Abstract
The N-terminal oligonucleotide/oligosaccharide-binding fold domain of the Schizosaccharomyces pombe protection of telomeres 1 (Pot1) protein, Pot1pN (residues 1–187 of full-length Pot1), specifically recognizes telomeric single-stranded DNA (ssDNA) via a complex series of molecular interactions that are punctuated by unusual internucleotide hydrogen bonds. While the structure of ssDNA-bound Pot1pN provides an initial model for understanding how the Pot1pN–ssDNA complex is assembled and how specific nucleotide recognition occurs, further refinement requires knowledge of the ssDNA-free state of Pot1pN and the dynamic changes that accompany the binding of ssDNA. Using NMR strategies, we found that ssDNA-free Pot1pN adopts a similar overall protein backbone topology as ssDNA-bound Pot1pN does. Although the backbone structure remained relatively unchanged, we observed unexpected differential dynamic changes within the ssDNA-binding pockets of Pot1pN upon binding of cognate ssDNA. These studies support a model in which conformational selection and induced fit play important roles in the recognition of ssDNA by Pot1pN. Furthermore, the studies presented here provide a more comprehensive understanding of how specific nucleotide recognition is achieved by the telomere-end protection family of essential proteins.
Keywords: NMR dynamics, OB fold, Pot1, single-stranded DNA binding, telomeres
Introduction
Telomeres are the nucleoprotein complexes that cap the ends of eukaryotic linear chromosomes and are responsible for maintaining the overall stability of genomic DNA and sustaining continued cellular proliferation (reviewed by Verdun and Karlseder1 and Aubert and Lansdorp2). Telomeres terminate in a conserved 3′ single-stranded overhang that, left uncapped, triggers damaging cellular responses, including inappropriate processing of the telomere (i.e., end-to-end chromosomal fusions and recombination events) caused by activation of the DNA-damage response pathway.3–8 Such events are circumvented through the capping of the single-stranded telomere ends by a family of highly specialized proteins called the telomere-end protection (TEP) proteins (reviewed by Croy et al.9).
TEP proteins bind telomeric single-stranded DNA (ssDNA) with high affinity and specificity, and our current understanding of how these proteins function arises from a combined structural and biochemical effort examining both full-length TEP proteins and their isolated ssDNA-binding domains (DBDs).10–15 Although functionally similar, the DBDs of TEP proteins display a surprising lack of sequence conservation.9 In contrast, structural studies, focused primarily on the ssDNA-bound states of these proteins, have identified a shared, highly conserved protein fold, the oligonucleotide/oligosaccharide-binding fold (OB fold). These structures reveal that ssDNA recognition by OB folds occurs primarily through interactions with the bases of bound oligonucleotides. The rich variety of amino acids present in TEP proteins in the context of a common structural framework has led to an assortment of chemically distinct ssDNA-binding interfaces.11,12,14,16,17 Despite this diversity, analysis of the ssDNA-bound structures of TEP proteins has revealed two primary conserved modes of interaction with the bound oligonucleotide: hydrogen-bonding and aromatic/hydrophobic base stacking interactions. Current models explain specific nucleotide recognition based on these bound structures, and rely most heavily on hydrogen-bonding interactions. However, biochemical data indicate that the specific nucleotide recognition can occur even in the absence of extensive hydrogen bonding between the protein and the nucleotide.13,18 Furthermore, some structurally identified hydrogen bonds that appear to be important for specific nucleotide recognition, when mutated, show no apparent effect on binding affinity.11 These results suggest that the specific recognition of telomeric ssDNA is more complex than indicated by simple interpretation of the interaction details available in these bound structures. Thus, the development of a more sophisticated understanding of specific recognition requires knowledge of the structural and dynamic changes that contribute to the thermodynamics of telomeric ssDNA binding.
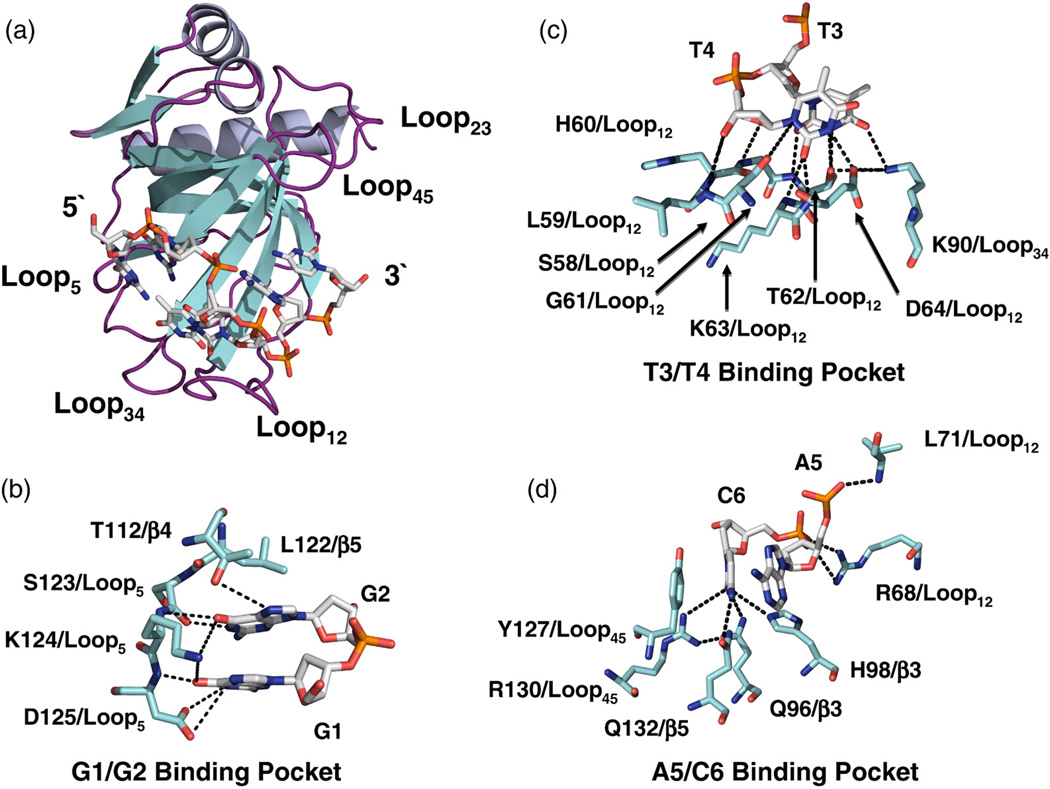
We chose to study the protection of telomeres 1 (Pot1) proteins, which represent the largest subfamily of TEP proteins and are found throughout nature in plants,5,19 fungi,7,20 and mammals,21 including humans.7,22 Pot1pN (residues 1–187 of Schizosaccharomyces pombe Pot1)7 is an autonomously folded DBD residing in the N-terminal region of full-length Pot1 that tightly binds a minimal oligonucleotide representing a single unit of the core repeating telomeric S. pombe sequence, d (GGTTAC) (specifically recognized nucleotides in boldface).10 The ssDNA-binding interface present in Pot1pN is formed from a single face of the OB fold β-barrel (consisting of β-strands β3 and β5) as well as Loop12 (connecting β1 and β2) and Loop45 (connecting β4 and β5) that together form the distinctive “taco-like” ssDNA-binding groove (Fig. 1). This ssDNA-binding interface constrains the inherently flexible oligonucleotide in an unusual, compact conformation punctuated by internucleotide hydrogen bonds connecting G1 to T3, G2 to T4, and A5 to the phosphodiester backbone of T3 and T410 (Fig. 1). In addition to these internucleotide interactions, a complex network of hydrogen bonds and aromatic stacking interactions is formed between the protein and the bound oligonucleotide. While the Pot1pN/d(GGTTAC) complex (Pot1pNB) structure provides an initial foundation for understanding the biochemically derived nucleotide specificity, it does not provide a complete picture of how the assembly of the complex occurs and its impact on specific nucleotide recognition.
Fig. 1.
(a) Ribbon (Pot1pN) and stick (ssDNA) representation of the high-resolution crystal structure of the Pot1pN/ssDNA complex11 highlighting the compact conformation of the bound oligonucleotide, d(GGTTAC). Pot1pN is shown in a ribbon representation with the α-helical elements shown in purple, the β-strand elements shown in yellow, and connecting loops shown in magenta. Each connecting loop found within the OB fold is labeled according to the β-strands that it connects (e.g., the loop connecting β−1 and −2 is designated as Loop12). d(GGTTAC) is shown in stick representation and colored according to standard atomic colors. (b–d) Stick representations detailing the molecular interactions that occur between individual residues in Pot1pN (blue sticks) and d(GGTTAC) (white sticks) in the three binding pockets—G1/G2, T3/T4, and A5/C6. Hydrogen-bonding interactions formed between Pot1pN and d(GGTTAC) are designated by black dashed lines. All figures were made using PyMOL version 1.0.23
Since structural and dynamic changes play important roles in the process of protein/ligand assembly and the thermodynamics of specific recognition, we used solution NMR-based relaxation, chemical shift perturbation (CSP), and residual dipolar coupling (RDC) experiments to probe the structural and dynamic changes that occur upon binding of Pot1pN to its minimal cognate ssDNA substrate, d (GGTTAC). Chemical shift indexing (CSI) and RDC experiments of ssDNA-free Pot1pN (Pot1pNF) and Pot1pNB indicate that the backbone structure of the Pot1pNF is highly similar to the OB fold present in the Pot1pNB crystal structure. Furthermore, these results indicate that the binding of ssDNA does not induce large global conformational changes in the structure of Pot1pN in solution. Rather, the binding of d(GGTTAC) to Pot1pN induces highly localized and varied structural and dynamic changes within the amide backbones of residues found within the loops comprising the G1/G2-, T3/T4-, and A5/C6-binding pockets. Taken together with previously reported structural and biochemical work, these results allow for the development of a more sophisticated mechanistic model for describing how the assembly of Pot1pNB contributes to the observed nucleotide sequence specificity.
Results
Backbone atom assignment of Pot1pNF
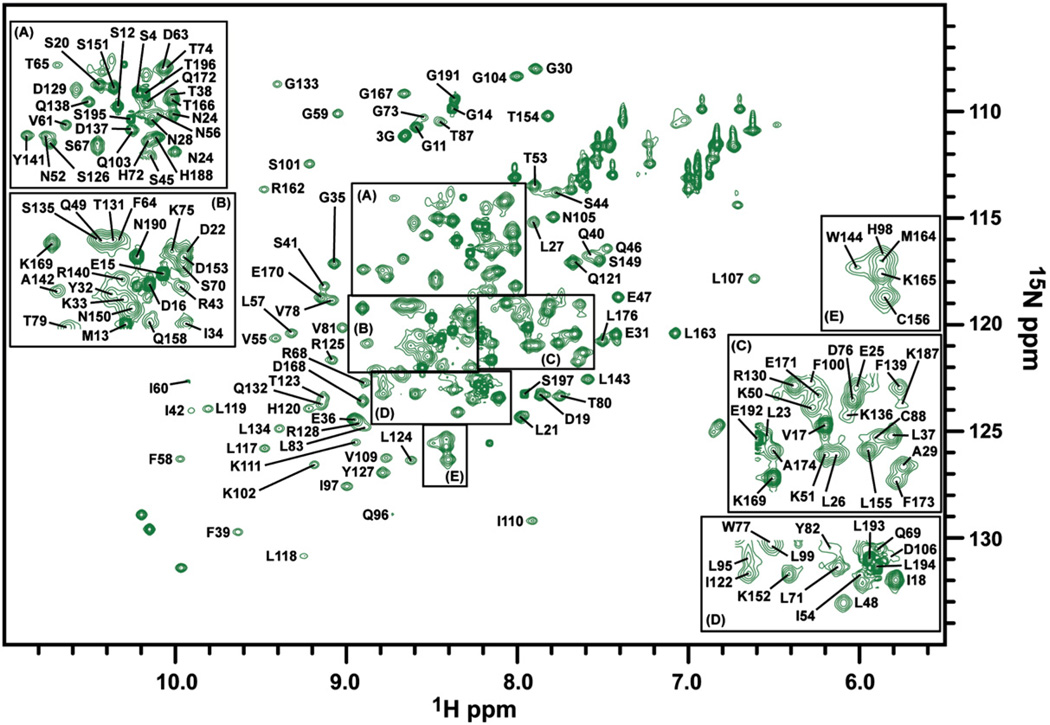
To gain insight into Pot1pNF in solution, we first assigned the backbone atoms (1HN, 15N, 13CO, 13Cβ, 13Cα) of Pot1pNF using standard triple-resonance NMR experiments (Supplemental Table 1) and annotated the 15N heteronuclear single-quantum coherence (HSQC) spectrum of Pot1pNF with completed assignments (Fig. 2). The construct of Pot1pN used in these experiments contains 189 observable residues, of which 155 (82%) have been completely assigned (in 1HN, 15N, 13CO, and 13Cα) and 8 (4%) have been partially assigned (only in 13CO, 13Cα, and, where applicable, 13Cβ). A total of five peaks could not be assigned in the 15N HSQC spectrum of Pot1pNF (Fig. 2) due to their absence in the triple-resonance data and/or intervening proline residues.
Fig. 2.
15N HSQC spectrum of the Pot1pNF collected on a Varian 500-MHz spectrophotometer equipped with a room temperature probe at 30 °C. Peaks representing the individual amino acids present in Pot1pN are labeled with their corresponding residue assignments. For clarity, selected regions of the 15N HSQC spectrum are enlarged [panels (a–e)] to display selected assignments.
We were able to almost fully recapitulate the assignment coverage observed in Pot1pNB with the exception of Ser8, Gly102, Leu165, and Asn166 (Supplemental Table 1), thus allowing for direct comparisons of almost all the residues present in each form of Pot1pN. Furthermore, we were able to assign Trp65, which was not visible in the 15N HSQC spectrum of Pot1pNB due to exchange broadening (Supplemental Table 1).24 The addition of Trp65 provides a complete residue assignment of the ssDNA-binding interface present in Pot1pN, as defined by the high-resolution crystal structure of Pot1pNB.11 Finally, as observed in the assignment of Pot1pNB, the remaining unassigned residues in Pot1pNF cluster either to the N- and C-termini or to a region of the protein directly adjacent to the ssDNA-binding interface (Supplemental Table 1).11
CSI, RDC measurements, and CSP mapping indicate minimal structural changes upon binding of ssDNA to Pot1pN
CSI analysis has been proven to be a reliable method25,26 for the identification of secondary structures. We used CSI analysis of 13Cα atoms to probe for changes that occur in the secondary structure elements present in Pot1pN upon binding of ssDNA. We found that the secondary structural elements predicted by CSI for both Pot1pNF and Pot1pNB are completely consistent with those found in the high-resolution structure of Pot1pNB (Supplemental Fig. 1). Moreover, differences between the 13Cα CSI values for Pot1pNF and Pot1pNB are quite modest, indicating that little perturbation of the overall secondary structure of Pot1pN occurs upon binding of ssDNA (Supplemental Fig. 1).
We also examined the perturbations in the backbone tertiary structure of Pot1pN that accompany the binding of d(GGTTAC) using RDC measurements. RDCs between 1H and 15N (DNH) atoms in weakly aligned liquid crystalline solutions provide structural restraints that correlate the overall orientation of individual bond vectors within a fixed molecular tensor axis system.27 Partial alignments of both Pot1pNF and Pot1pNB were obtained using a filamentous Pf1 bacteriophage liquid crystal system,28 resulting in DNH values ranging from 5 to 10 Hz (Supplemental Fig. 2 and Table 2). Determination of a self-consistent alignment tensor (principal component axes Sxx, Syy, and Szz) and respective Euler rotations (α, β, and γ) for Pot1pNF and that for Pot1pNB were done in an iterative fashion by removing DNH values with >3 Hz error that were rejected during the analysis of 1 million Monte Carlo simulated data sets (26 of 154 for Pot1pNB and 29 of 147 for Pot1pNF) (data not shown). The majority of residues that did not fit were found to be localized to nonregular secondary structure regions of the N-terminal non-OB-fold elements and loops connecting the regular secondary structure elements in the OB fold, as determined by the high-resolution crystal structure of Pot1pNB.11
Spectra collected to determine DNH values for Pot1pNF in low NaCl (50 mM) resulted in peak broadening and poor spectral quality. These spectral issues were alleviated by increasing the overall NaCl concentration to 400 mM (data not shown). In contrast, Pot1pNB exhibited excellent spectral quality at both salt concentrations, indicating that the free and bound states align via different mechanisms. Alignment by Pf1 bacteriophage occurs via electrostatic interactions,28 and since the electrostatic characters of Pot1pNF and Pot1pNB are significantly different, it is not surprising that they have unique alignment tensors (Supplemental Table 3). Therefore, to directly compare DNH values of Pot1pNF and Pot1pNB, we back-calculated DNH values based on the atomic coordinates present in the Pot1pNB crystal structure after rotation into each specific alignment tensor. Correlation plots of experimental versus calculated DNH values for Pot1pNF (Supplemental Fig. 3) and Pot1pNB (Supplemental Fig. 3) show Q-factors of ~0.42, indicating that both Pot1pNF and Pot1pNB are in good agreement with the 2.4-Å Pot1pNB crystal structure, with the potential exception of those residues located in nonstructured regions that were originally excluded from the analysis.29–32
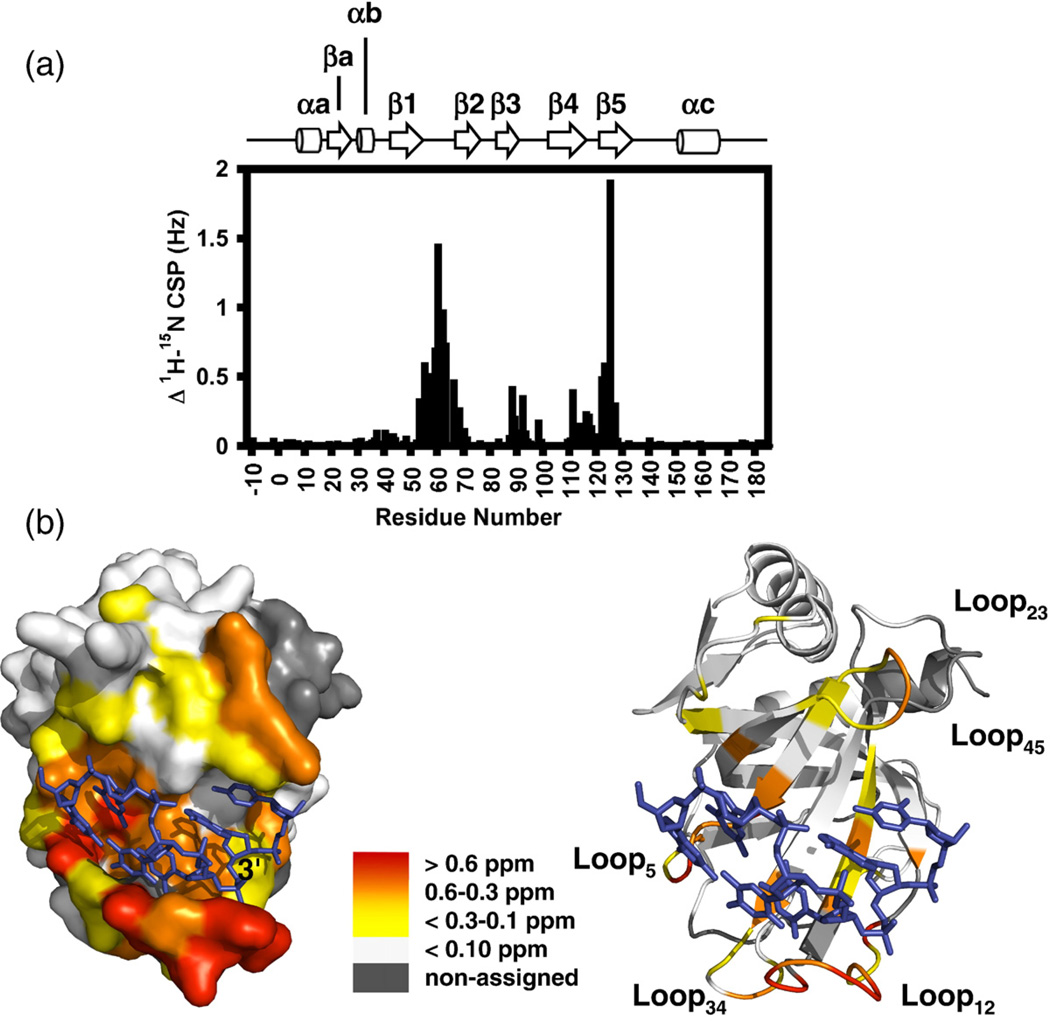
Finally, consistent with the minimal structural changes predicted by CSI and RDC analysis, CSP mapping of Pot1pNF and Pot1pNB indicates that the majority of residues (118 of 155) present in Pot1pN are unperturbed by the binding of cognate ssDNA (Fig. 3a and b). Rather, we observed perturbation of residues that are located within the ssDNA-binding interface as defined in the high-resolution crystal structure of Pot1pNB.11 Taken together, these three independent experiments provide ample evidence that the global backbone conformation of the OB-fold present in Pot1pN is preserved in the presence and in the absence of cognate ssDNA.
Fig. 3.
15N–1H CSP analyses of Pot1pNF and Pot1pNB. (a) Differences in the composite 15N–1H chemical shifts for Pot1pNF and Pot1pNB plotted as a function of residue number. The secondary structure elements of the high-resolution X-ray crystallographic structure of Pot1pNB11 are shown above for reference. (b) Surface (left) and ribbon (right) representation of the high-resolution X-ray crystallographic structure of Pot1pNB11 with the individual residues colored according to their respective chemical shift differences: >0.6 ppm in red; 0.6–0.3 ppm, orange; <.3–0.1 ppm, yellow; and <0.1 ppm, white. Nonassigned residues in either Pot1pNF or Pot1pNB are shown in gray. All figures were made using PyMOL version 1.0.23
Binding of d(GGTTAC) alters the fast-timescale dynamic behavior of Loop12 but not that of Loop45
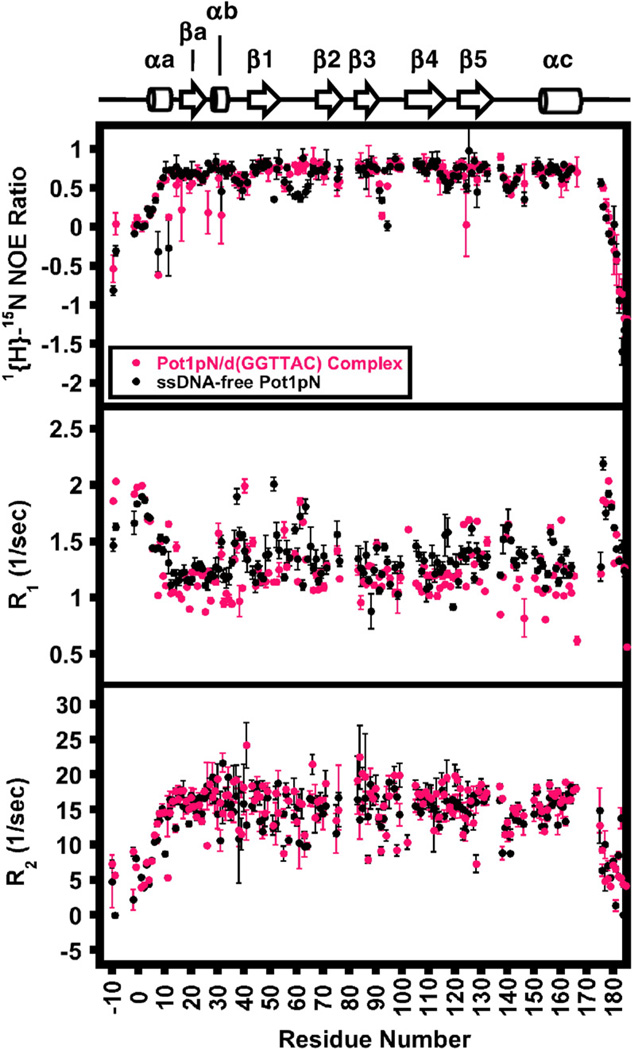
The dynamic behaviors of backbone amides in Pot1pNF and Pot1pNB on the picosecond–nanosecond timescale were assessed using standard two-dimensional 15N HSQC-based relaxation experiments [1{H}–15N steady-state heteronuclear Overhauser enhancement (HetNOE), T1, and T2] collected at two field strengths (Fig. 4 and Supplemental Table 4). We were able to obtain R1 (spin–lattice relaxation rate), R2 (spin–spin relaxation rate), and HetNOE values for all the backbone amides present in Pot1pNF except Ile54. In the case of Pot1pNB, R1, R2, and HetNOE values were obtained for all backbone amides except Leu23, Thr38, Arg48 (HetNOE only), Ile54, Ser70 (HetNOE only), Tyr82 (R2 only), Leu99 (R2 only), Lys136 (HetNOE only), and Lys187 (HetNOE only). In both cases, these select amide peaks exhibited poor peak dispersion and/or intensity and were excluded from our dynamics analysis. As expected, the terminal amides (1–30 and 188–197) and the majority of those present in the loops connecting individual β-strands in Pot1pNF and Pot1pNB experience dynamic motions on the fast timescale that are unaffected by the binding of d(GGTTAC) and are consistent with unstructured protein elements (Fig. 4 and Supplemental Fig. 4).
Fig. 4.
Parameters determined from 15N relaxation experiments. HetNOE (upper panel), R1 (middle panel), and R2 (lower panel) are plotted as a function of residue number for Pot1pNF (black) and Pot1pNB (magenta) collected at 500 MHz at 30 °C. Associated errors were derived as follows: HetNOE errors represent the standard deviation from triplicate experiments, while R1 and R2 errors were derived from the fitting of 300 simulated Monte Carlo data sets that included the noise errors estimated from duplicate points.33 The secondary structure elements of the high-resolution X-ray crystallographic structure of Pot1pNB11 are shown above for reference.
The majority of amides present in Pot1pNF and Pot1pNB were insensitive to changes in their respective picosecond–nanosecond dynamic motions upon binding of d(GGTTAC), with the notable exception of the ssDNA-binding Loop12 (Fig. 4). For Loop12, significant increases in the HetNOE ratios (Fig. 4) (average change of Loop12 amides from 0.54 in Pot1pNF to 0.73 in Pot1pNB) indicate a decrease in the dynamic behavior of these amides in response to ssDNA binding, moving from a partially disordered state to a highly ordered structure (Fig. 4 and Supplemental Fig. 4). Surprisingly, we found that the backbone amides in Loop45, which provides the other half of the ssDNA-binding interface (as described in Fig. 1a), undergo a modest change in the HetNOE ratios (average change from 0.61 in Pot1pNF to 0.67 in Pot1pNB) upon binding of d (GGTTAC) (Fig. 4 and Supplemental Fig. 4), suggesting that, unlike Loop12, Loop45 remains moderately flexible in the presence and in the absence of ssDNA (Fig. 4 and Supplemental Fig. 4). The dynamic characteristics of these amides in Loop12 and Loop45 in Pot1pNB observed in our studies correlate well with their respective B-factor values reported in the Pot1pNB crystal structure.11 These results indicate that the two ssDNA contacting loops, Loop12 and Loop45, which form the “taco-like” ssDNA-binding groove in Pot1pN, undergo differential dynamic and structural changes upon binding of d(GGTTAC).
Model-free analyses of Pot1pNF and Pot1pNB
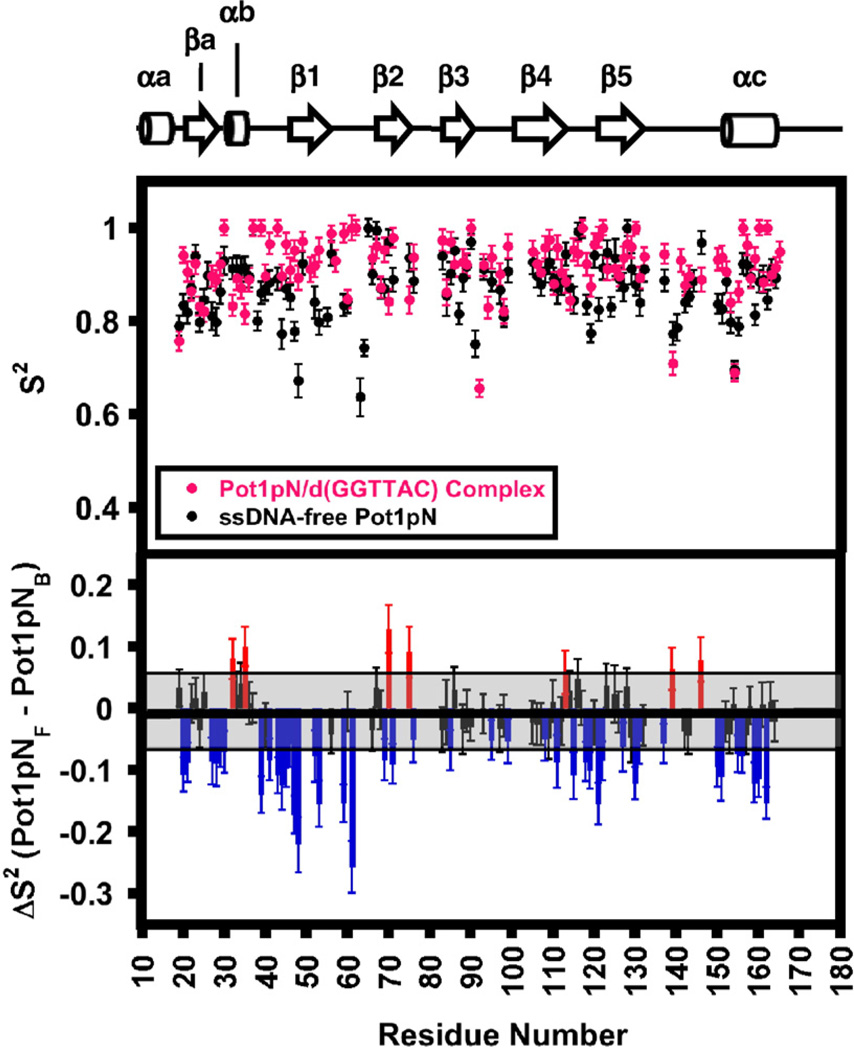
Model-free analyses of relaxation34,35 for Pot1pNF and Pot1pNB were performed using R1, R2, and HetNOE values collected at 500 and 600 MHz (Supplemental Table 5). Fitting of the data using ModelFree36 resulted in the majority of residues fitting to either model 1 or model 2 as described by Mandel et al.,36 indicating that the observed relaxation properties of Pot1pNF and Pot1pNB could be explained by simple motional models (Supplemental Table 5). The calculated overall molecular correlation times for Pot1pNF (12.5 ns) and Pot1pNB (13.4 ns) are consistent with the masses of free and bound Pot1pN (Supplemental Table 6). We observed a dramatic increase in the S2 (generalized order parameter) values for many of the backbone amides present in Loop12 and a less dramatic increase in the S2 values for those present in Loop45 (Fig. 5). We also detected dynamic changes in elements of the OB fold that do not make direct interactions with the bound oligonucleotide: β1, which directly precedes L12 (Val43, Asn44, Leu45, Phe46, Gly47, Ile48, Phe52, and Thr53), and the amides preceding and forming the C-terminal α-helix (Arg150, Leu151, Gly155, Asp156, Glu159, Gln160, and Ala162). Although these two regions do not directly interact with ssDNA, they do interact extensively with each other through predominantly hydrophobic interactions. Our relaxation data suggest that the loss of dynamic motion in Loop12 upon binding of d(GGTTAC) leads to a simultaneous loss of dynamic motion in β1 that is then propagated into the C-terminal helix (Fig. 5).
Fig. 5.
ModelFree S2 values for the backbone amides present in Pot1pNF (black) and Pot1pNB (magenta) derived from HetNOE, R1, and R2 experiments collected at 500 and 600 MHz at 30 °C. The upper panel contains S2 values calculated for Pot1pNF (black) and Pot1pNB (magenta) plotted as a function of residue number. Associated errors in S2 values were calculated by the fitting of 300 Monte Carlo simulated data sets with ModelFree.36 The lower panel is a bar graph showing the per-residue differences between the Pot1pNF and Pot1pNBS2 values plotted as a function of residue number. Residues falling within the average error of (shaded black box) the calculated S2 values were found to have no difference in the overall structure (black bars). Residues outside the average error were found to be less structured either in the absence (blue bars) or in the presence (red bars) of d(GGTTAC).
15N CPMG relaxation dispersion indicates a significant decrease in the overall dynamic behavior of Pot1pN on the microsecond–millisecond timescale upon binding d(GGTTAC)
Dynamic behavior on the microsecond–millisecond timescale can be associated with the conformational exchange of backbone amides between an NMR observed and often highly populated (visible) state and an invisible and generally sparsely populated (invisible) state of the protein (reviewed by Korzhnev and Kay37). Termed relaxation dispersion (Rex), molecular motions on these timescales directly increase the apparent R2 values of affected amides. We measured Rex directly using a modified R2 experiment in which R2 values of backbone amides in Pot1pNF and Pot1pNB were measured for a fixed period, during which CPMG (Carr–Purcell–Meiboom–Gill) pulses were applied at variable time delays (τCPMG) (20 µs to 1 ms in our experiment).38
Based on our R2-CPMG analysis (Supplemental Fig. 5 and Table 7 show data and fits for selected amides), we found that in the absence of bound oligonucleotide, there is a total of 40 backbone amides that experience microsecond–millisecond dynamic motion, of which 28 (70%) were found to cluster to the G1/G2- and T3/T4-binding pockets, with the remaining 12 located in the non-OB-fold structural elements found at the N- and C-termini of Pot1pN (Fig. 6a and Supplemental Fig. 6). The binding of d(GGTTAC) to Pot1pN results in a significant decrease in the overall dynamic behavior of Pot1pN on the microsecond–millisecond timescale, as only 26 backbone amides display microsecond–millisecond dynamic motions (Fig. 6b and Supplemental Fig. 6). While data collected for Pot1pNF and Pot1pNB at low field (600 MHz) were not of sufficient quality to perform an in-depth analysis of the “invisible” states sampled by these dynamically active backbone residues, mapping of these sites onto the Pot1pNB structure allows us to propose how such dynamic changes can contribute to specific nucleotide recognition. Specifically, we found that all the backbone amides present in the G1/G2-binding pocket that showed microsecond–millisecond dynamic behavior in Pot1pNF do not show the same motions in Pot1pNB. Additionally, we also observed a significant loss of dynamic motion in amides present within the T3/T4-binding pocket upon binding d(GGTTAC), specifically with respect to those found in Loop12 and Loop34. Finally, although the overall dynamic behavior of Pot1pNB was significantly decreased, we did observe a few amides that became dynamic upon binding d(GGTTAC) (Fig. 6b). Of these amides, Phe88 and Gln84 make contact with the bound oligonucleotide in the Pot1pNB crystal structure via aromatic stacking and hydrogen-bonding interactions, respectively. Taken together, these results suggest that the binding of d(GGTTAC) to Pot1pN directly impacts the overall dynamic behavior of backbone amides on the microsecond–millisecond timescale, with dynamic motion being quenched, especially in those loops that mediate specific nucleotide recognition.
Fig. 6.
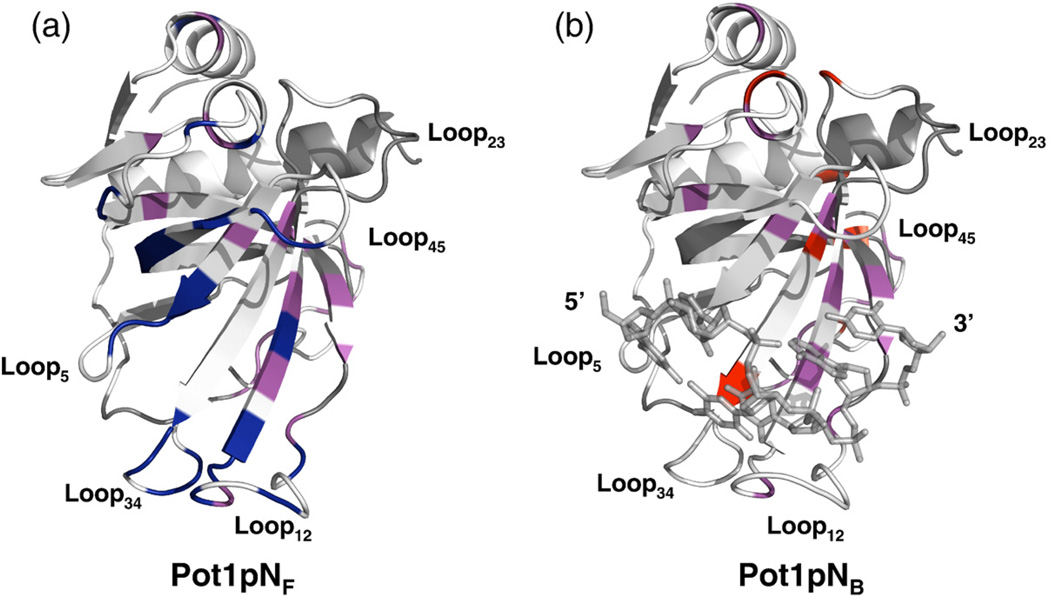
Ribbon (Pot1pN) and stick (ssDNA) representation of the high-resolution crystal structure of the Pot1pN–ssDNA complex11 highlighting the residues that undergo dynamic motion on the microsecond–millisecond timescale in both Pot1pNF and Pot1pNB. (a) Residues experiencing microsecond–millisecond dynamic motion in Pot1pNF only are shown in navy, those in both Pot1pNF and Pot1pNB are shown in magenta, and those that showed no motion are shown in white. (b) Residues experiencing microsecond–millisecond dynamic motion in Pot1pNB only are shown in navy, those in both Pot1pNF and Pot1pNB are shown in magenta, and those that showed no motion are shown in white. In both cases, residues shown in gray represent unassigned residues in both Pot1pNF and Pot1pNB. All figures were made using PyMOL version 1.0.23
Discussion
TEP proteins are a biologically important family that specifically recognizes the 3′ ssDNA overhangs present at telomere ends and are critical to proper telomere function (reviewed by Croy et al.9). Our current understanding of the underlying mechanisms that drive specific telomeric ssDNA recognition has been restricted due to the limited data sets addressing the ssDNA-free form of the TEP family. To date, two studies of ssDNA-free TEP proteins have been reported, but their overall ability to provide further insight into the process of ssDNA recognition has been hindered by limited data in the case of Cdc13-DBD39 and the presence of a heterogeneous TEBPα35-d(T4G4) complex in which an unbound protein co-crystallized with bound complexes.15 Therefore, we examined Pot1pNF by NMR using RDC experiments and CSI, which reveal that the overall backbone topology of the OB fold present in the Pot1pN bound state is essentially unchanged in the absence of bound oligonucleotide. These observations are further supported by our 1H–15N CSP data, which show that the binding of ssDNA does not lead to widespread perturbation of residues throughout Pot1pN. Rather, we found that changes are limited to residues present within the ssDNA-binding interface as defined by the Pot1pNB crystal structure.11 These results indicate that the binding of telomeric ssDNA to Pot1pN effects minimal change in the overall protein topology. In contrast, the ligand d(GGTTAC) is inherently disordered when free in solution25 and undergoes a substantial structural rearrangement to form the compact configuration observed in the Pot1pNB crystal structure.11 Taken together, these studies highlight an emerging shared mechanism for the recognition of telomeric ssDNA within the TEP family in which a disordered oligonucleotide undergoes a large structural rearrangement to fold into a largely ordered protein surface.
The absence of large protein conformational changes upon binding of ssDNA suggests that perhaps more subtle changes in dynamic behavior contribute to the observed nucleotide specificity conferred by Pot1pN. Two nonexclusive mechanisms described by these subtle dynamic motions have been implicated in mediating specific nucleotide recognition in RNA–protein interactions: conformational selection and induced fit (reviewed by Williamson,40 Leulliot and Varani,41 and Boehr and Wright42). Conformational selection involves the transient exchange of thermodynamically preferred structural conformations in the absence of ligand. Such selection involves dynamic motions that are generally thought to occur on a slower (microsecond–second) timescale. The structuring of disordered protein and/or ligand regions that occurs in induced fit can alter motions that occur on a faster (picosecond–nanosecond) timescale (reviewed by Mittermaier and Kay43). The use of NMR allows us to directly assess backbone motions occurring on both of these timescales, thus giving insight into the interaction mechanisms that are involved in the assembly of Pot1pNB.
Formation of the G1/G2-binding pocket
The G1/G2-binding pocket is formed by the coordinated interactions of β4 and Loop511 (Fig. 1b). In the absence and in the presence of ssDNA, the amide backbone of residues present within this binding pocket exhibits picosecond–nanosecond dynamic motions that are typical of well-ordered protein structural elements. Although we observed little change in picosecond–nanosecond motions, amides found within the G1/G2-binding pocket do experience microsecond–millisecond timescale motions in the absence of ssDNA that are quenched upon binding (Fig. 6a and b). Such dynamic attributes support a model in which the formation of the G1/G2-binding pocket occurs via conformational sampling, in which the final ssDNA-bound protein conformational state is selected from a dynamic ensemble of amide backbone conformations.44 In addition to conformational sampling, the binding of ssDNA to Pot1pN is also accompanied by a large structuring of the oligonucleotide, indicating that induced-fit mechanisms may also be employed.
Structural and mutagenesis data support a model in which the specific recognition of G1 arises in part from the complex hydrogen-bond network formed between the binding pocket and the oligonucleotide.10,11 Our dynamics data build upon these initial observations by suggesting that the well-ordered nature of the G1/G2-binding pocket in the absence of ssDNA is thermodynamically primed for optimum hydrogen-bonding interactions with guanosine at position 1. The presence of a guanosine-specific binding pocket also lends insight into previously published mutagenesis data that show the closely related inosine (which is identical with guanosine but lacks its 2′ exocyclic amine) is thermodynamically accommodated by Pot1pN, while cytosine is not.11 In contrast, the specific recognition of guanosine at position 2 arises from hydrogen-bonding interactions formed with thymine at position 4 and not from those provided by the G1/G2-binding pocket.11 Consistent with our dynamics studies, the guanosine-primed G1/G2-binding pocket may play an important role in the induced fitting of G2 by providing a well-ordered template that optimizes the positioning of G2 with hydrogen-bonding interactions with T4.
Assembly of the T3/T4-binding pocket
Unlike the G1/G2-binding pocket, which is well ordered in the presence and in the absence of ssDNA, picosecond–nanosecond motions show that backbone amides present in Loop12, which forms the centerpiece of the T3/T4-binding pocket, are significantly disordered in the absence of ssDNA and ordered upon binding (Fig. 4). This loop is well structured in Pot1pNB, suggesting that formation of the T3/T4-binding pocket proceeds via a “mutually” induced-fit type of mechanism that requires the coordinated co-folding of both Loop12 and Loop34 with the unstructured oligonucleotide40,45 (Fig. 4). The question then arises as to how the T3/T4-binding pocket and ssDNA oligonucleotide co-fold with one another. One possible mechanism is that the T3/T4-binding pocket and d(GGTTAC) fold into the conformation observed in the Pot1pNB crystal structure in an unbiased fashion based on a random sampling of an infinite number of conformational states. However, the microsecond–millisecond dynamic behavior exhibited by the backbone amides present in Loop12, β2, β3, and Loop34 supports a more complex mechanism in which the mutually induced fitting of oligonucleotide and protein is augmented by conformational sampling.44
Examination of the static crystal structure suggests that the specific recognition of T3 arises from hydrogen-bonding interactions offered solely by the T3/T4-binding pocket, while T4 arises from interactions formed with both G2 and the binding pocket.11 Our picosecond–nanosecond dynamics data indicate that, unlike the G1/G2-binding pocket, the important interactions that dictate the specific recognition of thymine at positions 3 and 4 are not presented within the context of a rigid structural template. Rather, microsecond–millisecond motions support a model in which these interactions are presented in the context of a transiently sampled ensemble of alternate conformations that are formed in the absence of ssDNA. Mechanistically, the formation of the T3/T4-binding pocket is similar to the formation of the G1/G2-binding pocket, as a thermodynamically preferred conformation, sampled from the ensemble, guides the correct induced fitting of the protein and oligonucleotide. This template may also provide positional regulation of T4 that results in an optimum orientation for forming the thermodynamically important interactions with G2.
Formation of the A5/C6-binding pocket
The binding pocket for A5/C6 is formed from the interactions of Loop12, β3, β4, Loop45, and β5 (Fig. 1d). Analysis of the picosecond–nanosecond timescale dynamics data shows that backbone amides present in Loop45 are partially disordered in Pot1pNF and that, unlike Loop12, Loop45 undergoes little change in dynamics upon binding of d (GGTTAC) (Fig. 4). In addition, Loop45 contains only a few backbone amides within the A5/C6-binding pocket that exhibit dynamic motion on the microsecond–millisecond timescale in the absence of ssDNA, Arg56, Arg113, Tyr115, and Gln120, all of which make contact with C6 (Figs. 1d and 6b). The binding of ssDNA leads to differential dynamic effects on the amide backbones of these residues. Arg56, Arg113, and Tyr115 undergo drastic dynamic losses, while the amide backbone of Gln120 remains dynamic. In addition, we found that the binding of d(GGTTAC) to Pot1pN induces a microsecond–millisecond dynamic behavior in the amide backbone of Gln84, which hydrogen bonds with C6.
Biochemical studies have shown a clear thermodynamic preference for adenosine at position 5 by Pot1pN, while preference for cytosine at position 6 is greatly reduced.10 Such thermodynamic characteristics are consistent with our dynamics data showing that Loop45, which directly contacts C6 in d(GGTTAC) (Fig. 1d), remains partially disordered and flexible in the presence and in the absence of bound oligonucleotide (Fig. 4). These dynamic motions reveal a malleable binding pocket that is able to accommodate nucleotides other than cytosine at position 6 with a lessened thermodynamic consequence. In addition to the retention of dynamic motion on the picosecond–nanosecond timescale, we also found that the binding of d (GGTTAC) induces the amide backbones of residues that hydrogen bond to C6 to become or remain dynamically active on the microsecond–millisecond timescale (Fig. 6b). These dynamic properties support a model for the specific recognition of A5 that arises from interactions not offered by the A5/C6-binding pocket. Instead, it is likely that the specific recognition of A5 results from the internucleotide interactions with the phosphodiester backbones of T3 and T4. This model is supported by the crystal structure of Pot1pN bound to d(GGTTA) (which does not contain cytosine at position 6) that shows minimal perturbation in the overall positioning of A5 relative to the Pot1pNB crystal structure.11 Such a mechanism for recognition of A5 requires exquisite control over the final bound conformation of the T3/T4-binding pocket and bound oligonucleotide.
The conformational selection and induced-fit models that we predicted to be important in the specific recognition of ssDNA by Pot1pN have also been proposed to be important in the context of the interaction of the human spliceosomal protein U1A with its cognate substrate, the polyadenylated inhibition element from a 3′ untranslated region of the U1A pre-mRNA.46 Specific recognition of mRNA by U1A is mediated in part by residues found within loop 3 (L3). In the absence of mRNA, L3 exhibits backbone dynamic motions that are consistent with a disordered loop undergoing conformational sampling. Upon binding mRNA, L3 becomes structured, effectively quenching the conformational sampling observed in the mRNA-free state. In addition to Pot1pN and U1A, similar dynamic observations have been ascribed to the recognition of RNA by a mouse RNA-binding protein, Musashi1.47 These studies together indicate that, like specific antigen–antibody interactions,48–50 the specific recognition of single-stranded nucleic acids is a complex and diverse event that incorporates physical interaction elements found in both the induced-fit model and the conformational selection model. This structural and dynamic analysis highlights the ability of NMR to complement and augment the currently available high-resolution X-ray crystallography structures, thus providing a more comprehensive interpretation of specific recognition of ssDNA by this important class of proteins.
Materials and Methods
Chemicals, reagents, and proteins
All chemicals and reagents were obtained from Fisher Scientific (Pittsburg, PA) unless otherwise indicated. HPLC-purified d(GGTTAC) was commercially synthesized (Operon, Huntsville, AL) and reverse-phase purified over a semipreparative C18 column (Grace Vydac, Hesperia, CA) in 1% triethanolamine acetate/H2O and eluted with a 5%–40% gradient of 1% triethanolamine acetate/19% H2O/80% acetonitrile. Chromatography materials were purchased from GE Healthcare (Piscat-away, NJ). Pf1 bacteriophage29 was a generous gift from Professor Arthur Pardi. Finally, the plasmid-encoding Pot1pN was graciously provided by Professor Thomas Cech.7
Expression and purification of Pot1pN
The 22-kDa (His)6-Pot1pN DBD from the S. pombe Pot1 protein was expressed in Escherichia coli, purified, and stored according to the protocols established by Lei et al.10 with final purified yields of 25 mg/L. Uniform 13C–15N or 15N labeling was performed by expressing each protein in modified minimal M9 media.39 Complexes of 15N-labeled Pot1pN and d(GGTTAC) at 1.25 mM in NMR sample buffer (50 mM KH2PO4, pH 6.15, 50 mM NaCl, 1 mM d-DTT, and 10% D2O) contained an excess of 0.25 molar equivalents of d(GGTTAC) to ensure that Pot1pN was present in a homogeneous ssDNA-bound complex. Due to the aggregation tendencies of Pot1pNF, NMR experiments were conducted at a final Pot1pN concentration of 350 µM. Finally, additional residues present in the N-terminal 6×His tag were identified with negative values to be consistent with the residue numbering scheme presented by Lei et al.11
NMR backbone assignment of Pot1pN
Standard Varian BioPack pulse sequence-derived two-and three-dimensional sensitivity-enhanced, gradient-selected non-TROSY assignment experiments [15N and 13C HSQC, HNCA, HN(CO)CA, HNCACB, CBCA(CO) NH, HN(CA)CO, and HNCO]51 were acquired at 30 °C on a Varian Inova 500-MHz spectrometer equipped with a Nalorac HCN warm probe containing single-axis Z-gradients. Collected data sets were processed using NMRPipe,33 and backbone resonance assignments were manually made using CCPNMR Analysis v1.5.52
CSI analyses of Pot1pNF and Pot1pNB
Secondary structural elements of both forms of Pot1pN were determined using per-residue 13Cα CSI methods previously published.25,26 Determination of CSI parameters for Pot1pNF and that for Pot1pNB were calculated using CCPNMR Analysis v1.5 software package-supplied macros.52 CSI values were determined by comparing the 13Cα values obtained for both Pot1pNF and Pot1pNB with those obtained for respective residues present in a random-coil pentapeptide sequence, GGXGG.53
RDC measurements of Pot1pNF and Pot1pNB
Measurements of RDCs (DNH) were determined using liquid crystal solutions containing 15 mg/mL of Pf1 bacteriophage in NMR buffer containing 400 mM NaCl.28,54 One-bond 1H–15N J-couplings in the absence (JHN) and in the presence (JNH+DNH) of alignment media for Pot1pNF and Pot1pNB were collected at 30 °C on a Varian Inova 500-MHz spectrometer equipped with a Nalorac HCN probe containing single-axis Z-gradients. Data were collected in an interleaved fashion using a two-dimensional doublet-selective, sensitivity-enhanced 1H–15N HSQC pulse sequence54 and processed using NMRPipe.33 Contributions of DNH to the couplings observed in the aligned state were calculated by subtracting the aligned-state couplings from the nonaligned-state couplings using the CCPNMR Analysis v1.5 software package.52 Principal component axes Sxx, Syy, and Szz of the alignment tensor and the protein orientation with respect to the molecular frame (Euler angles α, β, and γ) for Pot1pNB and Pot1pNF were determined using the atomic coordinates present in the Pot1pNB crystal structure using REDCAT.55
15N relaxation experiments
Picosecond-nanosecond dynamic (HetNOE, R1, and R256) and microsecond-millisecond dynamic (R2-CPMG experiments38) experiments were performed at 30 °C at two independent field strengths (500 and 600 MHz for HetNOE, R1, and R2 experiments and 600 and 900 MHZ for R2-CPMG experiments) on Varian Inova spectrometers equipped with a Varian HCN warm probe containing single-axis Z-gradients, except for R2-CPMG experiments, which were collected at 600 MHz using a Varian cold probe containing single-axis Z-gradients. However, due to the poor sensitivity for the R2-CPMG experiments collected at 600 MHz (data not shown), only 900-MHz data were analyzed.
HetNOE data were collected with and without a 3-s 1H saturation period with a constant recycle delay of 5 s. 15N R1 and R2 experiments were recorded in a pseudo-randomized, interleaved fashion with a constant recovery delay of 4 s with variable T1 relaxation delay periods of 0, 52.24, 104.48*, 208.96*, 417.92, 626.88, 835.84, 1253.76, 1606.78, and 1959 ms and variable T2 relaxation delays of 0, 30.93*, 46.89, 61.86, 77.32, 92.78*, 123.64, 154.64, and 170.10 ms (the asterisk represents repeated data points implemented at randomized positions). Heat-compensated R2-CPMG experiments were collected in a pseudo-randomized fashion with a constant T2 relaxation delay of 40 ms, a recovery delay of 2.5 s, and pseudo-randomized application of 180° refocusing CPMG pulses at frequencies of 0, 50*, 100, 200, 250, 400, 500, 550, 700, 750*, 800, and 950 Hz. Resultant spectra were processed using NMRPipe,33 and data were treated in an identical fashion using squared cosine-bell apodization functions, linear prediction to 2× points in the indirect dimension, and zero filled in both dimensions to 2× points. Data from the R2-CPMG experiments were treated in a similar fashion, with the exception that Gaussian apodization functions were used. Peak volumes were determined by nlinLS,34 with errors representing the Gaussian distribution of the root-mean-squared deviation of the peak volume to the noise threshold of the experiment.
Individual backbone amide HetNOE ratios were derived from peak volume data by taking the ratio of the non-1H saturated peak volume to the 1H saturated peak volume, with errors (1σ) derived from triplicate experiments collected at each magnetic field strength. R1 and R2 values were calculated using the peak analysis and quantification macros built into NMRPipe,34 with errors derived from a series of exponential fits of 300 synthetic data sets generated by Monte Carlo simulation.34 R2-CPMG experiments were processed using macros developed by Professor Lewis Kay, and normalized peak volumes were used to calculate the effective R2 (R2eff) values (1/s) as a function of CPMG frequency for each residue according to Eq. (1):
| (1) |
All data fitting was done using a hyperbolic tangent CPMG function embedded in CPMGFit†. Calculated errors for Rex and kex were derived from jacknife simulation of the individual data sets.
Model-free analyses of Pot1pNF and Pot1pNB
Relaxation parameters obtained for Pot1pNF and Pot1pNB collected at 500 and 600 MHz were subjected to the Lipari–Szabo model-free analysis34,35 using the Model-Free 4.15 and FAST-ModelFree software packages.36,57 Estimations of the global isotropic correlation time (τc) for each form of Pot1pN were derived from the 1σ adjusted mean R2/R1 values collected at 500 MHz. Initial estimations of the axially symmetric diffusion tensor were estimated from the Pot1pNB crystal structure11 and the measured 1σ adjusted mean R2/R1 ratios58 using pdbi-nertia and R2R1_diffusion‡. An axially symmetric diffusion tensor was chosen to describe diffusion of Pot1pNF and Pot1pNB and refined using data sets that eliminated the terminal highly dynamic residues (1–30 and 187–197) as well as residues for which all three of the 15N relaxation parameters (R1, R2, and HetNOE) could not be accurately determined. Calculation of ModelFree parameters was done by simultaneously fitting all 15N relaxation data collected at 500 and 600 MHz to one of five motional models that were selected based on the methods reported by Mandel et al.36 Errors reported in S2 values were derived from 300 synthetic data sets generated by Monte Carlo simulation.
Supplementary Material
Acknowledgements
This study was supported by a National Research Service Award Postdoctoral Fellowship (grant GM-071257 to J.E.C.), the National Institutes of Health (grant GM-059414 to D.S.W.), and the National Science Foundation (grant MCB-0617956 to D.S.W.). We thank Dr. Geoff Armstrong and the Rocky Mountain Regional 900-MHz NMR Facility of the University of Colorado at Denver Health Sciences Center (NIH-GM68928) for aiding in our NMR data collection at 900 MHz. We also thank Drs. Patrick Loria (Yale University), Arthur Palmer (Colombia University), Lewis Kay (University of Toronto), Elan Eisenmesser (University of Colorado at Denver Health Sciences Center), Michael Latham (University of Toronto), and Justin Douglas (University of Colorado at Boulder) for their thoughtful discussion, reading of the manuscript, suggestions, and contributions of software used in data analysis. Finally, we thank Dr. Art Pardi, Jennifer Roberts, and Lisa Warner (all from the University of Colorado at Boulder) for their helpful comments and suggestions in the preparation of this article.
Abbreviations used
- CSA
chemical shift anisotropy
- CSI
chemical shift indexing
- CSP
chemical shift perturbation
- CPMG
Carr-Purcell-Miboom-Gill
- DBD
ssDNA-binding domain
- DNH
residual dipolar coupling
- HSQC
heteronuclear singlequantum coherence
- JNH
J-coupling
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- OB
oligonucleotide/oligosaccharide binding
- Pot1
protection of telomeres 1
- Pot1pN
residues 1–187 of full-length Pot1
- Pot1pNF
ssDNA-free form of Pot1pN
- Pot1pNB
Pot1pN/d (GGTTAC) complex
- QF
Q-factor
- R1
spin–lattice relaxation
- R2
spin–spin relaxation
- RDC
residual dipolar coupling
- Rex
relaxation dispersion
- RMS
root mean squared difference
- ssDNA
single-stranded DNA
- HetNOE
1H15N steady state heteronuclear NOE
- S2
generalized order parameter
- SzzSyySzz
principal component axes of molecular alignment tensor
- τc
overall molecular correlation time
- τe
local correlation time
Footnotes
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2009.02.016
References
- 1.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 2.Aubert G, Lansdorp PM. Telomeres and aging. Physiol. Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 3.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunch JT, Bae NS, Leonardi J, Baumann P. Distinct requirements for Pot1 in limiting telomere length and maintaining chromosome stability. Mol. Cell. Biol. 2005;25:5567–5578. doi: 10.1128/MCB.25.13.5567-5578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shakirov EV, Surovtseva YV, Osbun N, Shippen DE. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinert TA, Hartwell LH. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 8.Veldman T, Etheridge KT, Counter CM. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr. Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11-389. J. Mol. Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe . Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 11.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 12.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 13.Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 14.Peersen OB, Ruggles JA, Schultz SC. Dimeric structure of the Oxytricha nova telomere end-binding protein alpha-subunit bound to ssDNA. Nat. Struct. Biol. 2002;9:182–187. doi: 10.1038/nsb761. [DOI] [PubMed] [Google Scholar]
- 15.Classen S, Ruggles JA, Schultz SC. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein alpha subunit both uncomplexed and complexed with telomeric ssDNA. J. Mol. Biol. 2001;314:1113–1125. doi: 10.1006/jmbi.2000.5191. [DOI] [PubMed] [Google Scholar]
- 16.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 17.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 18.Eldridge AM, Halsey WA, Wuttke DS. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry. 2006;45:871–879. doi: 10.1021/bi0512703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tani A, Murata M. Alternative splicing of Pot1 (protection of telomere)-like genes in Arabidopsis thaliana . Genes Genet. Syst. 2005;80:41–48. doi: 10.1266/ggs.80.41. [DOI] [PubMed] [Google Scholar]
- 20.Pitt CW, Moreau E, Lunness PA, Doonan JH. The pot1+ homologue in Aspergillus nidulans is required for ordering mitotic events. J. Cell Sci. 2004;117:199–209. doi: 10.1242/jcs.00844. [DOI] [PubMed] [Google Scholar]
- 21.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 22.Baumann P, Podell E, Cech TR. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol. Cell. Biol. 2002;22:8079–8087. doi: 10.1128/MCB.22.22.8079-8087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeLano WL. The PyMOL Molecular Graphics System DeLano Scientific. San Carlos, CA: 2002. [Google Scholar]
- 24.Croy JE, Fast JL, Grimm NE, Wuttke DS. Deciphering the mechanism of thermodynamic accommodation of telomeric oligonucleotide sequences by the Schizosaccharomyces pombe protection of telomeres 1 (Pot1pN) protein. Biochemistry. 2008;47:4345–4358. doi: 10.1021/bi701778x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wishart DS, Sykes BD. The 13C chemical shift index: a simple method for the identification of protein secondary structure using 13C chemical shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 26.Wishart DS, Skyes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 27.Lipsitz R, Tjandra N. Residual dipolar couplings in NMR structure analysis. Annu. Rev. Biophys. Biomol. Struct. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 28.Hansen MR, Hanson P, Pardi A. Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions. Methods Enzymol. 2000;317:220–240. doi: 10.1016/s0076-6879(00)17017-x. [DOI] [PubMed] [Google Scholar]
- 29.Alexandrescu A. Structure and disorder in the ribonuclease S-peptide probed by NMR residual dipolar couplings. Protein Sci. 2003;12:2132–2140. doi: 10.1110/ps.03164403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornilescu G, Marquardt JL, Ottiger M, Bax A. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J. Am. Chem. Soc. 1998;120:6836–6837. [Google Scholar]
- 31.Ottiger M, Bax A. Bicelle-based liquid crystals for NMR-measurement of dipolar couplings at acidic and basic pH values. J. Biomol. NMR. 1999;13:187–191. doi: 10.1023/a:1008395916985. [DOI] [PubMed] [Google Scholar]
- 32.Bax A. Weak alignment offers new NMR opportunites to study protein structure and dynamics. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delaglio E, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules: 1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 35.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules: 2. Analysis of experimental results. J. Am. Chem. Soc. 1982;104:4559–570. [Google Scholar]
- 36.Mandel AM, Akke M, Palmer AG. Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 37.Korzhnev DM, Kay LE. Probing invisible, low-populated states of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding. Acc. Chem. Res. 2008;41:442–451. doi: 10.1021/ar700189y. [DOI] [PubMed] [Google Scholar]
- 38.Loria JP, Ranee M, Palmer AG. ATROSY CPMG sequence for characterizing chemical exchange in large proteins. J. Biomol. NMR. 1999;15:151–155. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- 39.Eldridge AM, Wuttke DS. Probing the mechanism of recognition of ssDNA by the Cdc13-DBD. Nucleic Acids Res. 2008;36:1624–1633. doi: 10.1093/nar/gkn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson JR. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 41.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 42.Boehr DD, Wright PE. Biochemistry. How do proteins interact? Science. 2008;320:1429–1430. doi: 10.1126/science.1158818. [DOI] [PubMed] [Google Scholar]
- 43.Mittermaier A, Kay LE. New tools provide new insights in NMR studies of protein dynamics. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 44.Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- 45.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittermaier A, Varani L, Muhandiram DR, Kay LE, Varani G. Changes in side-chain and backbone dynamics identify determinants of specificity in RNA recognition by human U1A protein. J. Mol. Biol. 1999;294:967–979. doi: 10.1006/jmbi.1999.3311. [DOI] [PubMed] [Google Scholar]
- 47.Nagata T, Kanno R, Kurihara Y, Uesugi S, Imai T, Sakakibara S, et al. Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1. J. Mol. Biol. 1999;287:315–330. doi: 10.1006/jmbi.1999.2596. [DOI] [PubMed] [Google Scholar]
- 48.James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 49.Berger C, Weber-Bornhauser S, Eggenberger J, Hanes J, Pluckthun A, Bosshard HR. Antigen recognition by conformational selection. FEBS Lett. 1999;450:149–153. doi: 10.1016/s0014-5793(99)00458-5. [DOI] [PubMed] [Google Scholar]
- 50.Foote J, Milstein C. Conformational isomerism and the diversity of antibodies. Proc. Natl Acad. Sci. USA. 1994;91:10370–10374. doi: 10.1073/pnas.91.22.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett T, Farmer BT, Mueller L. Unambiguous resonance assignments in 13C, 15N-labelled nucleic acids by 3D triple resonance NMR. J. Am. Chem. Soc. 1993;115:11040–11041. [Google Scholar]
- 52.Vranken VE, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 53.Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 54.Cordier E, Dingley AJ, Grzesiek S. A doublet-separated sensitivity-enhanced HSQC for the determination of scalar and dipolar one-bond J-couplings. J. Biomol. NMR. 1999;13:175–180. doi: 10.1023/a:1008301415843. [DOI] [PubMed] [Google Scholar]
- 55.Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. J. Magn. Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 56.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 57.Cole R, Loria JP. FAST-ModelFree: a program for rapid automated analysis of solution NMR spin-relaxation data. J. Biomol. NMR. 2003;26:203–213. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 58.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.