Two mitochondrial translocases are essential for the division during early embryo and endosperm development in Arabidopsis.
Abstract
In the life cycle of flowering plants, the sporophytic generation takes up most of the time and plays a dominant role in influencing plant growth and development. The embryo cell and endosperm free nucleus divisions establish the critical initiation phase of early sporophyte development, which forms mature seeds through a series of cell growth and differentiation events. Here, we report on the biological functions of two Arabidopsis (Arabidopsis thaliana) mitochondrial proteins, TRANSLOCASE OF THE INNER MEMBRANE9 (TIM9) and TIM10. We found that dysfunction of either AtTIM9 or AtTIM10 led to an early sporophyte-lethal phenotype; the embryo and endosperm both arrest division when the embryo proper developed to 16 to 32 cells. The abortion of tim9-1 and tim10 embryos at the 16/32-cell stage was caused by the loss of cell viability and the cessation of division in the embryo proper region, and this inactivation was due to the collapse of the mitochondrial structure and activity. Our characterization of tim9-1 and tim10 showed that mitochondrial membrane permeability increased and that cytochrome c was released from mitochondria into the cytoplasm in the 16/32-cell embryo proper, indicating that mitochondrial dysfunction occurred in the early sporophytic cells, and thus caused the initiation of a necrosis-like programmed cell death, which was further proved by the evidence of reactive oxygen species and DNA fragmentation tests. Consequently, we verified that AtTIM9 and AtTIM10 are nonredundantly essential for maintaining the mitochondrial function of early embryo proper cells and endosperm-free nuclei; these proteins play critically important roles during sporophyte initiation and development in Arabidopsis.
Land plants have a remarkable life cycle that alternates between two distinct multicellular generations, the reduced gametophytes and the dominant sporophyte. The male and female gametophytes represent the gametophytic generation that alternates with the sporophytic generation (Kenrick and Crane, 1997; Niklas and Kutschera, 2010). In Arabidopsis (Arabidopsis thaliana), the sporophytic generation originates from the embryo and endosperm that are generated in the ovule after double fertilization. The embryo follows a predictable sequence of cell division and differentiation and develops from a zygote to a mature seed during embryogenesis. The endosperm grows rapidly as the syncytium and cellularizes, nourishing the developing embryo and determining the ovule size, and then disintegrates during the late stages of embryo development (Grossniklaus and Schneitz, 1998; Berger et al., 2006; Jenik et al., 2007).
Mitochondria play roles in a variety of fundamental and essential functions during the plant life cycle, including early sporophyte development. This has been confirmed through the identification of mitochondrial proteins with specific roles during Arabidopsis embryo and endosperm division (Millar et al., 2008; Hsu et al., 2010; Law et al., 2012). These processes are regulated by a large number of different mitochondrial proteins, and about 99% of these are encoded by nuclear DNA, synthesized in the cytosol, and imported into the mitochondria across one or both membranes (Burger et al., 2003; Elo et al., 2003). The mitochondrial protein import mechanism plays an important role in protein importation, including the TOM (for translocase of the outer membrane) complex and the TIM (for translocase of the inner membrane) complex on the mitochondrial outer and inner membranes, respectively (Chacinska et al., 2009). Between the double membranes, the mitochondrial intermembrane space (IMS) contains several tiny TIM proteins (TIM8–TIM10 and TIM13) that are known to be involved in the carrier transport pathway (Koehler et al., 1998a; Sirrenberg et al., 1998; Adam et al., 1999; Paschen et al., 2000). In yeast (Saccharomyces cerevisiae) and human, TIM9 and TIM10 had been proposed to exist in the form of a hexamer composed of three TIM9 and three TIM10 subunits (Webb et al., 2006; Baker et al., 2009). It was also revealed that in the carrier transport pathway, the TIM9:10 complex guided the precursor proteins through the IMS in yeast (Neupert and Herrmann, 2007). Once precursors translocate through the TOM channel into the IMS, they have to bind to the TIM9:10 complex and be transferred to the carrier translocase located on the inner membrane (TIM22 complex) to proceed to subsequent steps (Curran et al., 2002; Rehling et al., 2003; Wiedemann et al., 2006).
TIM9 and TIM10 are highly conserved proteins throughout the eukaryotic kingdom, including plants (Braun and Schmitz, 1999; Carrie et al., 2010). In potato (Solanum tuberosum), TIM9 and TIM10 were found to be two components of import proteins in the IMS, having functional roles in the efficient carrier proteins pathway (Lister et al., 2002). By searching yeast homologs, the genes encoding potential TIM9 and TIM10 homologs in Arabidopsis were found and defined to be components of the mitochondrial protein import apparatus (Bauer et al., 1999; Lister et al., 2003). Furthermore, the results of mitochondrial transport inhibitor treatment also suggested that AtTIM9 and AtTIM10 may participate in protein import (Lister et al., 2004). Recently, in addition to the basic import function, researchers have paid increasing attention to the function of TIM components during various phases of plant development, such as playing important roles in Arabidopsis seedling development (TIM21 and TIM23) and hypocotyl cell growth (TIM50; Hamasaki et al., 2012; Kumar et al., 2012; Wang et al., 2012). Although the AtTIM9 gene had been designated previously as EMB2474 and its mutation showed an embryo-lethal phenotype in Arabidopsis (Tzafrir et al., 2004), the mechanism of death during embryogenesis is unclear. Moreover, to date, little is known about the importance of TIM10 during Arabidopsis development, neither the relationship between AtTIM9 and AtTIM10 nor the potential functional roles of them in early sporophyte growth.
Here, we provide molecular and genetic evidence to demonstrate that loss of either AtTIM9 or AtTIM10 causes early sporophytic cells, including the embryo proper cells and endosperm-free nuclei, to stop dividing and arrest growth at the 16/32-cell stage of embryo development. The phenotypes we observed in the embryo proper and endosperm of the mutants were caused by initiation of necrosis-like programmed cell death (PCD) as defined by van Doorn and Woltering (2005), and this PCD process probably resulted from a collapse of mitochondrial structure and a decrease in mitochondrial activity. Our results indicated that AtTIM9 and AtTIM10 play nonredundant, essential roles in continuously dividing embryo proper and endosperm-free nuclei during early sporophyte development by maintaining normal mitochondrial function.
RESULTS
AtTIM9 and AtTIM10 Genes Are Both Essential for Sporophytic Progeny
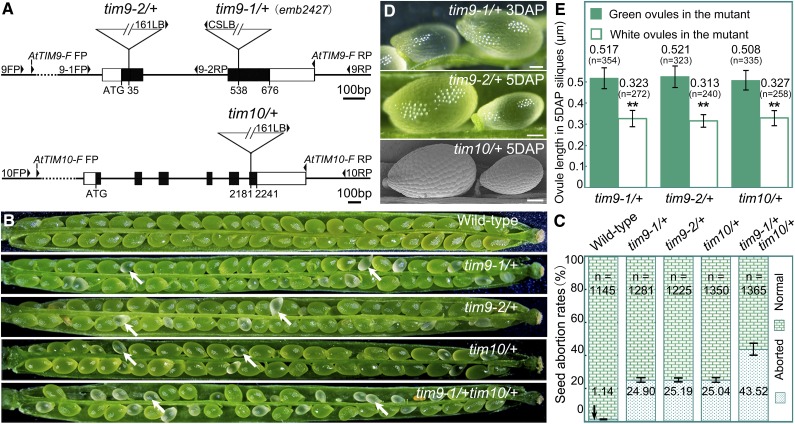
To investigate the function of the AtTIM9 and AtTIM10 genes during plant development, we obtained three Arabidopsis mutant lines from public mutant collections (Fig. 1A): tim9-1 (also known as emb2474; Tzafrir et al., 2004), tim9-2, and tim10. Segregation analysis of all three of the self-fertilized mutant progeny showed that the selective ratio was about 2:1 (resistant:sensitive), instead of the expected 3:1 (Table I). This result suggested that there was seed lethality in the resistant mutant progeny. PCR analysis also demonstrated that no viable homozygous mutant plants were found (Supplemental Fig. S1A). Therefore, each of the tim9-1, tim9-2, and tim10 mutants contains a single-copy transfer DNA (T-DNA) insertion in the genome and presents in the form of heterozygous tim9-1/+, tim9-2/+ and tim10/+, respectively, showing a homozygous seed-lethal phenotype.
Figure 1.
Characterization of the AtTIM9 and AtTIM10 mutations. A, Schematic diagrams of the AtTIM9 and AtTIM10 gene structures with the positions of the T-DNA insertions of three mutants. Exons are shown as black boxes, 5′ and 3′ regions as white boxes, and introns and untranslated regions as lines. FP, Forward primer; RP, reverse primer. B, Silique phenotypes of the wild type and mutants. The white arrows show the aborted white ovules. C, Seed abortion rates in the mutants compared with the wild type. The total number (n) of counted seeds is listed on the top of the histogram, and the seed abortion rate values are shown on the bottom. D, Ovule phenotypes in wild-type and mutant siliques. Bars = 1 mm. E, White ovule length compared with green ovules from heterozygous tim mutant parents. The total number (n) of counted ovules and the average ovule length values are shown above the histogram. [See online article for color version of this figure.]
Table I. Segregation of the tim9-1/+, tim9-2/+, and tim10/+ mutants.
| Cross (Female × Male)a | SMb | Resistant | Sensitive | R:S Ratec | Expected Rate |
|---|---|---|---|---|---|
| tim9-1/+ × tim9-1/+ | Bas | 2,008 | 1,052 | 1.91:1d | 3:1 |
| tim9-2/+ × tim9-2/+ | Sul | 1,595 | 796 | 2.03:1d | 3:1 |
| tim10/+ × tim10/+ | Sul | 1,454 | 718 | 2.00:1d | 3:1 |
Seeds of the selfing lines were collected and grown on selective plates to determine the segregation. bSelection marker (SM): BASTA (Bas) and sulfadiazine (Sul). cResistant (R):sensitive (S). dSignificantly different from the segregation ratio of 3:1 (P < 0.01).
We dissected maturing siliques from tim9-1/+, tim9-2/+, and tim10/+ plants to analyze the phenotypes of mutants and found that all of them contained a proportion of aborted white seeds (Fig. 1B, arrows). By contrast, the siliques from wild-type plants contained a well-ordered array of maturing green seeds (Fig. 1B). More than 2,000 selection marker-resistant progeny of each mutant displaying the aborted seed phenotype were observed, implying that the phenotypes are tightly linked to the T-DNA insertions in the respective mutants. We further calculated the percentage of aborted seeds in each mutant (Fig. 1C). The values were close to the expected value of 25%, reconfirming that mutations of AtTIM9 and AtTIM10 led to homozygous progeny lethality. Additional reciprocal crosses showed that the genetic transmission capacity of both females and males in the three mutants was similar to that of the wild type, indicating that the gametophyte fertilities were not affected due to a loss of function specifically of AtTIM9 or AtTIM10 (Supplemental Table S1). Together, these results indicate that the defective fertility phenotype of the mutants was only caused by aborted homozygous sporophytes after fertilization.
To determine whether AtTIM9 and AtTIM10 are required for early sporophytic development, we performed complementation tests (Supplemental Materials and Methods S1). The self-progeny of three independent T3 generation homozygous transgenic plants for each construct were each analyzed by genotyping (Supplemental Fig. S1A). Mutant tim9-1/+, tim9-2/+, and tim10/+ all could be successfully complemented to become fertile (Supplemental Fig. S1, B and C), indicating that the sporophyte-lethal phenotype was indeed caused by the loss of either AtTIM9 or AtTIM10 function. Thus, AtTIM9 and AtTIM10 both perform essential functions in early sporophyte development in Arabidopsis.
Mutations of AtTIM9 and AtTIM10 Arrest Development in Both Embryo and Endosperm When the Embryo Proper Grows to the 16/32-Cell Stage
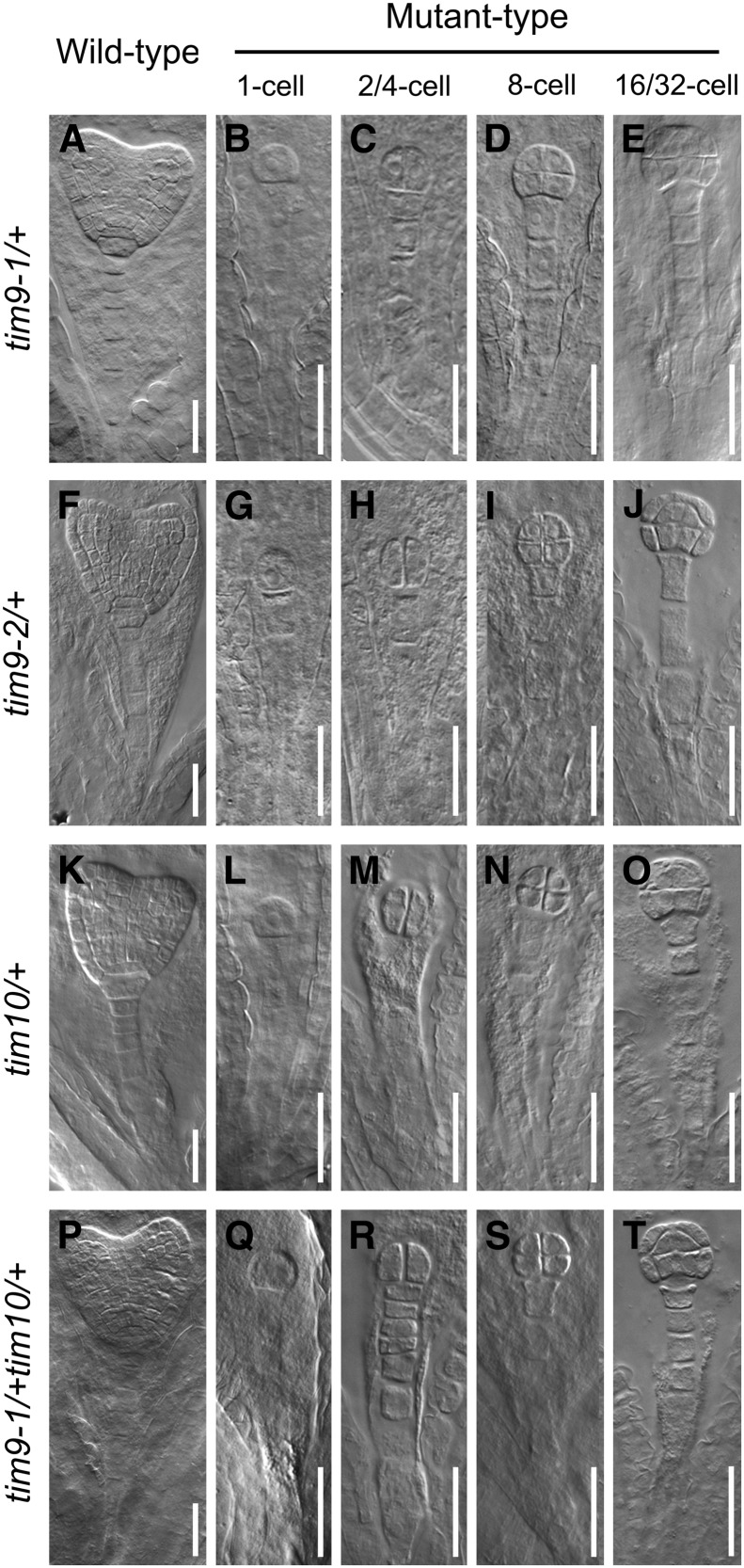
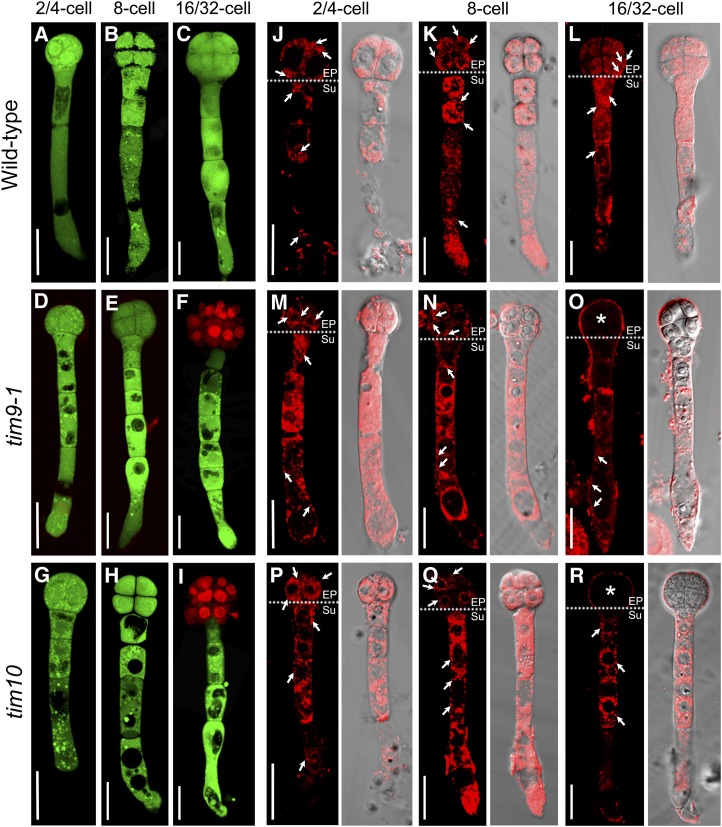
To precisely determine the developmental stage of the abortion of seeds of mutant plants, we examined embryos of immature ovules from tim9-1/+, tim9-2/+, and tim10/+. For controls, embryos in the normal ovules (mutant siliques) were developing into the early heart embryo stage at 4 d after pollination (DAP; Fig. 2, A, F, and K). Embryos in the aborted ovules were still delayed at the sequential stages containing one- to eight-cell embryo proper (Fig. 2, B–D, G–I, and L–N) and finally arrested at the 16/32-cell stage (Fig. 2, E, J, and O). No abnormalities in the suspensor suggested that the delayed growth was only present in the mutant embryo proper. Meanwhile, we found that the white ovules were generally smaller than the green ones, as the aborted seeds could be distinguished from normal ones in siliques at 3 DAP in each heterozygous mutant, according to the ovule size, and inferential statistical analysis of ovule lengths at 5 DAP showed a significant difference between green and white ovules (Fig. 1, D and E), implying that the development of the endosperm was also arrested. Next, we carefully counted representative numbers of each of the embryonic morphologies at different development stages in wild-type and heterozygote (tim9-1/+, tim9-2/+, and tim10/+) plants (Table II). About 25% of embryos in the mutants showed delayed growth throughout the developmental process and stagnated at the 16/32-cell stage, which was consistent with the segregation and abortion rates of the mutants (Table I; Fig. 1C). According to the tallied numbers of endosperm-free nuclei, the development of the endosperm also showed delayed growth in the early stages following fertilization (Table III). These data demonstrated that the embryos and syncytial endosperm of aborted ovules in tim9-1/+, tim9-2/+, and tim10/+ plants grew more slowly than did control ovules and finally arrested.
Figure 2.
Status of embryo development in the mutants. Normal and aborted embryos at the age of 4 DAP are observed within heterozygous siliques of tim9-1/+ (A–E), tim9-2/+ (F–J), tim10/+ (K–O), and tim9-1/+tim10/+ (P–T). As controls, normal embryos in heterozygous siliques are at the early heart stage (A, F, K, and P). Aborted embryos at the one-cell (B, G, L, and Q), two/four-cell (C, H, M, and R), eight-cell (D, I, N, and S), and 16/32-cell (E, J, O, and T) stages are shown for each heterozygous silique. Bars = 30 μm.
Table II. Distribution of embryo phenotypes in the wild type and mutants at sequential development stages.
Siliques were marked consecutively starting from 3 DAP, and the samples from days 3 to 6 were used for observation. For the siliques from the same line and at the same marking time, ovules in each silique were cleared and the embryos were counted at each stage, then the total and average of each stage embryo number were calculated and rounded to the whole number. Therefore, the number of each sample displays the embryo distribution of a whole silique. Nts/Nsi, Number of total ovules/number of siliques; Nas, number of average ovules in a single silique; EG, early globular; LG, later globular; EH, early heart; H, heart; ET, early torpedo; T, torpedo; C, cotyledon.
| Parents | DAP | Nts/Nsi | Nas | Arrested Stages |
Percentage Aborted | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| One Cell | Two/Four Cell | Eight Cell | 16/32 Cell | EG | LG | EH | H | ET | T | C | |||||
| Wild type | 3 | 226/6 | 38 | – | – | – | 2 | 15 | 15 | 6 | – | – | – | – | – |
| 4 | 286/7 | 41 | – | – | – | – | – | 5 | 16 | 20 | – | – | – | – | |
| 5 | 350/8 | 44 | – | – | – | – | – | – | – | 3 | 23 | 18 | – | – | |
| 6 | 231/5 | 46 | – | – | – | – | – | – | – | – | 28 | 10 | 8 | – | |
| tim9-1/+ | 3 | 233/5 | 47 | 1 | 2 | 3 | 6 | 14 | 12 | 9 | – | – | – | – | 25.53 |
| 4 | 262/6 | 44 | – | 1 | 2 | 8 | 4 | 6 | 17 | 6 | – | – | – | 25.00 | |
| 5 | 266/6 | 42 | – | 1 | 1 | 9 | – | 3 | 5 | 11 | 12 | – | – | 26.19 | |
| 6 | 235/5 | 47 | – | – | – | 12 | – | – | – | – | 3 | 25 | 7 | 25.53 | |
| tim9-2/+ | 3 | 280/6 | 49 | 1 | 1 | 4 | 7 | 15 | 11 | 10 | – | – | – | – | 26.09 |
| 4 | 303/6 | 50 | – | 1 | 3 | 9 | 5 | 7 | 18 | 7 | – | – | – | 26.00 | |
| 5 | 295/7 | 42 | – | 1 | 1 | 8 | – | 2 | 8 | 12 | 10 | – | – | 23.81 | |
| 6 | 257/6 | 43 | – | – | – | 11 | – | – | – | – | 2 | 23 | 7 | 25.58 | |
| tim10/+ | 3 | 295/6 | 49 | 2 | 2 | 3 | 5 | 14 | 13 | 10 | – | – | – | – | 26.00 |
| 4 | 289/7 | 41 | – | 1 | 3 | 7 | 3 | 6 | 18 | 3 | – | – | – | 26.83 | |
| 5 | 285/7 | 41 | – | 1 | 1 | 9 | – | – | 8 | 14 | 8 | – | – | 26.83 | |
| 6 | 224/5 | 45 | – | – | – | 11 | – | – | – | – | 4 | 25 | 5 | 24.44 | |
| tim9-1/+tim10/+ | 3 | 305/6 | 51 | 5 | 2 | 8 | 7 | 6 | 13 | 10 | – | – | – | – | 43.14 |
| 4 | 292/6 | 49 | 3 | 3 | 7 | 8 | 4 | 5 | 16 | 3 | – | – | – | 42.86 | |
| 5 | 298/6 | 50 | 4 | 2 | 6 | 10 | – | 2 | 5 | 14 | 7 | – | – | 44.00 | |
| 6 | 252/5 | 50 | 3 | 1 | 5 | 12 | – | – | – | – | 6 | 19 | 4 | 42.00 | |
Table III. Frequencies of embryo and endosperm in the wild type and mutants.
Ovules of the wild type and mutations were isolated and cleared to count the number of embryos in different stages and surrounding endosperm free nuclei. N, Number of total ovules; nuc, endosperm free nuclei.
| DAP | Parents | N | Embryo |
Endosperm |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Zygote | One Cell | Two/Four Cell | Eight Cell | 16/32 Cell | Less Than 30 nuc | 30 to 60 nuc | 60 to 120 nuc | More Than 120 nuc | |||
| % | |||||||||||
| 1 | Wild type | 156 | 15 | 27.9 | 55.8 | 1.3 | 0 | 16.4 | 53.7 | 22.4 | 7.5 |
| tim9-1/+ | 139 | 32.6 | 41.9 | 25.6 | 0 | 0 | 35.3 | 58.9 | 5.9 | 0 | |
| tim9-2/+ | 148 | 31.7 | 39 | 29.3 | 0 | 0 | 35.8 | 56.6 | 7.5 | 0 | |
| tim10/+ | 134 | 35.7 | 35.7 | 28.6 | 0 | 0 | 38.8 | 55.1 | 6.1 | 0 | |
| 2 | Wild type | 156 | 0 | 4.3 | 23.9 | 34.8 | 37 | 5.1 | 20.5 | 38.5 | 35.9 |
| tim9-1/+ | 139 | 2.1 | 11.8 | 46.7 | 35.2 | 4.2 | 20.9 | 38.8 | 28.8 | 11.5 | |
| tim9-2/+ | 148 | 2.2 | 11.1 | 48.9 | 33.3 | 4.5 | 21.6 | 40 | 27 | 11.4 | |
| tim10/+ | 134 | 2.1 | 10.9 | 50 | 32.7 | 4.3 | 20.1 | 39.6 | 27.6 | 12.7 | |
Considering that the lack of either AtTIM9 or AtTIM10 led to early sporophyte lethality, we crossed tim9-1/+ with tim10/+ to investigate if a more severe defective phenotype would occur in double mutant plants. In the F2 progeny, the tim9-1/+tim10/+ lines were identified and observed by dissection of siliques (Fig. 1B). The seed abortion rate of tim9-1/+tim10/+ was 43.52% (Fig. 1C), which conforms to the expected 43.75% rate according to Mendel’s second law; no more severe defective phenotype was observed in the double mutant, showing that the aborted embryos still arrested at the 16/32-cell stage (Fig. 2, P–T). These results indicated that AtTIM9 and AtTIM10 function nonredundantly during early sporophyte development to control embryo cell and endosperm-free nuclei division.
AtTIM9 and AtTIM10 Are Expressed Widely, Especially in the Embryo and Endosperm
Investigation of Arabidopsis Genevestigator expression data (Hruz et al., 2008) for AtTIM9 and AtTIM10 indicated that transcripts of both genes were present in all tissues (Supplemental Fig. S2). To further investigate the expression patterns of these two genes, we observed transgenic plants carrying ProAtTIM9::GUS and ProAtTIM10::GUS constructs and found that the two genes shared the same expression patterns (Supplemental Materials and Methods S1). In 10-d-old seedlings, GUS activity accumulated mainly in the tissues with actively dividing cells (Supplemental Fig. S3, A–C and G–I). During reproductive growth, GUS signals were evident in inflorescences (Supplemental Fig. S3, D and J), mature flowers (Supplemental Fig. S3, E and K), as well as embryos and endosperm at different stages (Supplemental Fig. S3, F and L, arrows and arrowheads), while they were not observed in the anthers or ovaries, hinting that loss of either AtTIM9 or AtTIM10 function would not result in gametophyte defects.
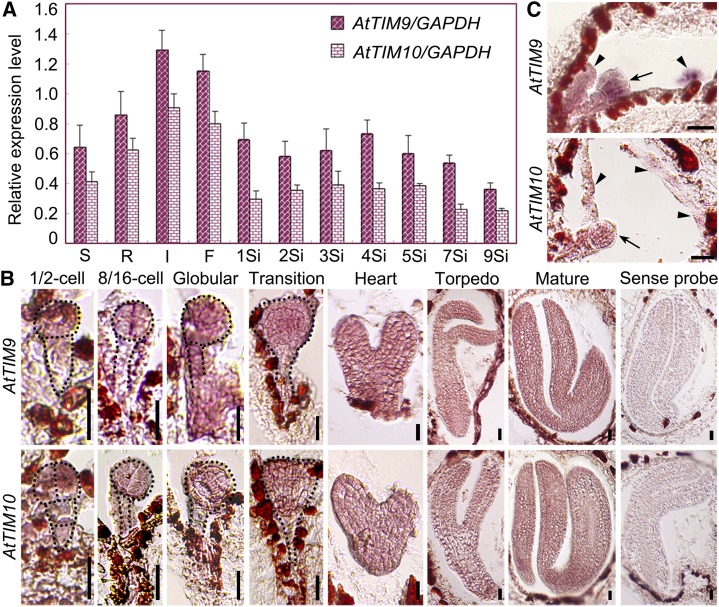
By using quantitative reverse transcription-PCR assays, we also observed that AtTIM9 and AtTIM10 transcripts were widely expressed in Arabidopsis tissues, including in siliques at different developmental stages (Fig. 3A). Since the mutant phenotypes were apparently associated with embryos and endosperm, we performed RNA in situ hybridization for wild-type ovules. AtTIM9 and AtTIM10 were detected both in the embryos and endosperm-free nuclei at different stages (Fig. 3, B and C, arrows and arrowheads). Thus, the expression of AtTIM9 and AtTIM10 is consistent with its functional relevance for early sporophyte, including embryo and endosperm development.
Figure 3.
Expression analysis of AtTIM9 and AtTIM10 genes at the transcriptional level. A, AtTIM9 and AtTIM10 temporal and spatial transcript levels as determined by quantitative real-time reverse transcription-PCR in wild-type plants. S, Seedlings; R, roots; I, inflorescences; F, flowers; 1Si to 9Si, siliques at 1 to 5 and 7, and 9 DAP. B and C, In situ hybridization of AtTIM9 and AtTIM10 transcripts (brown-red staining) accumulated in wild-type embryos and endosperm. Arrows and arrowheads show the embryos and endosperm in the ovules, respectively. Bars = 20 μm. [See online article for color version of this figure.]
AtTIM9 and AtTIM10 Proteins Are Highly Conserved and Localized in Mitochondria
In order to investigate the homology of AtTIM9 and AtTIM10 proteins, we performed alignments of the amino acid sequences (Supplemental Fig. S4) and modeled three-dimensional structures (Supplemental Fig. S5; Supplemental Materials and Methods S1) among TIM9 and TIM10 eukaryotic homologs. The results showed that both AtTIM9 and AtTIM10 have high homology at both the primary and secondary structure levels. Although the amino acid sequence identity between AtTIM9 and AtTIM10 is only 21.28% (Supplemental Fig. S4C), their predicted secondary structures are similar (Supplemental Fig. S5C), supporting the notion that TIM9 and TIM10 may have similar functions during embryonic development in Arabidopsis.
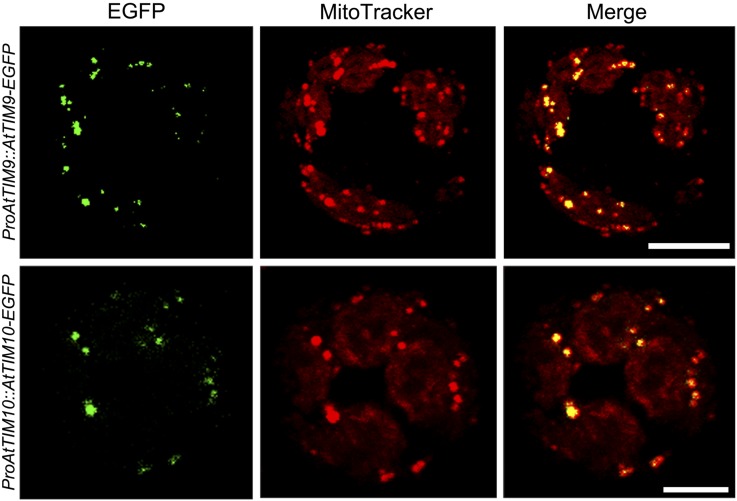
To further study the subcellular localization status of these proteins, transgenic lines were generated to express AtTIM9 or AtTIM10 in fusion with enhanced GFP (EGFP) at the N terminus under the control of their own promoters. The accumulation of AtTIM9-EGFP and AtTIM10-EGFP fusion proteins in stably transformed plantlets was verified (Supplemental Fig. S6), suggesting that the fusion proteins were present. Mesophyll protoplasts isolated from the transformants were examined in order to observe subcellular localization signals in living cells (Fig. 4). The results showed that protoplasts containing AtTIM9-EGFP and AtTIM10-EGFP both displayed dotted green fluorescence signals (Fig. 4, left), which overlapped with the mitochondrial indicator MitoTracker (Fig. 4, middle and right). These results confirmed the localization of AtTIM9-EGFP and AtTIM10-EGFP in mitochondria.
Figure 4.
Subcellular localization of AtTIM9 and AtTIM10. Either AtTIM9-GFP or AtTIM10-GFP is colocalized with mitochondria in Arabidopsis protoplasts. Bars = 10 μm. [See online article for color version of this figure.]
AtTIM9 and AtTIM10 Can Directly Interact with Each Other in Arabidopsis
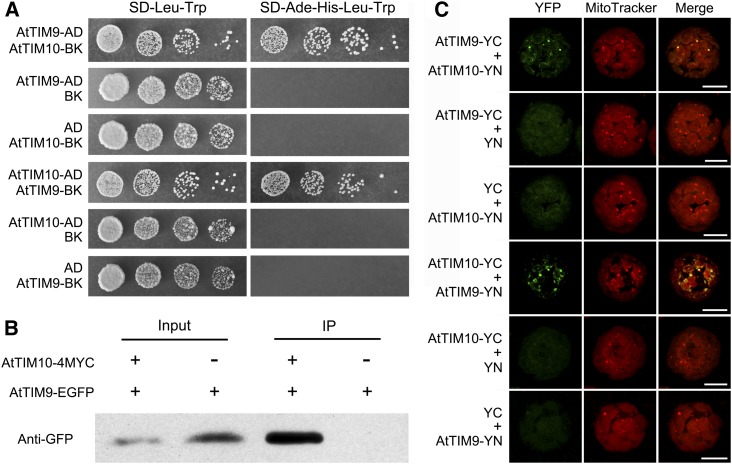
Initial confocal scanning of mesophyll protoplasts revealed that AtTIM9 and AtTIM10 are colocalized in the mitochondria of plants stably cotransformed with AtTIM9-EGFP and AtTIM10-enhanced cyan fluorescent protein (ECFP; Supplemental Fig. S7). This implied that AtTIM9 and AtTIM10 may possibly interact in some way. To investigate their interaction, we used yeast two-hybrid assays. Yeast cells that coexpressed the AtTIM10 bait and the AtTIM9 prey, or the AtTIM9 bait and the AtTIM10 prey, were allowed to grow on defective medium synthetic dextrose-adenine-His-Leu-Trp, showing that AtTIM9 and AtTIM10 strongly interact with each other on high-stringency selection plates (Fig. 5A). To confirm the interaction between AtTIM9 and AtTIM10 in vivo, we conducted a coimmunoprecipitation analysis. When the AtTIM9-EGFP and AtTIM10-4MYC fusion proteins were transiently coexpressed in Nicotiana benthamiana leaf cells, the AtTIM9-EGFP fusion protein could be coimmunoprecipitated with AtTIM10-4MYC (Fig. 5B). We further performed a bimolecular fluorescence complementation (BiFC) assay to verify their interaction in living plant cells. The results showed that the yellow fluorescent protein (YFP) fluorescence signal could be observed only in transformed Arabidopsis protoplasts that coexpressed the constructs AtTIM9-YC (aa156-239) and AtTIM10-YN (aa1-155) or AtTIM10-YC and AtTIM9-YN (Fig. 5C). Moreover, the YFP fluorescence signal accumulated exclusively in the mitochondria, reconfirming the subcellular localization of AtTIM9 and AtTIM10. Together, these results suggested that AtTIM9 and AtTIM10 may form a TIM9:10 complex and nonredundantly execute their functions in Arabidopsis, while lack of either of the subunits may lead to early embryo and endosperm lethality.
Figure 5.
Interaction between AtTIM9 and AtTIM10 proteins. A, Yeast two-hybrid assay showing interaction between AtTIM9 and AtTIM10. The 10-fold serial dilutions were plated onto lacking media. AD, pGADT7 vector; BK, pGBKT7 vector; Ade, adenine; SD, synthetic dextrose. B, Coimmunoprecipitation (IP) assay showing the interaction between AtTIM9 and AtTIM10 in N. benthamiana leaves in vivo. C, BiFC assay showing the interaction between AtTIM9 and AtTIM10 directly in Arabidopsis living protoplasts. YC, YFP C-terminal fragment (aa156-239); YN, YFP N-terminal fragment (aa1-155). Bars = 10 μm. [See online article for color version of this figure.]
Mutations of AtTIM9 and AtTIM10 Deleteriously Affect Cell Viability, Mitochondrial Activity, and Mitochondrial Structure in the Embryo Proper Cells and Surrounding Endosperm of Embryos at the 16/32-Cell Stage
To study how the lack of AtTIM9 or AtTIM10 affected early sporophytic cell division, we carefully isolated homozygous embryos from within aborted white ovules and examined the lethal embryos of tim9-1 and tim10 mutants. Double staining with fluorescein diacetate (FDA) and propidium iodide (PI) showed that all cells in the embryos maintained cell viability until the eight-cell stage (Fig. 6, A–E, G, and H). In tim9-1 and tim10 embryos, PI signals were shown in the nucleus of embryo proper cells at the 16/32-cell stage (Fig. 6, F and I), implying that cells in this region had died by this stage. Using MitoTracker as a marker, we detected active mitochondria throughout wild-type embryos, including in the embryo proper and in the suspensor cells (Fig. 6, J–L, arrows). Intriguingly, the active mitochondria of tim9-1 and tim10 homozygous embryos were distributed in the same manner as in wild-type embryos until the eight-cell stage (Fig. 6, M, N, P, and Q, arrows). However, mitochondria were not evident in embryo proper cells at the 16/32-cell stage (Fig. 6, O and R, asterisks), indicating that mitochondria were not viable in the embryo proper cells of aborted embryos.
Figure 6.
Cellular and mitochondrial viability in Arabidopsis embryos of the wild type, tim9-1, and tim10. A to I, Cell viability in the embryos of the wild type, tim9-1, and tim10 at different stages. Bars = 20 μm. J to R, Mitochondrial activity in the embryos of the wild type, tim9-1, and tim10 at different stages. Each plate consists of two group images: MitoTracker Red fluorescence images (left) and corresponding DIC images merged with the red fluorescence (right). Dotted lines in the left images divide the embryo into embryo proper (EP) and suspensor (Su) parts. Arrows show the active mitochondria in the embryos; asterisks indicate that no active mitochondria were found in the embryo proper cells. Bars = 20 μm.
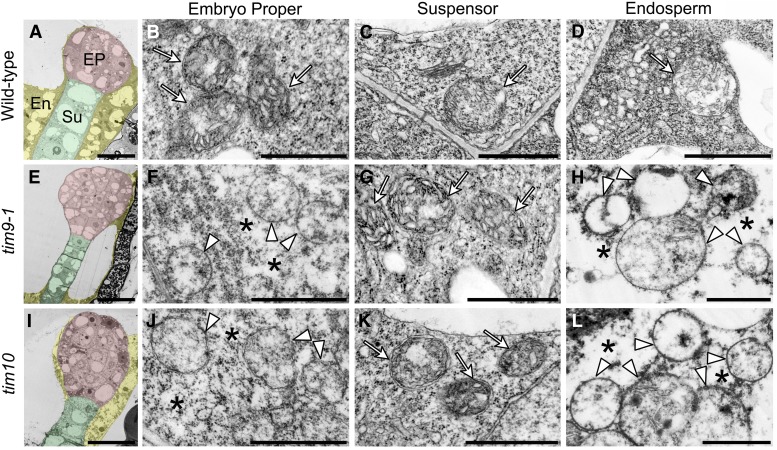
Furthermore, we analyzed the ultrastructure of mitochondria using transmission electron microscopy. The results showed that healthy mitochondria with abundant cristae were distributed everywhere in wild-type embryo cells and endosperm (Fig. 7, B–D, arrows), while embryo proper cells and syncytial endosperm in the tim9-1 and tim10 mutants contained swollen mitochondria with less intramitochondrial content and internal space lacking any obvious cristae structures (Fig. 7, F, H, J, and L, arrowheads). Also, we noticed that the cells with abnormal mitochondria showed shrunken cytoplasm, with the density of cytoplasmic materials becoming thin (Fig. 7, F, H, J, and L, asterisks) compared with the thick cytoplasm in normal cells (Fig. 7, B–D, G, and K). All of these features were consistent with other examples of necrotic cell death, which is a typical kind of PCD in plants (van Doorn et al., 2011). Consequently, the absence of AtTIM9 or AtTIM10 could lose the mitochondrial activity and destroy the mitochondrial structure, to influence cell viability in early embryo proper and endosperm.
Figure 7.
Mitochondrial structure in early embryo cells and endosperm of the wild type, tim9-1, and tim10. A, E, and I, Ultrastructure of the early embryo cells at the 16/32-cell stage from the wild type, tim9-1, and tim10. The embryo proper (EP), suspensor (Su), and endosperm (En) tissues are highlighted with red, green, and yellow, respectively. Bars = 20 μm. B to D, F to H, and J and L, Ultrastructural mitochondria of the embryo proper cells, suspensor cells, and endosperm-free nucleus in the wild type, tim9-1, and tim10. Arrows and arrowheads show the integral and degraded mitochondria, respectively; asterisks indicate the shrunken cytoplasm. Bars = 1 μm. [See online article for color version of this figure.]
Mitochondrial Membrane Permeability Increases in the Embryo Proper Cells of the tim9-1 and tim10 Mutants
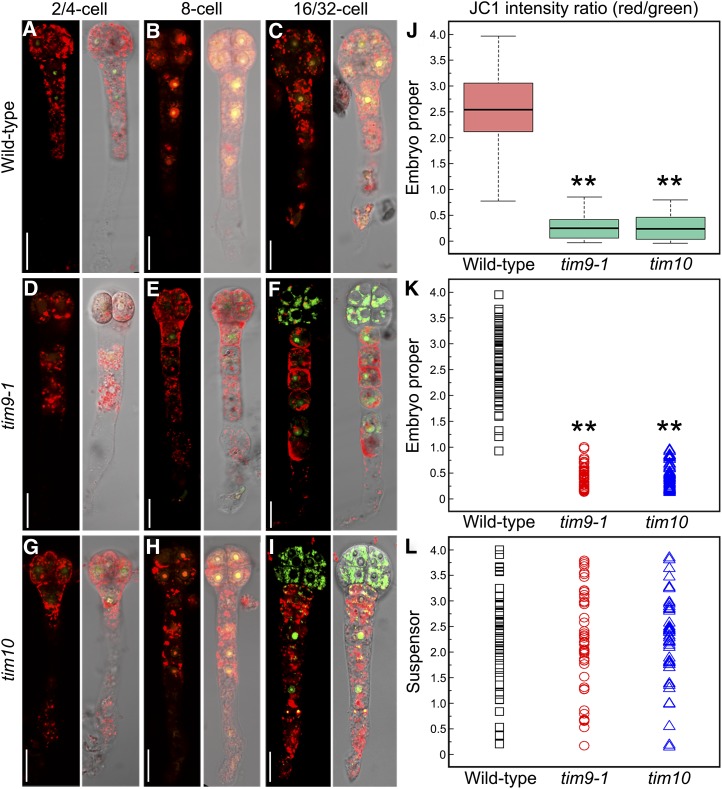
The abnormal mitochondrial activity and structure found in early embryo proper and endosperm may be related to mitochondrial dysfunction in tim9-1 and tim10. To explore this possibility, we investigated mitochondrial membrane permeability (MMP) level in tim9-1 and tim10 embryos by using the indicator 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1). Normally, JC-1 monomers could aggregate in healthy mitochondria with low membrane permeability and exhibit red fluorescence. When the membrane permeability increases in abnormal mitochondria, JC-1 monomers remain unaggregated and show green fluorescence (Zamzami et al., 1995; Martin et al., 2013). Our results showed that the fluorescence signals were more visibly red than green in embryo proper cells at the one- to eight-cell stages in the wild type, tim9-1, and tim10 (Fig. 8, A, B, D, E, G, and H). However, when the embryo proper grew to 16 to 32 cells, the green fluorescence signal was much stronger in tim9-1 and tim10 than in the wild type (Fig. 8, F and I). Quantitative analysis of red-to-green fluorescence ratios in embryo proper cells showed a wider range of dispersion in the wild type than in tim9-1 and tim10 (Fig. 8, J and K), indicating that the MMP level of the mutants was indeed increased in embryo proper samples at the 16/32-cell stage. In contrast, there were few differences in suspensor cells between the wild type and mutants (Fig. 8L). These results showed that the tim9-1 and tim10 mutants contained abnormal mitochondria with increased MMP levels in the 16/32-cell embryo proper.
Figure 8.
MMP in the embryos of the wild type, tim9-1, and tim10. A to I, MMP levels in the embryos of the wild type, tim9-1, and tim10 at different stages. Each plate consists of two group images: JC-1 aggregate red fluorescence merged with JC-1 monomer green fluorescence (left) and the red and green fluorescence merged with the DIC microscopy channel (right). Bars = 20 μm. J to L, The red/green JC-1 fluorescence ratios measured in wild-type (n = 56), tim9-1 (n = 50), and tim10 (n = 47) embryos at the 16/32-cell stage. Asterisks indicate statistically significant differences compared with the wild type (**P < 0.01).
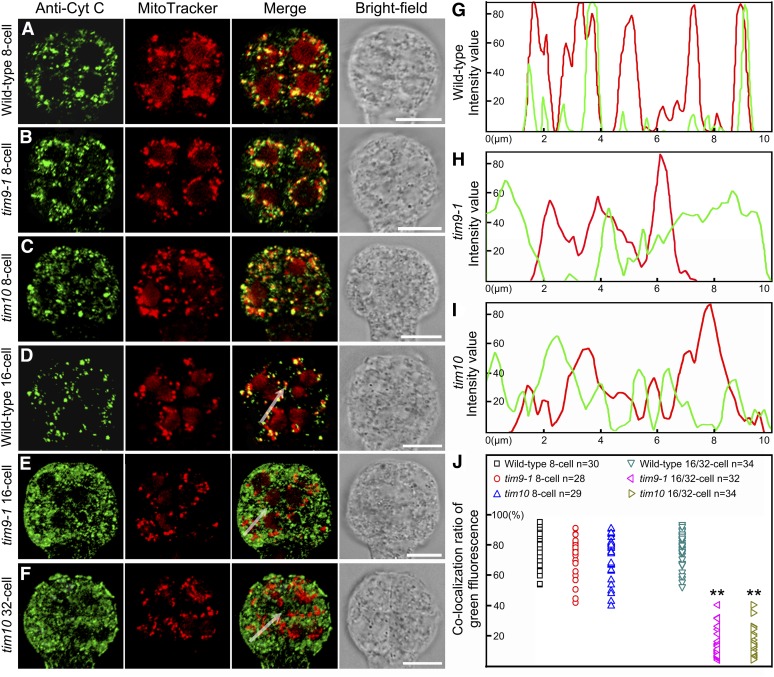
Cytochrome c Is Released from Mitochondria into the Cytoplasm in the Embryo Proper Cells of the tim9-1 and tim10 Mutants
The fact that 16/32-cell tim9-1 and tim10 embryo proper cells have increased MMP levels could conceivably lead to the release of cytochrome c (Cyt c) from mitochondria to the cytoplasm. The release of Cyt c at the onset of PCD is an evolutionarily conserved event appearing in organisms including plants and mammals (Martínez-Fábregas et al., 2013). To investigate whether this process occurred in the lethal cells of the tim9-1 and tim10 mutants, we used antibodies against Cyt c to detect its release in the embryo proper using immunofluorescence microscopy. At the eight-cell stage, most Cyt c was located in the mitochondria in both the wild type and mutants (Fig. 9, A–C). When tim9-1 and tim10 embryo proper reached the 16/32-cell stage, the green fluorescence signals of Cyt c observed in the cytoplasm hardly overlapped with the mitochondria director (Fig. 9, E and F), implying that Cyt c was released at this stage. Additional analysis of the fluorescence signal intensities produced along a randomly selected series of pixels revealed a correlation between Cyt c and mitochondria (Fig. 9, G–I). Further analysis of the ratio of overlapping green fluorescence also demonstrated that, at the 16/32-cell stage, most of the Cyt c was not colocalized with mitochondria but rather was released into the cytoplasm in the tim9-1 and tim10 mutants (Fig. 9J). These results suggest that the necrosis-like PCD was probably initiated in the embryo proper cells of the tim9-1 and tim10 mutants.
Figure 9.
Immunolocalization of Cyt c in the embryo proper of the wild type, tim9-1, and tim10. A to F, Localization of Cyt c in the embryo proper of the wild type, tim9-1, and tim10 at the eight- to 32-cell stages. Each plate consists of four group images from left to right: anti-Cyt c green fluorescence, MitoTracker Red fluorescence, green fluorescence merged with red fluorescence, and bright field. Bars = 10 μm. G to I, Intensity values of a plotted line by scanning through the merged regions in Figure 10, D to F. J, Overlapping ratio of green fluorescence measured in the embryo proper of the wild type, tim9-1, and tim10. Asterisks indicate statistically significant differences compared with the wild type (**P < 0.01).
PCD Occurs in Early Embryo Proper Cells and Endosperm-Free Nuclei of tim9-1 and tim10 Mutants
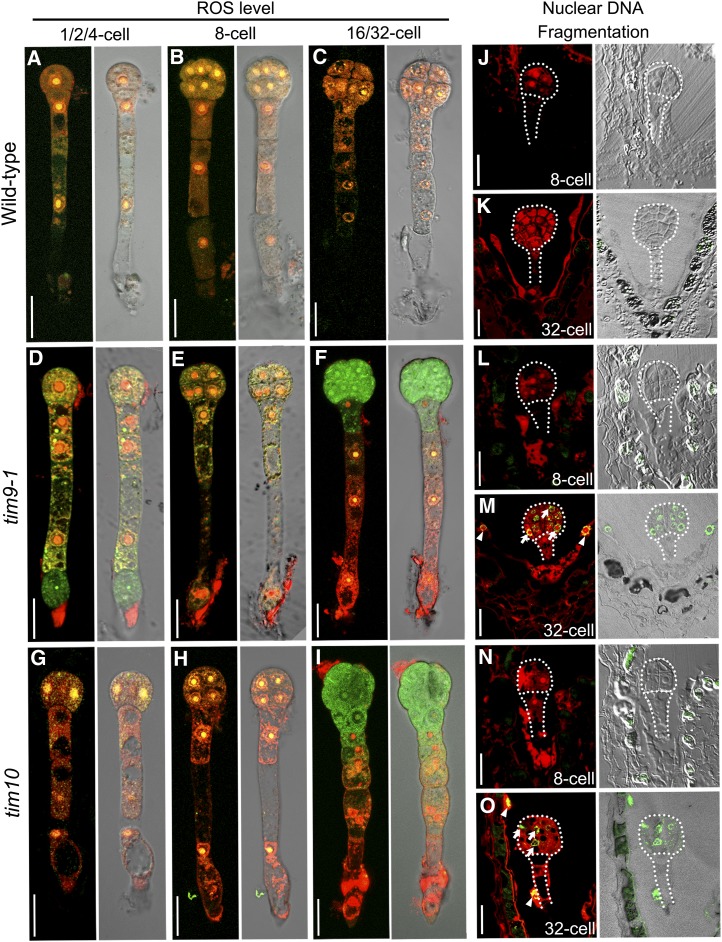
To explore whether the changes of the mitochondria in the tim9-1 and tim10 mutants caused the PCD event to happen, we used the fluorescent probes H2DCFDA (for 5-[and -6]-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester) and MitoSOX Red to selectively indicate reactive oxygen species (ROS) in the cytosol and mitochondria, respectively (Robinson et al., 2006; Bi et al., 2009). ROS are produced by mitochondria and can be involved in PCD, and the ROS levels in cytosol and mitochondria are normally considered to be an important indicator for PCD process (Fleury et al., 2002; Van Breusegem and Dat, 2006). In this assay, we found that ROS levels in tim9-1 and tim10 embryos before the eight-cell stage were similar to those in the wild type. However, when the embryo proper grew to the 16/32-cell stage, compared with the wild type (Fig. 10C), the ROS level in cytosol (green fluorescence) became higher in tim9-1 and tim10, while the mitochondrial ROS (red fluorescence) almost disappeared (Fig. 10, F and I). These results indicated that in tim9-1 and tim10 16/32-cell embryo proper cells, ROS as toxic compounds were not detoxified by the cytoplasm and accumulated in the cytosol; mitochondria also stopped producing the ROS source. These changes caused the ROS level balance between cytosol and mitochondria to be broken, which displayed a classical PCD feature in the aborted embryo proper of the two mutants.
Figure 10.
PCD occurs in early embryos and endosperm of the wild type, tim9-1, and tim10. A to I, ROS levels in the embryos at different stages. Each plate consists of two group images: cellular ROS green fluorescence merged with mitochondrial ROS red fluorescence (left) and the red and green fluorescence merged with the DIC microscopy channel (right). Bars = 20 μm. J to O, TUNEL-positive signal is indicated by the green fluorescence of fluorescein, and nuclei fluoresce deep red using the counterstain PI. Each plate consists of two group images: TUNEL-positive green fluorescence merged with DIC microscopy (left) and TUNEL-positive green fluorescence merged with nuclei red fluorescence (right). Arrows and arrowheads show the TUNEL signals in the embryonic nucleus and endosperm-free nuclei, respectively. Bars = 20 μm.
We further studied the PCD event under the control of the indicator terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay to test the nuclear DNA fragmentation in early sporophytic cells. Our results showed that the TUNEL signal was negative at the eight-cell stage in tim9-1 and tim10 ovules (Fig. 10, L and N). This was also the case in wild-type ovules (Fig. 10J). However, TUNEL-positive signals appeared in the tim9-1 (95%, n = 37) and tim10 (91%, n = 44) embryo proper and endosperm at the 32-cell stage (Fig. 10, H and M, arrows and arrowheads). These results indicated that nucleic acids degenerated and led to the eventual complete abortion of embryo proper cells and endosperm-free nucleus, potentially explaining the halted division and dead embryos and endosperm observed in the mutants. As such, AtTIM9 and AtTIM10 are both required for normal early sporophyte development in both embryo cells and syncytial endosperm.
DISCUSSION
AtTIM9 and AtTIM10 Are Both Essential for Maintaining Mitochondrial Function in Arabidopsis
Previous studies on yeast have reported that TIM9 and TIM10 play essential roles in mitochondrial biogenesis; the disruption of either protein causes cell lethality (Jarosch et al., 1997; Koehler et al., 1998b). In humans, among the Tim10/deafness/dystonia peptide protein family, only HmTIM8a has been investigated to be necessary during human neurological development to mediate the mitochondrial disease, deafness/dystonia syndrome (Tranebjaerg et al., 2001; Roesch et al., 2002), while the functions of human TIM9 and TIM10 remain unknown. Here, we found that mutants of AtTIM9 and AtTIM10 led to severe mitochondrial defects in early sporophytes, their encoding proteins AtTIM9 and AtTIM10 located in mitochondria, and play nonredundantly important roles in the division of early embryo cells and endosperm-free nuclei.,
Embryo cells contain a large number of mitochondria during development that enable a cell’s vigorous division and growth (Law et al., 2012). However, we found that mitochondria in the embryo proper and endosperm were not active and that mitochondrial cristae structure was lost when embryo arrest took place in the homozygous tim9-1 and tim10 mutants (Figs. 6 and 7). This destruction of mitochondrial structure could lead to various mitochondrial dysfunctions. As an example, ATP synthesis (oxidative phosphorylation) is critically dependent on the structural integrity of the mitochondrion (Sherratt, 1991; Frey and Mannella, 2000). Furthermore, we found the dysfunctional mitochondria increased MMP and released Cyt c into the cytoplasm in the mutant embryo proper cells (Figs. 8 and 9), suggesting that this is probably caused by the disruption of the mitochondrial import pathway in the absence of AtTIM9 or AtTIM10 (Lister et al., 2004). The TIM9:10 complex was reported to participate in the import of many important mitochondrial protein precursors, including ADP/ATP carrier in yeast and adenine nucleotide translocator in potato (Lister et al., 2002; Truscott et al., 2002). When TIM9 or TIM10 function is impaired, there may be an absence of ADP/ATP carrier proteins and a decrease of ATP level, which could abnormally increase MMP and release Cyt c (Pereira et al., 2007; Hasson et al., 2010). Accordingly, AtTIM9 and AtTIM10 play very important roles in maintaining mitochondrial function to control cell viability and division during early embryo and endosperm development.
The Early Sporophytic Abortion of the tim9-1 and tim10 Mutants through the Necrosis-Like PCD Pathway
A number of genes encoding proteins located in the mitochondria have been found to be essential for early embryo development in Arabidopsis; mutants of these genes show the phenotype of embryo arrest at the early globular stage (Okada et al., 2004; Berg et al., 2005; Vazzola et al., 2007; Jiang et al., 2012; Yu et al., 2012), indicating that their proteins involved in mitochondrial basal metabolic functions play many important roles during seed formation. However, the mechanism of death during early embryo lethality and how mitochondrial proteins are involved in the developmental process have been little studied. Therefore, in this study, we revealed that mutations of the AtTIM9 and AtTIM10 genes caused early embryo proper cell and syncytial endosperm lethality through the necrosis-like PCD pathway.
To date, there have only been a few reports about the relationship between mitochondrial import proteins and PCD. The small molecule MitoBloCK-6 was recently found to inhibit the import of TIM proteins and is further known to induce apoptosis via Cyt c release in human embryonic stem cells (Dabir et al., 2013). The mitochondrial import machinery also has been proven to contribute to the naturally occurring PCD during human pathologies (Sokol et al., 2014). In higher plants, PCD is an integral part during development and defense of the whole life, mainly including two distinguished classes: vacuolar cell death and necrosis (van Doorn et al., 2011). Vacuolar cell death in plants is also known as autophagy in animals and commonly happens during normal tissue and organ formation and elimination (Edinger and Thompson, 2004), whereas necrosis, as a programmed event, is typically an acute cell death response and always occurs under abiotic stresses (van Doorn and Woltering, 2005). To date, there are no molecular markers for the necrosis-like PCD, and it remains to be defined at the physiological level (van Doorn et al., 2011). Our evidence from transmission electron microscopy, ROS levels, and TUNEL signals indicated that when tim9-1 and tim10 embryos developed to the 16/32-cell stage, embryo proper cells and endosperm-free nuclei all display the features of necrosis-like PCD (Figs. 6, 9, and 10). During the PCD in plants, it has been reported that mitochondria play a central role (Jones, 2000; Vianello et al., 2007). Hence, the changes of MMP and Cyt c localization that we found in the lethal embryo proper of tim9-1 and tim10 (Figs. 7 and 8) confirmed that mitochondria significantly functioned in the necrosis-like PCD pathways in Arabidopsis.
Mitochondrial Dysfunction in the Early Sporophyte of tim9-1 and tim10 Mutants Is Only Found in Embryo Proper and Syncytial Endosperm, But Not in Suspensor Cells
Arabidopsis embryogenesis starts from the asymmetrical division of the zygote to form two cells of different sizes. The larger basal cell develops into a stalk-like support structure of six to eight cells called the suspensor, and the smaller apical cell is the founder of the embryo proper that contributes to most of the later seedling (Yeung and Meinke, 1993). In this study, mutations of AtTIM9 and AtTIM10 displayed the embryonic abortion phenotype, but with the well-developed suspensor, the lengths and cell numbers were similar to the wild type (Fig. 2). Furthermore, we found that the features of abnormal mitochondria and necrosis-like PCD only appeared in the mutant embryo proper cells and syncytial endosperm, while the suspensor cells remained normal (Figs. 6–10). As a result, we conclude that the mitochondrial dysfunction induced by the absence of AtTIM9 and AtTIM10 function only occurs in the embryo proper and endosperm, but not in suspensor cells.
Most flowering plants studied to date including Arabidopsis exhibit maternal mitochondrial inheritance (Mogensen, 1996; Nagata, 2010). The volume and density of mitochondria in zygote increase significantly as compared with the egg cell stage. This is followed by elongation of the zygote, which accumulates mitochondria in its apical region and subsequently starts asymmetrical division (Mansfield and Briarty, 1991; Russell, 1993). These findings indicate that mitochondria are required for the initial divisions of embryonic cells, and the ones that come from the female parent may play important roles during early sporophyte initiation and development. Our results showed that mitochondria in tim9-1 and tim10 mutants remained normal throughout the early sporophyte development stages until the eight-cell stage (Figs. 6–10), implying that the embryo proper and suspensor cells could divide into eight cells normally and that the endosperm-free nuclei also divide normally, with each region being sustained by their respective mitochondria. After the eight-cell stage in the tim9-1 and tim10 mutants, the suspensor developed completely to eight cells and stopped dividing (Mansfield and Briarty, 1991); therefore, the presence of active mitochondria in the suspensor cells of mutant embryos at the stage of arrest reflects the fact that division of the suspensor is completed first. On the other hand, the embryo proper and endosperm need to keep dividing; however, the dysfunctional mitochondria may no longer support natural cell growth and division and subsequently lead these early sporophytic cells to undergo the necrosis-like PCD.
In conclusion, AtTIM9 and AtTIM10 form a complex and function nonredundantly in early embryo and endosperm development in Arabidopsis. Our research into the functions of these two mitochondrial proteins opens a new horizon that the import proteins perform important roles in the maintenance of mitochondrial function in plants, thereby providing novel insight into the functions of import proteins in controlling the fundamental and essential process of cell division in early embryo proper cells and in endosperm-free nuclei.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used as the wild type in this study. The T-DNA insertion mutant lines were obtained from the Arabidopsis Biological Resource Center and the Nottingham Arabidopsis Stock Centre: CS16114 (tim9-1/+; also known as emb2474), CS442853 (tim9-2/+), and CS415161 (tim10/+). The double mutant tim9-1/+tim10/+ was generated by crossing tim9-1/+ and tim10/+. For the genetics experiment, seeds were surface sterilized and germinated on Murashige and Skoog medium with correspondent antibiotics for 7 to 10 d, then transplanted and grown on soil. All plants were grown in a growth chamber at Wuhan University at 22°C ± 2°C with a 16-h-light/8-h-dark photoperiod.
Mutant Verification, Complementation Analysis, and Phenotype Observation
Heterozygous lines were isolated by using a PCR screening approach with AtTIM9- and AtTIM10-specific primers and T-DNA primer (Supplemental Table S2), and the insertion positions were confirmed by sequencing with the T-DNA primer.
For complementation, the AtTIM9 and AtTIM10 full-length genomic fragments were amplified with KOD-Plus DNA polymerase (Toyobo) from wild-type Arabidopsis and cloned into pCambia1300 vector (Cambia), introduced into tim9-1/+, tim9-2/+, and tim10-1/+ heterozygote mutants via Agrobacterium tumefaciens (GV3101), and then transformed using the floral dip method. Primers used in the experiments are listed in Supplemental Table S2.
Ovules were isolated using two needles and cleared with chloral hydrate following the protocol described by Yadegari et al. (1994). Specimens were observed using the Olympus IX71 microscope under differential interference contrast (DIC) optics and photographed by a SPOT Xplorer Camera (Diagnostic).
Quantitative Real-Time PCR Assay
Quantitative real-time PCR was carried out using SYBR Green fluorescence and a Rotor-Gene Q real-time PCR machine (Qiagen). The expression of AtTIM9 and AtTIM10 in various RNA samples was normalized to the expression of an internal gene, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (Zhong and Simons, 1999). The relative expression levels were analyzed as described previously (Ma and Zhao, 2010). Three independent biological replicates and three technical replicates of each sample were made for quantitative PCR analysis. Primers used in the experiments are listed in Supplemental Table S2.
RNA in Situ Hybridization
The antisense and sense probes used in the experiments were all generated by PCR amplification with T7 promoters adding primers (Supplemental Table S2) and followed by in vitro transcription (DIG RNA Labeling Kit; Roche). Ovules at different developmental stages from wild-type Arabidopsis siliques were isolated. Fixating, embedding, and sectioning of ovules and procedures of RNA in situ hybridization were performed as described previously (Brewer et al., 2006). The section images were observed with the Olympus BX60 microscope and photographed by the Olympus DP72 CCD camera.
Subcellular Localizations
Western blotting of AtTIM9-EGFP and AtTIM10-EGFP fusion proteins was performed as described before (Liu et al., 2013), and detection was with anti-GFP rabbit antibody (1:2,000; Abclonal). Fluorescence was observed in Arabidopsis protoplasts. Mesophyll protoplasts isolated from the 4-week-old transformants were incubated with 2 mm MitoTracker Red FM (Molecular Probes) for 10 min and examined with a confocal microscope (Fluoview1000; Olympus). The images were taken under EGFP fluorescence (excitation, 488 nm; emission, 505–530 nm), the MitoTracker channel (excitation, 559 nm; emission, 530–560 nm), and ECFP fluorescence (excitation, 440 nm; emission, 505–530 nm). The merged images were generated using the Image Browser Fluoview version 2.7b software (Olympus).
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed according to the manufacturer’s instructions for the Matchmaker GAL4-based two-hybrid system 3 (Clontech). The full-length AtTIM9 open reading frame was subcloned into the pGADT7 or pGBKT7 vector and then transformed into yeast (Saccharomyces cerevisiae) strain AH109 by the lithium acetate method. The transformed colonies were plated onto the minimal synthetic dextrose base medium -Leu/-Trp or -Leu/-Trp/-His/-adenine (Clontech) at 30°C for 2 to 4 d to test for possible interactions. Positive yeast cells were grown to an optical density at 600 nm of 1, and then 5 μL of 10-fold serial dilutions of yeast cells was plated onto the two kinds of lacking media mentioned above. Primers used in this test are listed in Supplemental Table S2.
Coimmunoprecipitation Analysis
35S::AtTIM9-EGFP (pCambia1300) and 35S::AtTIM10-4MYC (pCambia2301) constructs were introduced into A. tumefaciens strain GV3101 and cotransformed into 6-week-old Nicotiana benthamiana leaves simultaneously using the agroinfiltration-mediated infiltration method. The proteins were extracted in the homogenization buffer adding 0.5 mm phenylmethylsulfonyl fluoride (Sigma-Aldrich) and incubated with 2 μg of anti-MYC mouse antibody (Abclonal) at 4°C for 4 h. After 10 μL of protein A sefinose (Bio Basic Inc.) was added and incubated at 4°C for 2 h, the beads were centrifuged and washed. The proteins were eluted with 40 μL of loading buffer and analyzed by immunoblotting using anti-GFP rabbit antibody (1:2,000; Abclonal). Primers used in this experiment are listed in Supplemental Table S2.
BiFC Assay
The full-length open reading frames of AtTIM9 and AtTIM10 were amplified and fused into the vectors pUC-SPYNE and pUC-SPYCE (Walter et al., 2004) to produce the C-terminal fusions AtTIM9-YFP1-174 aa (for amino acid), AtTIM10-YFP1-174 aa, AtTIM9-YFP-175-end aa, and AtTIM10-YFP-175-end aa. Mesophyll protoplasts isolated from Arabidopsis leaves were transformed with column-purified plasmid DNA (Yoo et al., 2007). After 12 h of incubation at 22°C in the dark, protoplasts were incubated with MitoTracker staining and analyzed and checked for the presence of fluorescence with confocal microscopy under the YFP fluorescence channel (excitation, 515 nm; emission, 505–530 nm) and the MitoTracker channel. Primers used in this experiment are listed in Supplemental Table S2.
Cell Viability, Mitochondrial Activity, and ROS Level Analysis
Isolation of Arabidopsis early embryos was performed according to method described by Hu et al. (2006). Early embryos isolated from the aborted tim9-1/+ and tim10/+ ovules were judged as homozygote, and their genotypes were defined as tim9-1 and tim10. All the isolated living early embryos were stained by 5 μg mL−1 FDA (Sigma) and 10 μg mL−1 PI (Sigma) mixed dyes to observe the activity of cells (for FDA: excitation, 488 nm; emission, 515–545; for PI: excitation, 559 nm; emission, 505–530 nm); by MitoTracker dye in 10% (w/v) mannitol to observe mitochondrial activity; and by 10 μm H2DCFDA (Molecular Probes) and 5 μm MitoSOX Red (Molecular Probes) mixed dyes to observe the ROS levels in cells and mitochondria, respectively (for H2DCFDA: excitation, 488 nm; emission, 515–545; for MitoSOX Red: excitation, 559 nm; emission, 505–530 nm). All images were obtained under confocal microscopy as described above.
Transmission Electron Microscopy
The method for ultrathin sections was carried out as described by Chen et al. (2012). Ovules (2 DAP) in wild-type siliques and aborted (white) ovules in tim9-1/+ and tim10/+ siliques were isolated for fixation and embedding, respectively. The ultrathin sections (60 nm) were cut with an RMC-MTX (Elexience) ultramicrotome. After staining with uranyl acetate and lead citrate, the samples were examined and photographed with a transmission electron microscope (Hitachi HT7700).
MMP Analysis
Isolated early embryos from wild-type ovules (2 DAP) and mutant white ovules were incubated in 10% (w/v) mannitol with 10 mg mL−1 JC-1 (Molecular Probes) for 30 min, then washed with 10% (w/v) mannitol, and their images were collected using the confocal microscope mentioned above. JC-1 aggregates were detected with red fluorescence (excitation, 559 nm; emission, 575–590 nm), and JC-1 monomers were detected with green fluorescence (excitation, 488 nm; emission, 515–545). The ratio of red to green fluorescence in JC-1 images was calculated by the Browser Fluoview version 2.7b software using R software (http://www.r-project.org/) for the box plot and Origin software (http://www.originlab.com/) for the dispersion graphs.
Fluorescence Immunolocalization Assay
For detecting Cyt c in the embryo proper, fluorescence immunolocalization was carried out as described previously (Zhang et al., 2008), analyzed using anti-Cyt c rabbit antibody (1:1,000; Agrisera), and observed using confocal microscopy as described above under the fluorescein isothiocyanate green fluorescence channel (excitation, 488 nm; emission, 505–530 nm) and the MitoTracker channel.
TUNEL Assay
Ovules in wild-type siliques (2 DAP) and aborted white ovules in tim9-1/+ and tim10/+ siliques were fixed in fixative (4% [w/v] paraformaldehyde) adding 0.1% (v/v) Triton X-100 on ice, embedded in the LR White (London Resin), and polymerized in 50°C for 24 h. Semithin sections of about 1 µm were cut using the MTX ultramicrotome (Elexience). The TUNEL assay was performed using the DeadEnd Fluorometric TUNEL system (Promega) according to the manufacturer’s instructions. Slides were immediately stained with 10 μg mL−1 PI and mounted with SlowFade Gold antifade reagent (Invitrogen). Samples were analyzed under confocal microscopy as described above, observing fluorescein isothiocyanate green fluorescence to find fragmented DNA signals and PI fluorescence to view the nuclear location.
Sequence data in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3g46560 (AtTIM9, also known as EMB2474; Tzafrir et al., 2004) and At2g29530 (AtTIM10).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Complementation assays of AtTIM9 and AtTIM10 mutants.
Supplemental Figure S2. Expression data of AtTIM9 and AtTIM10 from public databases.
Supplemental Figure S3. GUS staining in the ProAtTIM9::GUS and ProAtTIM10::GUS transgenic plants.
Supplemental Figure S4. AtTIM9 and AtTIM10 are conserved among eukaryotes.
Supplemental Figure S5. Homology modeling of AtTIM9 and AtTIM10.
Supplemental Figure S6. Immunoblot analysis of AtTIM9-EGFP and AtTIM10-EGFP fusion proteins.
Supplemental Figure S7. Colocalization of AtTIM9 and AtTIM10 in mitochondria.
Supplemental Table S1. Transmission of the tim9-1/+, tim9-2/+, and tim10/+ mutants.
Supplemental Table S2. Primers used in the experiments.
Supplemental Materials and Methods S1. Supplemental methods.
Supplementary Material
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre (University of Nottingham), the Arabidopsis Biological Resource Center (Ohio State University), and the SeedGene project (Meinke laboratory, Oklahoma State University) for kindly providing mutant seeds used in this study, Zhiyin Song (Wuhan University) for experimental instruction in mitochondria, Yingguo Zhu (Wuhan University) for providing BiFC vectors, and Zhiyong Gao (Wuhan University) for sharing coimmunoprecipitation methods.
Glossary
- IMS
intermembrane space
- PCD
programmed cell death
- T-DNA
transfer DNA
- DAP
days after pollination
- FDA
fluorescein diacetate
- PI
propidium iodide
- MMP
mitochondrial membrane permeability
- ROS
reactive oxygen species
- BiFC
bimolecular fluorescence complementation
- Cyt c
cytochrome c
- DIC
differential interference contrast
Footnotes
This work was supported by the National Basic Research Program of China (grant nos. 2012CB944801 and 2013CB126903) and the Key Grant Project of the Chinese Ministry of Education (grant no. 311026).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adam A, Endres M, Sirrenberg C, Lottspeich F, Neupert W, Brunner M. (1999) Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J 18: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MJ, Webb CT, Stroud DA, Palmer CS, Frazier AE, Guiard B, Chacinska A, Gulbis JM, Ryan MT. (2009) Structural and functional requirements for activity of the Tim9-Tim10 complex in mitochondrial protein import. Mol Biol Cell 20: 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer MF, Rothbauer U, Mühlenbein N, Smith RJ, Gerbitz K, Neupert W, Brunner M, Hofmann S. (1999) The mitochondrial TIM22 preprotein translocase is highly conserved throughout the eukaryotic kingdom. FEBS Lett 464: 41–47 [DOI] [PubMed] [Google Scholar]
- Berg M, Rogers R, Muralla R, Meinke D. (2005) Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J 44: 866–878 [DOI] [PubMed] [Google Scholar]
- Berger F, Grini PE, Schnittger A. (2006) Endosperm: an integrator of seed growth and development. Curr Opin Plant Biol 9: 664–670 [DOI] [PubMed] [Google Scholar]
- Bi Y, Chen W, Zhang W, Zhou Q, Yun L, Xing D. (2009) Production of reactive oxygen species, impairment of photosynthetic function and dynamic changes in mitochondria are early events in cadmium-induced cell death in Arabidopsis thaliana. Biol Cell 101: 629–643 [DOI] [PubMed] [Google Scholar]
- Braun HP, Schmitz UK. (1999) The protein-import apparatus of plant mitochondria. Planta 209: 267–274 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Heisler MG, Hejátko J, Friml J, Benková E. (2006) In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nat Protoc 1: 1462–1467 [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF. (2003) Mitochondrial genomes: anything goes. Trends Genet 19: 709–716 [DOI] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Whelan J. (2010) An in silico analysis of the mitochondrial protein import apparatus of plants. BMC Plant Biol 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138: 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Deng Y, Zhao J. (2012) Distribution and change patterns of free IAA, ABP 1 and PM H⁺-ATPase during ovary and ovule development of Nicotiana tabacum L. J Plant Physiol 169: 127–136 [DOI] [PubMed] [Google Scholar]
- Curran SP, Leuenberger D, Oppliger W, Koehler CM. (2002) The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J 21: 942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabir DV, Hasson SA, Setoguchi K, Johnson ME, Wongkongkathep P, Douglas CJ, Zimmerman J, Damoiseaux R, Teitell MA, Koehler CM. (2013) A small molecule inhibitor of redox-regulated protein translocation into mitochondria. Dev Cell 25: 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB. (2004) Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol 16: 663–669 [DOI] [PubMed] [Google Scholar]
- Elo A, Lyznik A, Gonzalez DO, Kachman SD, Mackenzie SA. (2003) Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 15: 1619–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury C, Mignotte B, Vayssière JL. (2002) Mitochondrial reactive oxygen species in cell death signaling. Biochimie 84: 131–141 [DOI] [PubMed] [Google Scholar]
- Frey TG, Mannella CA. (2000) The internal structure of mitochondria. Trends Biochem Sci 25: 319–324 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Schneitz K. (1998) The molecular and genetic basis of ovule and megagametophyte development. Semin Cell Dev Biol 9: 227–238 [DOI] [PubMed] [Google Scholar]
- Hamasaki H, Yoshizumi T, Takahashi N, Higuchi M, Kuromori T, Imura Y, Shimada H, Matsui M. (2012) SD3, an Arabidopsis thaliana homolog of TIM21, affects intracellular ATP levels and seedling development. Mol Plant 5: 461–471 [DOI] [PubMed] [Google Scholar]
- Hasson SA, Damoiseaux R, Glavin JD, Dabir DV, Walker SS, Koehler CM. (2010) Substrate specificity of the TIM22 mitochondrial import pathway revealed with small molecule inhibitor of protein translocation. Proc Natl Acad Sci USA 107: 9578–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SC, Belmonte MF, Harada JJ, Inoue K. (2010) Indispensable roles of plastids in Arabidopsis thaliana embryogenesis. Curr Genomics 11: 338–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Qin Y, Zhao J. (2006) Localization of an arabinogalactan protein epitope and the effects of Yariv phenylglycoside during zygotic embryo development of Arabidopsis thaliana. Protoplasma 229: 21–31 [DOI] [PubMed] [Google Scholar]
- Jarosch E, Rödel G, Schweyen RJ. (1997) A soluble 12-kDa protein of the mitochondrial intermembrane space, Mrs11p, is essential for mitochondrial biogenesis and viability of yeast cells. Mol Gen Genet 255: 157–165 [DOI] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W. (2007) Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol 23: 207–236 [DOI] [PubMed] [Google Scholar]
- Jiang L, Wang S, Li H, Zhang G, Li H. (2012) EMBRYONIC FACTOR 31 encodes a tyrosyl-tRNA synthetase that is essential for seed development. Mol Biol Rep 39: 8297–8305 [DOI] [PubMed] [Google Scholar]
- Jones A. (2000) Does the plant mitochondrion integrate cellular stress and regulate programmed cell death? Trends Plant Sci 5: 225–230 [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. (1997) The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Koehler CM, Jarosch E, Tokatlidis K, Schmid K, Schweyen RJ, Schatz G. (1998a) Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279: 369–373 [DOI] [PubMed] [Google Scholar]
- Koehler CM, Merchant S, Oppliger W, Schmid K, Jarosch E, Dolfini L, Junne T, Schatz G, Tokatlidis K. (1998b) Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J 17: 6477–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Yoshizumi T, Hongo H, Yoneda A, Hara H, Hamasaki H, Takahashi N, Nagata N, Shimada H, Matsui M. (2012) Arabidopsis mitochondrial protein TIM50 affects hypocotyl cell elongation through intracellular ATP level. Plant Sci 183: 212–217 [DOI] [PubMed] [Google Scholar]
- Law SR, Narsai R, Taylor NL, Delannoy E, Carrie C, Giraud E, Millar AH, Small I, Whelan J. (2012) Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol 158: 1610–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Chew O, Lee MN, Heazlewood JL, Clifton R, Parker KL, Millar AH, Whelan J. (2004) A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol 134: 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mowday B, Whelan J, Millar AH. (2002) Zinc-dependent intermembrane space proteins stimulate import of carrier proteins into plant mitochondria. Plant J 30: 555–566 [DOI] [PubMed] [Google Scholar]
- Lister R, Murcha MW, Whelan J. (2003) The mitochondrial protein import machinery of plants (MPIMP) database. Nucleic Acids Res 31: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Deng Y, Li G, Zhao J. (2013) Replication factor C1 (RFC1) is required for double-strand break repair during meiotic homologous recombination in Arabidopsis. Plant J 73: 154–165 [DOI] [PubMed] [Google Scholar]
- Ma H, Zhao J. (2010) Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J Exp Bot 61: 2647–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. (1991) Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69: 461–476 [Google Scholar]
- Martin MV, Fiol DF, Sundaresan V, Zabaleta EJ, Pagnussat GC. (2013) oiwa, a female gametophytic mutant impaired in a mitochondrial manganese-superoxide dismutase, reveals crucial roles for reactive oxygen species during embryo sac development and fertilization in Arabidopsis. Plant Cell 25: 1573–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Fábregas J, Díaz-Moreno I, González-Arzola K, Janocha S, Navarro JA, Hervás M, Bernhardt R, Díaz-Quintana A, De la Rosa MÁ. (2013) New Arabidopsis thaliana cytochrome c partners: a look into the elusive role of cytochrome c in programmed cell death in plants. Mol Cell Proteomics 12: 3666–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Small ID, Day DA, Whelan J. (2008) Mitochondrial biogenesis and function in Arabidopsis. The Arabidopsis Book 6: e0111, /10.1199/tab.0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen HL. (1996) The hows and whys of cytoplasmic inheritance in seed plants. Am J Bot 83: 383–404 [Google Scholar]
- Nagata N. (2010) Mechanisms for independent cytoplasmic inheritance of mitochondria and plastids in angiosperms. J Plant Res 123: 193–199 [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. (2007) Translocation of proteins into mitochondria. Annu Rev Biochem 76: 723–749 [DOI] [PubMed] [Google Scholar]
- Niklas KJ, Kutschera U. (2010) The evolution of the land plant life cycle. New Phytol 185: 27–41 [DOI] [PubMed] [Google Scholar]
- Okada K, Ohara K, Yazaki K, Nozaki K, Uchida N, Kawamukai M, Nojiri H, Yamane H. (2004) The AtPPT1 gene encoding 4-hydroxybenzoate polyprenyl diphosphate transferase in ubiquinone biosynthesis is required for embryo development in Arabidopsis thaliana. Plant Mol Biol 55: 567–577 [DOI] [PubMed] [Google Scholar]
- Paschen SA, Rothbauer U, Káldi K, Bauer MF, Neupert W, Brunner M. (2000) The role of the TIM8-13 complex in the import of Tim23 into mitochondria. EMBO J 19: 6392–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C, Camougrand N, Manon S, Sousa MJ, Côrte-Real M. (2007) ADP/ATP carrier is required for mitochondrial outer membrane permeabilization and cytochrome c release in yeast apoptosis. Mol Microbiol 66: 571–582 [DOI] [PubMed] [Google Scholar]
- Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kühlbrandt W, Wagner R, Truscott KN, Pfanner N. (2003) Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS. (2006) Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci USA 103: 15038–15043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch K, Curran SP, Tranebjaerg L, Koehler CM. (2002) Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum Mol Genet 11: 477–486 [DOI] [PubMed] [Google Scholar]
- Russell SD. (1993) The egg cell: development and role in fertilization and early embryogenesis. Plant Cell 5: 1349–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt HS. (1991) Mitochondria: structure and function. Rev Neurol (Paris) 147: 417–430 [PubMed] [Google Scholar]
- Sirrenberg C, Endres M, Fölsch H, Stuart RA, Neupert W, Brunner M. (1998) Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature 391: 912–915 [DOI] [PubMed] [Google Scholar]
- Sokol AM, Sztolsztener ME, Wasilewski M, Heinz E, Chacinska A. (2014) Mitochondrial protein translocases for survival and wellbeing. FEBS Lett 588: 2484–2495 [DOI] [PubMed] [Google Scholar]
- Tranebjaerg L, Jensen PK, Van Ghelue M, Vnencak-Jones CL, Sund S, Elgjo K, Jakobsen J, Lindal S, Warburg M, Fuglsang-Frederiksen A, et al. (2001) Neuronal cell death in the visual cortex is a prominent feature of the X-linked recessive mitochondrial deafness-dystonia syndrome caused by mutations in the TIMM8a gene. Ophthalmic Genet 22: 207–223 [DOI] [PubMed] [Google Scholar]
- Truscott KN, Wiedemann N, Rehling P, Müller H, Meisinger C, Pfanner N, Guiard B. (2002) Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol 22: 7780–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F, Dat JF. (2006) Reactive oxygen species in plant cell death. Plant Physiol 141: 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG, Beers EP, Dangl JL, Franklin-Tong VE, Gallois P, Hara-Nishimura I, Jones AM, Kawai-Yamada M, Lam E, Mundy J, et al. (2011) Morphological classification of plant cell deaths. Cell Death Differ 18: 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ. (2005) Many ways to exit? Cell death categories in plants. Trends Plant Sci 10: 117–122 [DOI] [PubMed] [Google Scholar]
- Vazzola V, Losa A, Soave C, Murgia I. (2007) Knockout of frataxin gene causes embryo lethality in Arabidopsis. FEBS Lett 581: 667–672 [DOI] [PubMed] [Google Scholar]
- Vianello A, Zancani M, Peresson C, Petrussa E, Casolo V, Krajňáková J, Patui S, Braidot E, Macrì F. (2007) Plant mitochondrial pathway leading to programmed cell death. Physiol Plant 129: 242–252 [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang Y, Carrie C, Giraud E, Elhafez D, Narsai R, Duncan O, Whelan J, Murcha MW. (2012) Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 24: 2675–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM. (2006) Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed α-propeller. Mol Cell 21: 123–133 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, Chacinska A. (2006) Chaperoning through the mitochondrial intermembrane space. Mol Cell 21: 145–148 [DOI] [PubMed] [Google Scholar]
- Yadegari R, Paiva G, Laux T, Koltunow AM, Apuya N, Zimmerman JL, Fischer RL, Harada JJ, Goldberg RB. (1994) Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell 6: 1713–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung EC, Meinke DW. (1993) Embryogenesis in angiosperms: development of the suspensor. Plant Cell 5: 1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu D, Jiang L, Gong H, Liu CM. (2012) EMBRYONIC FACTOR 19 encodes a pentatricopeptide repeat protein that is essential for the initiation of zygotic embryogenesis in Arabidopsis. J Integr Plant Biol 54: 55–64 [DOI] [PubMed] [Google Scholar]
- Zamzami N, Marchetti P, Castedo M, Decaudin D, Macho A, Hirsch T, Susin SA, Petit PX, Mignotte B, Kroemer G. (1995) Sequential reduction of mitochondrial transmembrane potential and generation of reactive oxygen species in early programmed cell death. J Exp Med 182: 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ren Y, Zhao J. (2008) Roles of extensins in cotyledon primordium formation and shoot apical meristem activity in Nicotiana tabacum. J Exp Bot 59: 4045–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Simons JW. (1999) Direct comparison of GAPDH, β-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem Biophys Res Commun 259: 523–526 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.