An ATP-dependent binding cassette transporter contributes to plant resistance to infection by Pseudomonas syringae and tolerance of abscisic acid.
Abstract
Plants have been shown previously to perceive bacteria on the leaf surface and respond by closing their stomata. The virulent bacterial pathogen Pseudomonas syringae pv tomato DC3000 (PstDC3000) responds by secreting a virulence factor, coronatine, which blocks the functioning of guard cells and forces stomata to reopen. After it is inside the leaf, PstDC3000 has been shown to up-regulate abscisic acid (ABA) signaling and thereby suppress salicylic acid-dependent resistance. Some wild plants exhibit resistance to PstDC3000, but the mechanisms by which they achieve this resistance remain unknown. Here, we used genome-wide association mapping to identify an ATP-dependent binding cassette transporter gene (ATP-dependent binding cassette transporter G family member16) in Arabidopsis (Arabidopsis thaliana) that contributes to wild plant resistance to PstDC3000. Through microarray analysis and β-glucuronidase reporter lines, we showed that the gene is up-regulated by ABA, bacterial infection, and coronatine. We also used a green fluorescent protein fusion protein and found that transporter is more likely to localize on plasma membranes than in cell walls. Transferred DNA insertion lines exhibited consistent defective tolerance of exogenous ABA and reduced resistance to infection by PstDC3000. Our conclusion is that ATP-dependent binding cassette transporter G family member16 is involved in ABA tolerance and contributes to plant resistance against PstDC3000. This is one of the first examples, to our knowledge, of ATP-dependent binding cassette transporter involvement in plant resistance to infection by a bacterial pathogen. It also suggests a possible mechanism by which plants reduce the deleterious effects of ABA hijacking during pathogen attack. Collectively, these results improve our understanding of basal resistance in Arabidopsis and offer unique ABA-related targets for improving the innate resistance of plants to bacterial infection.
Bacterial pathogens of plants disperse through the global hydrological cycle (Morris et al., 2008) and are likely to exert selection favoring defenses of wild plants at large spatial scales. In agriculture, these pathogens cause significant losses and necessitate extensive spraying of pesticides (Oerke, 2006). Although there is great potential to learn the mechanisms that underlie this resistance (Todesco et al., 2010), the genetic underpinnings of resistance in wild plants remain essentially unknown. Identification of natural mechanisms by which resistant wild plants prevent the growth of bacterial pathogens has the potential to improve crop yields and reduce exogenous pesticide use in agriculture.
The immune system of plants is sophisticated, involves the coordinated action of hundreds of genes (Jones and Dangl, 2006), and requires the presence of a single major hormone, salicylic acid (SA; Vlot et al., 2009). This pathway is cross regulated by other signaling pathways in the plant, including those controlled by jasmonic acid (Vlot et al., 2009) and abscisic acid (ABA; de Torres-Zabala et al., 2007). However, how and to what extent plants can regulate these hormonal-dependent pathways and cross talk is still not clear. SA-dependent defenses are effective against many bacterial pathogens, including the causal agents of bacterial spot disease, Pseudomonas syringae (Vlot et al., 2009). In some cases, plants perceive bacteria on the surface of the leaf and close the stomata, thereby preventing entry (de Torres-Zabala et al., 2007). However, at least one particularly virulent strain, P. syringae pv tomato (PstDC3000), has evolved a novel virulence mechanism, in which it secretes coronatine to force stomata to reopen (de Torres-Zabala et al., 2007; Montillet et al., 2013). After bacteria have entered the interior of a leaf, these virulent bacteria also use coronatine to hijack plant ABA biosynthesis, which elevates ABA levels and suppresses Isochorismate synthase1 expression, leading to down-regulation of SA-dependent defenses in the infected tissue (de Torres Zabala et al., 2009; Zheng et al., 2012). What has remained unknown is whether plants can counteract this suppression and if so, how.
ATP-dependent binding cassette (ABC) transporters have received particular attention recently for their importance in moving hormones, metal ions, and other compounds across membranes (Rea, 2007; Kang et al., 2011). The largest subfamily of ABC transporters in Arabidopsis (Arabidopsis thaliana), the Arabidopsis ATP-dependent binding cassette transporter G Family (AtABCG) group, contains 43 genes that share a common nucleotide-binding domain and transmembrane domain orientation (Verrier et al., 2008). To date, three members of this AtABCG family (AtABCG22, AtABCG25, and AtABCG40) have been associated with ABA transport directly. AtABCG22 and AtABCG40 are found on the guard cells specifically, and AtABCG25 is found on the vascular bundles (Kang et al., 2010; Kuromori et al., 2010, 2011). AtABCG36 Penetration Resistance3(PEN3)/Pleiotropic Drug Resistance8 (PDR8) is the only gene in AtABCG subfamily that has been reported for involvement in plant resistance to bacterial or fungal pathogens (Kobae et al., 2006; Stein et al., 2006; Xin et al., 2013). The functions of other AtABCGs and particularly, their possible roles in ABA transport or plant-pathogen interaction remain largely unknown. Here, we present evidence that one of the AtABCG transporters, AtABCG16, identified by genome-wide association (GWA) mapping is involved in ABA responses and plant resistance against PstDC3000.
RESULTS
GWA Mapping and Microarray Analysis Suggest the Involvement of AtABCG16 in Plant Resistance to Bacterial Pathogen PstDC3000
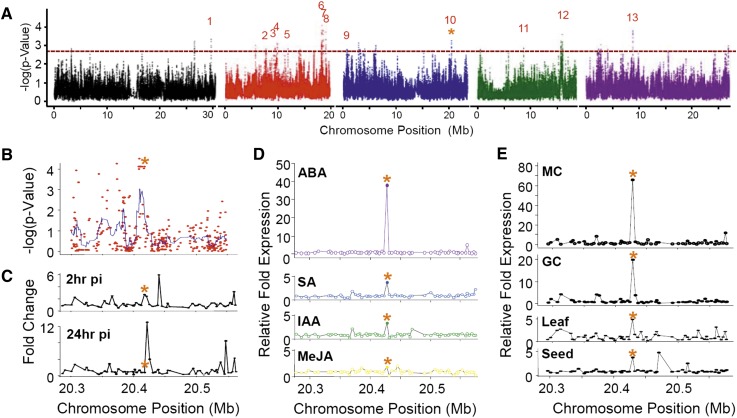
To identify candidate genes that contribute to differences in defense against bacterial infection in wild plants, we assessed a GWA map (Fig. 1A) constructed from 96 genotypes of the RegMap panel of Arabidopsis (Supplemental Table S1) consisting of a worldwide collection of plants for which over 200,000 single nucleotide polymorphisms (SNPs) have been identified (Atwell et al., 2010). GWA mapping has emerged as a particularly powerful method for identifying novel genes, because it uses genetic polymorphisms that are already present in wild populations and therefore, is likely to pinpoint genetic variation that is important in nature (Weigel, 2012). To assess resistance to infection, those plants were grown on soil and challenged by blunt syringe with a 104 colony-forming unit (cfu) mL−1 solution of PstDC3000 as described previously (Atwell et al., 2010). Our assessment of the GWA map revealed 13 major loci (Fig. 1A) and 54 candidate genes (Supplemental Table S2), of which one of the top candidates was an ABCG transporter (AtABCG16 at locus 10 on chromosome 3; Fig. 1B). This locus also included two additional gene candidates, At3g55070 and At3g55080, that contained equally significant SNPs (Supplemental Tables S3 and S4) but about which little is known.
Figure 1.
Genome-wide approaches identifying AtABCG16 as a candidate plant resistance gene through response to ABA. A, GWA map based on PstDC3000 growth in wild accessions of Arabidopsis (Supplemental Table S1) showing 13 loci (Supplemental Table S2), including locus 10, which contains AtABCG16. Locus 10 neighborhood covers 250 kb of chromosome 3 containing 30 genes upstream and 30 genes downstream of AtABCG16 (Supplemental Table S5) showing GWA scores for all SNPs in the interval with a smoothed peak (blue) containing three genes, including At3g55090 (B; Supplemental Tables S3 and S4), gene expression at 2 and 24 h postinoculation (pi) with PstDC3000 (C; Supplemental Table S5), exceptional expression response of AtABCG16 to exogenous 10 μm ABA (34th highest of 21,233 genes in the microarray) and moderate responses to 10 μm SA, IAA, or MeJA at 3 h (D), and tissue/location-specific expression at locus 10 interval in response to ABA in mesophyll cells (MCs), guard cells (GCs), leaves, and seeds (E). Microarray data were acquired from the eFP browser (Winter et al., 2007). *, Position of AtABCG16.
To better understand possible defense-related functions at this locus, we first mined published microarray data for the widely used ecotype Columbia-0 of Arabidopsis (Col-0) accession and found that AtABCG16 had a 2.76-fold up-regulation 2 h after exposure to PstDC3000 that diminished to 0.72-fold at 24 h after exposure (Fig. 1C). The early response to bacteria is consistent with functional role in the initial stages of plant responses to infection. Given the key roles of SA, ABA, indole-3-acetic acid (IAA), and methyl jasmonate (MeJA) in plant signaling and known transport roles of ABCG transporters in moving these compounds (Dean and Mills, 2004; Vlot et al., 2009; Kuromori et al., 2010), we then asked whether AtABCG16 expression was altered after exogenous application of these hormones to plants. We found that AtABCG16 was induced 3.8-fold and 3.5-fold by 10 µm exogenous SA and IAA, respectively, and not induced by MeJA (Fig. 1D). In contrast, AtABCG16 was up-regulated nearly 40-fold by ABA. Indeed, this was the 34th highest response of 22,591 Arabidopsis genes, and this gene was the only one within the 250-kb neighborhood to exhibit a response (Fig. 1D; Supplemental Table S5). Furthermore, tissue-specific microarrays showed that AtABCG16 was up-regulated 65-fold in mesophyll cells and nearly 20-fold in guard cells treated with exogenous ABA (Fig. 1E). Collectively, these data showed that this candidate gene responds to PstDC3000 and responds even more strongly to exogenous ABA.
Transferred DNA Insertion Mutants of AtABCG16 Show a Loss of ABA Tolerance in Plate Assays
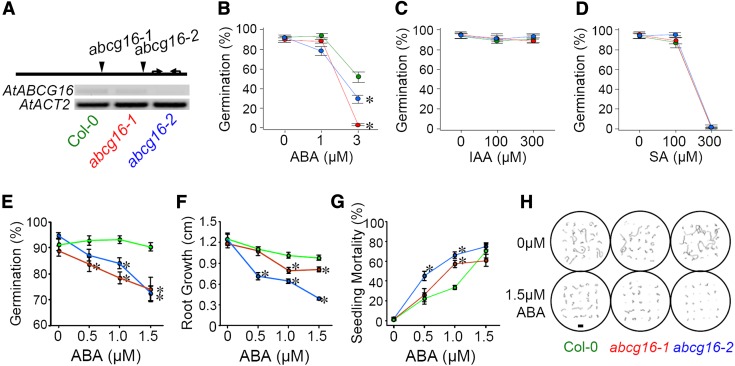
To assess experimentally whether AtABCG16 is required for plant tolerance of exogenous ABA, SA, or IAA, we used a standard hormone assay on Murashige and Skoog medium (MS) plates (Kuromori et al., 2010) to compare the tolerance of two transferred DNA (T-DNA) insertion knockout lines relative to the background (Col-0) in experiments where all three lines were grown simultaneously. To characterize the knockouts, we first ran quantitative reverse transcription (qRT) -PCR (Fig. 2A; Supplemental Tables S6 and S7) and found that they reduced the AtABCG16 gene expressions by 92% (SALK_087501; hereafter abcg16-1) and 84% (SALK_119868C; hereafter abcg16-2), respectively. Because SA and IAA likely operate at different concentrations from ABA in planta, we included both low-range (1 and 3 μm) and high-range (100 and 300 μm) tests of ABA, SA, and IAA. Through the plate assay, we found that both abcg16-1 and abcg16-2 had significantly reduced germination in the presence of exogenous ABA relative to the Col-0 background (F4,45 = 18.49, P < 0.0001; Fig. 2B; Supplemental Table S8), whereas no such response was detected in the presence of IAA (Fig. 2C; Supplemental Table S8) or SA (Fig. 2D; Supplemental Table S8). The loss of ABA tolerance was dramatic. Specifically, in the presence of 3 μm exogenous ABA, the abcg16-1 line had a 94.8% reduction in germination (Dunnett T = −8.7, P < 0.001) and the abcg16-2 line had a 43.5% reduction in germination (Dunnett T = −4.0, P = 0.002) relative to the background. Our conclusion from these assays was that AtABCG16 was necessary for successful germination in the presence of exogenous ABA but not SA or IAA at the concentrations tested.
Figure 2.
T-DNA insertion knockouts of AtABCG16 are less tolerant to exogenous ABA. A, T-DNA insertion sites (indicated by arrowheads) for each of two abcg16 knockout lines (abcg16-1 and abcg16-2) along the single exon of AtABCG16. Arrows indicate the positions of primers for detecting gene expression (Supplemental Table S6). Constitutive expression of AtABCG16 in the Col-0 wild-type background and two knockouts as determined by semiquantitative PCR (Supplemental Table S7) and shown here relative to a standard housekeeper (AtACT2). B to D, Plate assays comparing germination responses of abcg16-1 (red) and abcg16-2 (blue) relative to the Col-0 wild-type background (green) in the presence of exogenous 0 to 3 μm ABA (B), 0 to 300 μm SA (C), and 0 to 300 μm IAA (D) in water (Supplemental Table S8). Each mean (±1 se) represents the average of five replicate plates (each containing 25 seeds). E to G, Plate assays testing the effect of 0, 0.5, 1.0, or 1.5 μm exogenous ABA on germination on day 4 (F; percentage), root length on day 10 (G; centimeters), and mortality on day 30 (H; percentage). Each mean (±1 se) represents 10 replicate plates (each containing 25 seeds; Supplemental Fig. S1; Supplemental Table S9). *, Significant difference from wild-type line by Dunnett T contrast at P < 0.05. H, Representative tracings of roots for plates in 0 and 1.5 μm ABA treatments on day 10 (Supplemental Fig. S2). Bar = 1 cm.
To further assess the tolerance of abcg16-1 and abcg16-2 to exogenous ABA, we also tested these lines across a smaller range of 0 to 1.5 μm exogenous ABA using an established protocol (Kuromori et al., 2010), and again, we found highly significant defects in the tolerance of both knockout lines relative to the Col-0 background in terms of reduced germination (F6,108 = 3.71, P = 0.002; Fig. 2E; Supplemental Table S9), reduced root growth (F6,108 = 7.03, P < 0.001; Fig. 2, F and H; Supplemental Figs. S1 and S2; Supplemental Table S9), and elevated mortality of the knockouts (F6,103 = 3.92, P = 0.001; Fig. 2G; Supplemental Table S9). To assess the possibility of off-target effects of the T-DNA insertions in the abcg16-1 and abcg16-2 lines, we measured the expression of AtABCG17 and AtABCG18, which are directly adjacent to AtABCG16. We found no evidence of off-target effects of the T-DNA insertions (Supplemental Fig. S3), which was indicated by consistent expression of AtABCG17 and AtABCG18 in abcg16-1 and abcg16-2 relative to the Col-0 background line (CS60000). Collectively, these experiments showed that knockouts of AtABCG16 were defective in ABA tolerance, that the response was present at multiple stages of plant development, including germination and subsequent growth and survival of germinated seeds, and that the defects were likely to be caused specifically by the reduced expression of AtABCG16 (Fig. 2A) in these T-DNA insertion knockout lines.
AtABCG16 Potentially Localizes to the Plasma Membrane
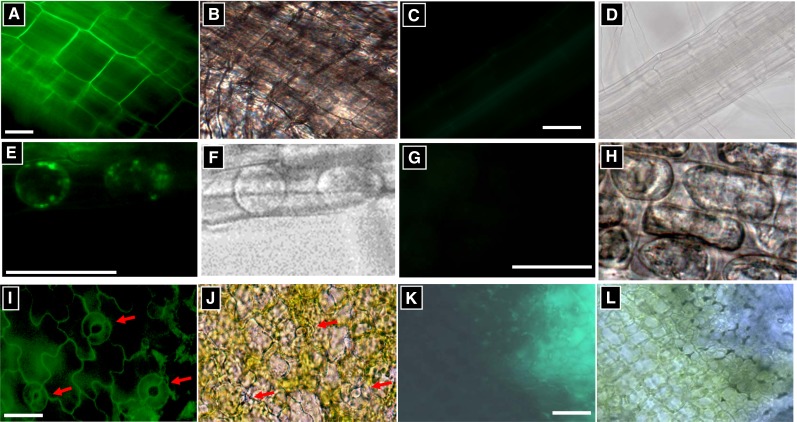
To localize AtABCG16, we use a construct consisting of the cauliflower mosaic virus 35S promoter driving the GFP fused to AtABCG16 (2X35S::GFP-AtABCG16). Subcellular localization of the fusion protein was observed using the filtered epifluorescence microscopy imaging of the green fluorescence signals in cells of the 2X35S::GFP-AtABCG16-transformed plants (Fig. 3). In root cells, fluorescence was highest on the cell surface, which was consistent with GFP-AtABCG16 localization in the plasma membrane or cell wall (Fig. 3A; Supplemental Fig. S4). To assess the hypothesis that the protein was in the cell wall, we treated the root tip cells with 20% Suc to create a high osmotic environment following a published protocol (Kuromori and Shinozaki, 2010). On the osmotic treatment cells, the fluorescence signal was observed to associate more with the cell interior and plasma membrane than on the cell wall (Fig. 3, E and F). In leaf cells, we observed fluorescence signals in both pavement cells and guard cells (Fig. 3, I and J). Collectively, these results are consistent with localization of AtABCG16 on the plasma membrane and seem to rule out localization on the cell wall but cannot rule out localization on vacuolar membranes or elsewhere in the cell interior.
Figure 3.
Plasma membrane localization of 2X35S::GFP-AtABCG16 fusion protein. A, C, E, G, I, and K show fluorescence levels under GFP expression conditions. B, D, F, H, J, and L were taken under white light. A to H show root cells. A, B, E, F, I, and J show plants transformed with 35S::GFP-AtABCG16. C, D, G, H, K, and L show untransformed plants with at least 2 times exposure time. E to H show plasmolyzed root cells after treatment with 20% Suc. I to L show leaf epidermal cells. Red arrows indicate stomata. Positive controls are shown in Supplemental Figure S4. Bars = 10 μm.
AtABCG16 Gene Expression Is Induced by Hormones and Bacteria
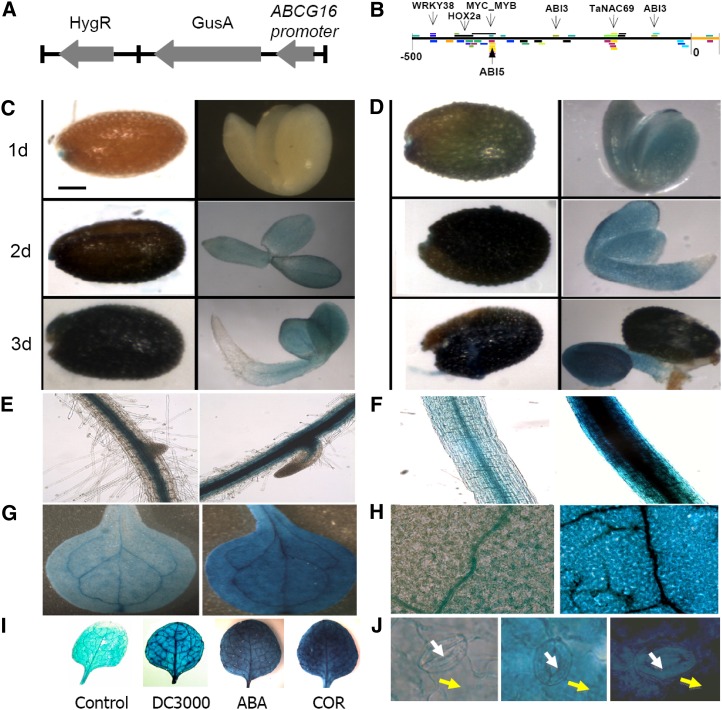
To further investigate how this gene is regulated, we created transgenic lines containing the GUS reporter gene driven by a native AtABCG16 promoter (Fig. 4A). This native promoter of AtABCG16 (Fig. 4B) contains important binding motifs for the ABA response factor, ABA Response Element, at −370, −250, and −80 as well as a binding site at −470 for WRKY38, a transcription factor directly linked to infection by PstDC3000 (Kim et al., 2008). Our AtABCG16 pro::GUS transgenic lines exhibited inducible GUS signals in control seeds during the germination process (Fig. 4C), which were significantly increased in seeds that were treated with 6 μm ABA (Fig. 4D). In AtABCG16 pro::GUS transgenic plants, in the absence of exogenous ABA, the GUS signals were observed in the center of the roots and hypocotyl (Fig. 4, E and F) and the veins of leaves (Fig. 4, G and H), consistent with association with vascular bundles. After treatment with 3 μm ABA for 24 h, GUS signals were observed ubiquitously in both roots and leaves (Fig. 4, E–H), indicating significant up-regulation in all types of cells. To see if AtABCG16 was up-regulated by bacterial pathogens, we tested the transgenic plants with the coronatine secreting strain (COR+) and PstDC3000 and separately tested them with a solution containing only the virulence factor coronatine. We found that PstDC3000 induced expression clearly in leaves and that coronatine up-regulated AtABCG16 even more strongly than ABA itself (Fig. 4I). Also, we observed elevated GUS levels in both guard cells and pavement cells under ABA induction (Fig. 4J), which confirmed our earlier microarray evidence that AtABCG16 was responding in multiple cell types within the leaf (Fig. 1E). Collectively, our GUS assay results showed that AtABCG16 responded to bacterial pathogen infection as well as to exogenous ABA and coronatine.
Figure 4.
AtABCG16 expression in seeds, roots, and leaves. A, Diagram of the AtABCG16pro::GUS construct including Hygromycin Resistance (HygR) selectable marker. B, The AtABCG16 promoter region includes three conserved motifs for the important ABA transcription factors ABA Insensitive3 (ABI3) and ABA Insensitive5 (ABI5) (Brady et al., 2003), one for WRKY38 (a known PstDC3000-induced transcription factor; Kim et al., 2008), and one for NAC-domain containing protein69 (NAC69) (a drought stress-related transcription factor; Xue et al., 2011) and positions of Homeobox protein 2a (Hox2a), myelocytomatosis oncogene_myeloblastosis oncogene (MYC_MYB) binding motifs as determined in Athamap. Expression of AtABCG16 on days 1, 2, and 3 in control seeds (C) or seeds treated with 6 μm ABA (D). E to H, GUS staining without (left) and with (right) ABA in root (E), hypocotyl (F), cotyledon (G), and rosette leaf (H) at 21 d. I, GUS staining of leaves in response to control solution (10 μm MgCl2) or 10 μm MgCl2 containing 108 cfu mL−1 of DC3000 (COR+), 3 μm ABA, and 1 μm COR (left to right). J, GUS staining at stomata: control (left), 3 μm ABA (center), and 1 μm coronatine (right) at 24 h after treatment. Guard and pavement cells are identified by white and yellow arrows, respectively.
AtABCG16 Reduces Bacterial Growth in Planta and Increases Plant Performance
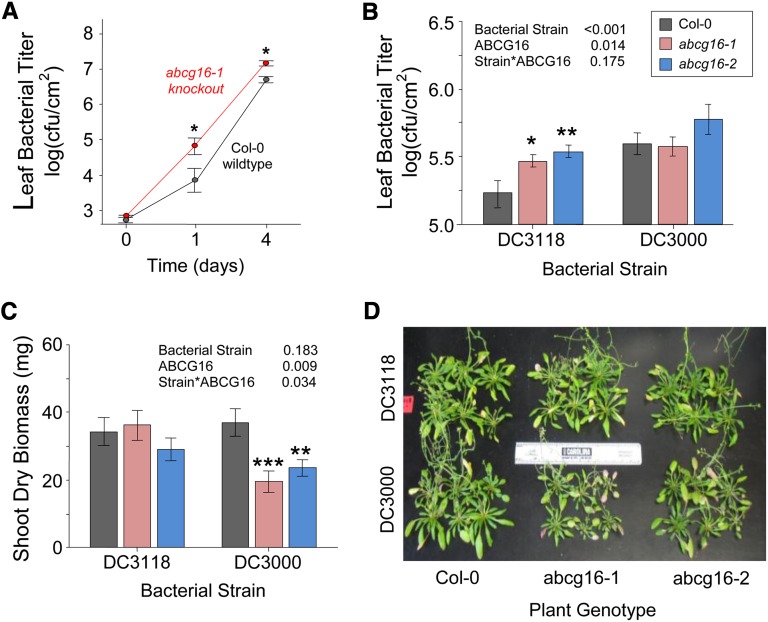
To assess the role of AtABCG16 in resistance to infection, we challenged plants with PstDC3000 using soil-grown plants of the Col-0 background and the T-DNA insertion knockout lines abcg16-1 and abcg16-2. In the first experiment, we compared growth of PstDC3000 in leaves of the Col-0 background and abcg16-1 after inoculation at 5 × 104 cfu mL−1 by blunt syringe. The PstDC3000 strain grew significantly more quickly and reached higher concentrations in the abcg16-1 mutant relative to the background line (F1,49 = 5.1, P = 0.028; Fig. 5A). To assess the possible effects on plant performance under more realistic conditions, we then performed a second experiment, where we spray inoculated soil-grown plants with bacteria and measured both bacterial and plant growth. To assess the possible role of AtABCG16, we sprayed both T-DNA insertion knockouts abcg16-1 and abcg16-2 with the COR+ and COR− bacterial strains and compared their responses relative to the Col-0 background. We found that the COR+ strain PstDC3000 was faster to colonize the interior of leaves and achieved higher titers (F1,31 = 15.4, P < 0.001; Fig. 5B; Supplemental Table S10), and it caused a greater reduction in shoot biomass than the COR− bacterial strain PstDC3118 but only in two T-DNA abcg16 knockout lines (F1,67 = 4.6, P = 0.034; Fig. 5, C and D; Supplemental Table S11). Together, these bacterial-challenge experiments suggested that AtABCG16 has an important role in the resistance of Arabidopsis against bacterial infection.
Figure 5.
Response of AtABCG16 knockout lines to bacterial treatments. Leaf bacterial titer means (±1 se) for wild-type plants (Col-0) and abcg16-1 knockout at 0, 1, and 4 d after blunt syringe inoculation of soil-grown plants with 5 × 104 cfu mL−1 of PstDC3000 in 10 mm MgSO4 buffer (A) and wild-type Col-0, abcg16-1 knockout, and abcg16-2 knockout plants titered at 18 d after spray infection with 1 × 108 cfu mL−1 of PstDC3118 or PstDC3000 in 10 mm MgSO4 buffer on day 36 of plant growth (B; Supplemental Table S10). C, Shoot dry biomass of 12 plants per treatment harvested on day 95 of plant growth (Supplemental Table S11). D, Reduced sizes of the two knockout lines in the presence of virulent PstDC3000 observed on day 95 of plant growth. *, P < 0.05. **, P < 0.01. ***, P < 0.001.
DISCUSSION
Although ABC transporters have been shown previously to transport ABA (Kang et al., 2010; Kuromori et al., 2010, 2011), this function has not been previously linked to resistance to bacterial infection. Here, we have shown that an ABCG transporter, AtABCG16, is important for ABA tolerance and also assists in plant basal resistance against PstDC3000. Both Atabcg16 T-DNA knockouts exhibited greater susceptibility to bacterial growth and reduced performance relative to untransformed plants. Together, these results suggest an important role of the gene in resistance to these bacteria. As such, the study provides one of the first demonstrations, to our knowledge, of a link between an ABC transporter and plant resistance to bacterial pathogens. Previously, only one other ABCG transporter, AtABCG36 (PEN3/PDR8), has been found to be involved in plant defense against pathogens (Kobae et al., 2006), but in that case, auxin transport was implicated (Strader and Bartel, 2009; Xin et al., 2013).
Although it seems clear that AtABCG16 is involved in both plant basal resistance to PstDC3000 and tolerance to exogenous ABA, we do not yet know whether these two effects are linked together. We have two reasons, however, for suspecting that a link does exist. First, our GUS assays indicated strong AtABCG16 expression in response to both the bacterial pathogen and hormone treatments. Indeed, AtABCG16 expression is induced by flagellin 22 amino acid fragment alone (C. Danna, personal communication). Interestingly, when we applied ABA to AtABCG16pro::GUS plants, the signal was uniformly detected across the leaf surface, including guard cells and nonguard cells. However, when we applied coronatine, we observed a dramatic response in nonguard cells but only moderate up-regulation in guard cells (shown by the arrows in Fig. 3I). This difference may reflect ABA movement into guard cells after synthesis in other cell types (Bauer et al., 2013; Boursiac et al., 2013). Second, the T-DNA insertion knockouts showed that AtABCG16 suppresses bacterial growth and increases tolerance to the exogenous ABA.
The bacterial virulence factor, coronatine, strongly induced expression of AtABCG16, but the exact nature of the relationship remains unclear. Most notably, we found that a bacterial strain that lacks coronatine, PstDC3118, had significantly higher growth in the T-DNA knockout lines, abcg16-1 and abcg16-2, than in the untransformed background plants. In other words, even the avirulent strain lacking COR secretion was able to achieve higher titers in the plants that lacked expression of this particular gene. These data suggest, therefore, that AtABCG16 has a role in resistance that is independent of mitigation of the effects of coronatine.
How the gene functions in ABA tolerance remains to be further clarified. AtABCG16 and two other ABC transporters implicated in ABA transport (AtABCG22 and AtABCG25) are half transporters, meaning that their products must form either homodimers or heterodimers to become functional transporters. Heterodimerization has been shown previously for ABC transporters (McFarlane et al., 2010) and provides a mechanism by which a large number of transporter structures could be assembled from a relatively small number of genes. It is possible that the transport function of AtABCG16 will involve the proteins from these transporters or other half ABCG transporters. Interestingly, the expression pattern of AtABCG16 is different from AtABCG25, AtABCG22, and AtABCG40 (Kang et al., 2010; Kuromori et al., 2010, 2011). In leaves, AtABCG25 is located on xylem, whereas AtABCG22 and AtABCG40 are expressed mostly on the membrane of guard cells. This would seem to suggest that AtABCG16 may not form heterodimers with these particular proteins and is consistent with the finding that AtABCG22 and AtABCG40 do not seem to interact in planta (Kuromori et al., 2011). Our analysis of ABCG family genes (Supplemental Fig. S5) indicates that AtABCG16 is most similar to AtABCG1, and our data suggest that the gene is most likely to have generalized expression across cell types, which may only be turned on under stress, whereas AtABCG25, AtABCG22, and AtABCG40 may have evolved more specialized functions with restricted expression that is cell-type specific (Ukitsu et al., 2007; Kuromori et al., 2011).
AtABCG16 exists on the third chromosome as a part of a tandem repeat of three ABC transporters that includes AtABCG17 and AtABCG18. There is no indication, however, that these three genes have overlapping function, and indeed, the differential responses of these genes to exogenous ABA, SA, and IAA are striking (Fig. 1D). However, because of the proximity of these three genes, it was particularly important to assess possible off-target effects of the T-DNA insertions in the abcg16-1 and abcg16-2 lines. Although both T-DNA insertion lines exhibited defectively low expression of AtABCG16 (Fig. 2A), they possessed normal expression of AtABCG17 and AtABCG18 relative to the Col-0 background (Supplemental Fig. S3). The strong defects that we observed in tolerance of abcg16-1 and abcg16-2 to ABA (Fig. 2, E–H) were, therefore, not caused by off-target effects on expression of AtABCG17 or AtABCG18. These data are consistent with a role of AtABCG16 specifically in tolerance of exogenous ABA.
To understand whether particular coding and promoter region differences among wild lines affect the function of AtABCG16, it will be important test the relative activities of alleles from susceptible and resistant wild lines in a common null background in the future. To assess more clearly whether AtABCG16 transports ABA, membrane transport assays (Kuromori et al., 2010) should also be performed. GWA mapping has attracted a great deal of interest in the literature (Atwell et al., 2010), but examples of successful use of this methodology remain very rare in plants. Our study is one of the first, to our knowledge, to identify a candidate gene directly from a GWA map and find supporting experimental evidence of relevant gene function.
In summary, we have provided evidence that AtABCG16 is involved in ABA tolerance and contributes to plant resistance against PstDC3000. We also confirm here the evidence from published microarrays that indicates very low levels of constitutive expression but dramatic induction of this gene by ABA and coronatine as well as its expression in both mesophyll and guard cells. Collectively, these results improve our understanding of basal resistance in Arabidopsis and offer unique ABA-related targets for improving the innate resistance of plants to bacterial infection.
MATERIALS AND METHODS
GWA Mapping
To identify candidates in plant resistance to PstDC3000, we created a GWA map for 96 accessions from the RegMap panel using data published previously (Atwell et al., 2010) of Arabidopsis (Arabidopsis thaliana) collected across the worldwide range. The dataset contained 205,803 SNPs derived from the 250K SNP data, version 3.06 (https://cynin.gmi.oeaw.ac.at/home/resources/atpolydb). All SNPs used in the analysis were diallelic and had the minor nucleotide represented in more than 5% of the accessions. For the GWA map, we used accession averages of leaf bacterial titer from three independent experiments where plants were infected by blunt syringe with a 1 × 104 cfu mL−1 solution of DC3000, and bacterial titers were measured at 0, 1, 4, and 7 d postinfection (Atwell et al., 2010; Supplemental Table S1). Wilcoxon rank sum tests were used to calculate the significance of association between each SNP and the phenotypic values using standard methods (Atwell et al., 2010). To handle confounding caused by population structure, we used the standard approach of mixed-model analysis known as Efficient Mixed-Model Association (Kang et al., 2008). We present log-transformed P values using the standard approach (Atwell et al., 2010) and refer to them as the GWA score. To control the bias of rare single extreme GWA score values, we smoothed the map by averaging the GWA score of every 10 adjacent SNPs. The 13 loci identified represented the subset common to both the Wilcoxon and Efficient Mixed-Model Association maps (Supplemental Table S2).
Microarray Data
Expression data for all genes in the 250-kb neighborhood of AtABCG16 (Supplemental Table S5) were extracted from the following published microarray datasets: PstDC3000 (NASCARRAYS-120; Numberger Laboratory), ABA (NASCARRAYS-176; Shimada Laboratory), MeJA (NASCARRAYS-174; Shimada Laboratory), IAA (NASCARRAYS-175; Shimada Laboratory), SA (NASCARRAYS-192; Shimada Laboratory), ABA leaf (Pandey et al., 2010), ABA seed (NASCARRAYS-183; Kamiya Laboratory), ABA guard cells (Pandey et al., 2010), and ABA mesophyll cells (Yang et al., 2008). All genes within the prescribed interval were included. Each of these published datasets included at least two biological replicates per treatment.
Plant Materials and Seed Sterilization
The wild-type Col-0 background (CS60000) and the two T-DNA insertion AtABCG16 knockouts, abcg16-1 (SALK_087501) and abcg16-2 (SALK_119868C), were acquired from the Arabidopsis Biological Resource Center at Ohio State. Seeds were sterilized before experiments using a standard bleach protocol consisting of 70% (v/v) ethanol for 1 min and 50% (v/v) bleach with 0.05% (v/v) Tween-20 (BP337-100; FisherBiotech) for 10 min followed by four washes with sterile distilled water before use.
Plate Assays with Exogenous ABA, IAA, and SA
To assess plant tolerances of exogenous ABA, IAA, and SA, we followed a standard plate assay (Kuromori et al., 2010) using 15-cm petri dishes containing 0.8% agar with one-half-strength MS (M5524; Sigma-Aldrich). ABA, IAA, and SA were added to cooling agar (45°C or less) to the specified concentrations just before pouring the plates. Seeds were sterilized and washed as described above. Each MS plate received 25 seeds followed by cold treatment at 4°C in the dark for 1 week to stratify the seeds. These plates were then moved to a plant growth cabinet at 22°C under a 12-h-light/12-h-dark photoperiod with 55% humidity, which was considered day 1 of the experiment. Plates were measured for percentage of germination on day 4, root length on day 10, and mortality on day 30. For measuring root length on day 10, all roots were traced on acetate paper, digitally scanned, and measured for pixel number by ImageJ. Root length was calculated as root pixel number divided by pixel number of a line of 1 cm in length. All experiments included 5, 6, or 10 independent replicate MS plates as indicated (Supplemental Tables S8 and S9).
Expression Analyses Using qRT-PCR
Total RNA was isolated from leaves, stems, flowers, and root tissues of 6-week-old plants using TriReagent according to the manufacturer’s protocol (MRC). Residual genomic DNA was removed first by treatment with RNase-free DNase I (Invitrogen). First-strand complementary DNA was synthesized using approximately 2 μg of total RNA, 0.5 μg of oligo(dT)18, and SuperScript III reverse transcriptase according to the manufacturer’s instructions (Invitrogen). Reverse transcription (RT) -PCR was carried out in an Eppendorf mastercycler using GoTaq Green Master Mix (Promega), which was programmed as follows: 2 min at 95°C for predenature and 25 to 40 cycles of 15 s at 94°C, 15 s at 55°C, and 20 s at 72°C for each gene. qRT-PCR was carried out in an ABI-7900 thermal cycling system using a Real-Time PCR Power Mix Kit (ABI) and programmed at the same condition. For control reactions, either no sample was added or RNA alone was added without RT to test if the RNA sample was contaminated with genomic DNA. Arabidopsis thaliana Actin protein2 was used as a reference gene. Oligonucleotide primer sets (Supplemental Table S6) were designed with the Primer Express 2.0 software (Applied Biosystems-Perkin-Elmer). To test the suitability of these primer sets, the specificity and identity of the RT-PCR products were monitored by melting curve analysis (65°C–99°C; 5°C s−1) of the reaction products, which can distinguish the gene-specific PCR products from nonspecific PCR products. All primers were synthesized by Integrated DNA Technologies. Data were analyzed by a standard method (Yuan et al., 2006).
Fluorescence Microscopy
To construct the GFP fusion lines, we first amplified the 2,211-bp AtABCG16 complementary DNA using PCR and cloned it into the pENTR/D-TOPO vector (Invitrogen). After sequence confirmation, we then integrated this construct (pENTR-AtABCG16) into the GFP fusion protein vector pMDC83 using LR clonase (Invitrogen). The construct of 2X35S::GFP-AtABCG16 was introduced into the recipient Col-0 plant through Agrobacterium sp.-mediated transformation using the floral-dip method (Clough and Bent, 1998). Hygromycin-resistant lines (T1) were screened and PCR confirmed. We used tissue from 1-week-old seedlings of T2 plants for study of the subcellular localization of the GFP fusion protein. Microscope images were taken using fluorescein isothiocyanate (FITC) -filtered epifluorescence microscopy (Olympus BX51 model) with blue light at 400× magnification. For plasmolysis, root sections were treated with 20% Suc for 10 min.
GUS Expression Assays with the AtABCG Promoter
For AtABCG16 promoter-driven GUS expression lines, a 1,388-bp AtABCG16 promoter region was amplified by using EX Taq (Takara) with the primers listed in Supplemental Table S6. Then, the PCR product was cloned into the PCR8/GW/TOPO vector (Invitrogen), sequence confirmed by Sanger sequencing, and integrated into the promoter analysis vector pMDC162 through the LR reaction (Curtis and Grossniklaus, 2003). Transgenic plants were made as described above. T2 lines were used for the GUS expression analysis. To test the expression profile of ABCG16 and the response to ABA, 1-week-old seedlings were soaked in 10 μm MgCl2 (control) or 3 μm ABA for 24 h. For other treatments, leaves of 3-week-old plants were soaked in 10 μm MgCl2 (control), 108 cfu mL−1 of DC3000, 3 μm ABA, and 1 μm coronatine for 24 h. The expression pattern of ABCG16 during seeds germination and the response to ABA were tested as described above. Seed germination assays were performed by soaking seeds in 10 μm MgCl2 (control) or 6 μm ABA for 1, 2, or 3 d. GUS staining was performed following standard procedures (Jefferson et al., 1987).
Bacterial Growth Assays
To assess the possible role of AtABCG16 on plant resistance, we performed bacterial growth assays using PstDC3000 alone and in conjunction with the COR− T-DNA insertion mutant bacterial strain PstDC3118 obtained from D. Cuppels and J. Tambong at Agriculture and Agri-Food. To generate the inoculation solution, we first streaked bacteria on a King’s B media agar plate with 50 μg L−1 of rifampicin (R3501-5G; Sigma-Aldrich), and after 3 d, we transferred several colonies to 50 mL of liquid King’s B media also containing 50 μg L−1 of rifampicin and shook this solution in an incubator for 24 h at 28°C. We then transferred an aliquot of 5 mL to a fresh tube of 50 mL of liquid King’s B media, shook this solution for 4 h at 28°C to achieve midlogarithmic stage growth, spun the tube for 5 min at 3,000 rpm to obtain a pellet, poured off the supernatant, resuspended the pellet in 1 mL of 10 mm MgSO4, vortexed, measured bacterial concentration by spectrophotometry, and diluted to the inoculation concentration of 5 × 104 cfu mL−1. For each plant, we inoculated two leaves with approximately 0.1 mL of 5 × 104 cfu mL−1 bacterial solution by blunt syringe. For the T-DNA insertion knockout lines, we sampled six independent plants per time point. We measured the knockouts and wild-type background plants simultaneously in each experiment. To measure leaf bacterial titers, we used a hole punch to collect a leaf disc (0.28 cm2) and then ground the leaf disc in 200 μL of 10 mm MgSO4 buffer; 40 μL from the diluted mixture were then plated on the agar plates with 50 μg mL−1 of rifampicin and incubated at 28°C for 3 d before counting the number of colonies.
Plant Growth Assay
To compare the effects of bacterial infection by COR+ and COR− bacteria on the performance of plants with and without functional AtABCG16, we grew the wild-type background line (Col-0) and both T-DNA insertion knockouts (abcg16-1 and abcg16-2) and then exposed them to either the COR+ strain (PstDC3000) or the COR− strain (PstDC3118) by repeated spray inoculation. Seeds were sown on soil (Promix BX) in standard 36-cell flats, cold treated for 1 week, and then placed in a growth room at 20°C under a 12-h-light/12-h-dark photoperiod. Seeds germinated the next day, which was considered day 1 of the experiment. Flats were redistributed at least one time per week to reduce positional effects. On day 11, we thinned seedlings to 5 plants per cell for a total of 120 plants per line. On day 35, we randomly assigned plants to receive either PstDC3000 or PstDC3118 by spray inoculation. On day 36, we sprayed the plants to saturation with 1 × 108 cfu mL−1 of PstDC3118 or PstDC3000 in 10 mm MgSO4 buffer. We then covered the flats with clear plastic covers. Given that Pseudomonas sp. infection is likely to be amplified by cold stress and high humidity, we then placed all plants in a 4°C cabinet for 48 h and sprayed the bacterial solution every 12 h for 2 d. After 48 h, we moved the flats back to the growth room but left the clear plastic covers on to maintain high humidity. On day 54 of plant growth (the 18th day after infection), we removed discs from plants in one-half of the cells for measurement of bacterial titer. The leaf disc was sterilized in 70% ethanol for 2 s, ground in 200 μL of 10 mm MgSO4 buffer, and then, diluted in 10 mm MgSO4 buffer and plated on King’s B media with 50 μg mL−1 of rifampicin. Plates were counted after 3 d at 28°C. On day 95, we harvested the remaining unsampled plants from each cell, photographed them, dried them at 60°C for 2 weeks, and weighed them to determine shoot dry biomass.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root tracings following ABA treatment.
Supplemental Figure S2. Photograph of roots shown in Fig. 2H.
Supplemental Figure S3. Expression of AtABCG17 and AtABCG18.
Supplemental Figure S4. 35S::GFP positive controls for root and leaf epidermis.
Supplemental Figure S5. Gene tree of ABC transporter G family group.
Supplemental Table S1. Titers of PstDC3000 in RegMap panel lines.
Supplemental Table S2. Candidate genes from genome-wide association map.
Supplemental Table S3. Base calls for SNP nos. 118594–118611.
Supplemental Table S4. Base positions and map scores for SNP nos. 118594–118611.
Supplemental Table S5. Expression response to bacterial infection, ABA, IAA, and SA.
Supplemental Table S6. Primers used for RT-PCR, gene and promoter cloning.
Supplemental Table S7. RT-PCR of AtABCG16.
Supplemental Table S8. Effects of ABA, IAA, and SA on germination.
Supplemental Table S9. Effects of ABA on germination, root growth, and mortality.
Supplemental Table S10. Bacterial titers following surface spray inoculation.
Supplemental Table S11. Shoot dry biomass following surface spray inoculation.
Supplementary Material
Acknowledgments
We thank Steve McCalley, Kaiyuan Pang, Darve Robinson, Justin Seaman, Andy Lariviere, Muhammad Saleem, Mahesh Basantani, and Rongjian Ye for assistance with data collection; Diane Cuppels and James Tambong for providing the DC3118 bacterial strain; Cristian Danna for sharing unpublished microarray data on AtABC16 expression response to flg22; Bjarni Vilhjalmsson, Christopher Toomajian, and Chang Lunching for providing helpful advice regarding R programming; Cristian Danna and Ram Dixit for valuable comments on the article; the Numberger, Shimada, and Kamiya Laboratories for providing microarray data to Nottingham Arabidopsis Stock Centre’s microarray database; and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants distributed by the Arabidopsis Biological Resource Center.
Glossary
- ABA
abscisic acid
- cfu
colony-forming unit
- Col-0
ecotype Columbia-0 of Arabidopsis
- GWA
genome-wide association
- IAA
indole-3-acetic acid
- MeJA
methyl jasmonate
- MS
Murashige and Skoog medium
- qRT
quantitative reverse transcription
- RT
reverse transcription
- SA
salicylic acid
- SNP
single nucleotide polymorphism
Footnotes
This work was supported by the U.S. National Science Foundation (grant nos. 1051581 to C.N.S. and 1050138 to M.B.T.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Ache P, Lautner S, Fromm J, Hartung W, Al-Rasheid KA, Sonnewald S, Sonnewald U, Kneitz S, Lachmann N, et al. (2013) The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr Biol 23: 53–57 [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Léran S, Corratgé-Faillie C, Gojon A, Krouk G, Lacombe B. (2013) ABA transport and transporters. Trends Plant Sci 18: 325–333 [DOI] [PubMed] [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P. (2003) The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J 34: 67–75 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala M, Bennett MH, Truman WH, Grant MR. (2009) Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J 59: 375–386 [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bögre L, Grant M. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26: 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JV, Mills JD. (2004) Uptake of salicylic acid 2-O-beta-D-glucose into soybean tonoplast vesicles by an ATP-binding cassette transporter-type mechanism. Physiol Plant 120: 603–612 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. (2008) Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y. (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107: 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Park J, Choi H, Burla B, Kretzschmar T, Lee Y, Martinoia E. (2011) Plant ABC transporters. Arabidopsis Book 9: e0153, /10.1199/tab.0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z. (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae Y, Sekino T, Yoshioka H, Nakagawa T, Martinoia E, Maeshima M. (2006) Loss of AtPDR8, a plasma membrane ABC transporter of Arabidopsis thaliana, causes hypersensitive cell death upon pathogen infection. Plant Cell Physiol 47: 309–318 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K. (2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA 107: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Shinozaki K. (2010) ABA transport factors found in Arabidopsis ABC transporters. Plant Signal Behav 5: 1124–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Sugimoto E, Shinozaki K. (2011) Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J 67: 885–894 [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Shin JJH, Bird DA, Samuels AL. (2010) Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22: 3066–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montillet JL, Leonhardt N, Mondy S, Tranchimand S, Rumeau D, Boudsocq M, Garcia AV, Douki T, Bigeard J, Laurière C, et al. (2013) An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol 11: e1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffière A, Yan S, Dominguez H, Thompson BM. (2008) The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J 2: 321–334 [DOI] [PubMed] [Google Scholar]
- Oerke EC. (2006) Crop losses to pests. J Agric Sci 144: 31–43 [Google Scholar]
- Pandey S, Wang RS, Wilson L, Li S, Zhao Z, Gookin TE, Assmann SM, Albert R. (2010) Boolean modeling of transcriptome data reveals novel modes of heterotrimeric G-protein action. Mol Syst Biol 6: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea PA. (2007) Plant ATP-binding cassette transporters. Annu Rev Plant Biol 58: 347–375 [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S. (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B. (2009) The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21: 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Balasubramanian S, Hu TT, Traw MB, Horton M, Epple P, Kuhns C, Sureshkumar S, Schwartz C, Lanz C, et al. (2010) Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465: 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukitsu H, Kuromori T, Toyooka K, Goto Y, Matsuoka K, Sakuradani E, Shimizu S, Kamiya A, Imura Y, Yuguchi M, et al. (2007) Cytological and biochemical analysis of COF1, an Arabidopsis mutant of an ABC transporter gene. Plant Cell Physiol 48: 1524–1533 [DOI] [PubMed] [Google Scholar]
- Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, Geisler M, Klein M, Kolukisaoglu U, Lee Y, Martinoia E, et al. (2008) Plant ABC proteins—a unified nomenclature and updated inventory. Trends Plant Sci 13: 151–159 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Weigel D. (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, Nomura K, Underwood W, He SY. (2013) Induction and suppression of PEN3 focal accumulation during Pseudomonas syringae pv. tomato DC3000 infection of Arabidopsis. Mol Plant Microbe Interact 26: 861–867 [DOI] [PubMed] [Google Scholar]
- Xue GP, Way HM, Richardson T, Drenth J, Joyce PA, McIntyre CL. (2011) Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4: 697–712 [DOI] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN., Jr (2006) Statistical analysis of real-time PCR data. BMC Bioinformatics 7: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X. (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.