant lines were screened for homozygous individuals carrying a T-DNA insertion using primers cPGI fwd5 GAGTCTGCTAAAGGACGCCAG, cPGI rev 10 GCAAGATTAGTGCTGACAGCAAC, and SALK left border primer LBaI TGGTTCACGTAGTGGGCCATCG. Although cpgi-1 (Salk_064423) was annotated as a SALK homozygous line, no homozygous individuals could be identified.
All plant lines were cultivated on one-half-strength MS medium (Murashige and Skoog, 1962) plus 0.8% (w/v) phyto agar and 2% (w/v) Suc. Seeds were stratified for 2 d at 4°C. Plants were grown in a growth chamber at a 16-h/8-h, 22°C/20°C day/night cycle, a photosynthetic photon flux density of 120 µmol m–2 s–1, and 60% humidity. Eleven-day-old seedlings were transferred to soil. Selection of transformed plants was performed on hygromycin-containing and/or kanamycin-containing (50 µg mL–1 each) plates.
Leaf Area and Fresh Weight Determination
At the time of harvest, pots with individual plants were photographed, and then whole rosettes were harvested and the fresh weight measured. Leaf area was determined from images using ImageJ 1.47m (http://imagej.nih.gov/ij/).
PAM Fluorometry
In vivo chlorophyll a fluorescence assay was performed by using PAM fluorometer of the IMAGING-PAM M-Series (Heinz Walz GmbH). Intact, 3-week-old plants were dark adapted for 30 min prior to fluorescence measurements. To investigate the photosynthetic apparatus, a dark-light induction curve was recorded by using the standard settings of the manufacturer’s software, i.e. actinic light 8 (186 µmol quanta m–2 s–1), with the following slow-induction parameters: delay, 40 s; clock, 20 s; and duration time, 315 s. At the start of every measurement, Fv/Fm was calculated. qP and qN at PSII were calculated according to Schreiber et al. (1986) and ETR according to Genty et al. (1989) as summarized by Maxwell and Johnson (2000).
Cloning and Yellow Fluorescent Protein Localization Studies
PGI and cPGI open reading frames without stop codons were amplified from Arabidopsis Col-0 leaf tissue cDNA adding XbaI/XmaI (PGI fwd XbaI TCTAGAAAATGGCCTCTCTCTCAGGCC, PGI rev XmaI CCCGGGTGCGTACAGGTCATCCACATT) and BamHI/XmaI (cPGI fwd BamHI GGATCCAAAATGGCGTCATCAACCGC, cPGI rev XmaI CCCGGGCATCTGGGGCTCGGAAC) restriction sites to the PGI or cPGI reading frame, respectively. PCR products were subcloned into pJet1.2 vector (Thermo Scientific Life Science), sequenced, and subsequently cloned in frame into either SpeI/XmaI or BamHI/XmaI opened pHygII-UT-c-term-Venus. The pHygII-UT-c-term-Venus consists of the pUBQ10 promoter (Norris et al., 1993; Krebs et al., 2012) with mutated SacI restriction site-Multiple Cloning Site (MCS)-Venus-heat shock protein18.2 terminator (Nagaya et al., 2010) cassette and was kindly provided by Rainer Waadt and cloned via HindIII/EcoRI into hygII-MCS plasmid (kindly provided by Jörg Kudla’s laboratory; Walter et al., 2004).
Constructs were transformed into Agrobacterium tumefaciens GV3101 and infiltrated into Nicotiana benthamiana leaves. After 4 d, subcellular localization of PGI, cPGI, and an empty vector control in leaf cells was analyzed in leaf epidermal cell by spinning-disc confocal microscopy (QLC100 confocal scanning unit from Solamere Technology Group attached to a Nikon Eclipse TE 2000-U bright-field microscope) using an argon laser (500M Select, Laserphysics; excitation wavelength filter at 514 nm and emission filter 500–550 nm). Images were captured by a CCD camera (CoolSnap-HQ, Photometrics) using the Metamorph software (Universal Imaging Corporation).
The two cPGI-specific amiRNAs (amiRNA1 TGTACTGTTAATATGCTCCCG, amiRNA2 TATCTAGAACGTTCCAGACTT) were identified, and appropriate oligos were designed with the online tool at http://wmd3.weigelworld.org (Schwab et al., 2006). Neither of the two amiRNA sequences returned PGI1 as a hit when the BLAST on the National Center for Biotechnology Information Web site (http://blast.ncbi.nlm.nih.gov/Blast.cgi; database: Refseq_rna; organism: Arabidopsis) was used. The closest hits covered a maximum of 61% and 66% of the 21 nucleotides at E values of 4.7 and 1.2, respectively, indicating that the amiRNA sequences are specific for cPGI transcript. The CaMV35S:amiRNA cPGI1 and two constructs were cloned using the TOPO and Gateway system (Life Technologies). The final PCR product including a cPGI-specific amiRNA was cloned into pENTR/D-TOPO vector to give the entry clones. Subsequently, pGWB2 (Nakagawa et al., 2007) was used in LR-reactions (Life Technologies) to obtain destination clones. Constructs were transformed into A. tumefaciens GV3101 and used to transform Col-0 plants with the floral dip method (Clough and Bent, 1998). Positive T1 individuals were selected based on kanamycin and hygromycin resistance conferred by pGWB2. After transfer to soil and another 3 weeks of growth in long-day conditions, cPGI enzyme activity was determined compared to a wild-type control. In total, five amiRNA lines were analyzed in detail that showed almost no detectable cPGI activity in leaf extracts. For amiRNA1, two lines designated amiR-cpgi6 and amiR-cpgi10 and, for amiRNA2, three lines designated amiR2-cpgi8, amiR2-cpgi9, and amiR2-cpgi10 were selected.
Attempts to isolate homozygous amiR-cpgi plants revealed that despite selection on antibiotic medium, the high nonphotochemical quenching phenotype was lost in subsequent generations, indicating that amiRNA expression might be silenced in later generations. Hence, for all analyses, amiR-cpgi plants were selected from a segregating T3 population based on occurrence of the high nonphotochemical quenching phenotype (Fig. 4, B and C).
Plant Protein Extraction and PGI Activity Assay
Total protein was extracted by grinding 100 mg of leaf tissue in liquid nitrogen, resuspending the material in extraction buffer containing 100 mm HEPES-NaOH (pH 7.4), 1 mm EDTA, 5 mm β-mercaptoethanol (Carl Roth GmbH and Co KG), and 50 µg mL–1 phenylmethylsulfonyl fluoride in 100% ethanol (AppliChem GmbH) followed by a 10-min incubation on ice. Total protein of the centrifugation-cleared solution was determined by standard Bradford assay (Roti-Quant, Carl Roth GmbH).
PGI activity was measured in a TECAN Infinite M200 multiplate reader (Tecan Austria GmbH) as described previously (Kunz et al., 2010). Briefly, 190 µL of assay buffer (50 mm HEPES-NaOH, pH 7.4, 1 mm EDTA, 3 mm MgCl2, 3 mm Fru6P, 1 mm NAD+, and 0.4 units mL–1 Fru6P dehydrogenase from Leuconostoc mesenteroides, Roche Applied Science) per sample was added to each well, and absorption was measured at 366 nm. Then, 10 µL of protein extract (10 µg of total protein per well) was added, and absorption at 366 nm was monitored. PGI activity was calculated from the slope of NADH production and normalized to total protein. Inactivation of the plastidic PGI isoform by heating an aliquot of each extract to 50°C for 10 min (Jones et al., 1986b) was used to distinguish between cytosolic and plastidic PGI contribution to total PGI activity.
Carbohydrate Measurements
Starch and Suc leaf content were determined as described previously (Kunz et al., 2010) with minor modifications. Briefly, liquid nitrogen-ground, frozen leaf material (80–100 mg) was used for extraction in 900 µL of ethanol (80% [v/v]; 60 min at 75°C). Suc and starch were determined from supernatant and pellet of the same extract after centrifugation (5 min at 14,000 rpm), respectively. Isolation and measurement of water-soluble heteroglycans were performed as described in Fettke et al. (2004).
For whole-leaf starch staining, chlorophyll of freshly harvested leaf rosettes was first removed in 70% (v/v) ethanol at 60°C for 20 min and subsequently stained in iodine-potassium iodide solution (Lugol’s solution, AppliChem). Water-rinsed leaves were mounted and photographed.
Vitality Staining of Pollen Grains
The viability of the pollen grains was assessed using the staining method according to Alexander (1969). Whole filaments were mounted on glass slides in staining solution and incubated for 10 min. Viable pollen grains were able to absorb the dye and would stain red.
Sequence data of loci investigated in this study can be found in the Arabidopsis Genome Initiative under the following accession numbers: At4g24620 (PGI1), At5g42740 (cPGI), At5g48300 (ADG1), At5g50950 (FUM2), and At2g40840 (DPE2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Plant growth of mutants and the corresponding wild types in long- and short-day conditions.
Supplemental Figure S2. Reduced PGI activity in amiR-cpgi plants.
Supplemental Figure S3. Photosynthetic and metabolic data of three independent lines expressing the second amiRNA targeting cPGI transcript (amiR2-cpgi).
Supplemental Figure S4. qN and qP in amiR-cpgi, dpe2, and wild-type plants.
Supplemental Figure S5. Phenotype of a homozygous cpgi-1 plant expressing cPGI driven by the CaMV 35 promoter.
Supplemental Figure S6. USP promoter activity monitored histochemically as GUS activity and analyzed microscopically in flowers and developing seeds of transgenic pUSP:uidA plants.
Supplemental Figure S7. Diagram illustrating the calculation of the expected genotype distribution in the self-progeny of heterozygous cPGI/cpgi plants.
Supplemental Table S1. Observed and expected distribution of genotypes from the progeny of heterozygous cPGI T-DNA mutants.
Supplemental Table S2. Germination rate of segregating seeds collected from heterozygous cPGI/cpgi plants and the Col-0 wild type.
Supplementary Material
Acknowledgments
We thank Dr. Frank Ludewig (University of Cologne) for critical reading and comments on the manuscript, Dr. Rainer Waadt (University of California, San Diego) for providing the UBQ10-MCS-Venus-heat shock protein18.2 cassette, Dr. Joerg Kudla (University of Muenster) for hygII-MCS plasmid, Drs. Thomas Sharkey (University of Wisconsin, Madison) and Junshi Yazaki (The Salk Institute) for providing dpe2 lines and SALK_149466, respectively, and Sonja Lott (University of Cologne) and Sophie Wolf (Free University of Berlin) for excellent technical assistance.
Glossary
- Fru6P
Fru 6-phosphate
- Glc6P
Glc 6-phosphate
- Glc1P
Glc 1-phosphate
- T-DNA
transfer DNA
- Col-0
ecotype Columbia
- amiRNA
artificial microRNA
- CaMV
Cauliflower mosaic virus
- PAM
pulse amplitude modulation
- Fv/Fm
maximum photochemical efficiency of PSII in the dark-adapted state
- qN
nonphotochemical quenching coefficient
- qP
photochemical quenching coefficient
- ETR
electron transport rate
- MS
Murashige and Skoog
- cDNA
complementary DNA
- SHG
soluble heteroglycan
Footnotes
This work was supported by the International Max Planck Research School Cologne (to S.Z.-N.), the Deutsche Forschungsgemeinschaft (Schwerpunktprogramm 1108 and FL 126/23–1 to U.-I.F. and R.E.H.), the Human Frontiers Science Program, the Alexander von Humboldt Foundation, the Deutscher Akademischer Austauschdienst Rise program (to H.-H.K.), and the National Science Foundation (MCB0918220 and MCB1414339). The fusion protein localization was funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy (grant no. DE–FG02–03ER15449 to J.I.S.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bäumlein H, Boerjan W, Nagy I, Bassüner R, Van Montagu M, Inzé D, Wobus U. (1991) A novel seed protein gene from Vicia faba is developmentally regulated in transgenic tobacco and Arabidopsis plants. Mol Gen Genet 225: 459–467 [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C. (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB. (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 [DOI] [PubMed] [Google Scholar]
- Chia T, Thorneycroft D, Chapple A, Messerli G, Chen J, Zeeman SC, Smith SM, Smith AM. (2004) A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J 37: 853–863 [DOI] [PubMed] [Google Scholar]
- Cho MH, Lim H, Shin DH, Jeon JS, Bhoo SH, Park YI, Hahn TR. (2011) Role of the plastidic glucose translocator in the export of starch degradation products from the chloroplasts in Arabidopsis thaliana. New Phytol 190: 101–112 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cséke C, Weeden NF, Buchanan BB, Uyeda K. (1982) A special fructose bisphosphate functions as a cytoplasmic regulatory metabolite in green leaves. Proc Natl Acad Sci USA 79: 4322–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custers JBM, Snepvangers SCHJ, Jansen HJ, Zhang L, Lookeren Campagne MM. (1999) The 35S-CaMV promoter is silent during early embryogenesis but activated during nonembryogenic sporophytic development in microspore culture. Protoplasma 208: 257–264 [Google Scholar]
- Egli B, Kölling K, Köhler C, Zeeman SC, Streb S. (2010) Loss of cytosolic phosphoglucomutase compromises gametophyte development in Arabidopsis. Plant Physiol 154: 1659–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettke J, Chia T, Eckermann N, Smith A, Steup M. (2006) A transglucosidase necessary for starch degradation and maltose metabolism in leaves at night acts on cytosolic heteroglycans (SHG). Plant J 46: 668–684 [DOI] [PubMed] [Google Scholar]
- Fettke J, Eckermann N, Poeste S, Pauly M, Steup M. (2004) The glycan substrate of the cytosolic (Pho 2) phosphorylase isozyme from Pisum sativum L.: identification, linkage analysis and subcellular localization. Plant J 39: 933–946 [DOI] [PubMed] [Google Scholar]
- Fettke J, Eckermann N, Tiessen A, Geigenberger P, Steup M. (2005) Identification, subcellular localization and biochemical characterization of water-soluble heteroglycans (SHG) in leaves of Arabidopsis thaliana L.: distinct SHG reside in the cytosol and in the apoplast. Plant J 43: 568–585 [DOI] [PubMed] [Google Scholar]
- Fettke J, Malinova I, Albrecht T, Hejazi M, Steup M. (2011) Glucose-1-phosphate transport into protoplasts and chloroplasts from leaves of Arabidopsis. Plant Physiol 155: 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Dietrich D, Fait A, Fernie AR, Holdsworth MJ, Baker A, Theodoulou FL. (2007) The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol 144: 1467–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR. (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990: 87–92 [Google Scholar]
- Gibon Y, Pyl ET, Sulpice R, Lunn JE, Höhne M, Günther M, Stitt M. (2009) Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ 32: 859–874 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97: 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison-Lowe NJ, Olsen LJ. (2008) Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 4: 339–348 [DOI] [PubMed] [Google Scholar]
- Howden R, Park SK, Moore JM, Orme J, Grossniklaus U, Twell D. (1998) Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Batth TS, Petzold CJ, Redding-Johanson AM, Mukhopadhyay A, Verboom R, Meyer EH, Millar AH, Heazlewood JL. (2011) Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. J Proteome Res 10: 1571–1582 [DOI] [PubMed] [Google Scholar]
- Izumi M, Hidema J, Makino A, Ishida H. (2013) Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol 161: 1682–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TWA, Gottlieb LD, Pichersky E. (1986b) Reduced enzyme activity and starch level in an induced mutant of chloroplast phosphoglucose isomerase. Plant Physiol 81: 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TWA, Pichersky E, Gottlieb LD. (1986a) Enzyme activity in ems-induced null mutations of duplicated genes encoding phosphoglucose isomerases in Clarkia. Genetics 113: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A, Yamane K, Miyashita NT. (2000) DNA polymorphism at the cytosolic phosphoglucose isomerase (PgiC) locus of the wild plant Arabidopsis thaliana. Genetics 156: 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler H, Häusler RE, Schulz B, Gröner F, Flügge UI, Weber A. (2000) Molecular characterisation of a new mutant allele of the plastid phosphoglucomutase in Arabidopsis, and complementation of the mutant with the wild-type cDNA. Mol Gen Genet 263: 978–986 [DOI] [PubMed] [Google Scholar]
- Krebs M, Held K, Binder A, Hashimoto K, Den Herder G, Parniske M, Kudla J, Schumacher K. (2012) FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J 69: 181–192 [DOI] [PubMed] [Google Scholar]
- Kruckeberg AL, Neuhaus HE, Feil R, Gottlieb LD, Stitt M. (1989) Decreased-activity mutants of phosphoglucose isomerase in the cytosol and chloroplast of Clarkia xantiana. Impact on mass-action ratios and fluxes to sucrose and starch, and estimation of Flux Control Coefficients and Elasticity Coefficients. Biochem J 261: 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz HH, Häusler RE, Fettke J, Herbst K, Niewiadomski P, Gierth M, Bell K, Steup M, Flügge UI, Schneider A. (2010) The role of plastidial glucose-6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol (Stuttg) 12: 115–128 [DOI] [PubMed] [Google Scholar]
- Lin TP, Caspar T, Somerville C, Preiss J. (1988) Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADPglucose pyrophosphorylase activity. Plant Physiol 86: 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Sharkey TD. (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218: 466–473 [DOI] [PubMed] [Google Scholar]
- Lu Y, Steichen JM, Yao J, Sharkey TD. (2006) The role of cytosolic α-glucan phosphorylase in maltose metabolism and the comparison of amylomaltase in Arabidopsis and Escherichia coli. Plant Physiol 142: 878–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig F, Flügge UI. (2013) Role of metabolite transporters in source-sink carbon allocation. Front Plant Sci 4: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinova I, Steup M, Fettke J. (2013) Carbon transitions from either Calvin cycle or transitory starch to heteroglycans as revealed by 14C-labeling experiments using protoplasts from Arabidopsis. Physiol Plant 149: 25–44 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Irles-Segura A, Mateu I, Nunes-Nesi A, Fernie AR, Segura J, Ros R. (2010) The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol 152: 1830–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nagaya S, Kawamura K, Shinmyo A, Kato K. (2010) The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol 51: 328–332 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Neuhaus HE, Kruckeberg AL, Feil R, Stitt M. (1989) Reduced-activity mutants of phosphoglucose isomerase in the cytosol and chloroplast of Clarkia xantiana. II. Study of the mechanisms which regulate photosynthate partitioning. Planta 178: 110–122 [DOI] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge UI, Schneider A. (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17: 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC. (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89 [DOI] [PubMed] [Google Scholar]
- Norris SR, Meyer SE, Callis J. (1993) The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol 21: 895–906 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Zhou W, Keech O, Francisco PB, Udomchalothorn T, Tschoep H, Stitt M, Gibon Y, Smith SM. (2010) Arabidopsis has a cytosolic fumarase required for the massive allocation of photosynthate into fumaric acid and for rapid plant growth on high nitrogen. Plant J 62: 785–795 [DOI] [PubMed] [Google Scholar]
- Rasse DP, Tocquin P. (2006) Leaf carbohydrate controls over Arabidopsis growth and response to elevated CO2: an experimentally based model. New Phytol 172: 500–513 [DOI] [PubMed] [Google Scholar]
- Schmitz J, Schöttler MA, Krueger S, Geimer S, Schneider A, Kleine T, Leister D, Bell K, Flügge UI, Häusler RE. (2012) Defects in leaf carbohydrate metabolism compromise acclimation to high light and lead to a high chlorophyll fluorescence phenotype in Arabidopsis thaliana. BMC Plant Biol 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Häusler RE, Kolukisaoglu U, Kunze R, van der Graaff E, Schwacke R, Catoni E, Desimone M, Flügge UI. (2002) An Arabidopsis thaliana knock-out mutant of the chloroplast triose phosphate/phosphate translocator is severely compromised only when starch synthesis, but not starch mobilisation is abolished. Plant J 32: 685–699 [DOI] [PubMed] [Google Scholar]
- Schreiber U, Schliwa U, Bilger W. (1986) Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth Res 10: 51–62 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Warthmann N, Weigel D. (2010) Directed gene silencing with artificial microRNAs. Methods Mol Biol 592: 71–88 [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R. (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG. (2008) Functional characterization of the Arabidopsis AtSUC2 Sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol 148: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M. (1990) Fructose-2,6-bisphosphate as a regulatory molecule in plants. Annu Rev Plant Physiol Plant Mol Biol 41: 153–185 [Google Scholar]
- Stitt M, Wirtz W, Gerhardt R, Heldt HW, Spencer C, Walker D, Foyer C. (1985) A comparative study of metabolite levels in plant leaf material in the dark. Planta 166: 354–364 [DOI] [PubMed] [Google Scholar]
- Stitt M, Zeeman SC. (2012) Starch turnover: pathways, regulation and role in growth. Curr Opin Plant Biol 15: 282–292 [DOI] [PubMed] [Google Scholar]
- Streb S, Eicke S, Zeeman SC. (2012) The simultaneous abolition of three starch hydrolases blocks transient starch breakdown in Arabidopsis. J Biol Chem 287: 41745–41756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M. (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Walters RG, Ibrahim DG, Horton P, Kruger NJ. (2004) A mutant of Arabidopsis lacking the triose-phosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiol 135: 891–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Singer SD, Liu Z. (2012) Silencing of meiosis-critical genes for engineering male sterility in plants. Plant Cell Rep 31: 747–756 [DOI] [PubMed] [Google Scholar]
- Weber A, Servaites JC, Geiger DR, Kofler H, Hille D, Gröner F, Hebbeker U, Flügge UI. (2000) Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12: 787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise SE, Weber AP, Sharkey TD. (2004) Maltose is the major form of carbon exported from the chloroplast at night. Planta 218: 474–482 [DOI] [PubMed] [Google Scholar]
- Wilkinson JE, Twell D, Lindsey K. (1997) Activities of CaMV 35S and nos promoters in pollen: implications for field release of transgenic plants. J Exp Bot 48: 265–275 [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Wang X, Hong Z. (2010) Precocious pollen germination in Arabidopsis plants with altered callose deposition during microsporogenesis. Planta 231: 809–823 [DOI] [PubMed] [Google Scholar]
- Yu TS, Lue WL, Wang SM, Chen J. (2000) Mutation of Arabidopsis plastid phosphoglucose isomerase affects leaf starch synthesis and floral initiation. Plant Physiol 123: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharov A, Giersberg M, Hosein F, Melzer M, Müntz K, Saalbach I. (2004) Seed-specific promoters direct gene expression in non-seed tissue. J Exp Bot 55: 1463–1471 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. (2007) The diurnal metabolism of leaf starch. Biochem J 401: 13–28 [DOI] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB. (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.