Alteration of the intracellular redox state during endoplasmic reticulum stress contributes to autophagy activation in Chlamydomonas reinhardtii.
Abstract
The accumulation of unfolded/misfolded proteins in the endoplasmic reticulum (ER) results in the activation of stress responses, such as the unfolded protein response or the catabolic process of autophagy to ultimately recover cellular homeostasis. ER stress also promotes the production of reactive oxygen species, which play an important role in autophagy regulation. However, it remains unknown whether reactive oxygen species are involved in ER stress-induced autophagy. In this study, we provide evidence connecting redox imbalance caused by ER stress and autophagy activation in the model unicellular green alga Chlamydomonas reinhardtii. Treatment of C. reinhardtii cells with the ER stressors tunicamycin or dithiothreitol resulted in up-regulation of the expression of genes encoding ER resident endoplasmic reticulum oxidoreductin1 oxidoreductase and protein disulfide isomerases. ER stress also triggered autophagy in C. reinhardtii based on the protein abundance, lipidation, cellular distribution, and mRNA levels of the autophagy marker ATG8. Moreover, increases in the oxidation of the glutathione pool and the expression of oxidative stress-related genes were detected in tunicamycin-treated cells. Our results revealed that the antioxidant glutathione partially suppressed ER stress-induced autophagy and decreased the toxicity of tunicamycin, suggesting that oxidative stress participates in the control of autophagy in response to ER stress in C. reinhardtii In close agreement, we also found that autophagy activation by tunicamycin was more pronounced in the C. reinhardtii sor1 mutant, which shows increased expression of oxidative stress-related genes.
All living organisms have evolved sophisticated mechanisms to efficiently respond and adapt their growth and metabolism to different types of stress. A well-documented example of such stress-induced responses is the process of autophagy or self-degradation, which is structurally and functionally conserved in all eukaryotes. During autophagy (also known as macroautophagy), cytoplasmic components, including proteins, membranes, and even organelles, are nonselectively enclosed within a double-membrane vesicle known as autophagosome and delivered to the vacuole/lysosome for degradation of toxic or damaged components and recycling of needed nutrients (Xie and Klionsky, 2007; Nakatogawa et al., 2009; Li and Vierstra, 2012; Liu and Bassham, 2012).
Autophagy is mediated by a set of proteins coded by ATG (autophagy-related) genes that are widely conserved from yeast (Saccharomyces cerevisiae) to humans. Homologs of ATG genes have been reported in plant and algal genomes, indicating that autophagy is also conserved in photosynthetic organisms (Thompson and Vierstra, 2005; Bassham et al., 2006; Diaz-Troya et al., 2008b; Avin-Wittenberg et al., 2012). Some ATG proteins play a structural role in autophagy and are essential for the formation of the autophagosome. For instance, the ATG8 protein anchors to the autophagosome membrane through its covalent binding to phosphatidylethanolamine, and the ATG8-phosphatidylethanolamine conjugate is essential for the formation and completion of the autophagosome (Ichimura et al., 2000). ATG genes are conserved in Chlamydomonas reinhardtii (algae; Diaz-Troya et al., 2008b; Pérez-Pérez et al., 2010), and autophagy has been investigated in this model system by monitoring the abundance, lipidation state, and cellular distribution of the ATG8 protein under several growth and stress conditions. As reported for other systems, nitrogen or carbon depletion triggers autophagy in C. reinhardtii (Pérez-Pérez et al., 2010). Moreover, entry of C. reinhardtii cells into stationary growth phase activates autophagy in a reversible manner, because the process is quickly down-regulated when cells return to exponential growth (Pérez-Pérez et al., 2010). Autophagy is also induced in C. reinhardtii cells treated with hydrogen peroxide or methyl viologen (MV), indicating that oxidative stress triggers this process in algae (Pérez-Pérez et al., 2010, 2012a). Reactive oxygen species (ROS) are potent inducers of autophagy in C. reinhardtii and plants (Liu and Bassham, 2012; Pérez-Pérez et al., 2012b). Indeed, a link between photo-oxidative damage, ROS accumulation, and autophagy activation has been shown in C. reinhardtii cells with a decreased carotenoid content caused by either the mutation of phytoene synthase or the inhibition of phytoene desaturase by the herbicide norflurazon (Pérez-Pérez et al., 2012a). Moreover, ROS generated in the chloroplast of carotenoid-deficient cells or the chloroplast of wild-type cells subjected to high light stress activate autophagy (Pérez-Pérez et al., 2012a).
The accumulation of unfolded/misfolded proteins in the endoplasmic reticulum (ER) is known to trigger autophagy in yeast (Bernales et al., 2006; Yorimitsu et al., 2006) and mammals (Ogata et al., 2006). More recently, induction of autophagy by ER stress has also been reported in land plants and algae (Pérez-Pérez et al., 2010; Liu et al., 2012), indicating that the signaling pathways controlling autophagy activation in response to this intracellular stress might be conserved in photosynthetic organisms. In C. reinhardtii, tunicamycin, which induces ER stress by inhibition of N-linked glycosylation, strongly increases the abundance and lipidation of ATG8 and modifies its cellular distribution (Pérez-Pérez et al., 2010). The accumulation of misfolded proteins in the ER is a potent stress signal that induces the expression of chaperones and other proteins required for the reestablishment of cell homeostasis, a signaling process known as the unfolded protein response (UPR; Walter and Ron, 2011). ER stress is perceived in the cell by key signaling proteins, such as the highly conserved inositol-requiring enzyme1 kinase, which transduces stress signals to the nucleus by promoting the splicing of basic leucine zipper (bZIP)-like transcription factors (Walter and Ron, 2011). Eukaryotic cells use a quality control mechanism that recognizes aberrantly folded proteins in the ER for their degradation through the proteasome, a process that is known as ER-associated degradation (Walter and Ron, 2011). Prolonged ER stress also triggers autophagy to remove unfolded proteins and counterbalance ER expansion caused by UPR (Bernales et al., 2006).
Unlike other cellular compartments, the ER provides, through ER-resident oxidoreductases, an oxidative environment that facilitates the oxidation of cysteines and thereby, the formation of disulfide bonds (Tu and Weissman, 2004). In yeasts and mammals, it has been shown that endoplasmic reticulum oxidoreductin1 (ERO1) is a major source of ROS in the ER and the cell (Haynes et al., 2004; Tu and Weissman, 2004). Oxidative protein folding in the ER occurs, in part, through the formation of disulfide bonds by protein disulfide isomerases (PDIs). To introduce disulfides into client proteins, PDIs must be maintained in an oxidized state, and ERO1 is mainly responsible for keeping PDI oxidized and active in the ER (Frand and Kaiser, 1999). ERO1 uses molecular oxygen as the final electron acceptor and hence, forms one molecule of hydrogen peroxide for every disulfide that it introduces (Tu and Weissman, 2004). Several studies have established an association between ER stress and ROS generation (Malhotra and Kaufman, 2007; Rutkowski and Kaufman, 2007; Ozgur et al., 2014), and the molecular mechanisms by which ROS are produced during UPR have been thoroughly reviewed (Santos et al., 2009).
ROS have been proposed to play an important role in the mechanisms of autophagy induction in response to various stress conditions in mammals, plants, and algae (Huang et al., 2011; Scherz-Shouval and Elazar, 2011; Szumiel, 2011; Li et al., 2012; Pérez-Pérez et al., 2012b). Although ER stress promotes ROS production and induces autophagy, it remains unknown whether ROS are involved in ER stress-induced autophagy. In this study, we provide evidence connecting autophagy activation in ER-stressed cells with redox imbalance generated from the UPR-regulated oxidative folding machinery in C. reinhardtii and propose that oxidative stress contributes to the induction of autophagy by ER stress.
RESULTS
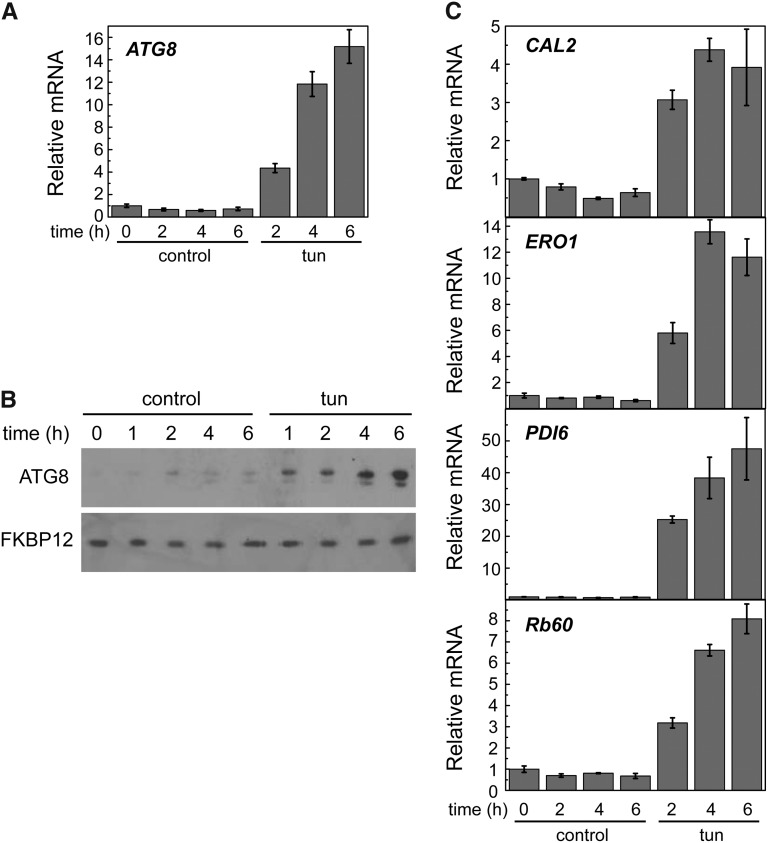
Tunicamycin Triggers ER Stress and Autophagy in C. reinhardtii
In a previous study, we showed that tunicamycin triggers autophagy in C. reinhardtii (Pérez-Pérez et al., 2010). To further characterize the effect of tunicamycin on autophagy, we analyzed the expression of the ATG8 gene by quantitative real-time PCR (qPCR). Tunicamycin treatment resulted in a progressive increase of ATG8 transcript levels (Fig. 1A). In close agreement, an increase in ATG8 protein abundance and the appearance of modified ATG8 forms were observed in ER-stressed cells (Fig. 1B). No effect was observed on the amount of FK506 Binding Protein12 (FKBP12), an ER stress unrelated protein (Crespo et al., 2005). Transcription of ATG3, encoding an E2-like enzyme involved in ATG8 lipidation (Ichimura et al., 2000), was also analyzed in tunicamycin-treated cells to investigate the participation of other ATG genes in ER stress. qPCR analysis revealed that, similar to ATG8, ATG3 expression was up-regulated with tunicamycin as well as other stressors previously shown to induce autophagy, such as hydrogen peroxide, MV, or norflurazon (Pérez-Pérez et al., 2010, 2012a; Supplemental Fig. S1). As expected, tunicamycin had no effect on the expression of FKBP12 (Supplemental Fig. S2).
Figure 1.
Tunicamycin triggers autophagy and ER stress in C. reinhardtii Log-phase cells grown in TAP medium were treated with 5 µg mL−1 tunicamycin, and samples were taken at the indicated times and processed for expression analysis of ATG8 (A) and CAL2, ERO1, PDI6, and Rb60 (C) by qPCR (A and C) or western blot (B). Thirty micrograms of total extracts were resolved by 15% SDS-PAGE followed by western blotting with anti-ATG8 and anti-FKBP12 antibodies. Values are means of three independent experiments. tun, Tunicamycin.
The cellular response to ER stress in C. reinhardtii is still poorly characterized, and no markers have been established to investigate this process in this model alga. In Arabidopsis (Arabidopsis thaliana), it has been reported that expression of some UPR genes, such as the ER-resident chaperone Calreticulin2 (CAL2; At1g09210), increases in response to tunicamycin (Martínez and Chrispeels, 2003). We identified a member of the calreticulin family in the C. reinhardtii nuclear genome (Supplemental Fig. S3), which we denoted as CAL2. To investigate whether the expression of CAL2 is subject to UPR regulation in C. reinhardtii, transcription of this gene was determined by qPCR analysis in cells treated with tunicamycin or other stressors that should not trigger ER stress. CAL2 mRNA levels were increased with tunicamycin (Fig. 1C) but not hydrogen peroxide, MV, or norflurazon (Supplemental Fig. S1A), showing that this gene can be used to monitor ER stress in C. reinhardtii.
ERO1 has not been previously described in algae, but based on its high evolutionary conservation, we identified an ERO1 homolog in the C. reinhardtii genome (Supplemental Fig. S3). To investigate the participation of C. reinhardtii ERO1 in ER stress, ERO1 expression was analyzed by qPCR in cells treated with tunicamycin. We found a strong induction of ERO1 expression in ER-stressed cells (Fig. 1C). In yeast, ERO1 and PDI expressions are coordinately regulated and subjected to UPR regulation (Frand and Kaiser, 1998; Pollard et al., 1998). Rb60/PDI1A is the only canonical PDI that has been studied in C. reinhardtii, although other PDI-like proteins seem to be conserved in the C. reinhardtii genome (Lemaire and Miginiac-Maslow, 2004; Filonova et al., 2013). Rb60 seems to have a dual localization in the ER and the chloroplast (Levitan et al., 2005), but its role in ER stress has not been investigated. We analyzed the expression of Rb60 in response to tunicamycin treatment and found that, similar to ERO1, this gene was strongly up-regulated (Fig. 1C). Searching for PDI proteins other than Rb60 in C. reinhardtii, we identified a PDI-like protein that we denoted PDI6 containing an N-terminal J domain (Supplemental Fig. S3). This domain structure is conserved in ER proteins involved in the recognition and binding of misfolded proteins that fail to achieve their correct conformation in the ER (Schroda, 2004). We investigated the participation of PDI6 in ER stress by analyzing the expression of this gene in tunicamycin-treated cells. PDI6 mRNA level dramatically increased in ER-stressed cells (Fig. 1C), strongly suggesting that this gene is involved in the cellular response to this stress. Taken together, these results indicated that CAL2, ERO1, Rb60, and PDI6 expression can be used to monitor ER stress in C. reinhardtii.
In addition to tunicamycin, we tested the effect of thapsigargin, an inhibitor of the ER Ca2+-ATPase that triggers ER stress by depletion of luminal calcium stores (Urano et al., 2000), on autophagy and ER stress markers. Like tunicamycin treatment, thapsigargin led to ER stress and autophagy activation in C. reinhardtii (Supplemental Fig. S4).
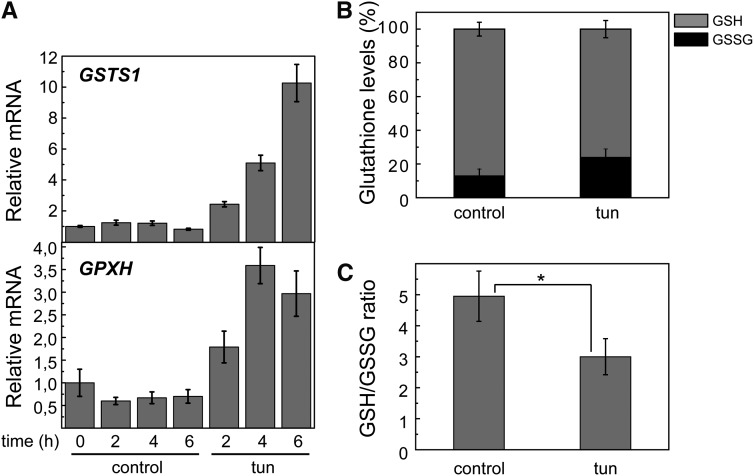
Tunicamycin Triggers Oxidative Stress
We explored the possible induction of oxidative stress caused by ER stress through different approaches. We analyzed the expression of Glutathione Peroxidase Homologous (GPXH) and Glutathione-S-transferase1 (GSTS1) genes in ER-stressed cells. Both genes can be induced by several ROS, although GPXH is more significantly induced by singlet oxygen (Ledford et al., 2007; Fischer et al., 2007, 2012). Our results revealed a high induction of GSTS1 and a moderate increase of GPXH expression in response to tunicamycin treatment (Fig. 2B), suggesting that ER stress may result in the activation of oxidative stress signaling in C. reinhardtii However, the intracellular pool of total glutathione, comprising reduced glutathione (GSH) and oxidized glutathione (GSSG), was determined in cells treated with tunicamycin. Our results indicated that ER stress raised the intracellular concentration of GSSG (Fig. 2B) and decreased the GSH-to-GSSG ratio (Fig. 2C), a hallmark of redox imbalance (Foyer and Noctor, 2011). Taken together, these results indicated that ER stress triggers oxidative stress in C. reinhardtii.
Figure 2.
Tunicamycin triggers oxidative stress. A, Expression analysis of GSTS1 and GPXH genes by qPCR in cells treated with 5 µg mL−1 tunicamycin at the indicated times. B, Intracellular pools of GSH and GSSG were determined in C. reinhardtii cells treated with 5 µg mL−1 tunicamycin or drug vehicle (control); a 100% glutathione pool corresponds to 837 ± 58 and 744 ± 64 pmol per 1 million cells for control and tunicamycin-treated cells, respectively. C, GSH-to-GSSG ratio in tunicamycin-treated cells compared with untreated cells. Values are means of three independent experiments. tun, Tunicamycin. *, Differences were significant at P < 0.05 according to Student’s t test.
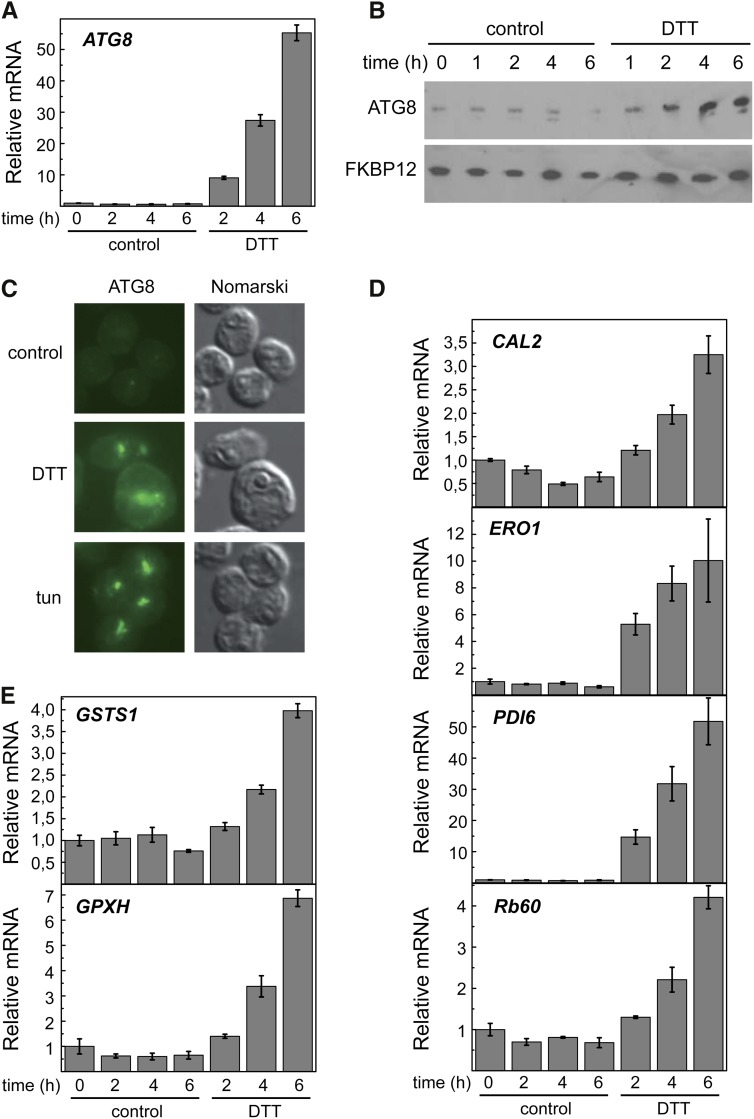
Dithiothreitol Causes ER Stress and Autophagy Activation
Disruption of disulfide bonds by dithiothreitol (DTT) results in accumulation of unfolded proteins in the ER and consequently, triggers ER stress. Massive aggregation of misfolded proteins caused by DTT has been shown to induce autophagy in various systems, including yeasts, mammals, and more recently, plants (Bernales et al., 2006; Yorimitsu et al., 2006; Liu et al., 2012). We investigated the effect of DTT on ER stress and autophagy in C. reinhardtii by different approaches. Autophagy was investigated in DTT-treated cells by examining ATG8 expression. Our results showed that DTT induces ATG8 expression (Fig. 3A), suggesting that DTT efficiently triggers autophagy in C. reinhardtii Activation of autophagy by DTT was confirmed by western-blot and immunofluorescence analyses of ATG8. The abundance of this protein progressively increased in response to DTT, which was shown by western blot (Fig. 3B). In agreement with this result, the cellular distribution of ATG8 drastically changed on DTT treatment, which was observed by immunofluorescence microscopy. In untreated cells, the ATG8 signal was very weak and localized in a single spot in some cells, whereas this signal was much stronger and detected as multiple spots in response to DTT or tunicamycin treatment (Fig. 3C).
Figure 3.
DTT triggers autophagy and ER stress in C. reinhardtii. Log-phase cells grown in TAP medium were treated with 2.5 mm DTT, and samples were taken at the indicated times for qPCR (A), western-blot (B), or immunofluorescence (C) analyses of ATG8. Immunofluorescence images correspond to 6 h of treatment. In addition to ATG8, expressions of the UPR-regulated genes CAL2, ERO1, PDI6, and Rb60 (D) or the oxidative stress-related genes GSTS1 and GPXH (E) were analyzed by qPCR. Values are means of three independent experiments.
ER stress was monitored in DTT-treated cells by qPCR analysis of CAL2, ERO1, PDI6, and Rb60 genes. DTT treatment resulted in the up-regulation of these four genes (Fig. 3D). This result confirmed that CAL2, ERO1, PDI6, and Rb60 genes might be used as ER stress markers in C. reinhardtii Given the tight correlation found between ER and oxidative stresses in tunicamycin-treated cells (Fig. 2), we also analyzed the expression of the oxidative stress-related genes GSTS1 and GPXH. Interestingly, reductive stress induced by DTT may indirectly lead to oxidative stress in the cell, because transcription of both GSTS1 and GPXH genes was enhanced (Fig. 3E).
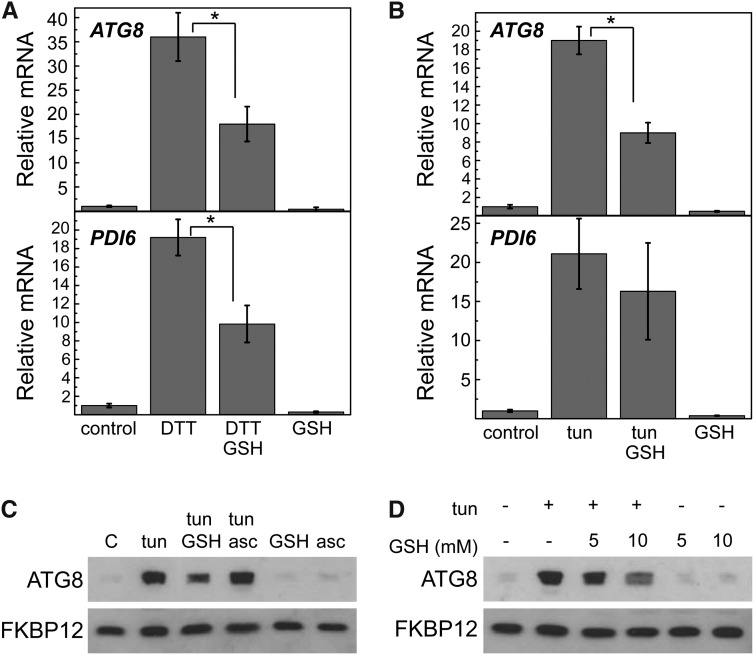
Glutathione Partially Suppresses ER Stress-Induced Autophagy
To test whether redox imbalance contributes to autophagy activation during ER stress, autophagy and ER stress markers were analyzed in cells treated with DTT, which massively reduces disulfide bonds. DTT was used alone or combined with GSH, an abundant antioxidant in the cell that plays an important role in ROS scavenging and maintenance of cellular redox homeostasis (Foyer and Noctor, 2011). Our results show that ATG8 induction caused by DTT is decreased by 50% in the presence of GSH (Fig. 4A), indicating that autophagy might be partially suppressed by GSH. Interestingly, ER stress was also decreased by exogenous GSH in DTT-treated cells, which was revealed by the lower expression of PDI6 (Fig. 4A) and other ER stress-induced genes (Supplemental Fig. S5A). Because GSH decreased both ER stress and autophagy, it was not possible to separate these two processes, and therefore, down-regulation of autophagy might be caused by the partial suppression of ER stress or might be caused by ROS scavenging or redox signaling mechanisms dependent on GSH. Thus, additional investigation of the role of oxidative stress in the control of ER stress-induced autophagy should be performed under experimental conditions, where ER stress and oxidative stress can be independently tested. We found that GSH was also able to partially suppress tunicamycin-induced expression of ATG8. However, unlike DTT, GSH did not significantly decrease the tunicamycin-induced expression of ER stress markers (Fig. 4B; Supplemental Fig. S5B). In agreement with decreased ATG8 transcription, the abundance of the ATG8 protein was also down-regulated in cells treated with both tunicamycin and GSH compared with cells treated only with tunicamycin (Fig. 4C). Moreover, when the GSH concentration was increased from 5 to 10 mm, a stronger decrease of ATG8 protein level was observed (Fig. 4D). To analyze the specificity of the response to GSH, we also investigated whether ascorbate, an abundant metabolite in plant cells with antioxidant properties (Foyer and Noctor, 2011), is able to mitigate the increase of ATG8 protein level caused by tunicamycin, but unlike GSH, no effect was observed on ATG8 abundance (Fig. 4C). We found, however, that ascorbate potently and rapidly increased the expression of GSTS1, indicating that this antioxidant is acting on the expression of some genes (Supplemental Fig. S6).
Figure 4.
GSH partially prevents ER stress-induced autophagy. Log-phase cells grown in TAP medium were treated with 2.5 mm DTT (A) or 5 µg mL−1 tunicamycin (B) alone or combined with 10 mm GSH. After 8 h of treatment, cells were processed for expression analysis of ATG8 and PDI6 genes by qPCR. Values are means of three independent experiments. *, Differences were significant at P < 0.05 according to Student’s t test between DTT and DTT-GSH or tunicamycin and tunicamycin-GSH. C and D, Western-blot analyses of ATG8 and FKBP12 proteins in log-phase cells treated with 5 µg mL−1 tunicamycin, 5 to 10 mm GSH, 2 mm ascorbate (asc), or combinations of these compounds. Thirty micrograms of total extracts were resolved by 15% SDS-PAGE followed by western blotting with anti-CrATG8 and anti-CrFKBP12 antibodies. tun, Tunicamycin.
The effect of GSH on ER stress-induced autophagy was also examined by immunolocalization assays of ATG8 in C. reinhardtii cells. As previously shown (Pérez-Pérez et al., 2010), tunicamycin treatment strongly increased the ATG8 signal, which accumulated in a few intense spots (Fig. 5A). However, the presence of GSH in the culture medium significantly decreased the intensity of the ATG8 signal, although some spots were still clearly visible, indicating that autophagy was reduced but not fully abolished in these cells (Fig. 5A). Quantification of the immunofluorescence signal from individual cells confirmed the inhibitory effect of GSH on autophagy induced by ER stress (Fig. 5B). These results are in close agreement with qPCR and western-blot assays of ATG8 performed in tunicamycin-treated cells in the presence of exogenous GSH (Fig. 4) and strongly suggest that oxidative stress might be involved in the induction of autophagy in ER-stressed cells.
Figure 5.
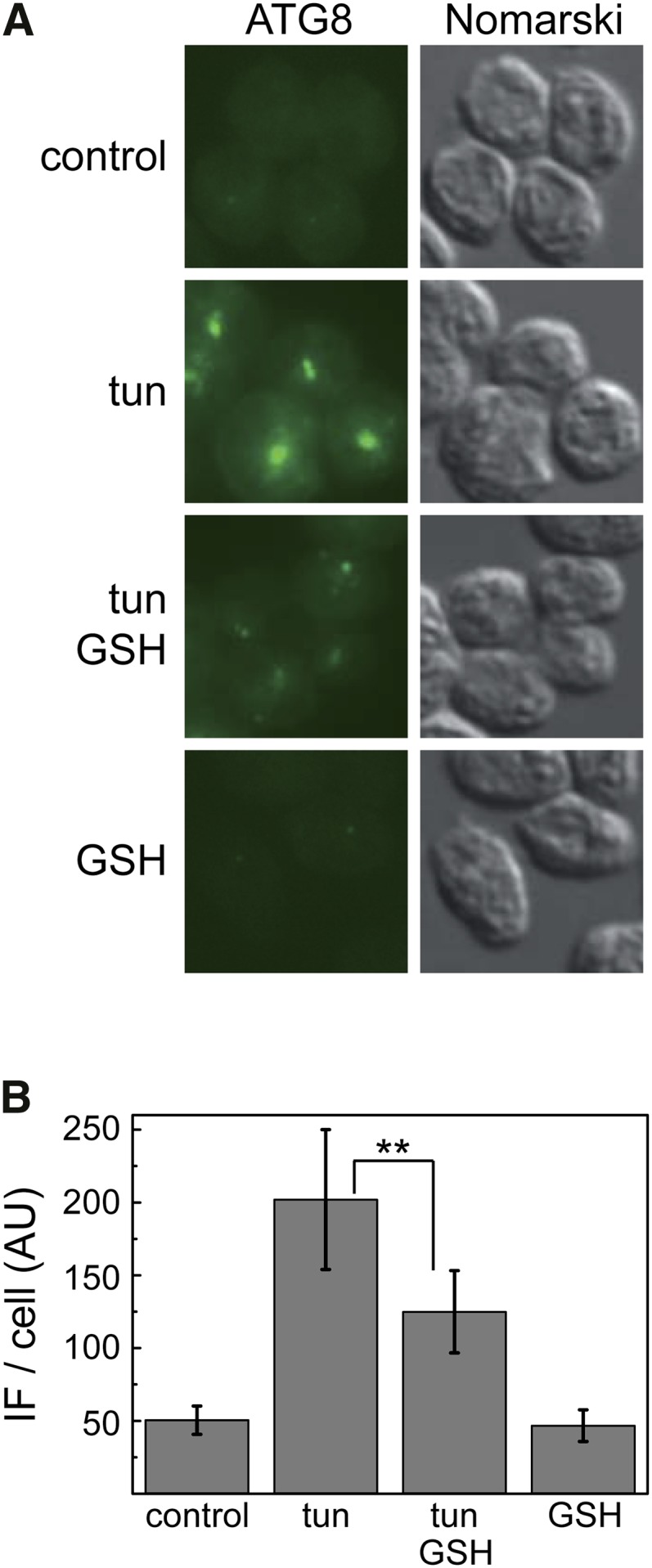
GSH decreases tunicamycin-induced cellular accumulation of ATG8 in C. reinhardtii. A, Immunolocalization of ATG8 in C. reinhardtii cells treated for 8 h with 5 µg mL−1 tunicamycin alone or combined with 10 mm GSH. Bar = 5 µm. B, Quantification of the immunofluorescence (IF) signal detected in individual cells from the experiment described in A. For each condition, a minimum of 100 individual cells was analyzed using ImageJ software. tun, Tunicamycin. **, Differences were significant at P < 0.01 according to Student’s t test between tunicamycin and tunicamycin GSH.
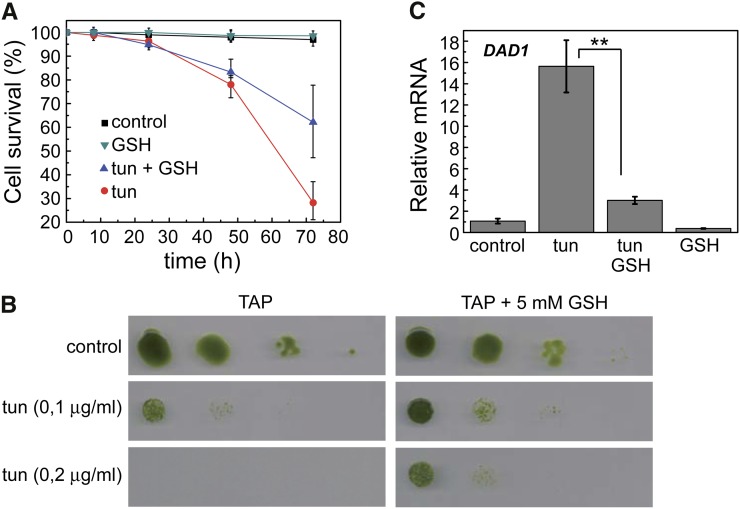
Glutathione Decreases the Toxicity of Tunicamycin in C. reinhardtii
Protein glycosylation in the ER is essential for cell growth, and thus, its inhibition by tunicamycin is toxic for the cell. Indeed, we found that tunicamycin reduced cell viability in C. reinhardtii after 24 h of treatment (Fig. 6A). Our results indicated that the presence of exogenous GSH in the medium decreased the toxicity of tunicamycin, which was revealed by Evans blue staining (Fig. 6A) or serial spot dilution assays (Fig. 6B). Interestingly, cells were still fully viable at the time of autophagy activation by tunicamycin (6–8 h; Fig. 6A), supporting a prosurvival role of this catabolic process in response to ER stress. In relation to the effect of tunicamycin on cell viability, we analyzed the expression of the DAD1 gene, a homolog of defender against apoptotic cell death1 that is conserved in many eukaryotes, including C. reinhardtii (Kelleher and Gilmore, 1994, 1997; Gallois et al., 1997; Moharikar et al., 2007). The DAD1 protein is part of the oligosaccharyltransferase complex and seems to play an important role in N-linked glycosylation in the ER (Kelleher and Gilmore, 1994), a process specifically inhibited by tunicamycin. Our results revealed that DAD1 expression was up-regulated on prolonged exposure (48 h) to tunicamycin (Fig. 6C), when cell viability is significantly compromised (Fig. 6A). No induction of DAD1 was observed in cells treated with tunicamycin for 6 h (Supplemental Fig. S7), indicating that DAD1 expression is associated to tunicamycin-induced loss of cell viability. In agreement with the protective role of GSH on cell growth in ER-stressed cells, the presence of this antioxidant decreased tunicamycin-mediated DAD1 expression (Fig. 6C).
Figure 6.
GSH reduces the toxicity of tunicamycin in C. reinhardtii. A, Log-phase cells grown in TAP medium were treated with tunicamycin (5 µg mL−1), GSH (10 mm), or both compounds combined. Untreated cells were used as control. Cell viability was determined by Evans blue staining at the indicated times. Results are means of four independent experiments. B, Cells were subjected to 10-fold serial dilutions and spotted onto TAP plates containing the indicated concentrations of tunicamycin and GSH. Plates were grown at 25°C under continuous illumination for 5 d. C, Expression analysis of the DAD1 gene by qPCR. Log-phase cells were treated with 5 µg mL−1 tunicamycin alone or combined with 10 mm GSH for 48 h and then processed for RNA isolation and analysis. Values are means of three independent experiments. tun, Tunicamycin. **, Differences were significant at P < 0.01 according to Student’s t test between tunicamycin and tunicamycin-GSH.
The sor1 Mutant Displays Increased Autophagy in Response to ER Stress
To further investigate the redox regulation of ER stress-induced autophagy in C. reinhardtii, we analyzed the activation of this degradative pathway by tunicamycin in the sor1 mutant. This strain lacks the Singlet Oxygen Resistant1 (SOR1) protein, a putative bZIP transcription factor that controls the expression of a large number of oxidative stress response and detoxification genes, including GSTS1 (Fischer et al., 2012). Treatment of sor1 cells with tunicamycin revealed a more pronounced accumulation of the ATG8 protein in the sor1 mutant compared with wild-type cells (Fig. 7A), suggesting a higher induction of autophagy in this mutant in response to ER stress. Expression analysis of the ATG8 gene also showed a higher response to tunicamycin in sor1 compared with cw15 cells (Fig. 7B). As previously reported (Fischer et al., 2012), we found that sor1 mutant cells show a constitutively higher expression of GSTS1 (Fig. 7B). Interestingly, despite this elevated expression, tunicamycin further increased GSTS1 mRNA abundance in the sor1 mutant (Fig. 7B). Taken together, these results strongly suggested that autophagic activity triggered by ER stress is higher in cells with anomalous expression of oxidative stress-related genes.
Figure 7.
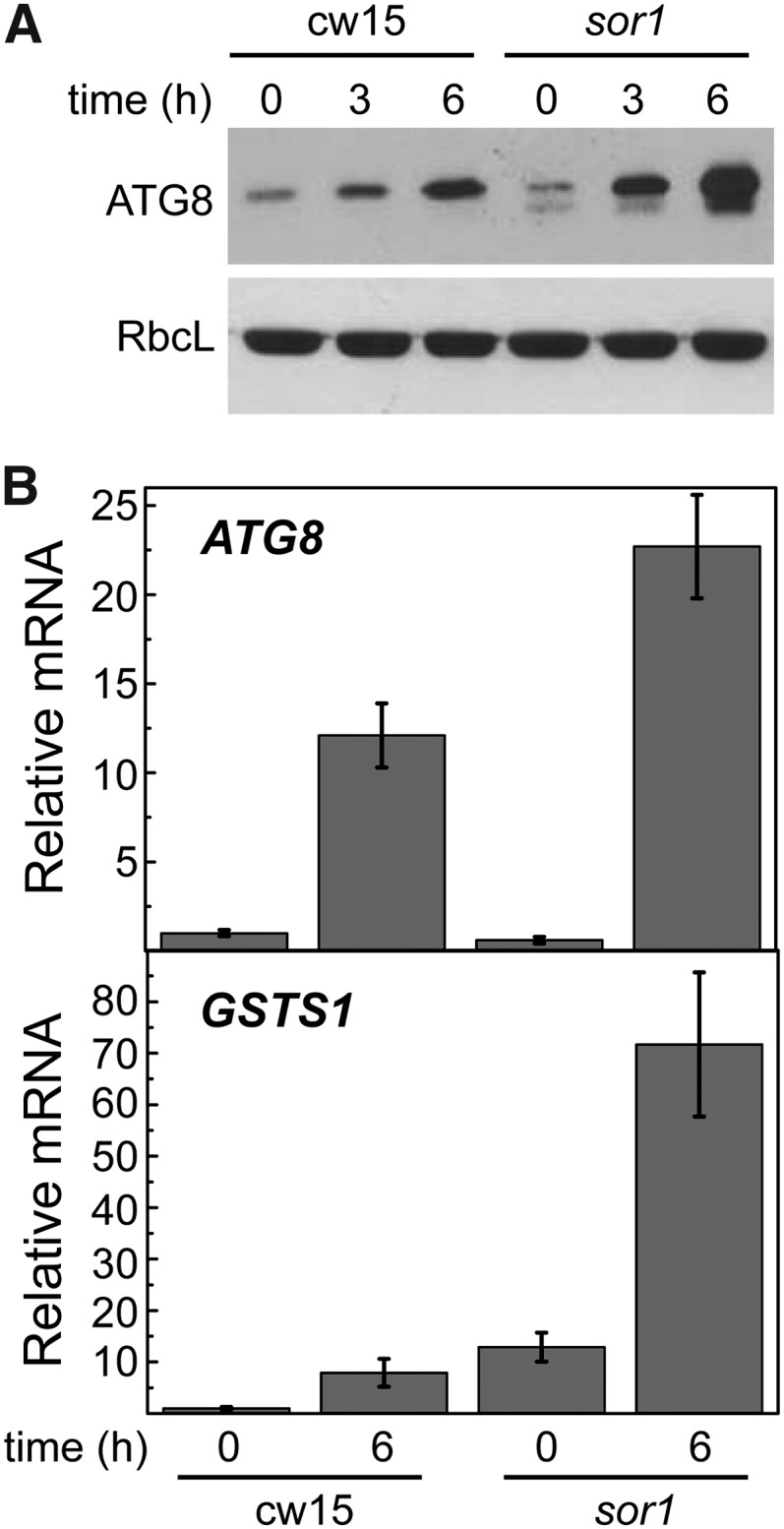
The sor1 mutant displays increased autophagy in response to ER stress. A, Log-phase cw15 and sor1 cells were treated with tunicamycin (5 µg mL−1), and at the indicated times, samples were taken and processed for western-blot analysis. B, Expression analysis of ATG8 and GSTS1 genes by qPCR. Log-phase cw15 and sor1 cells were treated with tunicamycin (5 µg mL−1) for 6 h and subsequently subjected to RNA isolation and analysis. Values are means of three independent experiments. RbcL, Rubisco large subunit.
DISCUSSION
In a previous report, we showed that ER stress induces autophagy in C. reinhardtii (Pérez-Pérez et al., 2010). In this study, we show that ER stress mediated by tunicamycin or DTT treatment strongly increased the expression of the ATG8 gene, which is in agreement with the pronounced accumulation of the ATG8 protein and its modified forms detected in ER-stressed cells (Figs. 1 and 3). Induction of autophagy by ER stress is evolutionarily conserved and has been reported in yeast (Bernales et al., 2006; Yorimitsu et al., 2006), mammals (Ogata et al., 2006), and more recently, plants (Liu et al., 2012). Studies performed in yeast strongly suggest that autophagy serves to counterbalance ER expansion that occurs as a consequence of UPR signaling (Bernales et al., 2006). Degradation of ER membranes by autophagy during ER stress is conserved in plants, because this type of membrane has been detected inside autophagic bodies in Arabidopsis plants treated with tunicamycin (Liu et al., 2012).
This study also showed that the accumulation of unfolded proteins in the ER triggers oxidative stress in C. reinhardtii Formation of disulfide bonds in the ER and associated oxidative protein folding are linked to the generation of hydrogen peroxide by the activity of the ERO1 oxidoreductase that catalyzes the reoxidation and activation of PDI (Tu and Weissman, 2004). Originally identified in yeast (Frand and Kaiser, 1998; Pollard et al., 1998), ERO1 is widely conserved in all eukaryotes, including plants (Aller and Meyer, 2013). In this study, we found that ERO1 is conserved in C. reinhardtii and that its expression is highly induced by the ER stressors tunicamycin and DTT (Figs. 1 and 3). ERO1 is a hydrogen peroxide-generating enzyme, and its high expression may lead to increased levels of hydrogen peroxide under ER stress. Indeed, our results indicated that C. reinhardtii cells subjected to ER stress displayed elevated expression of oxidative stress-related genes and higher levels of the intracellular pool of GSSG (Figs. 2, B and C and 3E). Thus, different lines of evidence indicate that ER stress triggers oxidative stress in C. reinhardtii. In yeast, ROS, such as hydrogen peroxide, are generated during ER stress, and overexpression of ERO1 causes a significant increase in ROS and GSSG pool, suggesting that up-regulation of the oxidative protein folding machinery by the UPR contributes to ROS accumulation (Haynes et al., 2004). In a recent report, it has also been shown that ER stress triggers ROS signaling in plants (Ozgur et al., 2014), indicating that ROS production on ER stress is widely conserved.
In addition to ERO1, we identified other genes subjected to UPR regulation in C. reinhardtii. The expression levels of two PDI genes, Rb60 and PDI6, and the CAL2 chaperone were increased in response to ER stress (Figs. 2 and 4). Rb60 is a canonical PDI with an atypical dual localization in the ER and the chloroplast (Levitan et al., 2005), where it regulates psbA translation by light (Danon and Mayfield, 1991). The function of Rb60 in the ER is unclear, although based on its high identity to classical PDI and its localization, it likely participates in oxidative protein folding in the ER. PDI6 encodes a PDI-like protein containing an N-terminal J domain that is conserved among ER cochaperones (Schroda, 2004) and a C-terminal Arg-rich domain involved in ER stress quality control in mammals (Mizobuchi et al., 2007). The domain architecture of PDI6 seems to be well conserved in microalgae and less represented in plants, where the J domain-containing protein ATERDJ3A from Arabidopsis seems to be the closest homolog (Supplemental Fig. S3). No clear homologs of this protein have been found in yeasts or humans. Our results revealed that PDI6 mRNA levels are particularly sensitive to ER stress, and a massive increase in the expression of this gene was observed with tunicamycin or DTT (Figs. 1, 3, and 4). Expression of the calreticulin CAL2 gene was also found to be up-regulated by the UPR in C. reinhardtii (Figs. 1, 3, and 4). Similar to ERO1, CAL2 is conserved in plants and other systems (Supplemental Fig. S3), and microarray analysis of Arabidopsis seedlings treated with tunicamycin or DTT suggested that this gene is up-regulated under ER stress conditions (Martínez and Chrispeels, 2003). Our results indicate that the abundance of ERO1, Rb60, PDI6, or CAL2 mRNAs can be used as markers of the UPR in C. reinhardtii and strongly suggest that the mechanisms of oxidative protein folding described in other eukaryotes are conserved in green algae.
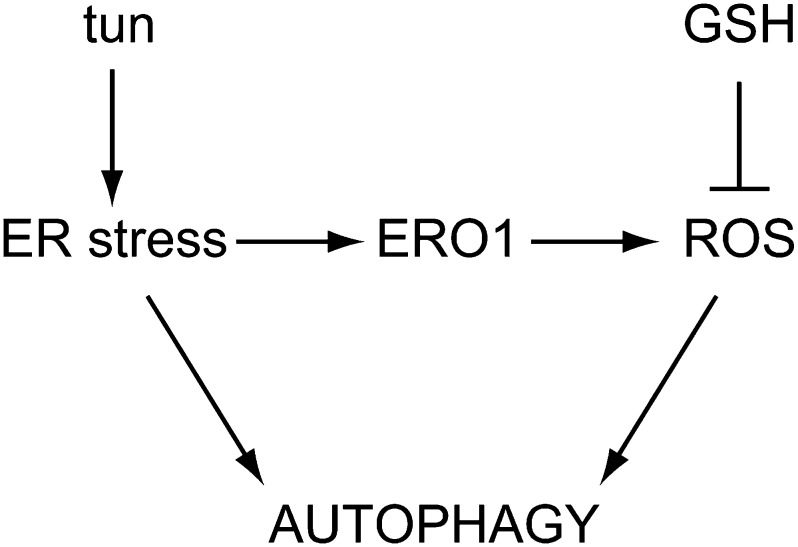
Given that autophagy is regulated by ROS (Pérez-Pérez et al., 2012b) and ER stress triggers oxidative stress responses in C. reinhardtii (Fig. 2), ER stress-derived ROS may likely contribute to autophagy regulation in response to this stress (Fig. 8). In close agreement with this hypothesis, we found that the antioxidant GSH down-regulates autophagy in ER-stressed cells (Figs. 4 and 5). Glutathione is the main free soluble thiol in the cell and acts as a redox buffer by maintaining the intracellular environment in a reduced state (Foyer and Noctor, 2011). Moreover, glutathione has also been shown to play a protective role from ER-generated oxidative stress. Prolonged UPR activation in a yeast strain that is genetically predisposed to sustained ER stress causes oxidative stress, ultimately leading to cell death (Haynes et al., 2004). In this strain, the presence of GSH in the culture medium relieved oxidative stress and prevented cell death without diminishing UPR activation, indicating that ROS accumulate during ER stress and are toxic for the cell (Haynes et al., 2004). The effect of GSH in alleviating oxidative stress and promoting growth of ER-stressed cells might be conserved in C. reinhardtii, because our results showed that GSH decreases the toxicity of tunicamycin (Fig. 6). The protective role of GSH was emphasized by the finding that this antioxidant decreased the expression of the DAD1 gene, which is specifically up-regulated when cell viability is compromised because of prolonged ER stress (Fig. 6C). DAD1 is a putative antiapoptotic gene widely conserved in eukaryotes, including humans, plants, yeasts, and algae (Nakashima et al., 1993; Kelleher and Gilmore, 1994, 1997; Gallois et al., 1997; Moharikar et al., 2007). The DAD1 protein is localized in the ER and forms part of the oligosaccharyltransferase complex, which catalyzes N-linked glycosylation (Kelleher and Gilmore, 1994). Although the precise function of this protein is not clear, DAD1 seems to be involved in preventing cell death and regulating N-linked glycosylation. Our results are in agreement with these putative roles, because expression of the C. reinhardtii DAD1 gene is induced by tunicamycin treatment, which is toxic for the cell and specifically inhibits N-linked glycosylation.
Figure 8.
Control of autophagy by ER stress and ER-derived ROS in C. reinhardtii. Tunicamycin (tun) treatment results in toxic accumulation of unfolded proteins in the ER, leading to autophagy activation as a defensive mechanism. ER stress also increases ROS production in the ER by de oxidoreductase ERO1, which contributes to the up-regulation of autophagy. The antioxidant properties of GSH counterbalance ROS signaling to the autophagic machinery but do not abrogate ER stress, resulting in partial inactivation of autophagy.
The hypothesis that ER stress-induced autophagy and redox signaling are linked was strengthened by the finding that tunicamycin triggers autophagy more potently in sor1 mutant cells (Fig. 7). The sor1 strain was isolated in a screen for mutants with increased tolerance to singlet oxygen and shows a constitutively higher expression of a large number of oxidative stress response and detoxification genes (Fischer et al., 2012). Mapping of the sor1 mutation identified a putative bZIP transcription factor denoted as SOR1 that controls, among other things, the expression of the glutathione S-transferase gene GSTS1 (Fischer et al., 2012), which is up-regulated in ER-stressed cells (Fig. 2B). The high autophagic activity observed in sor1 mutant cells treated with tunicamycin might be related to the enhanced expression of ROS-induced genes reported in this mutant, because our results indicated that ER stress-induced autophagy is linked to redox signaling in C. reinhardtii (Figs. 2, 4, and 5). Given the role of SOR1 in the control of glutathione-based ROS scavenging by regulating the expression of genes, such as GSTS1 or GPXH (Fischer et al., 2012), and the regulatory function of this antioxidant in ER stress-induced autophagy (Figs. 4 and 5), it is possible that the up-regulation of autophagy observed in the sor1 mutant is related to the high expression of these genes, which may result in reduced intracellular levels of glutathione in sor1 cells.
Overall, our results indicate that ER stress triggers autophagy and oxidative stress in C. reinhardtii. Under ER stress triggered by tunicamycin, GSH was found to decrease autophagy induction, whereas UPR markers remained unaffected. This uncoupling of UPR and autophagy in the presence of an antioxidant participating in diverse ROS scavenging mechanisms suggests that ROS production may participate in the control of ER stress-induced autophagy in C. reinhardtii (Fig. 8). However, GSH effects on ER stress may involve other mechanisms not associated with ROS scavenging. For example, given the protective role that glutathione plays in the ER (Haynes et al., 2004), it may also be possible that glutathione specifically regulates autophagy by restoring the GSH-to-GSSG ratio in the ER, which is critical for maintaining the functionality and redox balance at this cellular compartment. Glutathione or redox imbalance may also play a signaling role during ER stress (e.g. through glutathionylation of specific target proteins; Zaffagnini et al., 2012). In this work, we have established the importance of the intracellular redox state and glutathione during ER stress-induced autophagy. Additional studies will be required to unravel how redox active molecules control autophagy induction and examine the interplay between these mechanisms and other ER stress signaling pathways.
MATERIALS AND METHODS
Strains and Growth Conditions
Chlamydomonas reinhardtii cell wall-deficient cw15 4B+ and sor1 mutant strains were obtained from the Chlamydomonas Culture Collection. C. reinhardtii cells were grown under continuous illumination at 25°C in Tris-acetate phosphate (TAP) medium as described (Harris, 1989). When required, cells in exponential growth phase (106 cells per milliliter) were treated with 5 µg mL−1 tunicamycin (654380; Calbiochem) from 5 mg mL−1 stock in dimethylformamide, 2.5 mm DTT (A2948; Applichem), 1 mm hydrogen peroxide (H1009; Sigma-Aldrich), 1 µm MV (85617-7; Sigma-Aldrich), or 20 µm norflurazon (PS1044; Sigma-Aldrich).
Gene Accession Numbers
C. reinhardtii genes analyzed in this study were identified at the Phytozome Web site (http://www.phytozome.net/cgi-bin/gbrowse/chlamy/) under the following accession numbers: ATG3, Cre02.g102350.t1.2; ATG8, Cre16.g689650.t1.2; CAL2, Cre01.g038400.t1.2; ERO1, Cre17.g723150.t1.3; PDI6, Cre12.g518200.t1.3; Rb60, Cre02.g088200.t1.2; GPXH, Cre10.g458450.t1.3; GSTS1, Cre16.g688550.t1.2; and DAD1, Cre02.g108400.t1.2.
RNA Isolation and Quantification
C. reinhardtii total RNA was isolated from frozen cell pellets as previously described (Crespo et al., 2005). First strand complementary DNA was produced using 2 µg of total RNA, oligo(dT) primer, and 100 units of SuperScript II RNase H reverse transcription (18064-014; Invitrogen) in a 50-µL reaction. Reverse transcription qPCR was performed with a StepOne Real-Time PCR System (Applied Biosystems). PCR reactions, in a final volume of 20 µL, contained 10 µL of FastStart Universal SYBR Green Master (04913850001; Roche), 1 μL of complementary DNA dilution, 250 nm each primer, and distilled water up to 20 µL. Conditions used for amplification in the thermocycler were preincubation at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s and annealing and elongation at 55°C to 58°C (depending on the gene analyzed) for 60 s. All reactions were performed in triplicate with two of four biological replicates. The CBLP gene was used as a control constitutively expressed gene (Pootakham et al., 2010). The primer pairs used for qPCR were 5′-CTTCTCGCCCATGACCAC-3′ and 5′-CCCACCAGGTTGTTCTTCAG-3′ for CBLP, 5′-CGAGTTCAAGGTCGAGCAGT-3′ and 5′-CCACCCACAGACATGGTGTA-3′ for ATG3, 5′-TCCCCGATATCGACAAGAAG-3′ and 5′-TGCGGATGACGTACACAAAT-3′ for ATG8, 5′-ACCCTGACTACGTCCACGAC-3′ and 5′-GTCCTCAGCGAACTTCTTGG-3′ for CAL2, 5′-TGTCAACCTGCTCATCAACC-3′ and 5′-CTGCTGCTGCTACTGCTGTC-3′ for ERO1, 5′-GGTGTGGCTGGTTGAGTTCT-3′ and 5′-CTCTTTGGCGTCCTCACAGT-3′ for PDI6, 5′-CCAAGCGCTTTAAGAAGGTG-3′ and 5′-GTAGGGAAGCCCTTGACCTC-3′ for Rb60, 5′-AGGTTCTGGATGCGTTCCTA-3′ and 5′-ACACAGTCAGGGCGAAGAAG-3′ for DAD1, 5′-GCGGTCGCCAATAACCAAT-3′ and 5′-AAGGGCTGTCCCGAAAGC-3′ for GPXH (Fischer et al., 2009), 5′-CTGACCATCAGCCACGACT-3′ and 5′-ACATCGAACACCAGGGTAGC-3′ for FKBP12, and 5′-CAGAGGTGAAAGGCGGATAC-3′ and 5′-GTGTTGCAATGGACTTCAGC-3′ for GSTS1 (Fischer et al., 2012).
Protein Preparation and Immunoblot Analysis
C. reinhardtii cells from liquid cultures were collected by centrifugation (4,000g for 5 min), washed one time in 50 mm Tris-HCl (pH 7.5) buffer, and resuspended in a minimal volume of the same solution. Cells were lysed by two cycles of slow freezing to −80°C followed by thawing at room temperature. The soluble cell extract was separated from the insoluble fraction by centrifugation (15,000g for 15 min) in a microcentrifuge at 4°C. For immunoblot analyses, total protein extracts (30 µg) were subjected to 15% SDS-PAGE and then transferred to nitrocellulose membranes (162-0115; Bio-Rad). Anti-CrATG8 (Pérez-Pérez et al., 2010) and secondary antibodies were diluted 1:2,500 and 1:10,000, respectively, in PBS containing 0.1% (v/v) Tween 20 (A4974; Applichem) and 5% (w/v) milk powder. The ECL-Plus immunoblotting detection system (RPN2132; GE Healthcare) was used to detect the proteins with horseradish peroxidase-conjugated anti-rabbit secondary antibodies (A6154; Sigma-Aldrich). Anti-FKBP12 antibody was diluted 1:3,000 and used as the loading control as described previously (Crespo et al., 2005). Proteins were quantified with the Coomassie Brilliant Blue dye-binding method as described by the manufacturer (500-0006; Bio-Rad).
Glutathione Determination
C. reinhardtii cells were collected by centrifugation (5,000g for 5 min), washed one time in 50 mm sodium phosphate (pH 7.5) buffer, resuspended in 0.2 n HCl, and lysed by two cycles of frozen-thawed at −80°C. Crude extracts were cleared by centrifugation at 15,000g for 20 min at 4°C, and 500 μL of sample was neutralized by adding 50 μL of 50 mm NaH2PO4 (pH 7.5) and 0.2 n NaOH to a final pH between 5 and 6. The neutralized sample was directly used for measuring total glutathione (GSH plus GSSG) by the recycling assay initially described by Tietze (1969) and adapted by Queval and Noctor (2007). The method relies on the glutathione reductase-dependent reduction of 5,5′-dithiobis(2-nitro-benzoic acid) (DTNB; D8130; Sigma). GSSG was measured after treatment of the neutralized sample with 10 mm 4-vinylpyridine (V320-4; Sigma) for 30 min at 25°C. To remove excess 4-vinylpyridine, the derivatized sample was centrifuged two times at 15,000g for 20 min at 4°C. To measure total glutathione or GSSG, sample was added to a mix containing 120 mm NaH2PO4 (pH 7.5), 300 μm DTNB, 500 μm NADPH, 1 mm EDTA (pH 8), and 1 units mL−1 glutathione reductase (G3664; Sigma), and DTNB reduction was measured at 412 nm. Different GSH (G4251; Sigma) concentrations ranging from 0 to 5 μm were used as standards.
Viability Assay
Cell viability was estimated by determining the percentage of C. reinhardtii cells that excluded Evans blue dye (E2129; Sigma), which only stains nonviable cells; 450 µL of C. reinhardtii cells was incubated with 0.1% (w/v) Evans blue for 5 min, washed one time with 500 µL of TAP medium, and resuspended in an equal volume of TAP medium. Cells were examined in a phase contrast microscope to visualize uptake of the dye.
Fluorescence Microscopy
C. reinhardtii cells were fixed and stained for immunofluorescence microscopy as previously described (Diaz-Troya et al., 2008a; Pérez-Pérez et al., 2010). Affinity-purified polyclonal anti-ATG8 was used as the primary antibody at 1:500 dilution. For signal detection, a fluorescein isothiocyanate-labeled goat anti-rabbit antibody (1:500; F4890; Sigma-Aldrich) was used. Preparations were photographed on a DM6000B microscope (Leica) with an ORCA-ER camera (Hamamatsu) and processed with the Leica Application Suite Advanced Fluorescence software package (Leica). For the comparative analysis of the fluorescein isothiocyanate signal from different samples, the same acquisition time was fixed. Immunofluorescence signals in individual cells were quantified using the ImageJ software.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of autophagy induction by different stress conditions in C. reinhardtii.
Supplemental Figure S2. Expression analysis of the FRKB12 gene by qPCR.
Supplemental Figure S3. Domain structure and amino acid sequence analysis of CAL2, ERO1, and PD16 proteins from C. reinhardtii.
Supplemental Figure S4. Effect of thapsigargin on autophagy and ER stress.
Supplemental Figure S5. Expression analysis of CAL2, ERO1, and Rb60 genes by qPCR.
Supplemental Figure S6. Expression analysis of the GSTS1 gene by qPCR in cells treated with ascorbate.
Supplemental Figure S7. DAD1 expression is up-regulated upon prolonged ER stress.
Supplementary Material
Acknowledgments
We thank Francisco J. Florencio for helpful discussions and advice.
Glossary
- DTNB
5,5′-dithiobis(2-nitro-benzoic acid)
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- MV
methyl viologen
- PDI
protein disulfide isomerase
- qPCR
quantitative real-time PCR
- ROS
reactive oxygen species
- TAP
Tris-acetate phosphate
- UPR
unfolded protein response
Footnotes
This work was supported, in part, by the Intra European Fellowship (European Union Marie Curie Fellowship no. PIEF–GA–2011–298652–REDOXDYNAMICS to M.E.P.-P.), by LABEX DYNAMO (grant no. ANR–11–LABX–0011 to M.E.P.-P. and S.D.L.), by the Spanish Ministry of Economy and Competitiveness (grant nos. BFU–2009–07368 to J.L.C. and BFU–2012–35913 to J.L.C.), and by Junta de Andalucía (grant no. CVI-7336 to J.L.C.).
The online version of this article contains Web-only data.
References
- Aller I, Meyer AJ. (2013) The oxidative protein folding machinery in plant cells. Protoplasma 250: 799–816 [DOI] [PubMed] [Google Scholar]
- Avin-Wittenberg T, Honig A, Galili G. (2012) Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 249: 285–299 [DOI] [PubMed] [Google Scholar]
- Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, Yoshimoto K. (2006) Autophagy in development and stress responses of plants. Autophagy 2: 2–11 [DOI] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. (2006) Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol 4: e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo JL, Díaz-Troya S, Florencio FJ. (2005) Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol 139: 1736–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. (1991) Light regulated translational activators: identification of chloroplast gene specific mRNA binding proteins. EMBO J 10: 3993–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Troya S, Florencio FJ, Crespo JL. (2008a) Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii. Eukaryot Cell 7: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. (2008b) The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4: 851–865 [DOI] [PubMed] [Google Scholar]
- Filonova A, Haemsch P, Gebauer C, Weisheit W, Wagner V. (2013) Protein disulfide isomerase 2 of Chlamydomonas reinhardtii is involved in circadian rhythm regulation. Mol Plant 6: 1503–1517 [DOI] [PubMed] [Google Scholar]
- Fischer BB, Dayer R, Schwarzenbach Y, Lemaire SD, Behra R, Liedtke A, Eggen RI. (2009) Function and regulation of the glutathione peroxidase homologous gene GPXH/GPX5 in Chlamydomonas reinhardtii. Plant Mol Biol 71: 569–583 [DOI] [PubMed] [Google Scholar]
- Fischer BB, Dayer R, Wiesendanger M, Eggen RIL. (2007) Independent regulation of the GPXH gene expression by the primary and secondary effects of high light stress in Chlamydomonas reinhardtii. Physiol Plant 130: 195–206 [Google Scholar]
- Fischer BB, Ledford HK, Wakao S, Huang SG, Casero D, Pellegrini M, Merchant SS, Koller A, Eggen RI, Niyogi KK. (2012) SINGLET OXYGEN RESISTANT 1 links reactive electrophile signaling to singlet oxygen acclimation in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 109: E1302–E1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA. (1998) The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell 1: 161–170 [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA. (1999) Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell 4: 469–477 [DOI] [PubMed] [Google Scholar]
- Gallois P, Makishima T, Hecht V, Despres B, Laudié M, Nishimoto T, Cooke R. (1997) An Arabidopsis thaliana cDNA complementing a hamster apoptosis suppressor mutant. Plant J 11: 1325–1331 [DOI] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- Haynes CM, Titus EA, Cooper AA. (2004) Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell 15: 767–776 [DOI] [PubMed] [Google Scholar]
- Huang J, Lam GY, Brumell JH. (2011) Autophagy signaling through reactive oxygen species. Antioxid Redox Signal 14: 2215–2231 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. (1994) The Saccharomyces cerevisiae oligosaccharyltransferase is a protein complex composed of Wbp1p, Swp1p, and four additional polypeptides. J Biol Chem 269: 12908–12917 [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. (1997) DAD1, the defender against apoptotic cell death, is a subunit of the mammalian oligosaccharyltransferase. Proc Natl Acad Sci USA 94: 4994–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford HK, Chin BL, Niyogi KK. (2007) Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot Cell 6: 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire SD, Miginiac-Maslow M. (2004) The thioredoxin superfamily in Chlamydomonas reinhardtii. Photosynth Res 82: 203–220 [DOI] [PubMed] [Google Scholar]
- Levitan A, Trebitsh T, Kiss V, Pereg Y, Dangoor I, Danon A. (2005) Dual targeting of the protein disulfide isomerase RB60 to the chloroplast and the endoplasmic reticulum. Proc Natl Acad Sci USA 102: 6225–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD. (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Li L, Ishdorj G, Gibson SB. (2012) Reactive oxygen species regulation of autophagy in cancer: implications for cancer treatment. Free Radic Biol Med 53: 1399–1410 [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC. (2012) Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ. (2007) Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal 9: 2277–2293 [DOI] [PubMed] [Google Scholar]
- Martínez IM, Chrispeels MJ. (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizobuchi N, Hoseki J, Kubota H, Toyokuni S, Nozaki J, Naitoh M, Koizumi A, Nagata K. (2007) ARMET is a soluble ER protein induced by the unfolded protein response via ERSE-II element. Cell Struct Funct 32: 41–50 [DOI] [PubMed] [Google Scholar]
- Moharikar S, D’Souza JS, Rao BJ. (2007) A homologue of the defender against the apoptotic death gene (dad1 )in UV-exposed Chlamydomonas cells is downregulated with the onset of programmed cell death. J Biosci 32: 261–270 [DOI] [PubMed] [Google Scholar]
- Nakashima T, Sekiguchi T, Kuraoka A, Fukushima K, Shibata Y, Komiyama S, Nishimoto T. (1993) Molecular cloning of a human cDNA encoding a novel protein, DAD1, whose defect causes apoptotic cell death in hamster BHK21 cells. Mol Cell Biol 13: 6367–6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26: 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur R, Turkan I, Uzilday B, Sekmen AH. (2014) Endoplasmic reticulum stress triggers ROS signalling, changes the redox state, and regulates the antioxidant defence of Arabidopsis thaliana. J Exp Bot 65: 1377–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Couso I, Crespo JL. (2012a) Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 8: 376–388 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Florencio FJ, Crespo JL. (2010) Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 152: 1874–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Lemaire SD, Crespo JL. (2012b) Reactive oxygen species and autophagy in plants and algae. Plant Physiol 160: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard MG, Travers KJ, Weissman JS. (1998) Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell 1: 171–182 [DOI] [PubMed] [Google Scholar]
- Pootakham W, Gonzalez-Ballester D, Grossman AR. (2010) Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol 153: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G, Noctor G. (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ. (2007) That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci 32: 469–476 [DOI] [PubMed] [Google Scholar]
- Santos CX, Tanaka LY, Wosniak J, Laurindo FR. (2009) Mechanisms and implications of reactive oxygen species generation during the unfolded protein response: roles of endoplasmic reticulum oxidoreductases, mitochondrial electron transport, and NADPH oxidase. Antioxid Redox Signal 11: 2409–2427 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Elazar Z. (2011) Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci 36: 30–38 [DOI] [PubMed] [Google Scholar]
- Schroda M. (2004) The Chlamydomonas genome reveals its secrets: chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth Res 82: 221–240 [DOI] [PubMed] [Google Scholar]
- Szumiel I. (2011) Autophagy, reactive oxygen species and the fate of mammalian cells. Free Radic Res 45: 253–265 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD. (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Tietze F. (1969) Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal Biochem 27: 502–522 [DOI] [PubMed] [Google Scholar]
- Tu BP, Weissman JS. (2004) Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164: 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287: 664–666 [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334: 1081–1086 [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ. (2006) Endoplasmic reticulum stress triggers autophagy. J Biol Chem 281: 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, Bedhomme M, Marchand CH, Morisse S, Trost P, Lemaire SD. (2012) Redox regulation in photosynthetic organisms: focus on glutathionylation. Antioxid Redox Signal 16: 567–586 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.