The discrete localization of laccases to secondary cell walls directs lignification in protoxylem.
Abstract
Plants precisely control lignin deposition in spiral or annular secondary cell wall domains during protoxylem tracheary element (TE) development. Because protoxylem TEs function to transport water within rapidly elongating tissues, it is important that lignin deposition is restricted to the secondary cell walls in order to preserve the plasticity of adjacent primary wall domains. The Arabidopsis (Arabidopsis thaliana) inducible VASCULAR NAC DOMAIN7 (VND7) protoxylem TE differentiation system permits the use of mutant backgrounds, fluorescent protein tagging, and high-resolution live-cell imaging of xylem cells during secondary cell wall development. Enzymes synthesizing monolignols, as well as putative monolignol transporters, showed a uniform distribution during protoxylem TE differentiation. By contrast, the oxidative enzymes LACCASE4 (LAC4) and LAC17 were spatially localized to secondary cell walls throughout protoxylem TE differentiation. These data support the hypothesis that precise delivery of oxidative enzymes determines the pattern of cell wall lignification. This view was supported by lac4lac17 mutant analysis demonstrating that laccases are necessary for protoxylem TE lignification. Overexpression studies showed that laccases are sufficient to catalyze ectopic lignin polymerization in primary cell walls when exogenous monolignols are supplied. Our data support a model of protoxylem TE lignification in which monolignols are highly mobile once exported to the cell wall, and in which precise targeting of laccases to secondary cell wall domains directs lignin deposition.
During all stages of a land plant’s life cycle, lignified secondary cell walls provide critical mechanical properties for water transport and the upright growth habit. Water conduction in xylem tissue occurs in tracheary elements (TEs), whose secondary cell walls are composed of cellulose, hemicelluloses, and the phenolic lignin polymer. Lignification of the secondary cell wall imparts strength, rigidity, and water impermeability to the polysaccharide components. The deposition of lignin in secondary cell walls is developmentally regulated, and different cell types generate distinctive secondary cell wall patterns. Protoxylem TEs, for example, form annular or helical secondary wall thickenings, whereas metaxylem TEs deposit secondary cell walls in a reticulated or pitted pattern (Esau, 1965). Protoxylem TEs form in young elongating plant tissues and therefore the restriction of lignin deposition to the annular or helical secondary cell wall thickenings, as opposed to the intervening primary cell walls, is crucial to allow continued axial elongation. The mechanisms restricting lignin deposition specifically to secondary cell wall thickenings have not been identified.
Monolignol (lignin monomer) biosynthesis occurs in the cytosol in close proximity to the endoplasmic reticulum (ER), because the pathway includes both cytosolic- and ER-localized enzymes (Bonawitz and Chapple, 2010). Multienzyme complexes, potentially anchored at specialized subdomains on the ER surface, have been postulated to channel phenolic metabolite production during lignification (Chen et al., 2011; Bassard et al., 2012). Both ATP-binding cassette (ABC) transporters of monolignols and proton-dependent transporters of monolignol glucosides have been proposed as monolignol export mechanisms (Ehlting et al., 2005; Miao and Liu, 2010; Liu, C.J., 2012; Tsuyama et al., 2013). However, genetic evidence does not support a role for monolignol glucosides as direct precursors of lignin in Arabidopsis (Arabidopsis thaliana; Chapelle et al., 2012). Monolignol transport activity has been reported for heterologously expressed AtABCG29, which was capable of transporting p-coumaryl alcohol in yeast (Saccharomyces cerevisiae) cells and isolated plasma membrane vesicles (Alejandro et al., 2012). Localized monolignol biosynthesis and/or export are hypothesized to be mechanisms for concentrating monolignols in secondary, but not primary, cell wall domains in protoxylem TEs.
An alternative mechanism for controlling the pattern of lignin deposition could be the site-specific localization of the enzymes required for monolignol polymerization within the cell wall. Monolignol polymerization occurs by oxidative combinatorial coupling of monolignols, catalyzed by cell wall-localized laccases and/or peroxidases (Vanholme et al., 2012). A peroxidase, originally isolated from xylogenic Zinnia elegans cell cultures, has been localized to secondary cell walls, which demonstrates the spatial association of oxidative enzymes with lignin deposition (Sato et al., 2006), but the function of this peroxidase in vivo has not been demonstrated. Recent analysis of the Arabidopsis LACCASE4 (LAC4), LAC11, and LAC17 genes showed that loss of function of these genes had a dramatic effect on lignification of metaxylem and fiber cells in inflorescence stems (Berthet et al., 2011; Zhao et al., 2013). However, the roles of either laccases or peroxidases in controlling the spatial pattern of cell wall lignification in protoxylem TEs have not been investigated.
Studying protoxylem TE development is challenging because they are located deep within root or shoot tissues, but the identification of key transcriptional regulators that activate differentiation of protoxylem TEs has led to the development of a genetically tractable experimental system. In this system, the activity of VASCULAR NAC DOMAIN 7 (VND7), a NAC domain transcription factor that regulates protoxylem TE cell fate (Kubo et al., 2005), is paired with an inducible transcriptional activator, viral protein16 (VP16) coupled to a glucocorticoid receptor (GR; Yamaguchi et al., 2010). Upon induction with dexamethasone (DEX), nearly all cells in Arabidopsis seedlings transdifferentiate into protoxylem TE-like cells (Yamaguchi et al., 2010). This system allows protoxylem development to be studied in diverse mutant backgrounds, and permits the expression and high-resolution imaging of proteins tagged with GFP during xylogenesis.
This study examines the cellular mechanisms underlying the discrete lignification of spiral or annular secondary cell walls in protoxylem TEs. With GFP tagging and live-cell monitoring in the VND7-VP16-GR lines, we tested whether monolignol biosynthetic enzymes, putative monolignol transporters, or laccases were specifically localized in lignifying cell wall domains. Fluorescently tagged monolignols were used to assay whether monolignols would polymerize in secondary cell wall domains in laccase loss-of-function mutants, as well as in primary cell walls of overexpression lines. Whereas earlier studies identified which members of the laccase gene family were active in lignification of metaxylem vessel and fiber cell types in the Arabidopsis stem (Berthet et al., 2011; Zhao et al., 2013), this work directly addresses the role of these oxidative enzymes in directing lignin polymerization in lignified secondary cell wall domains adjacent to unlignified primary cell walls of the protoxylem TE.
RESULTS
Live-Cell Imaging of Lignification of Induced Protoxylem TEs
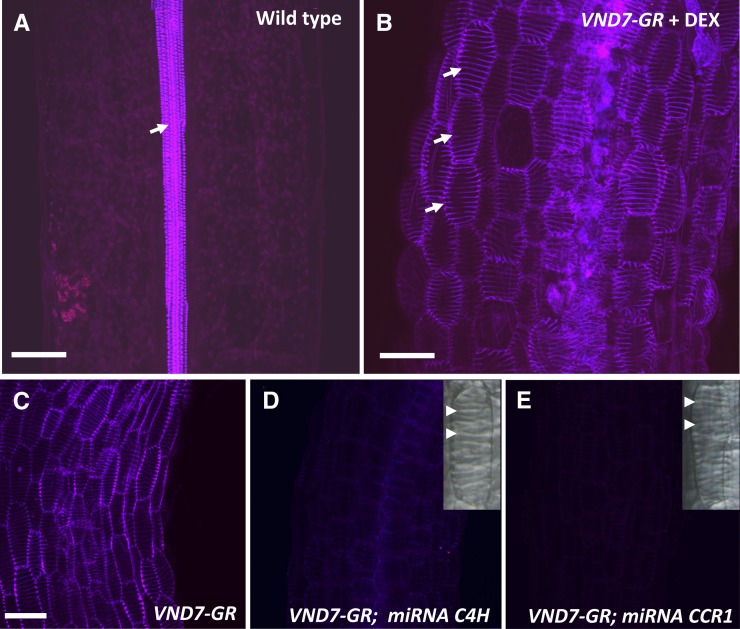
Although the genes encoding lignin biosynthetic enzymes are up-regulated in plant lines expressing the master transcription factor VND7, the presence of lignin in the induced protoxylem TEs has not been established. To examine lignification in induced protoxylem TEs, cell wall phenolic autofluorescence was profiled using two-photon excitation microscopy, with UV excitation (350–370 nm) and emission (420–540 nm; Fig. 1). In hypocotyls from uninduced control seedlings, the only intrinsic fluorescence from lignified secondary cell walls came from endogenous protoxylem TEs in the central vascular cylinder (Fig. 1A). However, within 48 h of VND7 induction, nearly 100% of hypocotyl epidermal cells of VND7-VP16-GR seedlings transdifferentiated into protoxylem TEs with highly fluorescent secondary cell walls (Fig. 1, B and C).
Figure 1.
Lignin autofluorescence in secondary cell walls of endogenous and VND7-induced Arabidopsis seedling TEs. A, In a control, uninduced, 10-d-old Arabidopsis hypocotyl, only protoxylem TEs (arrow) emit autofluorescence when excited by UV light. B, In an Arabidopsis hypocotyl carrying VND7-VP16-GR, after induction with DEX, epidermal cells transdifferentiate into protoxylem TEs (arrows) and emit autofluorescence. C to E, UV autofluorescence of protoxylem TEs imaged under identical conditions in VND7-VP16-GR alone or in plants expressing both VND7-VP16-GR and artificial microRNAs targeting monolignol biosynthetic genes C4H (D) or CCR1 (E). Protoxylem TE differentiation and secondary cell wall (arrowheads) formation was not inhibited by constitutive expression of artificial microRNAs (insets in D and E). Maximum projection images of z-stacks are shown. Bars = 50 μm.
In order to verify that the UV autofluorescence observed in VND7-induced TEs is dependent on monolignol biosynthesis, the expression of two key monolignol biosynthetic genes, CINNAMATE-4-HYDROXYLASE (C4H) or CINNAMOYL-CoA REDUCTASE1 (CCR1), was suppressed through the use of targeted artificial microRNAs (Schwab et al., 2006). Plants constitutively overexpressing the C4H or CCR1 microRNA recapitulated the range of phenotypes previously described in c4h and ccr1 mutants (Jones et al., 2001; Ruegger and Chapple, 2001; Schilmiller et al., 2009; Thévenin et al., 2011), including irregular xylem and a marked reduction in lignin deposition (Pro-35S:miRNA C4H lines, Supplemental Fig. S1; and Pro-35S:miRNA CCR lines, Smith et al., 2013). Constitutive expression of the C4H or CCR1 microRNA during VND7-induced TE formation did not inhibit secondary cell wall formation (Fig. 1, D and E, insets) but two-photon microscopy revealed severely reduced UV autofluorescence in these cell wall domains (Fig. 1, D and E), compared with the wild type (Fig. 1C). Together, these results indicate that the autofluorescence in secondary cell walls of VND7-induced protoxylem TEs is from lignin, because it is dependent not only on the function of the general phenylpropanoid pathway enzyme C4H but also on the function of the monolignol-specific pathway enzyme CCR1. This establishes the VND7-inducible protoxylem system as a valid model for analysis of cell wall lignification.
Dynamic Rearrangement of Monolignol Biosynthetic Enzymes during Protoxylem TE Differentiation
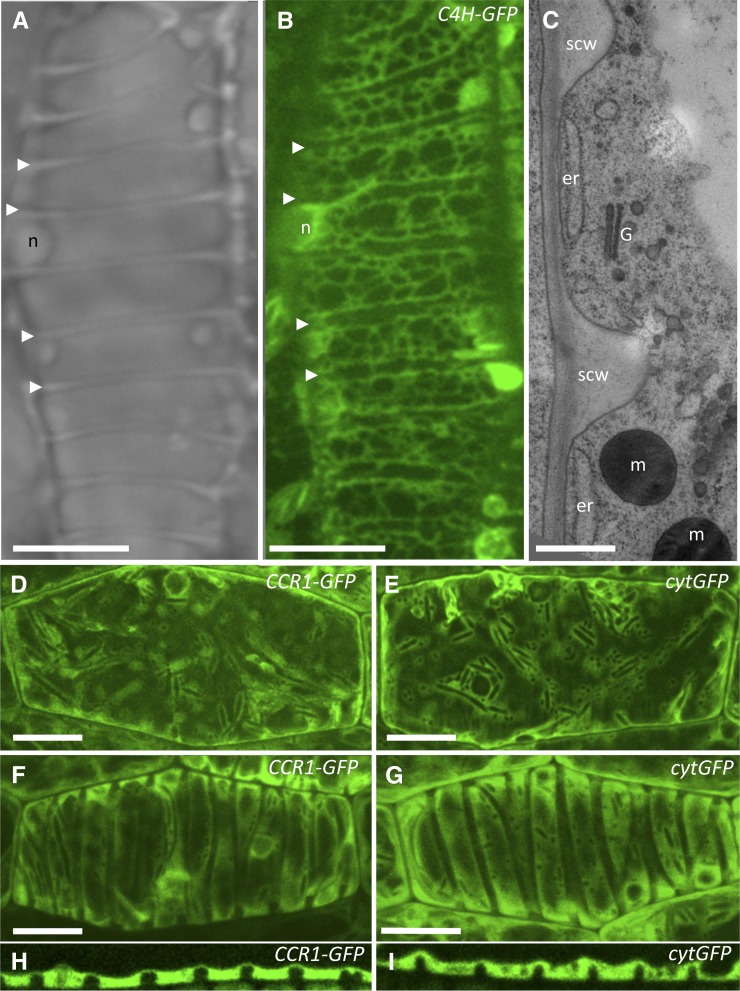
One hypothesis to explain specific deposition of lignin only in spiral cell wall thickenings of protoxylem TEs is that monolignol biosynthetic enzymes in these cells may be preferentially organized around sites of secondary cell wall deposition. To directly image the subcellular distribution of C4H during protoxylem TE differentiation, a C4H-GFP translational fusion was stably transformed into VND7-VP16-GR plants. Similar C4H-GFP fusions were shown to be functional in plants and localized to the ER when transiently expressed in tobacco (Nicotiana tabacum) leaf cells (Ro et al., 2001; Bassard et al., 2012). In epidermal cells of uninduced C4H-GFP-transformed VND7-VP16-GR plants, C4H-GFP was clearly localized to the reticulate network of tubular ER, as confirmed by colocalization with an ER-localized red fluorescent protein (RFP)-HDEL marker (Supplemental Fig. S2). Induction of VND7-mediated protoxylem TE differentiation had a profound impact on the pattern of C4H-GFP distribution. Live-cell imaging was used to visualize the C4H-GFP-labeled ER pattern, as it changed from an open tubular ER network early in development (Supplemental Fig. S2, A–C) to a denser lamellar pattern concentrated between developing secondary cell wall domains (Fig. 2B; Supplemental Fig. S2, D–G). Continuous ER strands were parallel to, and generally excluded from, domains directly below developing secondary cell wall thickenings, which were observed in bright-field microscopy (Fig. 2A). Evidence of ER exclusion below secondary cell wall domains was also observed in endogenous root protoxylem TEs using transmission electron microscopy (TEM) of high-pressure frozen/freeze-substituted samples (Fig. 2C). The changes of C4H-GFP distribution during VND7-induced TE differentiation are therefore associated with overall changes in ER distribution and morphology, but do not appear to be attributable to relocalization of C4H-GFP to specific regions of the ER.
Figure 2.
C4H-GFP and CCR1-GFP are localized between secondary cell wall domains during VND7-induced protoxylem TE differentiation. A, Bright-field image of a differentiating protoxylem TE induced in VND7-VP16-GR plants, showing secondary cell wall thickenings (arrowheads). B, Maximum projection image of optical sections through the same cell shown in A, in which C4H-GFP localization is observed in the ER network that is excluded below developing secondary cell walls (arrowheads) and is adjacent to primary cell wall domains. C, Transmission electron micrograph showing laminar ER, Golgi, and mitochondria in relation to secondary cell walls of root protoxylem TEs. D and E, Maximum projection images of optical sections showing cytoplasmic localization of CCR1-GFP (D) and cytGFP (E) in control hypocotyl epidermal cells of VND7-VP16-GR seedlings without DEX induction. F to I, After VND7-VP16-GR protoxylem TE induction for 24 h, both CCR1-GFP (F) and cytGFP (G) were predominately localized in the cytoplasm between the secondary cell wall thickenings and as shown in a single median optical slice in CCR1-GFP (H) and cytGFP (I). er, Endoplasmic reticulum; G, Golgi; m, mitochondria; n, nucleus; scw, secondary cell wall. Bars = 12 μm in A, B, and D to G; 500 nm in C.
Because CCR1 is specific to monolignol biosynthesis, its subcellular distribution was tracked during VND7-mediated TE lignification. First, the function of the CCR1-GFP fusion protein was tested by complementing ccr1irx4 loss-of-function mutants, in which CCR1-GFP rescued the irregular xylem phenotype of ccr1irx4 mutants (Supplemental Fig. S3). CCR1 is predicted to be a cytosolic enzyme and CCR1-GFP localization exactly matched cytosolic-localized (cyt) GFP (Fig. 2, D and E). During all stages of VND7-mediated TE differentiation, both CCR1-GFP and cytGFP signals were observed throughout the cytoplasm, with no bias toward secondary cell wall domains (Fig. 2, F–I). Thus, spiral secondary cell wall lignification is not correlated with the spatial distribution of the C4H and CCR1 monolignol biosynthetic enzymes.
ABC Transporters Localize to All Plasma Membrane Domains during Protoxylem TE Differentiation
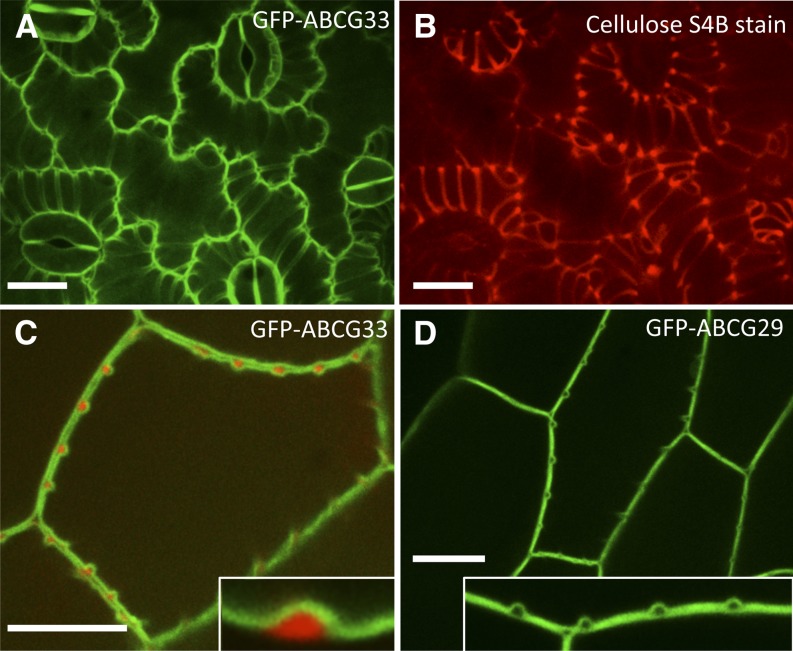
ABCB11, ABCG29, and ABCG33 transporters were previously identified as candidate monolignol exporters (Ehlting et al., 2005; Kaneda et al., 2011; Alejandro et al., 2012). To test the hypothesis that these putative monolignol transporters could have a restricted plasma membrane localization adjacent to secondary cell wall domains, GFP translational fusions of ABCB11, ABCG29, and ABCG33 were transformed into VND7-VP16-GR plants. When VND7-mediated protoxylem TE differentiation was induced, all fluorescently tagged ABC transporters were localized to plasma membranes (Fig. 3; Supplemental Fig. S4). Yellow Fluorescent Protein (YFP)-tagged ABCG11, which was previously shown to function as a plasma membrane-localized cuticular wax and cutin exporter (Bird et al., 2007), was included as a positive control for plasma membrane localization (Supplemental Fig. S4). Secondary cell wall domains were counterstained with the cellulose-specific Pontamine Fast Scarlet 4B (S4B) dye (Fig. 3, B and C; Anderson et al., 2010). When the GFP channel signal was merged with that of the cellulose counterstain, it was clear that transporters were equally distributed along both secondary and adjacent primary wall regions (Fig. 3, A and C). This uniform plasma membrane distribution of candidate monolignol ABC transporters persisted until the initiation of programmed cell death, when fragmentation of the GFP-labeled plasma membranes was observed (Supplemental Fig. S5; Supplemental Movie S1). These results demonstrate that the candidate monolignol ABC transporters, including ABCG29 (Fig. 3D), are not preferentially excluded from, nor localized to, the plasma membrane domains facing developing secondary cell walls.
Figure 3.
ABC transporters are evenly localized in plasma membranes during protoxylem TE differentiation in VND7-VP16-GR seedlings. A, Maximum projection image of cotyledon epidermal cells undergoing protoxylem TE differentiation, expressing GFP-ABCG33. B, Cellulose stain, Pontamine S4B, counterstained image of cells shown in A, highlighting secondary cell wall domains. C, Median optical slice of induced protoxylem TE, showing invaginations of GFP-ABCG33 (green), counterstained with Pontamine S4B (red) demonstrating secondary cell wall domains, with inset image of the optical cross section of one secondary wall thickening. D, Induced protoxylem TE from plants expressing GFP-ABCG29, with inset showing plasma membrane localization over primary and secondary wall domains. Bars = 15 μm.
Laccase-Dependent Monolignol Polymerization in Secondary Cell Wall Domains
The results of the preceding experiments suggest that monolignols are likely produced and exported into the cell wall in a diffuse, rather than targeted, manner. If this model is correct, then exogenously supplied monolignols are predicted to be highly mobile throughout the cell wall matrix, and should only become polymerized within the secondary cell wall domains. The tools to assess this prediction were recently developed, in the form of fluorescently tagged monolignol analogs, such as γ-nitrobenzofuran (NBD)-tagged coniferyl alcohol (CA), that become polymerized into lignin via oxidase-mediated radical coupling (Tobimatsu et al., 2011). When applied in planta, these NBD-tagged probes were incorporated into the lignifying tissues in Arabidopsis and Pinus radiata plants (Tobimatsu et al., 2013).
The availability of NBD-tagged monolignols together with the VND7-induced protoxylem system provided the opportunity to document the lignification of discrete cell wall domains in individual protoxylem TEs. NBD-CA and untagged CA were applied together to induced VND7-VP16-GR seedlings and labeling of the differentiating TEs was monitored. To ensure that free monolignols were not simply being trapped in the polymer matrix after treatment with fluorescent monolignols, and that only the insoluble lignin polymer was being imaged, treated seedlings were thoroughly washed in 49% methanol:1% acetic acid at 45°C for up to 72 h to remove excess NBD-CA, prior to imaging.
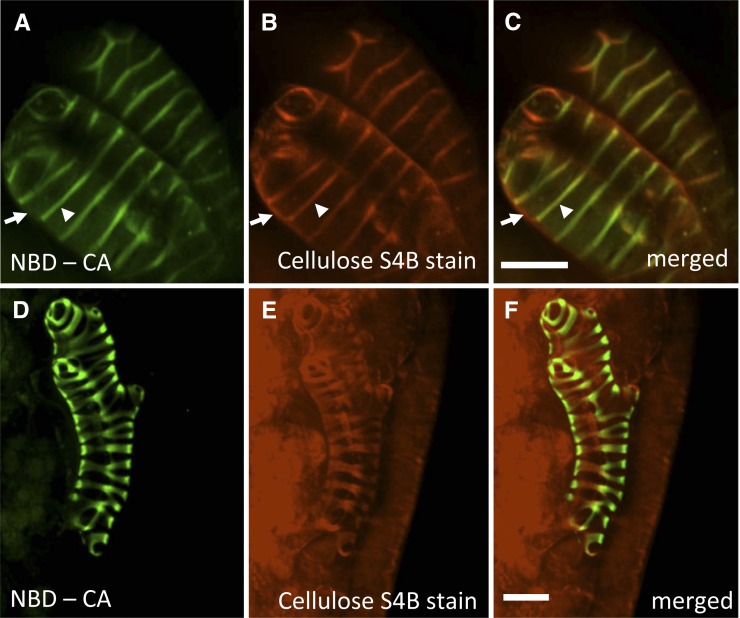
Secondary cell wall domains, as identified by S4B staining for cellulose, were strongly labeled by NBD-CA treatment, whereas adjacent primary cell walls were not (Fig. 4, A–C, arrowheads and arrows, respectively). Similarly, in leaf tissue in which only a single leaf mesophyll cell had transdifferentiated into a protoxylem TE, incorporation of NBD-CA was restricted to the secondary cell walls of the transdifferentiated cell (Fig. 4, D–F). Collectively, these results demonstrate that monolignols can diffuse freely throughout the apoplast and become polymerized into lignin only in the secondary cell walls of differentiating protoxylem TEs.
Figure 4.
Mobility of monolignols in the apoplast: Fluorescently tagged monolignols are specifically incorporated into the secondary cell walls of VND7-induced protoxylem TEs. A, Green fluorescence of incorporated NBD-CA specifically in secondary cell walls (arrowhead) and not primary cell walls (arrows) of epidermal cells induced to transdifferentiate into protoxylem TEs. B, Cellulose-specific Pontamine S4B counterstained image of cells shown in A, demonstrating secondary cell walls (arrowhead) and primary cell walls (arrow). C, Merged image of A and B showing colocalization of signal in secondary cell walls. D, In an area in which only one leaf mesophyll cell differentiated into a protoxylem TE, exogenously added NBD-CA was specifically incorporated in secondary wall thickenings and not in the surrounding neighbors. E, Pontamine S4B counterstained image of protoxylem TE shown in D, demonstrating background stain of primary cell wall domains of surrounding leaf cells, and strong secondary cell wall domains in lone protoxylem TE. F, Merged image of D and E. Bars = 15 μm.
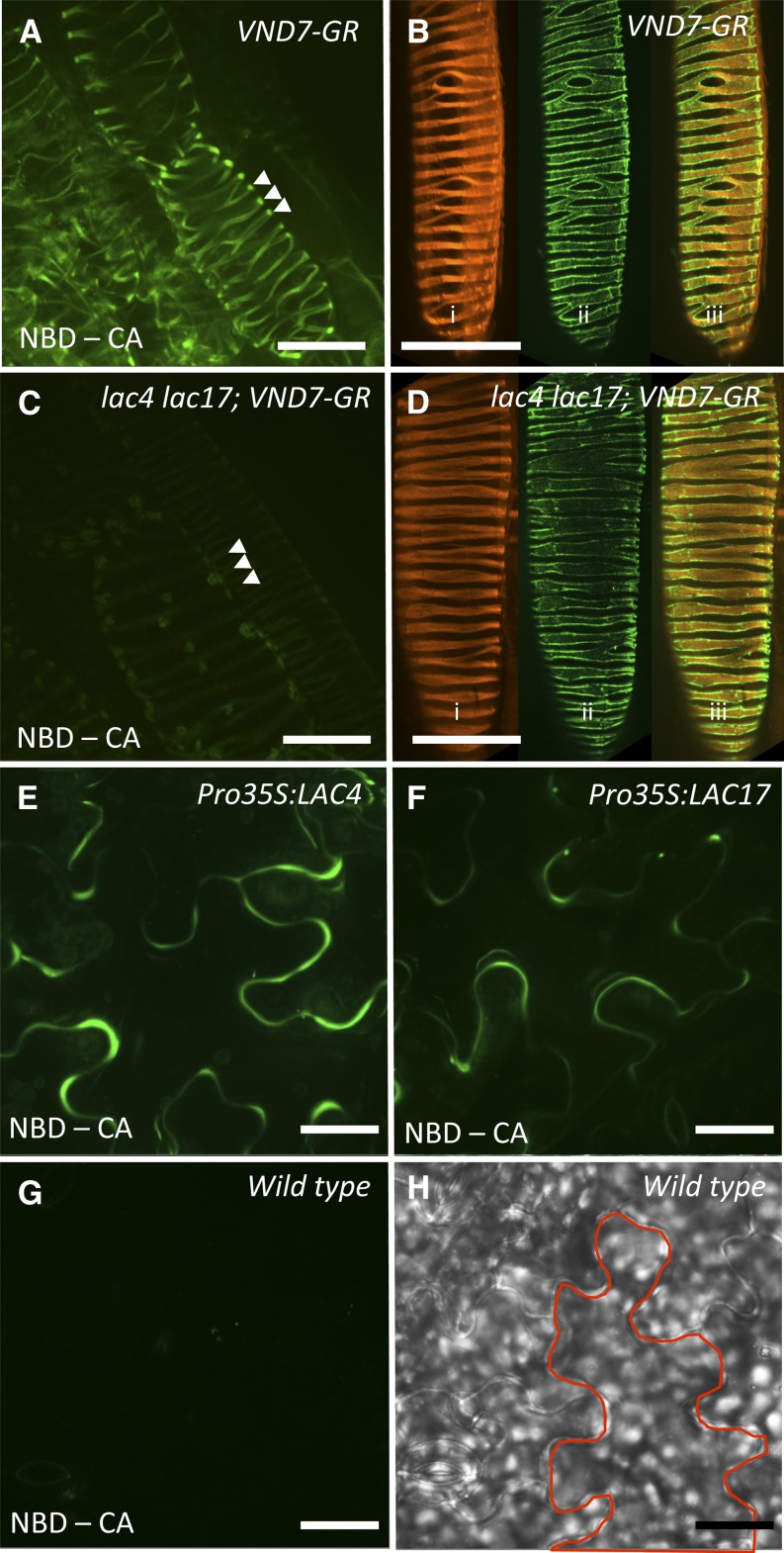
If monolignols are exported to, and are mobile in, all cell wall domains, but become polymerized only in discrete secondary cell wall thickenings, then the presence of laccases and/or peroxidases in those spiral wall thickenings could determine the sites of lignin polymerization. The LAC4 and LAC17 genes are two of the most strongly differentially expressed genes in plant cells responding to increased VND7 or VND6 activity (Ohashi-Ito et al., 2010; Yamaguchi et al., 2010, 2011). In order to directly test the contribution of these laccases to the lignification patterns of protoxylem TE cell walls, the inducible VND7 construct was transformed into lac4 lac17 double mutants. In contrast with strong NBD-CA incorporation in the secondary cell walls of induced protoxylem in the wild-type VND7-VP16-GR seedlings (Fig. 5A, arrowheads), lac4 lac17 double mutants did not incorporate the fluorescent monolignols after induction of VND7 (Fig. 5C, arrowheads). At the same time, VND7-mediated secondary cell wall formation was not affected in lac4 lac17 mutants because secondary wall thickenings in both wild-type (Fig. 5B) and lac4 lac17 double mutants (Fig. 5D) were identical, as assessed by cellulose staining with Pontamine S4B, and immunofluorescent labeling of xylan using LM10 antibodies. These results demonstrate that although other secondary cell wall components such as glucuronoxylan or dense deposits of cellulose were present in the lac4 lac17 mutants, the polysaccharides were not sufficient to trap free monolignols in secondary cell walls in the absence of the required laccases.
Figure 5.
LACs are necessary and sufficient to direct lignin polymerization in Arabidopsis cell wall domains. A, Maximum projection image of hypocotyl epidermal cells induced to transdifferentiate into protoxylem TEs in VND7-VP16-GR seedlings, showing NBD-CA fluorescence in the secondary cell walls (arrowheads). B, VND7-induced protoxylem TE showing the following: i, cellulose deposition (Pontamine S4B stain); ii, whole mount immunolocalization of xylan (LM10 antibody); and iii, merged images. C, Maximum projection image of epidermal cells induced to transdifferentiate into protoxylem TE, in VND7-VP16-GR seedlings in lac4 lac17 double-mutant backgrounds showing that NBD-CA was not incorporated into the secondary cell walls (arrowheads). D, VND7-VP16-GR induced protoxylem TE in lac4 lac17 double mutants showing identical deposition patterns for the following: i, cellulose (Pontamine S4B stain); ii, xylan (LM10 immunolocalization); and iii, merged images, as observed in VND7-VP16-GR alone shown in B. E and F, Constitutive expression of LAC4 (E) or LAC17 (F) catalyzes lignin polymerization of NBD-CA in primary cell walls of cotyledon epidermal cells. G, Control wild-type cotyledon epidermal cells do not incorporate NBD-CA into their cell walls. H, Corresponding bright-field image of control wild-type cotyledons with one cell outlined. Bars = 25 μm.
The loss-of-function mutant analysis demonstrated that LAC4 and LAC17 were necessary for protoxylem lignification. In order to evaluate whether these laccase proteins are sufficient for lignin polymerization, we generated plant lines constitutively overexpressing LAC4 (Pro-35S:LAC4) or LAC17 (Pro-35S:LAC17) and tested whether primary cell walled cells in Arabidopsis seedlings would lignify. We predicted that the presence of LAC4 or LAC17 in a cell wall, even a primary cell wall, would be sufficient to enable cell wall lignification when monolignols were exogenously applied. When Pro-35S:LAC4 or Pro-35S:LAC17 seedlings were treated with exogenously supplied, fluorescently tagged monolignols, primary cell walls in cotyledon epidermal cells had strong fluorescence, thus indicating cell wall lignification (Fig. 5, E and F). By contrast, wild-type cotyledons did not incorporate the fluorescently tagged monolignols (Fig. 5, G and H). Taken together, it is evident that laccases are both necessary and capable of directing lignification in cell walls when monolignols are available.
LAC4 and LAC17 Localize to Secondary Cell Walls in Protoxylem TEs
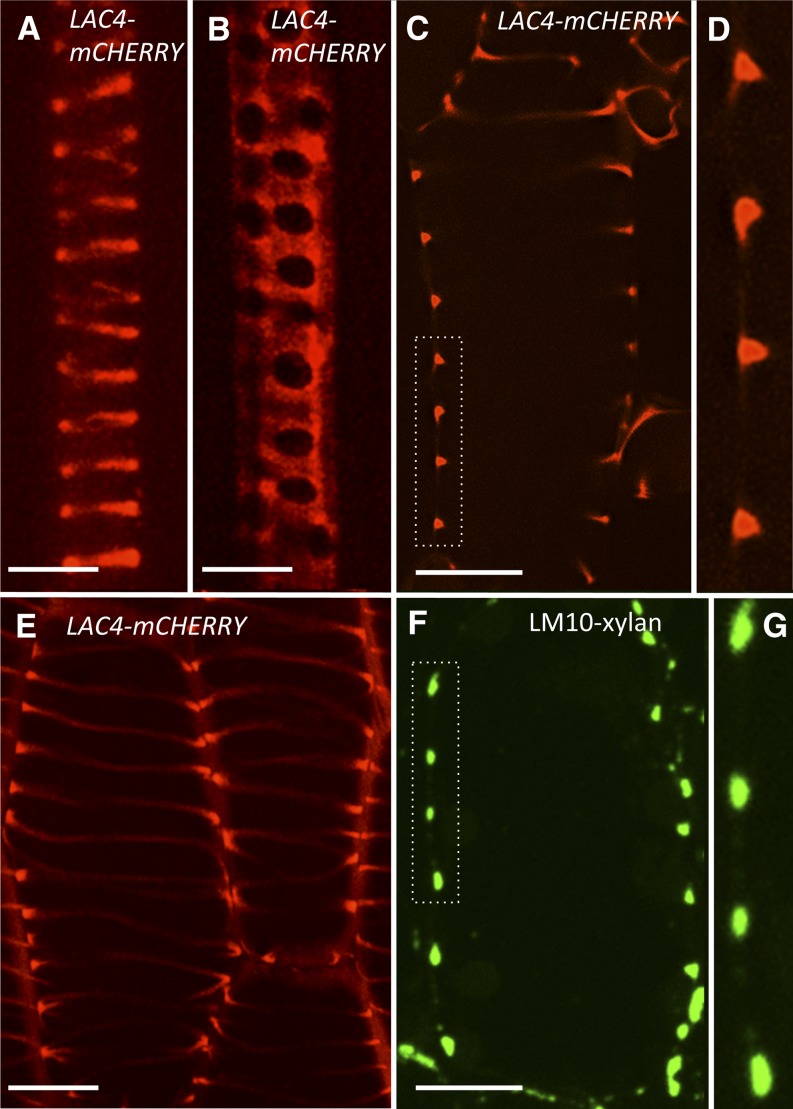
If the presence of the enzymes responsible for oxidative coupling of monolignols is the key factor determining where lignin is deposited, then the spatial organization of these apoplastic proteins is predicted to be tightly controlled. To test this hypothesis, LAC4 and LAC17 were tagged with mCherry (ProLAC4:LAC4-mCherry and ProLAC17:LAC17-mCherry), a pH-stable monomeric RFP that can function in the acidic environment of the apoplast. The irregular xylem phenotype of lac4lac17 double-mutant inflorescence stems was complemented by expression of the LAC4-mCherry construct, thus demonstrating that the LAC4-mCherry protein is functional in planta (Supplemental Fig. S3). In primary roots from lac4lac17 seedlings expressing ProLAC4:LAC4-mCherry, the fluorescent signal was specifically detected in endogenous TEs and was localized to the helical and pitted secondary cell wall thickenings of protoxylem and metaxylem TEs, respectively (Fig. 6, A and B). The expression and subcellular localization of LAC17-mCherry was identical to those observed for LAC4-mCherry, and is shown in Supplemental Figure S6.
Figure 6.
LAC4 localizes specifically to protoxylem TE secondary cell walls. A and B, LAC4 (LAC4-mCherry) localization in endogenous protoxylem (A) and metaxylem (B) TE from primary control roots. C, Median optical slice of an induced protoxylem TE, in hypocotyl epidermis, after 18 h of VND7-VP16-GR induction showing LAC4-mCherry localization, specifically in developing secondary cell walls. D, Higher magnification of developing secondary cell wall outlined in C. E, Maximum projection image showing that LAC4-mCherry localization persists in secondary cell walls after extended exposure to DEX and programed cell death. F, Immunolocalization of xylan (LM10 antibody) on a sectioned differentiating protoxylem TE showing identical deposition patterns as observed for LAC4-mCherry. G, Higher magnification of the developing secondary cell wall outlined in F. Bars = 5 μm in A and B; 12 μm in C to F.
To visualize the LAC4-mCherry pattern at high resolution, protoxylem was induced on the plant surface using VND7-VP16-GR plants transformed with the ProLAC4:LAC4-mCherry construct. The LAC4-mCherry signal was specifically localized to the developing secondary cell wall domains during the early stages of VND7-induced TE differentiation, as shown in a single optical section (Fig. 6, C and D). The concentration of LAC4-mCherry within developing secondary cell walls continued throughout TE differentiation and persisted after programmed cell death (Fig. 6E, maximum projection image). The pattern of LAC4-mCherry distribution was identical to the patterns of LM10 immunofluorescence labeling of glucuronoxylan in secondary cell walls (Fig. 6, F and G, immunolabeled sections and inset). The localization of LAC4 and LAC17 exclusively to secondary cell wall domains, and loss of monolignol incorporation in the lac4 lac17 double mutants, indicate that apoplastic targeting of these laccases plays a key role in the precise pattern of lignin deposition in protoxylem TEs.
DISCUSSION
The goal of this study was to identify mechanisms that allow protoxylem TEs to specifically deposit lignin in spiral cell wall thickenings, while the intervening primary cell walls remain lignin free. This pattern of secondary cell wall lignification is crucial for the ability of protoxylem TEs to facilitate water transport to young and rapidly expanding plant organs. The combination of two technologies (fluorescently tagged monolignols and the VND7-inducible protoxylem system in Arabidopsis) provided a platform to directly visualize the lignification process in an experimental system with rich transcriptomic and molecular genetic resources. The precise spatial association of fluorescent protein-tagged LAC (LAC4-mCherry and LAC17-mCherry) with the spiral wall domains demonstrated that these oxidative enzymes are localized to secondary cell walls. The loss of lignification in lac4lac17 mutants, and ectopic lignification in overexpression lines of LAC4 and LAC17, demonstrated that laccases are necessary and sufficient to direct lignin polymerization. By contrast, localization of GFP-tagged monolignol biosynthetic enzymes and putative monolignol transporter proteins in developing protoxylem TEs demonstrated that the spatial distribution of these components was not correlated with the lignified cell wall pattern.
The relevant laccases in this protoxylem system were predicted from transcriptomic studies that identified LAC4 and LAC17 as genes that were strongly differentially expressed in response to the master regulator transcription factor VND7 (Yamaguchi et al., 2010, 2011). The dramatic reduction of fluorescent lignin incorporation in protoxylem TEs from lac4 lac17 mutants was more severe than the 40% reduction in total lignin reported earlier in lac4 lac17 double-mutant inflorescence stems (Berthet et al., 2011). This difference may reflect the different types of lignified cells being examined in each study (i.e. the abundance of fibers and metaxylem TEs in the Arabidopsis inflorescence stem versus protoxylem TEs). Although lac4 lac17 double-mutant phenotypes in stems included hypolignified fibers and irregular metaxylem vessels (Berthet et al., 2011), loss of function of a third laccase, in the lac4 lac17 lac11 triple mutants, led to severe growth defects and a striking loss of lignified cells in the stem (Zhao et al., 2013). These data make the case that LAC11 is a third redundant component required for lignification in the fiber-rich stem, and this is consistent with the preferential activation of LAC11 expression by the fiber-specific transcription factor SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 (Ohashi-Ito et al., 2010; Zhao et al., 2013). Thus, analyses of both gene expression and mutant phenotypes indicate that although the combination of LAC4, LAC17, and LAC11 support lignification in stems, LAC4 and LAC17 are the major players in protoxylem TE lignification.
Although clarification of the functional members of the laccase gene family is interesting, it is the precise localization of these laccases to the spiral secondary cell walls of protoxylem TEs that is most relevant for the targeted lignification of these secondary cell wall domains. During protoxylem development, there are many Golgi bodies that pause at the microtubule-lined plasma membrane domain underlying the secondary cell wall (Wightman and Turner, 2008), presumably to deliver xylans, cellulose synthases, and cell wall proteins. LAC4-mCherry fluorescence was detected in nascent secondary cell wall thickenings from the earliest stages of secondary cell wall formation, suggesting a testable hypothesis that laccases are secreted simultaneously with secondary cell wall matrix polysaccharides. The molecular mechanisms underlying the specific targeting of secondary cell wall cellulose synthases, xylans, and laccases to the plasma membrane domains below developing secondary cell walls are still unknown. The high flux of vesicle delivery to the microtubule-lined plasma membrane domains may represent a transient reorientation of secretory vesicle flow during this stage of differentiation. A similar phenomenon was hypothesized to account for the directed delivery of plasma membrane proteins to the periarbuscular membrane during arbuscular mycorrhizal symbiosis in Medicago truncatula (Pumplin et al., 2012). Although a transient reorientation of secretory vesicle traffic is the simplest explanation, an alternative hypothesis of uniform secretion of laccases coupled to rapid remobilization toward secondary cell walls, resulting in the observed pattern of LAC4 and LAC17 localization, cannot be ruled out. The development of LAC4-mCherry and LAC17-mCherry proteins provides important tools to study these hypothesized targeting mechanisms.
The model that monolignols are exported out of the cell into all areas of the apoplast, rather than being selectively exported into secondary versus primary cell walls, is in agreement with the recent characterization of Casparian strip lignification in root endodermal cells. Lee et al. (2013) demonstrated that although plasma membrane-bound Casparian strip membrane domain proteins (CASPs) function to organize the extracellular oxidative peroxidases specifically to lignifying Casparian strips, other plasma membrane proteins, including the monolignol transporter ABCG29, are excluded from Casparian strip domains by the CASP complexes (Roppolo et al., 2011; Alejandro et al., 2012). Given the exclusion of ABCG29 from the lignifying domain of the Casparian strip, we hypothesized that a similar, or inverse, pattern might be observed in induced protoxylem TE expressing a variety of GFP-tagged putative monolignol transporters (Alejandro et al., 2012). However, the uniform localization of several different ABC transporters during protoxylem differentiation was not consistent with the hypothesis that localized monolignol export facilitates specific lignification patterns. A careful examination of the transcriptome of the VND7-induced seedlings (Yamaguchi et al., 2010, 2011) did not reveal increased expression of CASP, CASP-like proteins, or peroxidases. Instead, which cell domains lignify appears to be determined by the oxidative machinery and is independent of the source of monolignols, an observation compatible with the good neighbor hypothesis that cells surrounding lignifying TEs may actively contribute to TE lignification (Hosokawa et al., 2001; Tokunaga et al., 2005; Pesquet et al., 2013; Smith et al., 2013). However, future research may identify other components that may be limiting for the lignification process during protoxylem TE differentiation in addition to LAC4 and LAC17.
In lac4lac17 mutants, the secondary cell wall polysaccharide environment remained intact, but this was not sufficient to create the secondary cell wall lignification pattern. These data indicate that differences in nanoscale cell wall polysaccharide environments between the pectin/xyloglucan-rich primary cell wall and the xylan-rich secondary cell wall do not contribute to trapping monolignols in protoxylem secondary cell wall domains. Instead, the presence of laccases in secondary cell wall domains appears to be a key feature, because LAC4 and LAC17 were both necessary for, and capable of, directing cell wall lignification.
MATERIALS AND METHODS
Plant Growth
All Arabidopsis (Arabidopsis thaliana) plants were grown under 16-h-light/21°C and 8-h-dark/16°C conditions. Seeds were surface sterilized using chlorine gas generated in a closed container using 100 mL of bleach and 3 mL of concentrated HCl for 3 to 6 h. Seeds were subsequently plated on one-half-strength Murashige and Skoog (MS) medium (Sigma), kept in the dark at 4°C for 2 to 3 d, and subsequently moved to growth conditions. Pro-35S:RFP-HDEL plants and plasmid (Nelson et al., 2007) were obtained from the Arabidopsis Biological Resource Center (stock no. CD3 960). The lac4-2 lac17 lines were obtained from Richard Sibout and Lise Jouanin and were originally described in Berthet et al. (2011). Pro-35S:VND7-VP16-GR seedlings and plasmid were originally described in Yamaguchi et al. (2010). VND7 activity in 7- to 10-d-old seedlings was induced as described in Yamaguchi et al. (2010).
All transgenic plant lines were generated using ecotype Columbia-0 of Arabidopsis plants, Agrobacterium tumefaciens (strain GV3101), and the floral dip method. Primary transformants were plated on one-half-strength MS medium and selected for survival on 50 μg mL–1 kanamycin, 25 μg mL–1 hygromycin, and 25 μg mL–1 Basta (DL-phosphinothricin; Bayer), or by spraying 10-d-old seedlings grown on soil with 120 μg mL–1 Basta.
Cell Wall Labeling
Cellulose staining using Pontamine S4B (Sigma) was performed by incubating seedlings in 10 mg mL–1 S4B solution in one-half-strength MS medium for 10 to 20 min and washing once in one-half-strength MS medium prior to imaging. Labeling cell wall lignin using NBD-CA was carried out as follows: a mixture of 0.5 mm NBD-CA (Tobimatsu et al., 2011, 2013) and 5 mm CA (Sigma) was added to liquid one-half-strength MS medium to a final concentration of 1 and 10 μm, respectively, together with 10 μm dexamethasone (Sigma) for VND7 induction. VND7-VP16-GR seedlings were incubated in this induction solution for 4 to 48 h, and they were gently shaken under regular growth conditions (listed above). No differences in NBD-CA incorporation were observed when preinducing VND7 activity for 24 h prior to adding the NBD-CA mix. Seedlings were subsequently rinsed twice for 15 min in one-half-strength MS medium and fixed in 4% paraformaldehyde, 50 mm PIPES, 5 mm MgSO4, and 5 mm EGTA solution overnight. Seedlings were subsequently washed twice in TBS plus Tween 20 (TBST; 10 mm Tris, pH 7, 0.25 m NaCl, 0.1% Tween 20) solution. Seedlings were heated to 45°C for 4 to 72 h in 49% (v/v) methanol:1% (v/v) acetic acid solution with gentle shaking to wash away unincorporated NBD-CA. Seedlings were subsequently washed and rehydrated in a graded methanol series, mounted in one-half-strength MS medium, and imaged. For whole-mount immunolabeling, VND7-induced seedlings were fixed as described above and stored in TBST buffer at 4°C until hybridization. Fixed seedlings were incubated in 5% bovine serum albumin (Sigma) in TBST for 1 to 2 h at room temperature and were subsequently incubated with LM10 primary antibody (McCartney et al., 2005) at 1:50 dilution overnight at 4°C. Seedlings were rinsed three times in TBST solution for 5 min and were subsequently incubated in a 1:100 dilution of secondary anti-rat Alexa 488 antibody (Invitrogen) at room temperature for 1 h. After washing twice with TBST, samples were mounted onto slides and imaged. For immunolocalization of thick sections, the above procedure was performed on sectioned material from LR white resin-infiltrated seedlings after high-pressure freezing and freeze substitution in glutaraldehyde and uranyl acetate. Sections were generated using a Leica Ultracut UCT Ultramicrotome and were mounted on Teflon-coated glass slides (Electron Microscopy Sciences).
Microscopy and Image Analysis
For live-cell imaging, a Perkin-Elmer UltraView VoX spinning disk confocal mounted on a Leica DMI6000 inverted microscope and a Hamamatsu 9100-02 CCD camera were used with the following excitation and emission filters: GFP (488 and 525), YFP (514 and 540), and RFP (561 and 595). Samples were mounted in water and imaged using a Leica oil immersion 63× objective (1.4 numerical aperture, Plan-Apo). All images were processed using Volocity image analysis software (Improvision). GFP settings were used for imaging lignin with NBD-CA and RFP settings were used for imaging cellulose after treatment with S4B.
For visualizing lignin autofluorescence, an Olympus FV1000 Multiphoton Laser Scanning Microscope with a tunable MaiTai BB DeepSee (710–990 nm) laser adjusted to 730 nm was used. Simultaneous absorption of two 730-nm photons results in excitation of lignin at approximately 350 to 370 nm. During imaging, two emission channels were simultaneous collected using 420- to 460-nm and 495- to 540-nm filter sets. Samples were fixed in 6:1 ethanol:acetic acid solution for 24 h, washed twice with 95% ethanol, and gradually rehydrated using 70% ethanol, 30% ethanol, and water solutions. Samples were subsequently mounted in chloral hydrate solution (9:1:3 (w/v/v) chloral hydrate:glycerol:water) and were imaged using an Olympus XL Plan N25X objective. All images were processed using Volocity image analysis software (Improvision) and Photoshop software (Adobe Systems). For Supplemental Figures S1 and S3, hand sections from mature Arabidopsis inflorescence stems were stained with toluidine blue (Ted Pella) or freshly mounted and imaged for lignin autofluorescence using a Leica DMR microscope equipped with a standard mercury arc lamp with excitation (340–380 nm) and emission (450 nm) filter sets. For TEM analysis of TEs, 7-d-old seedling roots were high-pressure frozen, freeze substituted, sectioned, and prepared for TEM as previously described (Smith et al., 2013).
Molecular Biology
All DNA sequences were cloned into pDONR221 or pDONRzeo using Gateway cloning methodology (Invitrogen) and were subsequently transferred into specific expression vectors as indicated. All PCR amplifications were carried out using Phusion High Fidelity DNA polymerase (New England Biolabs) and two-step adapter PCR was used to incorporate full-length Gateway attb sequences into the PCR products.
Genomic regions containing the open reading frames (ORFs) for ABCB11, ABCG29, ABCG33, C4H, and CCR1 were subsequently cloned into the vectors driven by the UBIQUITIN10 (UBQ10) promoter (Grefen et al., 2010) and were transformed into VND7-VP16-GR plants. C-terminal GFP fusions (ProUBQ10:ABCB11-GFP, ProUBQ10:C4H-GFP, and ProUBQ10:CCR1-GFP), and N-terminal GFP fusions (ProUBQ10:GFP-ABCG29 and ProUBQ10:GFP-ABCG33). Pro-35S:YFP-ABCG11 plants (Bird et al., 2007) were crossed with Pro-35:VND7-VP16-GR plants using standard crossing techniques. A genomic LAC4 and LAC17 fragment containing 2 kb of the 5′ promoter and an ORF with no stop codon was amplified and cloned into a modified pMDC111 vector (Curtis and Grossniklaus, 2003), in which the GFP coding region was excised by digesting with the AscI and SacI restriction enzymes, and replaced with the mCherry coding sequence. Pro-35S:LAC4 and Pro-35S:LAC17 constructs were generated by cloning the respective genomic ORFs into the pK2GW7 vector (VIB Department of Plant Systems Biology, Ghent University) and subsequent transformation into wild-type plants.
Artificial microRNA targeting C4H and CCR1 was designed using WMD2 (http://wmd2.weigelworld.org), generated as described in Schwab et al. (2006), and was subsequently cloned into the pK2GW7 vector (VIB Department of Plant Systems Biology, Ghent University).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers LAC4 (AT2G38080), LAC17 (AT5G60020), CCR1 (AT1G15950), C4H (AT2G30490), VND7 (AT1G71930), ABCG11 (AT1G17840), ABCB11 (AT1G02520), ABCG29 (AT3G16340), and ABCG33 (AT2G37280).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypic characterization of Pro35S:miRNA C4H plants.
Supplemental Figure S2. C4H-GFP localization and ER structure during VND7-induced protoxylem TE differentiation.
Supplemental Figure S3. Complementation of irregular xylem phenotypes of ccr1irx4 and lac4 lac17 mutants with GFP or mCherry fusion proteins.
Supplemental Figure S4. ABC transporters are evenly localized in plasma membranes during protoxylem TE differentiation in VND7-VP16-GR seedlings.
Supplemental Figure S5. C4H-YFP and YFP-ABCG11 localization during protoxylem programmed cell death of transdifferentiated Arabidopsis epidermal cells.
Supplemental Figure S6. LAC17 localizes specifically to protoxylem TE secondary cell walls.
Supplemental Movie S1. Z-stack animation of YFP-ABCG11 localization in numerous plasma membrane-derived vesicles during protoxylem TE programmed cell death.
Supplementary Material
Acknowledgments
We thank the University of British Columbia Bioimaging Facility for technical support and Etienne Grienenberger for technical advice and comments on this article.
Glossary
- TE
tracheary element
- ER
endoplasmic reticulum
- ABC
ATP-binding cassette
- RFP
red fluorescent protein
- TEM
transmission electron microscopy
- cyt
cytosolic-localized
- YFP
yellow fluorescent protein
- S4B
Fast Scarlet 4B
- NBD
γ-nitrobenzofuran
- CA
coniferyl alcohol
- MS
Murashige and Skoog
- TBST
TBS plus Tween 20
- ORF
open reading frame
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (Discovery and Collaborative Research and Training Experience Program grants to B.E. and A.L.S.), the Japan Society for the Promotion of Science (Kakenhi grant nos. 24114002 and 25291062 to T.D.), the Nara Institute of Science and Technology Global Initiative Program (to T.D.), the U.S. Department of Energy Office of Science (grant no. DE–SC0006930 to Y.T. and J.R.), and the U.S. Department of Energy Great Lakes Bioenergy Research Center (grant no. BER DE–FC02–07ER64494 to Y.T. and J.R.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alejandro S, Lee Y, Tohge T, Sudre D, Osorio S, Park J, Bovet L, Lee Y, Geldner N, Fernie AR, et al. (2012) AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol 22: 1207–1212 [DOI] [PubMed] [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C. (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassard JE, Richert L, Geerinck J, Renault H, Duval F, Ullmann P, Schmitt M, Meyer E, Mutterer J, Boerjan W, et al. (2012) Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 24: 4465–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L. (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52: 485–498 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C. (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Chapelle A, Morreel K, Vanholme R, Le-Bris P, Morin H, Lapierre C, Boerjan W, Jouanin L, Demont-Caulet N. (2012) Impact of the absence of stem-specific β-glucosidases on lignin and monolignols. Plant Physiol 160: 1204–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC, Li Q, Shuford CM, Liu J, Muddiman DC, Sederoff RR, Chiang VL. (2011) Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc Natl Acad Sci USA 108: 21253–21258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al. (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42: 618–640 [DOI] [PubMed] [Google Scholar]
- Esau K. (1965). Vascular Differentiation in Plants. Holt, Rinehart and Winston, New York [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR. (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Suzuki S, Umezawa T, Sato Y. (2001) Progress of lignification mediated by intercellular transportation of monolignols during tracheary element differentiation of isolated Zinnia mesophyll cells. Plant Cell Physiol 42: 959–968 [DOI] [PubMed] [Google Scholar]
- Jones L, Ennos AR, Turner SR. (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Kaneda M, Schuetz M, Lin BS, Chanis C, Hamberger B, Western TL, Ehlting J, Samuels AL. (2011) ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J Exp Bot 62: 2063–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N. (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412 [DOI] [PubMed] [Google Scholar]
- Liu CJ. (2012) Deciphering the enigma of lignification: precursor transport, oxidation, and the topochemistry of lignin assembly. Mol Plant 5: 304–317 [DOI] [PubMed] [Google Scholar]
- McCartney L, Marcus SE, Knox JP. (2005) Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem 53: 543–546 [DOI] [PubMed] [Google Scholar]
- Miao YC, Liu CJ. (2010) ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc Natl Acad Sci USA 107: 22728–22733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Oda Y, Fukuda H. (2010) Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22: 3461–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E, Zhang B, Gorzsás A, Puhakainen T, Serk H, Escamez S, Barbier O, Gerber L, Courtois-Moreau C, Alatalo E, et al. (2013) Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. Plant Cell 25: 1314–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Zhang X, Noar RD, Harrison MJ. (2012) Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc Natl Acad Sci USA 109: E665–E672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro DK, Mah N, Ellis BE, Douglas CJ. (2001) Functional characterization and subcellular localization of poplar (Populus trichocarpa x Populus deltoides) cinnamate 4-hydroxylase. Plant Physiol 126: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roppolo D, De Rybel B, Tendon VD, Pfister A, Alassimone J, Vermeer JE, Yamazaki M, Stierhof YD, Beeckman T, Geldner N. (2011) A novel protein family mediates Casparian strip formation in the endodermis. Nature 473: 380–383 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Chapple C. (2001) Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159: 1741–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Demura T, Yamawaki K, Inoue Y, Sato S, Sugiyama M, Fukuda H. (2006) Isolation and characterization of a novel peroxidase gene ZPO-C whose expression and function are closely associated with lignification during tracheary element differentiation. Plant Cell Physiol 47: 493–503 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C. (2009) Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Roach M, Mansfield SD, Ellis B, Samuels L. (2013) Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 25: 3988–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenin J, Pollet B, Letarnec B, Saulnier L, Gissot L, Maia-Grondard A, Lapierre C, Jouanin L. (2011) The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol Plant 4: 70–82 [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y, Davidson CL, Grabber JH, Ralph J. (2011) Fluorescence-tagged monolignols: synthesis, and application to studying in vitro lignification. Biomacromolecules 12: 1752–1761 [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y, Wagner A, Donaldson L, Mitra P, Niculaes C, Dima O, Kim JI, Anderson N, Loque D, Boerjan W, et al. (2013) Visualization of plant cell wall lignification using fluorescence-tagged monolignols. Plant J 76: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga N, Sakakibara N, Umezawa T, Ito Y, Fukuda H, Sato Y. (2005) Involvement of extracellular dilignols in lignification during tracheary element differentiation of isolated Zinnia mesophyll cells. Plant Cell Physiol 46: 224–232 [DOI] [PubMed] [Google Scholar]
- Tsuyama T, Kawai R, Shitan N, Matoh T, Sugiyama J, Yoshinaga A, Takabe K, Fujita M, Yazaki K. (2013) Proton-dependent coniferin transport, a common major transport event in differentiating xylem tissue of woody plants. Plant Physiol 162: 918–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W. (2012) Metabolic engineering of novel lignin in biomass crops. New Phytol 196: 978–1000 [DOI] [PubMed] [Google Scholar]
- Wightman R, Turner SR. (2008) The roles of the cytoskeleton during cellulose deposition at the secondary cell wall. Plant J 54: 794–805 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Goué N, Igarashi H, Ohtani M, Nakano Y, Mortimer JC, Nishikubo N, Kubo M, Katayama Y, Kakegawa K, et al. (2010) VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol 153: 906–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T. (2011) VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J 66: 579–590 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA. (2013) Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.