Cadmium inhibits controlled nitrate uptake by roots, but this has the beneficial effect of reducing cadmium entry into roots.
Abstract
Identification of mechanisms that decrease cadmium accumulation in plants is a prerequisite for minimizing dietary uptake of cadmium from contaminated crops. Here, we show that cadmium inhibits nitrate transporter 1.1 (NRT1.1)-mediated nitrate (NO3−) uptake in Arabidopsis (Arabidopsis thaliana) and impairs NO3− homeostasis in roots. In NO3−-containing medium, loss of NRT1.1 function in nrt1.1 mutants leads to decreased levels of cadmium and several other metals in both roots and shoots and results in better biomass production in the presence of cadmium, whereas in NO3−-free medium, no difference is seen between nrt1.1 mutants and wild-type plants. These results suggest that inhibition of NRT1.1 activity reduces cadmium uptake, thus enhancing cadmium tolerance in an NO3− uptake-dependent manner. Furthermore, using a treatment rotation system allowing synchronous uptake of NO3− and nutrient cations and asynchronous uptake of cadmium, the nrt1.1 mutants had similar cadmium levels to wild-type plants but lower levels of nutrient metals, whereas the opposite effect was seen using treatment rotation allowing synchronous uptake of NO3− and cadmium and asynchronous uptake of nutrient cations. We conclude that, although inhibition of NRT1.1-mediated NO3− uptake by cadmium might have negative effects on nitrogen nutrition in plants, it has a positive effect on cadmium detoxification by reducing cadmium entry into roots. NRT1.1 may regulate the uptake of cadmium and other cations by a common mechanism.
Cadmium is highly toxic to humans (Nicholson et al., 1983), and its primary route of entry into the body is through crops grown in cadmium-contaminated soil (Clemens et al., 2013). A recent survey indicated that vegetables and rice (Oryza sativa) account for approximately 40% and 38%, respectively, of total cadmium exposure in residents of Shanghai, China’s largest city (He et al., 2013). However, cadmium contamination of agricultural soils as a result of rapid industrial development and release of agrochemicals into the environment is an increasingly serious problem. Many strategies have been proposed for remediating cadmium-contaminated soil to prevent cadmium uptake by crops. These strategies include the dig-and-dump method or encapsulation of the contaminated soil, chemical immobilization or extraction of cadmium, and phytoremediation by cadmium-hyperaccumulating plants (Pulford and Watson, 2003). However, the dig-and-dump and chemical methods are expensive, whereas phytoremediation requires several growing seasons to be effective, making it impractical in regions where farmland is limited and food supply insufficient.
The shortfalls of these strategies have prompted researchers to develop alternative techniques that are cost-effective and interfere less with crop production. Use of nitrogen fertilizers is one of the most important agronomic practices and it has been suggested that their appropriate use might provide a relatively inexpensive, time-saving, and effective strategy for reducing cadmium entry into, and accumulation in, crops because NO3− facilitates cadmium uptake in hydroponically grown plants (Sarwar et al., 2010; Luo et al., 2012). However, in a preliminary study, we found that, in plants grown in soil, the effect of the nitrogen form on cadmium accumulation was strongly associated with the pH-buffering capacity of the soil. In soil with a lower pH-buffering capacity, application of ammonium (NH4+) resulted in higher cadmium levels in plants than application of NO3−, probably as a result of soil acidification by NH4+, and the opposite effect was seen when plants were grown in soil with higher pH-buffering capacity (S.K. Fan, S.T. Du, and C.W. Jin, unpublished data). This suggests that management of the use of nitrogen fertilizers to prevent cadmium entry into crops might be difficult because of the wide variation in soil pH-buffering capacity.
Because NO3− facilitates cadmium uptake in hydroponically grown plants as described above, modification of NO3− uptake pathways in plants might also affect cadmium uptake, in which case modifying these pathways to reduce cadmium entry into crops could circumvent the risks and the difficulties involved in nitrogen fertilizer management. Exposure to cadmium has been shown to reduce NO3− uptake by roots (Hernández et al., 1997; Gouia et al., 2000; Rizzardo et al., 2012), but this has been assumed to be deleterious to plant growth (Finkemeier et al., 2003; Rizzardo et al., 2012). The process by which NO3− is taken up across the root plasma membrane is complex, and several nitrate transporters (NRTs) involved in NO3− uptake from the growth medium have been characterized. In Arabidopsis (Arabidopsis thaliana), NRT1.1 is a dual-affinity transporter involved in both high- and low-affinity uptake, NRT1.2 is involved only in low-affinity NO3− uptake, whereas NRT2.1, NRT2.2, and NRT2.4 are only involved in high-affinity NO3− uptake (Wang et al., 2012; Léran et al., 2014). However, the transporter responsible for the cadmium-induced decrease in NO3− uptake remains unknown. Given the presumed association between NO3− uptake and cadmium uptake, it is important to identify the molecular mechanism involved in this process, and it is particularly important to determine whether the modulation of relevant NO3− transporters affects cadmium entry into plants.
In this study, we investigated the relationship between NO3− uptake and cadmium uptake in Arabidopsis roots. To our knowledge, our results reveal a new mechanism for resisting cadmium toxicity: Cadmium reduces NO3− uptake by inhibiting NRT1.1 activity, which in turn reduces cadmium entry into root cells. As a result, cadmium levels in plants are lower and plant growth is improved. Our findings may provide a strategy for minimizing cadmium accumulation in crops grown in contaminated soil using biotechnological pathways to decrease NO3− uptake.
RESULTS
Cadmium Inhibits NO3− Uptake and Impairs NO3− Homeostasis in Roots
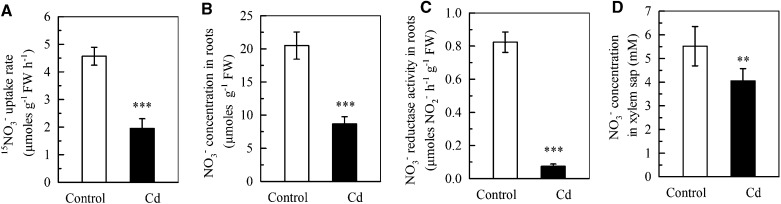
As described above, cadmium has been shown to inhibit NO3− uptake in several plant species (Hernández et al., 1997; Gouia et al., 2000; Rizzardo et al., 2012). NO3− uptake in the roots of ecotype Columbia-0 of Arabidopsis (Col-0) was evaluated using 15NO3− at the same concentration as the unlabeled form used for plant growth. As shown in Figure 1A, the rate of NO3− uptake was significantly decreased by more than 50% after 7 d of exposure to 10 μm cadmium. Because a reduced rate of NO3− uptake might negatively affect NO3− homeostasis in plants, we measured the NO3− level in the roots and found that it was also significantly reduced by approximately 50% (Fig. 1B). The NO3− level in roots is controlled by three integrated processes: uptake from the growth medium, assimilation by nitrate reductase (NR), and translocation from the roots to the shoots. An Illumina mRNA-sequencing (Seq) analysis showed that 7 d of treatment with 10 μm cadmium differentially affected the expression of the two NR-encoding genes NITRATE REDUCTASE1 (NIA1) and NIA2, with the transcript level of NIA1 being significantly decreased and transcript level of NIA2 slightly, but not significantly, increased (Table I). Expression of genes related to NO3− translocation was also differentially affected by cadmium. The transcript level of NRT1.5, a gene that encodes a transporter involved in xylem loading for root-to-shoot transport of NO3− (Lin et al., 2008), was reduced, whereas that of NRT1.8, a gene encoding a transporter that functions in removal of NO3− from the xylem sap (Li et al., 2010), was markedly increased (Table I). We therefore measured the NR activity in the roots and the NO3− level in the xylem sap and found that both were significantly decreased by cadmium exposure (Fig. 1, C and D). Because inhibition of NO3− assimilation and translocation would theoretically lead to an increased NO3− level in roots, we concluded that the observed reduction in NO3− accumulation in cadmium-exposed roots was due to reduced NO3− uptake. The rate of NO3− uptake evaluated above was measured using 2.25 mm 15NO3− and therefore included both high- and low-affinity uptake. We then measured high-affinity NO3− uptake using 200 μm NO3− and found that it was significantly increased by cadmium (Supplemental Fig. S1). This suggests that the decreased NO3− uptake seen in cadmium-exposed Col-0 plants grown under our normal growth conditions (2.25 mm NO3−) results from a dynamic interaction between increased high-affinity uptake and decreased low-affinity uptake.
Figure 1.
Effect of cadmium on NO3− uptake and concentration and NO3− reductase activity in the roots and the NO3− concentration in xylem sap in Col-0 plants. Plants were precultured in complete nutrient solution using 2.25 mm NO3− and 750 μm NH4+ as the nitrogen source for 5 weeks, and then were transferred to complete nutrient solution alone (control) or containing 10 μm CdCl2 (Cd) for 7 d, after which the analyses were performed. A, Rate of NO3− uptake by the roots. B, NO3− concentration in the roots. C, NO3− reductase activity in the roots. D, NO3− concentration in the xylem sap. NO3− uptake by Col-0 plants was measured using 2.25 mm 15NO3− for 5 min. Bars represent the sd (n = 4–5 biological replicates). Asterisks indicate significant differences compared with the controls (**P < 0.01, ***P < 0.001; two-tailed Student’s t test). FW, Fresh weight.
Table I. Effects of cadmium treatment on expression in roots of genes related to NO3− uptake, assimilation, and translocation.
Gene expression was analyzed by Illumina mRNA-Seq. A statistical cutoff of P < 0.05 after Bonferroni correction was used to determine which genes were differentially expressed. Positive ratios indicate gene induction; negative ratios indicate gene repression.
| Gene ID | Annotation | Control | Cadmium | Cadmium to Control Ratio | P Value |
|---|---|---|---|---|---|
| FPKM | log2 | ||||
| AT1G08090 | NRT2.1 | 59.49 | 425.22 | 2.84 | 7.11E-08a |
| AT1G08100 | NRT2.2 | 0.11 | 1.6302 | 3.90 | 7.89E-04b |
| AT1G18880 | NRT1.9 | 57.63 | 47.42 | −0.28 | 6.60E-01d |
| AT1G12110 | NRT1.1 | 718.32 | 242.21 | −1.57 | 1.66E-04b |
| AT1G32450 | NRT1.5 | 630.92 | 330.96 | −0.93 | 1.60E-02c |
| AT1G37130 | NIA2 | 195.46 | 307.52 | 0.65 | 2.87E-01d |
| AT1G69850 | NRT1.2 | 109.58 | 141.10 | 0.36 | 4.42E-01d |
| AT1G77760 | NIA1 | 629.52 | 230.05 | −1.45 | 4.19E-02c |
| AT4G21680 | NRT1.8 | 0.44 | 35.98 | 6.37 | 2.74E-03c |
| AT5G50200 | High-affinity NRT3.1 | 287.74 | 723.25 | 1.33 | 1.24E-02c |
| AT5G60770 | NRT2.4 | 0.12 | 0.07 | −0.87 | 6.51E-01d |
P < 1.00E-04. bP < 1.00E-03. cP < 5.00E-02. dP < 1.
Inhibition of NRT1.1 Activity Results in Decreased NO3− Uptake
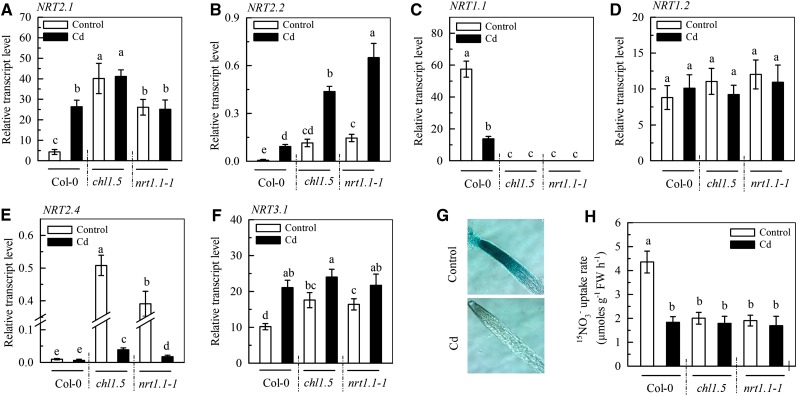
In Arabidopsis, NRT1.1, NRT1.2, NRT2.1, NRT2.2, and NRT2.4 are involved in root uptake of NO3− from the growth medium (Wang et al., 2012; Léran et al., 2014). To investigate the molecular basis underlying the inhibition of NO3− uptake in cadmium-exposed Col-0 plants under our growth conditions, we examined the expression of these five NRT genes in roots using Illumina mRNA-Seq analysis and found that only the expression of NRT1.1 was significantly decreased by 7 d of treatment with 10 μm cadmium, expression of the other NRT genes either being increased (NRT2.1 and NRT2.2) or not affected (NRT1.2 and NRT2.4; Table I). The expression of NRT3.1, which encodes a protein required for NRT2.1-mediated transport activity (Tsay et al., 2007), was also significantly increased by cadmium (Table I). These results were confirmed by real-time quantitative PCR (Col-0 in Fig. 2, A–F). These results, together with the finding of significantly increased high-affinity uptake in cadmium-exposed Col-0 roots (Supplemental Fig. S1), suggested that the inhibition of NO3− uptake by cadmium measured at 2.25 mm resulted from inhibition of NRT1.1 activity, rather than changes in other NRTs. Consistent with this notion, GUS staining of the roots of pNRT1.1::NRT1.1-GUS transgenic plants showed that 7 d of treatment with 10 μm cadmium caused a large decrease in the NRT1.1-GUS protein level (Fig. 2G). We then compared the rate of NO3− uptake by the roots of Col-0 plants and two NRT1.1-null mutants, chlorate-resistant1.5 (chl1.5) and nrt1.1-1, using 2.25 mm 15NO3−. In cadmium-free medium, the rate of NO3− uptake by Col-0 plants was more than double that in the nrt1.1 mutants, and cadmium had little effect on the NO3− uptake of the nrt1.1 mutants but decreased the rate of uptake by Col-0 plants to the same level as in the mutants (Fig. 2H). These results demonstrate that inhibition of NRT1.1 activity was responsible for the reduction in the rate of NO3− uptake measured at 2.25 mm in the presence of cadmium.
Figure 2.
Role of NRT1.1 in the cadmium-induced inhibition of NO3− uptake. A to F, The treatments of the Col-0 plants, chl1.5 and nrt.1-1 mutants, and pNRT1.1::NRT1.1-GUS transgenic plants were the same as those in Figure 1, A to F. For real-time quantitative PCR analysis of expression of NRT2.1, NRT2.2, NRT1.1, NRT1.2, NRT2.4, and NRT3.1, transcript levels were normalized to those of UBIQUITIN10 mRNA (100%). G, GUS staining of the root of pNRT1.1::NRT1.1-GUS transgenic plants. H, NO3− uptake rate in Col-0, chl1.5, and nrt1.1-1 plants measured using 2.25 mm 15NO3– for 5 min. The bars in A to F and H represent the sd (n = 4–8 biological replicates). Different lowercase letters above bars indicate significant differences at P < 0.05 (lsd test). FW, Fresh weight.
The nrt1.1 Mutants Have Increased Cadmium Tolerance and Lower Cadmium Levels
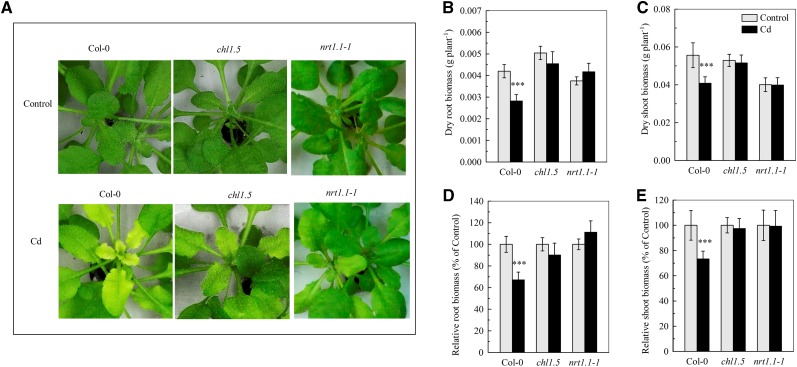
We then investigated the association between NRT1.1 and cadmium tolerance in Arabidopsis plants. After 7 d of exposure to 10 μm cadmium, the newly formed leaves of Col-0 plants developed severe chlorosis, whereas this effect was clearly less pronounced in the chl1.5 and nrt1.1-1 mutants (Fig. 3A). Furthermore, the root and shoot biomasses of Col-0 plants exposed to cadmium were significantly reduced by approximately 40% and 30%, respectively, whereas there was no significant effect in the chl1.5 and nrt1.1-1 mutants (Fig. 3, B–E). Similar results were obtained after 7 d of exposure to 20 μm cadmium (Supplemental Fig. S2A), conditions used in latter two studies on cadmium uptake in NO3−-free medium. In addition, after exposure to 10 μm cadmium for 7 d, the root and shoot biomasses of a third NRT1.1-null mutant, chl1.6, were not affected by cadmium, whereas those of the corresponding wild type, Landsberg erecta (Ler), were significantly decreased by cadmium (Supplemental Fig. S3A). These results suggest that inhibition of NRT1.1 activity could be a means of defense against cadmium toxicity in plants.
Figure 3.
Sensitivity of Col-0, chl1.5, and nrt1.1-1 plants to cadmium. The plants were treated in the same way as in Figure 1. A, Chlorosis of the newly formed leaves. B, Dry root biomass. C, Dry shoot biomass. D, Relative root biomass. E, Relative shoot biomass. The relative biomass was calculated as the mean dry weight expressed as a percentage of the control dry weight in the same plant line. Bars represent the sd (n = 8 biological replicates). Asterisks indicate a significant difference from the control value (***P < 0.001; two-tailed Student’s t test).
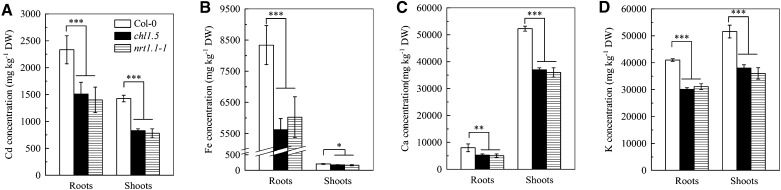
We next measured cadmium levels and found that both the roots and shoots of the chl1.5 and nrt1.1-1 mutants contained significantly lower levels than those of Col-0 plants after 7 d of exposure to either 10 μm cadmium (Fig. 4A) or 20 μm cadmium (Supplemental Fig. S2B). In addition, after 7 d of exposure to 10 μm cadmium, cadmium levels in the shoots and roots of the chl1.6 mutant were significantly lower than those in Ler plants (Supplemental Fig. S3B). These results indicate that lack of NRT1.1 function reduces cadmium entry into plants, validating our previous assumption that modification of the NO3− uptake pathways might affect cadmium uptake in plants. Interestingly, iron, calcium, and potassium levels were also significantly lower in the two nrt1.1 mutants than in the Col-0 plants after exposure to 10 μm cadmium for 7 d (Fig. 4, B–D).
Figure 4.
Metal concentrations in Col-0, chl1.5, and nrt1.1-1 plants. Plants were precultured as described in Figure 1 for 5 weeks, and then were transferred to complete nutrient solution containing 10 μm CdCl2 for 7 d, after which the metal concentrations in the roots and shoots were measured. A, Cadmium concentration. B, Iron concentration. C, Calcium concentration. D, Potassium concentration. Bars represent the sd (n = 5 biological replicates). Asterisks indicate significant differences compared with the Col-0 plants (*P < 0.05, **P < 0.01, ***P < 0.001; two-tailed Student’s t test). DW, Dry weight.
The nrt1.1 Mutants Do Not Show Decreased Cadmium Uptake in NO3−-Free Growth Medium
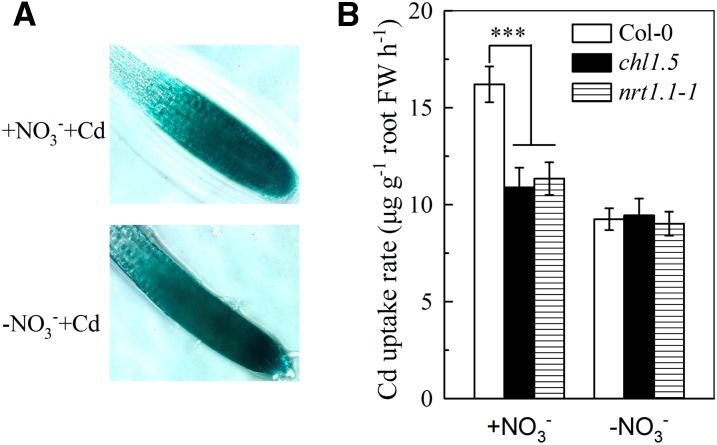
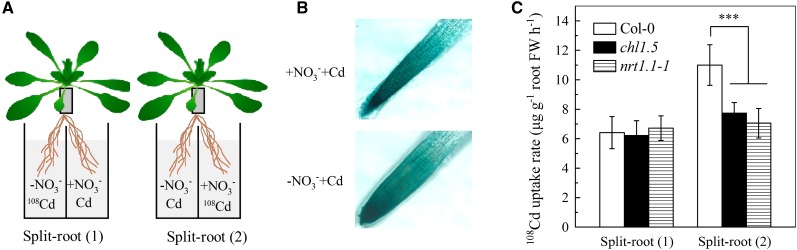
Because NRT1.1 is also involved in numerous physiological processes in addition to NO3− uptake (Ho et al., 2009), it was important to clarify whether regulation of cadmium uptake by NRT1.1 was associated with NO3− uptake. When 6-week-old pNRT1.1::NRT1.1-GUS plants were transferred for 1 h to complete nutrient solution or NO3−-free nutrient solution (both containing 20 μm cadmium), GUS staining of the two sets of roots was similar (Fig. 5A), indicating that short-term removal of NO3− from the growth medium had little effect on NRT1.1 activity. We then measured cadmium uptake by the roots in 1 h in NO3−-free medium and found no difference between Col-0 plants and the chl1.5 and nrt1.1-1 mutants. By contrast, in NO3−-containing growth medium, cadmium uptake was significantly higher in Col-0 plants than in the chl1.5 or nrt1.1-1 mutant (Fig. 5B). These results show that regulation of cadmium uptake by NRT1.1 is NO3− uptake dependent. To further verify this conclusion, a split-root experiment was designed. As shown in Figure 6A, one-half of the roots of each plant were bathed for 1 h in NO3−-free medium containing 20 μm 108Cd and the other one-half in NO3−-containing medium containing 20 μm unlabeled cadmium (split-root system 1) or in NO3−-free medium containing 20 μm unlabeled cadmium and the other one-half in NO3−-containing medium containing 20 μm 108Cd (split-root system 2). This split-root experiment allows the same NO3− supply to be provided to all plants regardless of labeling treatments. Figure 6B shows that comparable GUS staining was seen in pNRT1.1::NRT1.1-GUS roots in the NO3−-containing side of split-root system 1 and the NO3−-free side of split-root system 2, showing NRT1.1 activity was not affected by the 1 h of localized NO3− removal. As expected, the roots of Col-0 plants in the NO3−-containing side of split-root system 2 showed significantly higher 108Cd uptake than those of the chl1.5 or nrt1.1-1 mutant, whereas in the NO3−-free side of side of split-root system 1, there was no difference in 108Cd uptake between the three types of plant (Fig. 6C), providing further evidence that NO3− uptake is necessary for regulation of cadmium uptake by NRT1.1. We then grew Col-0 plants and the two nrt1.1 mutants in NO3−-free medium containing 10 μm cadmium for 7 d and found no significant difference between them in terms of the reduction in the biomass caused by cadmium (Supplemental Fig. S4A) or in cadmium levels (Supplemental Fig. S4B) in the roots or shoots. These results clearly contrast with those obtained for plants grown in NO3−-containing medium (Figs. 3, D and E, and 4A), and again indicate that the reduction in cadmium uptake by inhibition of NRT1.1 is dependent on NO3− uptake.
Figure 5.
Effects of short-term NO3− removal on cadmium uptake in Col-0, chl1.5, and nrt1.1-1 plants. Plants were precultured as described in Figure 1 for 6 weeks and then were transferred to either complete (+NO3−) or NO3−-free (−NO3−) nutrient solution containing 20 μm CdCl2 for 1 h. A, GUS staining of the roots of pNRT1.1::NRT1.1-GUS transgenic plants. B, Rate of cadmium uptake by roots of Col-0, chl1.5, and nrt1.1-1 plants. Bars represent the sd (n = 5 biological replicates). Asterisks indicate significant differences compared with Col-0 plants (***P < 0.001; two-tailed Student’s t test). FW, Fresh weight.
Figure 6.
108Cd uptake by roots grown in a split-root system. Plants were precultured as described in Figure 1 for 5 weeks, and then the root system of each plant was divided into two approximately equal parts, each of which was transferred to a different container in complete nutrient solution. The plant was left undisturbed for 1 week, and then the roots were immersed in either complete (+NO3−) or NO3−-free (−NO3−) nutrient solution containing either 20 μm CdCl2 or 20 μm 108CdCl2 for 1 h, as indicated. A, Schematic representation of the split-root experimental protocol. B, GUS staining of the roots of pNRT1.1::NRT1.1-GUS transgenic plants. C, 108Cd uptake rates by Col-0, chl1.5, and nrt1.1-1 roots. The bars represent the sd (n = 5 biological replicates). Asterisks indicate significant differences compared with Col-0 plants (***P < 0.001; two-tailed Student’s t test). FW, Fresh weight.
The irt1/nrt1.1 Double Mutant Has Lower Cadmium Levels than irt1 Mutants
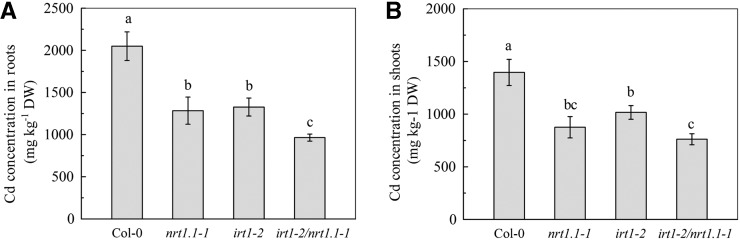
To investigate how cadmium uptake was reduced by inhibition of NRT1.1-mediated NO3− uptake, we measured mRNA levels in the roots of Col-0 plants and the chl1.5 mutant using Illumina mRNA-Seq. Because cadmium can enter root cells via various transporters/channels for various bivalent nutrient cations (Verbruggen et al., 2009; Lux et al., 2011; Clemens et al., 2013), we first focused on the expression of these transporter genes in roots (Supplemental Table S1). After 7 d of exposure to 10 μm cadmium, the chl1.5 mutant did not show any significant decrease in expression of any of these genes compared with Col-0 plants, whereas in the absence of cadmium, only IRT1, the gene coding for iron-regulated transporter1 (Vert et al., 2002), showed significantly lower expression in chl1.5 mutants than in Col-0 plants. These findings implied that the decrease in cadmium uptake caused by inhibition of NRT1.1 might be a result of lower IRT1 activity. We therefore generated an irt1/nrt1.1 double mutant by crossing the IRT1-null mutant irt1-2 with the nrt1.1-1 mutant and measured root and shoot cadmium concentrations. As shown in Figure 7, cadmium levels in the nrt1.1-1 and irt1-2 mutants were significantly lower than those in Col-0 plants, but were even lower in the irt1-2/nrt1.1-1 double mutant. Because the NO3− status of the growth medium affects IRT1 expression (Zhao and Ling, 2007), we also cultivated the plants in medium containing a higher concentration of NO3− (10 mm instead of the normal 2.25 mm) and measured IRT1 mRNA levels. Interestingly, in the absence of cadmium, similar IRT1 mRNA levels were seen in Col-0 plants and the chl1.5 and nrt1.1-1 mutants, but in the presence of cadmium, levels were significantly lower in the chl1.5 and nrt1.1-1 plants than in the Col-0 plants (Supplemental Fig. S5). However, cadmium levels in both the roots and shoots were also significantly lower in the irt1-2/nrt1.1-1 double mutant than in the irt1-2 mutant (Supplemental Fig. S6). These results indicate that inhibition of IRT1 activity does not explain, or at least does not fully explain, why blocking NO3− uptake by NRT1.1 reduces cadmium entry into roots.
Figure 7.
Cadmium concentrations in Col-0, nrt1.1-1, irt1-2, and irt1-2/nrt1.1-1 plants after 7 d of cadmium exposure. The indicated plants were treated as in Figure 4. A, Root cadmium concentration. B, Shoot cadmium concentration. Bars represent the sd (n = 5 biological replicates). Different lowercase letters above bars indicate significant differences at P < 0.05 (lsd test). DW, Dry weight.
The nrt1.1 Mutants Do Not Show Reduced Metal Contents when Metal Ions and NO3− Are Provided Synchronously
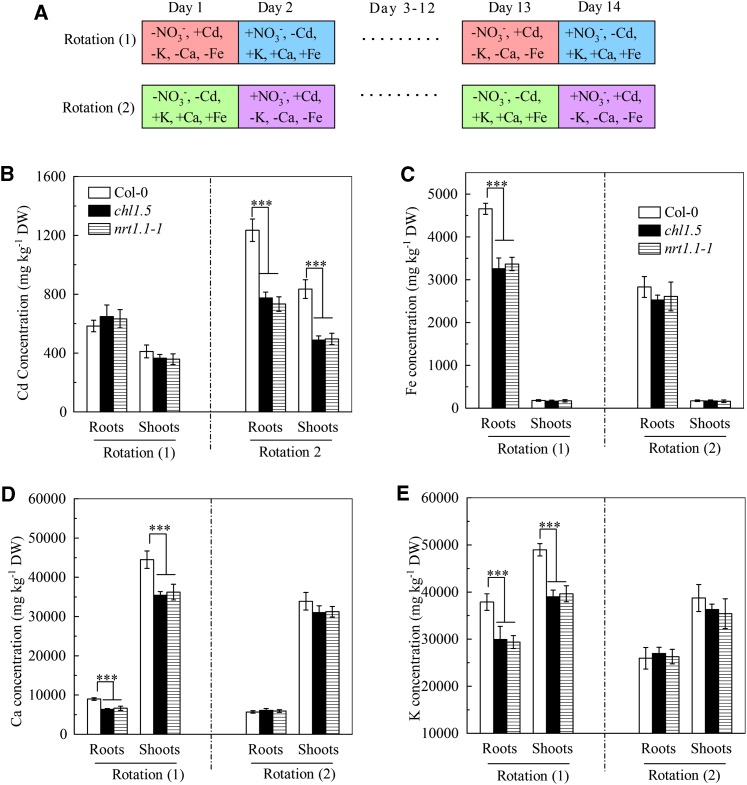
Because the nrt1.1 mutants contained lower levels of cadmium and of several other metals than Col-0 plants (Fig. 4, B–D), we thought that uptake of cadmium and other cations might be regulated by NRT1.1 by a common mechanism and therefore designed two NO3−-cation treatment rotation schemes (seven cycles of two treatments on alternate days) to test this hypothesis. As shown in Figure 8A, using the rotation system 1, the plants were incubated in medium lacking Fe2+, Ca2+, K+, and NO3−, but containing Cd2+ on one day and in medium containing Fe2+, Ca2+, K+, and NO3−, but no Cd2+ the next day. Using the rotation system 2, the plants were incubated with medium containing Fe2+, Ca2+, and K+ but no NO3− or Cd2+ on one day and with medium lacking Fe2+, Ca2+, and K+, but containing NO3− and Cd2+ the next day. As shown in Figure 8, B to E, after 14 d of growth using rotation system 1, both the chl1.5 and nrt1.1-1 mutants had significantly lower iron, calcium, and potassium levels, but not cadmium levels, than Col-0 plants. Using rotation system 2, the two nrt1.1 mutants showed significantly lower cadmium levels, but not iron, calcium, and potassium levels, than Col-0 plants. These results support the above idea that the mechanism by which NRT1.1 regulates uptake of the other cations may be the same as that by which it regulates cadmium uptake (i.e. both processes require the simultaneous uptake of NO3−).
Figure 8.
Effects of NO3−-cation treatment rotation on metal concentrations in Col-0, chl1.5, and nrt1.1-1 plants. A, Scheme showing the protocol of NO3−-cation treatment rotations. Plants were precultured in NO3−-free medium with 3 mm NH4+ as the sole nitrogen source for 4 weeks and then underwent two treatment rotation schemes as follows. In rotation system 1, the plants were grown on alternate days for 14 d in complete nutrient solution lacking NO3−, potassium, calcium, and iron, but containing 10 μm cadmium (−NO3−, +Cd, −K, −Ca, and −Fe; 1), or complete nutrient solution (+NO3−, −Cd, +K, +Ca, and +Fe; 2). In rotation system 2, the plants were grown on alternate days for 14 d in complete nutrient solution lacking NO3− (−NO3−, −Cd, +K, +Ca, and +Fe; 1), or complete nutrient solution lacking potassium, calcium, and iron but containing 10 μm cadmium (+NO3−, +Cd, −K, −Ca, and −Fe; 2). B to E, Metal concentrations. Bars represent the sd (n = 5–8 biological replicates). Asterisks indicate significant differences compared with Col-0 plants (***P < 0.001; two-tailed Student’s t test). DW, Dry weight. [See online article for color version of this figure.]
DISCUSSION
Plants can employ a number of strategies to minimize cadmium toxicity, including immobilization (e.g. binding to the cell wall), compartmentalization (e.g. vacuolar segregation), and chelation (e.g. cadmium-phytochelatin and cadmium-metallothionein complexation: Verbruggen et al., 2009; Lux et al., 2011; Mendoza-Cózatl et al., 2011; Clemens et al., 2013). However, these strategies do not improve the safety of crops for human consumption because they do not reduce cadmium accumulation in the plant. It has been assumed that prevention of cadmium entry into roots might be an additional strategy by which plants could resist cadmium toxicity (Sanità di Toppi and Gabbrielli, 1999), but little evidence is available to support this idea. In this study, we revealed a mechanism of plant resistance to cadmium toxicity, namely that cadmium inhibits NRT1.1-mediated NO3− uptake in roots, which in turn reduces cadmium entry into roots, thus facilitating cadmium detoxification in plants. It may therefore be possible to design other more practical methods for inhibiting NRT1.1-mediated NO3− uptake and reducing cadmium contamination of food.
Although several NRTs are involved in NO3− uptake in Arabidopsis, under our growth conditions, cadmium only inhibited the NO3− uptake controlled by NRT1.1 (Fig. 2; Table I). NRT1.1 is a dual-affinity transporter involved in both high- and low-affinity uptake (Liu et al., 1999). However, high-affinity NO3− uptake in Col-0 plants was unexpectedly found to be increased after cadmium exposure (Supplemental Fig. S1), probably as a result of induction of NRT2.1, NRT2.2, and NRT3.1 (Fig. 2, A, B, and F; Table I), suggesting that the inhibitory effect of cadmium on NRT1.1-mediated NO3− uptake in Col-0 plants may have been underestimated under our growth conditions. This inhibition of NO3− uptake resulted in impaired NO3− homeostasis in the plants (Fig. 1B), which might result in insufficient nitrogen supply and reduced growth. In addition, several nitrogen-containing compounds, including phytochelatins, reduced glutathione, and metallothionein, are involved in cadmium detoxification (Verbruggen et al., 2009), and insufficient uptake of NO3− is thought to be detrimental to cadmium detoxification (Finkemeier et al., 2003; Rizzardo et al., 2012). However, this assumption was not supported by our study, because the three nrt1.1 mutants chl1.5, nrt1.1-1, and chl1.6 showed higher cadmium tolerance and lower cadmium levels than the corresponding wild-type plants (Figs. 3 and 4; Supplemental Figs. S2 and S3), providing evidence that inhibition of NRT1.1 helps to prevent cadmium toxicity.
In addition to NO3− uptake, NRT1.1 functions in many other physiological processes (Ho et al., 2009), such as regulation of primary root growth (Guo et al., 2001), triggering of root colonization of NO3−-rich patches (Remans et al., 2006), regulation of another NO3− transporter NRT2.1 (Muños et al., 2004), auxin transport (Krouk et al., 2010), and regulation of tolerance to NH4+ toxicity (Hachiya et al., 2011). Some functions of NRT1.1 are independent of NO3− uptake. However, our findings that nrt1.1 mutants did not have lower cadmium levels or show better growth than wild-type plants when NH4+ was the sole nitrogen source (Supplemental Fig. S4) show that NO3− uptake is necessary for regulation of cadmium uptake by NRT1.1. Further support for this conclusion was provided by measurements of cadmium uptake during short-term removal of NO3− in a single growth medium or in split-root studies, which showed that the higher cadmium uptake in Col-0 plants compared with nrt1.1 mutants was abolished by removing NO3− from the growth medium (Figs. 5 and 6).
Recent studies have shown that induction of NRT1.8 or inhibition of NRT1.5 favors cadmium tolerance in plants, probably by reducing the amount of cadmium translocated from the roots to the shoots (Li et al., 2010; Chen et al., 2012). However, neither NRT1.5 nor NRT1.8 appeared to play a role in NRT1.1-regulated cadmium uptake, because expression of these genes in the nrt1.1 mutants was affected by cadmium in a similar manner to that in Col-0 plants (Supplemental Fig. S7). NRT1.1 was recently proposed to function similarly to NRT1.5 in root-to-shoot NO3− translocation (Léran et al., 2013). However, the results of our short-term NO3− removal study did not support a role of this recently proposed NRT1.1 function in regulating cadmium uptake by roots. During short-term removal of NO3−, NRT1.1-controlled NO3− translocation probably continued normally in Col-0 plants (GUS staining in pNRT1.1::NRT1.1-GUS plants was barely affected; Figs. 5A and 6B), but no difference in cadmium uptake was seen between Col-0 plants and the nrt1.1 mutants (Figs. 5B and 6C). However, we cannot exclude the possibility that inhibition of NRT1.1 might act in parallel with inhibition of NRT1.5 to enhance cadmium tolerance by reducing cadmium translocation from the roots to the shoots. Under our growth conditions, this effect may have been concealed by the decrease in cadmium uptake caused by inhibition of NO3− uptake by NRT1.1. Future studies need to distinguish between the effects of NO3− translocation and NO3− uptake to determine whether this is the case.
It is worth noting that the high-affinity NO3− uptake in the nrt1.1 mutants in both the presence and absence of cadmium was higher than that in Col-0 plants in the absence of cadmium (Supplemental Fig. S1), probably due to the higher expression of NRT2.1, NRT2.2, NRT2.4, and NRT3.1 in the mutants than in the wild-type plants (Fig. 2, A–F). Furthermore, the high-affinity NO3− uptake in Col-0 plants was increased after cadmium exposure (Supplemental Fig. S1). It was therefore necessary to clarify whether the increase in high-affinity NO3− uptake was involved in the regulation of cadmium uptake by NRT1.1. We used an NRT2.1-null mutant nrt2.1 to clarify this issue. Because NRT2.1 is a high-affinity NO3− transporter (Cerezo et al., 2001), we grew the plants in low NO3− (0.2 mm) medium and found that shoot and root cadmium levels in the nrt2.1 mutant were significantly lower than those in Col-0 plants (Supplemental Fig. S8). This suggests that the increase in high-affinity NO3− uptake, or at least the increase in NRT2.1-mediated high-affinity NO3− uptake, does not play a role in NRT1.1-regulated cadmium uptake and may hinder attempts at preventing cadmium uptake by the roots. The question remains regarding why high-affinity NO3− uptake in Col-0 plants is increased in the presence of cadmium. One possible explanation is that it is increased to compensate for the decreased NRT1.1-mediated NO3− uptake so as to minimize the impairment of NO3− homeostasis in cadmium-treated plants.
Because cadmium is a nonessential element for plants, entry of cadmium into root cells may rely on transporters/channels for various bivalent nutrient cations (Verbruggen et al., 2009; Lux et al., 2011; Clemens et al., 2013). Transcript analysis suggested that the reduced cadmium uptake associated with blocking of NRT1.1 activity might be a result of inhibition of IRT1 activity (Supplemental Fig. S5; Supplemental Table S1). However, this speculation was refuted by the observation that cadmium levels in the irt1-2 mutant were significantly higher than those in the irt1-2/nrt1.1-1 double mutant (Fig. 7; Supplemental Fig. S6). The mechanism by which NRT1.1 regulates cadmium uptake therefore remains unknown. Our NO3−-cation treatment rotation study showed that NRT1.1 might regulate the uptake of cadmium and other cations by a common mechanism involving the simultaneous uptake of NO3− (Fig. 8), but this needs to be verified directly. Theoretically, decreased uptake of the anion NO3− should be accompanied by decreased cation uptake so as to maintain the ionic balance in the roots. Previous studies have shown that plants fed the cation NH4+ contain lower concentrations of metal nutrients than plants fed the anion NO3− (Kirkby and Mengel, 1967; Kirkby and Knight, 1977; Van Beusichem et al., 1988). Thus, an ion-balancing process may be the common mechanism by which NRT1.1-mediated NO3− uptake regulates cadmium uptake and the uptake of the other cations. If this were the case, inhibition of NO3− uptake controlled by other NRTs may also decrease cadmium uptake. The observation of lower cadmium levels in the nrt2.1 mutant than in the Col-0 plants supports this idea.
In conclusion, although inhibition of NO3− uptake by cadmium may be detrimental to nitrogen nutrition in plants, it facilitates cadmium detoxification. Most previously identified cadmium detoxification mechanisms rely primarily on changes in the form or distribution of cadmium in plant tissues, but not on the exclusion of cadmium from plants. Here, we describe such a mechanism. Modification of NO3− uptake in crops by modulating NRT1.1 activity might provide a biological engineering approach to reducing accumulation of cadmium in edible organs, thus improving food safety.
MATERIALS AND METHODS
Plant Material
The mutants chl1.5 (Huang et al., 1996), nrt1.1-1 (salk_097431), nrt2.1 (cs859604), and irt1-2 (salk_054554; Nishida et al., 2011) and the pNRT1.1::NRT1.1-GUS transgenic plant line (Guo et al., 2001) were on the Arabidopsis (Arabidopsis thaliana) Col-0 background, whereas the chl1.6 (cs6154) mutant was on the Ler background (Tsay et al., 1993). The chl1.5 and irt1-2 seeds were a kind gift from Dr. Philippe Nacry (Biochimie et Physiologie Moléculaire des Plantes) and Dr. Takafumi Mizuno (Mie University), respectively. The salk_097431, cs6154, cs859604, and pNRT1.1::NRT1.1-GUS (cs6513) seeds were purchased from the Arabidopsis Biological Resource Center; the seeds for the last two plant lines were donated to the Arabidopsis Biological Resource Center by Dr. Nigel Crawford. The insertions in these lines were verified using the primers listed in Supplemental Table S2. The irt1-2/nrt1.1-1 double mutant was obtained by crossing nrt1.1-1 with irt1-2, and the homozygous line irt1-2/nrt1.1-1 was isolated and confirmed by PCR using the gene-specific primers listed in Supplemental Table S2.
Hydroponic Culture
Seeds were germinated on a nylon net floating in complete nutrient solution [750 μm NaH2PO4, 500 μm MgSO4, 375 μm K2SO4, 2.25 mm KNO3, 375 μm (NH4)2SO4, 1 mm CaCl2, 10 μm H3BO3, 0.5 μm MnSO4, 0.5 μm ZnSO4, 0.1 μm CuSO4, 0.1 μm (NH4)6Mo7O24, and 25 μm Fe-EDTA, pH 5.8]. On d 7, the seedlings were transferred to sand supplemented with fresh complete nutrient solution. After 14 d, batches of four seedlings were transplanted into 0.4-L pots filled with complete nutrient solution, which was renewed every other day. At 5 or 6 weeks of age (as indicated in the text), these plants were used in studies and treated as indicated in the figure legends, except in the case of the NO3−-cation treatment rotation study described below. In NO3− removal studies, the K+ equilibrium of the nutrient solution was maintained by replacing KNO3 with K2SO4.
Treatment Rotation Study
Because precultivation in NO3−-containing medium might result in lower iron, calcium, and potassium levels in the nrt1.1 mutants than in Col-0 plants, the plants were precultured for 4 weeks in NO3−-free medium using 3 mm NH4+ as the sole nitrogen source prior to NO3−-cation treatment rotation studies. At 4 weeks, the concentrations of iron, calcium, and potassium in the chl1.5 and nrt1.1-1 mutants were similar to those in Col-0 plants (data not shown). The 4-week-old plants were then exposed to NO3−-cation treatment rotations as indicated in Figure 8A using the following four media, some of which were prepared by modifying the complete nutrient solution as follows: (1) removal of K2SO4, KNO3, CaCl2, and Fe-EDTA and addition of 10 μm CdCl2 (−NO3−, +Cd, −K, −Ca, and −Fe); (2) complete nutrient solution (+NO3−, −Cd, +K, +Ca, and +Fe); (3) replacement of KNO3 with 1125 μm K2SO4 (−NO3−, −Cd, +K, +Ca, and +Fe); and (4) replacement of KNO3 with 2.25 mm NaNO3, removal of K2SO4, CaCl2, and Fe-EDTA, and addition of 10 μm CdCl2 (+NO3−, +Cd, −K, −Ca, and −Fe). Addition of 3 mm NaCl to complete nutrient solution containing 10 μm CdCl2 had little effect on cadmium uptake or growth of Col-0 plants and nrt1.1 mutants (data not shown), so the effect of Na+ addition in the fourth growth medium on plant cadmium uptake should be negligible.
Measurement of 15NO3− Uptake Rate, NR Activity, and NO3− Concentrations
The plants were precultured as described above for 5 weeks and then were transferred to complete nutrient solution with or without 10 μm cadmium for 7 d, after which the following analyses were performed. To measure the rate of 15NO3− uptake, the plants were washed for 1 min in 0.1 mm CaSO4 and then transferred for 5 min to cadmium-free or cadmium-containing complete nutrient solution in which KNO3 was replaced by either 2.25 mm K15NO3 (atom % 15N: 99%) or 200 μm K15NO3 and 1.025 mm K2SO4. They were then washed for 1 min in 0.1 mm CaSO4, after which the roots were dried for 72 h at 80°C and analyzed using an isotope ratio mass spectrometer (Finnigan MAT Delta S), 15NO3– influx being calculated from the total nitrogen and 15N content. NR activity was measured as described by Jin et al. (2009b). NO3− concentrations in the roots and xylem sap were measured as described by Li et al. (2010).
Measurement of Short-Term Cadmium Uptake
For measurements in a single growth medium, plants were precultured as described above for 6 weeks, transferred to either complete or NO3−-free nutrient solution containing 20 μm CdCl2 for 1 h, and then quickly rinsed with complete nutrient solution and transferred to a solution of 2 mm MES and 5 mm CaCl2 for 15 min. The fresh weight of the roots was then recorded and cadmium levels measured by inductively coupled plasma-mass spectrometry.
For measurements in split-root growth media, the root system of each 5-week-old plant was gently separated into two approximately equal portions, which were placed in separate containers (Fig. 6A) and supplied with complete nutrient solution for 1 week to allow the plant to adapt to split-root conditions. Different nutrient mixtures (complete nutrient solution or NO3−-free nutrient solution containing either 20 μm CdCl2 or 20 μm 108CdCl2 [atom % 108Cd: 71%]) were then added for 1 h to the two containers (Fig. 6A), and then both sides of the root system were rinsed as described above and the fresh root weight of the 108Cd-treated side was recorded and 108Cd levels were measured by inductively coupled plasma-mass spectrometry.
GUS Expression Analysis
Histochemical assay of GUS gene expression in the roots of pNRT1.1::NRT1.1-GUS transgenic plants was performed as described by Guo et al. (2001) and the distribution and intensity of the blue product were observed under a microscope (Nikon Eclipse E600; Nikon) and photographed using a camera attached to the microscope.
Illumina mRNA-Seq and Real-Time Reverse Transcription-PCR Analyses
Total RNA in roots was extracted using TRIzol (Invitrogen). RNA quality was checked using a Bioanalyzer 2100 (Agilent Technologies), and high-quality RNA (RNA integrity number > 8) was treated with DNase I to completely remove any genomic DNA contamination. About 10 μg of total RNA was converted to complementary DNA using mRNA-Seq kits from Illumina, and the barcoded complementary DNA library was sequenced on an Illumina HiSeq 2000 by the Shanghai Majorbio Bio-pharm Technology Corporation. We used TopHat software (Trapnell et al., 2009) to align the sequence reads to the reference genome and Cufflink (Roberts et al., 2011) to call the expression values (fragments per kilobase of exon model per million mapped fragments [FPKM]) of annotated genes.
The real-time reverse transcription-PCR analyses were performed as previously described (Jin et al., 2009a).
Measurement of Metal Concentrations
Plant tissues were dried at 80°C for 48 h, and then the dried samples were wet digested as previously described (Jin et al., 2009a), the digests diluted with ultrapure water, and the concentrations of metals, including 108Cd, were analyzed using inductively coupled plasma-mass spectrometry.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT1G08090 (NRT2.1), AT1G08100 (NRT2.2), AT1G18880 (NRT1.9), AT1G12110 (NRT1.1), AT1G32450 (NRT1.5), AT1G37130 (NIA2), AT1G69850 (NRT1.2), AT1G77760 (NIA1), AT4G19690 (IRT1), AT4G21680 (NRT1.8), AT5G50200 (NRT3.1), and AT5G60770 (NRT2.4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of 10 μm cadmium on high-affinity NO3− uptake by Col-0, chl1.5, or nrt1.1-1 plants.
Supplemental Figure S2. Effect of 20 μm cadmium on growth of, and cadmium levels in, Col-0, chl1.5, and nrt1.1-1 plants.
Supplemental Figure S3. Effect of 10 μm cadmium on growth of, and cadmium levels in, Ler and chl1.6 plants.
Supplemental Figure S4. Effect of NO3− removal on growth of, and cadmium uptake by, Col-0, chl1.5, and nrt1.1-1 plants.
Supplemental Figure S5. Effect of 10 μm cadmium on IRT1 expression in Col-0, chl1.5, and nrt1.1-1 plants in growth medium containing 10 mm NO3−.
Supplemental Figure S6. Cadmium levels in Col-0, nrt1.1-1, irt1-2, and irt1-2/nrt1.1-1 plants after 7 d of exposure to cadmium in 10 mm NO3− medium.
Supplemental Figure S7. Effect of 10 μm cadmium on expression of NRT1.5 and NRT1.8 in Col-0, chl1.5, and nrt1.1-1 plants.
Supplemental Figure S8. Cadmium levels in Col-0 plants and nrt2.1 mutants grown for 7 d in 200 μm NO3− medium.
Supplemental Table S1. Effect of 10 μm cadmium on the expression of genes related to metal cation uptake.
Supplemental Table S2. Primers used in this work.
Supplementary Material
Glossary
- NRT
nitrate transporter
- Col-0
ecotype Columbia-0 of Arabidopsis
- NR
nitrate reductase
- Seq
sequencing
- Ler
Landsberg erecta
- FPKM
fragments per kilobase of exon model per million mapped fragments
Footnotes
This work was supported by the Natural Science Foundation of China (grant no. 31270041), the Zhejiang Province Natural Science Foundation (grant nos. LR13C130001 and LY14C130001), the Fundamental Research Funds for the Central Universities (grant no. 2014QNA6006), and the Natural Science Foundation of Ningbo City (grant no. 2011A610002).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Cerezo M, Tillard P, Filleur S, Muños S, Daniel-Vedele F, Gojon A. (2001) Major alterations of the regulation of root NO(− uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol 127: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Lv XF, Li JY, Yi HY, Gong JM. (2012) Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol 159: 1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Aarts MGM, Thomine S, Verbruggen N. (2013) Plant science: the key to preventing slow cadmium poisoning. Trends Plant Sci 18: 92–99 [DOI] [PubMed] [Google Scholar]
- Finkemeier I, Kluge C, Metwally A, Georgi M, Grotjohann N, Dietz KJ. (2003) Alterations in Cd-induced gene expression under nitrogen deficiency in Hordeum vulgare. Plant Cell Environ 26: 821–833 [DOI] [PubMed] [Google Scholar]
- Gouia H, Gorbel MH, Meyer C. (2000) Effects of cadmium on activity of nitrate reductase and on other enzymes of the nitrate assimilation pathway in bean. Plant Physiol Biochem 38: 629–638 [Google Scholar]
- Guo FQ, Wang R, Chen M, Crawford NM. (2001) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13: 1761–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya T, Mizokami Y, Miyata K, Tholen D, Watanabe CK, Noguchi K. (2011) Evidence for a nitrate-independent function of the nitrate sensor NRT1.1 in Arabidopsis thaliana. J Plant Res 124: 425–430 [DOI] [PubMed] [Google Scholar]
- He P, Lu Y, Liang Y, Chen B, Wu M, Li S, He G, Jin T. (2013) Exposure assessment of dietary cadmium: findings from Shanghainese over 40 years, China. BMC Public Health 13: 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández LE, Gárate A, Carpena-Ruiz R. (1997) Effects of cadmium on the uptake, distribution and assimilation of nitrate in Pisum sativum. Plant Soil 189: 97–106 [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Huang NC, Chiang CS, Crawford NM, Tsay YF. (1996) CHL1 encodes a component of the low-affinity nitrate uptake system in Arabidopsis and shows cell type-specific expression in roots. Plant Cell 8: 2183–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin CW, Du ST, Chen WW, Li GX, Zhang YS, Zheng SJ. (2009a) Elevated carbon dioxide improves plant iron nutrition through enhancing the iron-deficiency-induced responses under iron-limited conditions in tomato. Plant Physiol 150: 272–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin CW, Du ST, Zhang YS, Lin XY, Tang CX. (2009b) Differential regulatory role of nitric oxide in mediating nitrate reductase activity in roots of tomato (Solanum lycocarpum). Ann Bot (Lond) 104: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkby EA, Knight AH. (1977) Influence of the level of nitrate nutrition on ion uptake and assimilation, organic acid accumulation, and cation-anion balance in whole tomato plants. Plant Physiol 60: 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkby EA, Mengel K. (1967) Ionic balance in different tissues of the tomato plant in relation to nitrate, urea, or ammonium nutrition. Plant Physiol 42: 6–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Léran S, Muños S, Brachet C, Tillard P, Gojon A, Lacombe B. (2013) Arabidopsis NRT1.1 is a bidirectional transporter involved in root-to-shoot nitrate translocation. Mol Plant 6: 1984–1987 [DOI] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, et al. (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Li JY, Fu YL, Pike SM, Bao J, Tian W, Zhang Y, Chen CZ, Zhang Y, Li HM, Huang J, et al. (2010) The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, et al. (2008) Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF. (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo BF, Du ST, Lu KX, Liu WJ, Lin XY, Jin CW. (2012) Iron uptake system mediates nitrate-facilitated cadmium accumulation in tomato (Solanum lycopersicum) plants. J Exp Bot 63: 3127–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux A, Martinka M, Vaculík M, White PJ. (2011) Root responses to cadmium in the rhizosphere: a review. J Exp Bot 62: 21–37 [DOI] [PubMed] [Google Scholar]
- Mendoza-Cózatl DG, Jobe TO, Hauser F, Schroeder JI. (2011) Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic. Curr Opin Plant Biol 14: 554–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16: 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Kendall MD, Osborn D. (1983) Cadmium and mercury nephrotoxicity. Nature 304: 633–635 [DOI] [PubMed] [Google Scholar]
- Nishida S, Tsuzuki C, Kato A, Aisu A, Yoshida J, Mizuno T. (2011) AtIRT1, the primary iron uptake transporter in the root, mediates excess nickel accumulation in Arabidopsis thaliana. Plant Cell Physiol 52: 1433–1442 [DOI] [PubMed] [Google Scholar]
- Pulford ID, Watson C. (2003) Phytoremediation of heavy metal-contaminated land by trees—a review. Environ Int 29: 529–540 [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103: 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardo C, Tomasi N, Monte R, Varanini Z, Nocito FF, Cesco S, Pinton R. (2012) Cadmium inhibits the induction of high-affinity nitrate uptake in maize (Zea mays L.) roots. Planta 236: 1701–1712 [DOI] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27: 2325–2329 [DOI] [PubMed] [Google Scholar]
- Sanità di Toppi LS, Gabbrielli R. (1999) Response to cadmium in higher plants. Environ Exp Bot 41: 105–130 [Google Scholar]
- Sarwar N, Saifullah, Malhi SS, Zia MH, Naeem A, Bibi S, Farid G. (2010) Role of mineral nutrition in minimizing cadmium accumulation by plants. J Sci Food Agric 90: 925–937 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. (2007) Nitrate transporters and peptide transporters. FEBS Lett 581: 2290–2300 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Van Beusichem ML, Kirkby EA, Baas R. (1988) Influence of nitrate and ammonium nutrition on the uptake, assimilation, and distribution of nutrients in Ricinus communis. Plant Physiol 86: 914–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C, Schat H. (2009) Mechanisms to cope with arsenic or cadmium excess in plants. Curr Opin Plant Biol 12: 364–372 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Hsu PK, Tsay YF. (2012) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17: 458–467 [DOI] [PubMed] [Google Scholar]
- Zhao T, Ling HQ. (2007) Effects of pH and nitrogen forms on expression profiles of genes involved in iron homeostasis in tomato. Plant Cell Environ 30: 518–527 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.