Arabidopsis lines lacking either light-harvesting complex (LHC) protein Lhcb1 or Lhcb2 were produced, revealing that Lhcb1 and Lhcb2 play different roles during state transitions and that both are necessary but neither is sufficient. Plant lacking Lhcb1 lacked most LHCII trimers but plants lacking Lhcb2 contained LHCII similar to the wild type but could not perform state transitions.
Abstract
Photosynthetic light harvesting in plants is regulated by phosphorylation-driven state transitions: functional redistributions of the major trimeric light-harvesting complex II (LHCII) to balance the relative excitation of photosystem I and photosystem II. State transitions are driven by reversible LHCII phosphorylation by the STN7 kinase and PPH1/TAP38 phosphatase. LHCII trimers are composed of Lhcb1, Lhcb2, and Lhcb3 proteins in various trimeric configurations. Here, we show that despite their nearly identical amino acid composition, the functional roles of Lhcb1 and Lhcb2 are different but complementary. Arabidopsis thaliana plants lacking only Lhcb2 contain thylakoid protein complexes similar to wild-type plants, where Lhcb2 has been replaced by Lhcb1. However, these do not perform state transitions, so phosphorylation of Lhcb2 seems to be a critical step. In contrast, plants lacking Lhcb1 had a more profound antenna remodeling due to a decrease in the amount of LHCII trimers influencing thylakoid membrane structure and, more indirectly, state transitions. Although state transitions are also found in green algae, the detailed architecture of the extant seed plant light-harvesting antenna can now be dated back to a time after the divergence of the bryophyte and spermatophyte lineages, but before the split of the angiosperm and gymnosperm lineages more than 300 million years ago.
INTRODUCTION
Most of the photons that are converted to biochemical energy and biomass through photosynthesis are harvested by the major light-harvesting chlorophyll a/b binding antenna complex light-harvesting complex II (LHCII), which is one of the most abundant proteins on earth. The LHC proteins are synthesized in the cytosol, posttranslationally imported into the chloroplasts, and inserted into the thylakoid membranes. The apoproteins of LHCII contain three membrane-spanning regions and bind several xanthophyll molecules in addition to chlorophyll a and b. The nuclear genes coding for the components of this complex, originally denoted CAB (for chlorophyll a/b binding) genes, were among the first plant genes ever to be sequenced (Bedbrook et al., 1980). Angiosperm plant LHCII trimers are made up of three very similar, but distinct, forms of the protein encoded by the Lhcb1, Lhcb2, and Lhcb3 genes (Jansson et al., 1992). Lhcb1 gene products are the most abundant; the products of both Lhcb1 and Lhcb2 accumulate under low-light conditions when photosynthetic light harvesting limits photosynthesis (Jansson, 1994). Lhcb4, Lhcb5, and Lhcb6 are monomeric proteins associated with photosystem II (PSII), while the Lhca polypeptides make up the antenna of photosystem I (PSI).
The Lhcb1 and Lhcb2 proteins can be phosphorylated at a Thr residue close to the N terminus, in a process dependent on the chloroplast serine-threonine protein kinase STN7 (Bellafiore et al., 2005). STN7 may itself be the kinase although the possibility that another kinase(s) may be involved as well has not been formally excluded. LHC phosphorylation drives state transitions, a process of partial functional redistribution of a mobile pool of LHCII between PSII and PSI (Bennett, 1977), triggered by changes in the redox state of the plastoquinone (PQ) pool. A second homologous kinase, STN8, also has the capacity to phosphorylate LHCII; however, its activity toward the complex is much lower than that of STN7 (Bonardi et al., 2005; Leoni et al., 2013). LHCII is also phosphorylated at other sites (Michel et al., 1991; Vener et al., 2001; Vainonen et al., 2005; Rinalducci et al., 2006), but this seems not to play any role in state transitions. The thylakoid-associated phosphatase 38 (TAP38/PPH1) catalyzes the reverse reaction (Pribil et al., 2010; Shapiguzov et al., 2010), a process that seems to be independent of the redox state, and the balance between the activities of STN7 and PPH1/TAP38 determines the phosphorylation state of LHCII. It was recently shown that upon long-term acclimation to a variety of natural light conditions, LHCII is phosphorylated, and part of the (phosphorylated) LHCII serves as an effective antenna for PSI (Wientjes et al., 2013a).
State transitions can be triggered experimentally by supplementing the light with far-red light (which only excites PSI and is termed “state 1 light”), leading to LHCII dephosphorylation and driving the mobile LHCII toward PSII, or by turning off the far-red light, inducing phosphorylation and attachment of LHCII to PSI (state 2). In nature, large and sudden changes in light quality are less common than changes in light quantity, and changes in the balance of PSI and PSII excitation induced by alterations in light intensity are therefore probably the most important drivers of LHCII phosphorylation and dephosphorylation (Rintamäki et al., 2000). Phosphorylation-driven changes in state transitions provide the flexibility to maintain the redox balance under conditions of changing light quality and quantity. Lhcb3 lacks the phosphorylation site and is not directly involved in state transitions, but as plants lacking Lhcb3 slightly increase the amounts of Lhcb1 and Lhcb2, the rate of state transitions is slightly higher in Lhcb3 knockout (KO) mutants, probably because more substrate is available for STN7 (Damkjaer et al., 2009).
Lhcb1 and Lhcb2 show an extraordinary degree of evolutionary conservation. Green algae and the moss Physcomitrella patens have many proteins of the Lhcb1/2/3 clade, yet no direct counterparts of Lhcb1 and Lhcb2 exist in these organisms, although Lhcb3-like proteins are found in Physcomitrella and other non-seed plants (Elrad and Grossman, 2004; Alboresi et al., 2008). This dates the separation of the Lhcb3 and Lhcb1/Lhcb2 clades to ∼400 million years ago (MYA). However, Lhcb1 and Lhcb2, proteins differing only by 14 diagnostic amino acids, can be distinguished in both the conifer and the angiosperm lineage (Jansson and Gustafsson, 1990). Thus, the last common ancestor of conifers and angiosperms, an as yet unidentified primitive gymnosperm living ∼300 MYA, must have possessed both the Lhcb1 and Lhcb2 proteins. Not all LHCII trimers seem to be engaged in state transitions; some are always associated with PSII, corresponding to the so-called S and M trimers of the PSII supercomplex (Boekema et al., 1999). Lhcb3 seems to be mainly confined to M trimers, where it forms a heterotrimer with two Lhcb1 subunits (Hankamer et al., 1997; Damkjaer et al., 2009). Very little or no Lhcb3 is found in LHCII-PSI, suggesting that M trimers do not participate in state transitions (Jansson et al., 1997; Galka et al., 2012). It has been demonstrated that the trimers performing state transitions are peripherally attached to PSII. These trimers have been shown to associate more stably with, and transfer the energy more efficiently to, PSI than PSII when forming LHCII-PSI complexes (Galka et al., 2012; Wientjes et al., 2013a).
In Arabidopsis thaliana, Lhcb1 is encoded by five genes, three of which are closely linked on chromosome 1, and Lhcb2 is encoded by three genes (Jansson, 1999), two of which are closely linked on chromosome 2. This close genetic linkage means that loss-of-function (quintuple Lhcb1 or triple Lhcb2) T-DNA KO mutants are almost impossible to generate. Introducing an antisense construct against one of these genes (Lhcb2) results in simultaneous—partial or complete—inhibition of expression of Lhcb1 as well as of Lhcb2 (Andersson et al., 2003), not surprisingly since the genes have such a high degree of DNA sequence identity that cross-inhibition is expected. Therefore, genetic approach cannot be utilized to address the functional differences between Lhcb1 and Lhcb2, differences that must exist since they have both been in existence for hundreds of millions of years. We have recently shown that phosphorylation of Arabidopsis Lhcb2 is more rapid than that of Lhcb1, suggesting that the first phase of state transition is mediated by Lhcb2 (Leoni et al., 2013), but why do seed plants need both Lhcb1 and Lhcb2? This is the question that we address in this contribution.
RESULTS
Lhcb1 or Lhcb2 Can Be Specifically Depleted with Artificial MicroRNAs
To address the individual roles of Lhcb1 and Lhcb2, we generated artificial microRNA (amiRNA) Arabidopsis lines deficient in either Lhcb1 or Lhcb2, the amiLhcb1 and amiLhcb2 lines, respectively. We tested two amiRNA constructs both for Lhcb1 and for Lhcb2. For Lhcb1 repression, only amiLhcb1v1 had the capacity to silence Lhcb1 gene expression and the effect was variable between lines. We screened the progeny of 30 individual transformation events, and the two lines (amiLhcb1v1.14 and amiLhcb1v1.3) that had the lowest Lhcb1 levels were used for further analysis. For Lhcb2, both amiLhcb2v1 and amiLhcb2v2 constructs efficiently silenced Lhcb2 gene expression. It is possible that one, or more, of the Lhcb1 genes partially escaped inhibition by the amiRNA constructs; alternatively, the higher expression levels of Lhcb1 made full inhibition hard to achieve. Whatever the explanation for this finding, it does not affect the analysis described in this article.
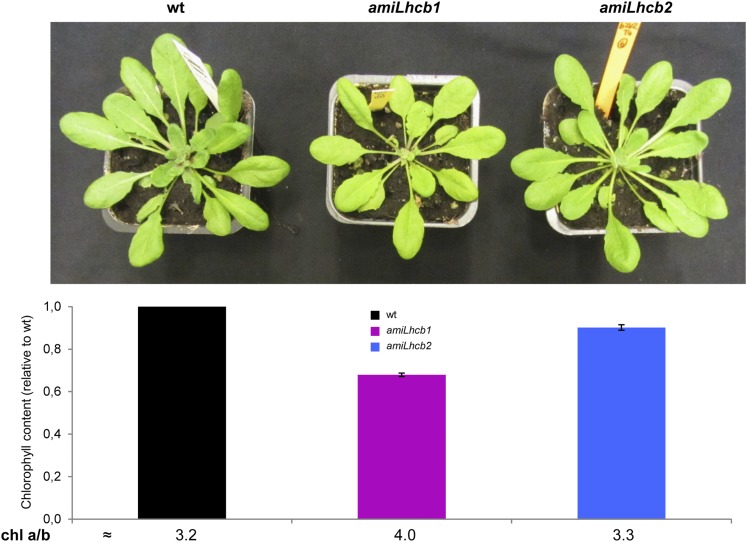
When grown under controlled conditions, amiLhcb1 plants grew more slowly than the wild type and the leaves were slightly smaller and paler, while the growth rate and pigment levels in amiLhcb2 were similar to those of wild-type plants (Figure 1). The chlorophyll content of amiLhcb1 was reduced by ∼30% compared with the wild type, and the chlorophyll a/b ratio was 4, compared with 3.2 in the wild type. The growth and pigmentation phenotype of the amiLhcb1 plants thus resembled that of asLhcb2 (which lacks both Lhcb1 and Lhcb2), showing that the growth and pigmentation phenotype of asLhcb2 (Andersson et al., 2003) was, not surprisingly, caused mainly by depletion of Lhcb1. Fluctuating light on the other hand, low-light intensity (50 μmol photons m−2 s−1) repeatedly interrupted by a high light peak (500 μmol photons m−2 s−1), resulted in stunted growth of both amiLhcb1 (142.55 ± 19.34 mg) and amiLhcb2 (154.72 ± 22.54 mg) in comparison to the wild type (386.43 ± 37.49 mg).
Figure 1.
Phenotypic Analysis of Examined Lines.
Visual appearance, chlorophyll levels, and chlorophyll a/b ratios in leaves of wild-type, amiLhcb1, and amiLhcb2 Arabidopsis lines grown under normal light conditions. Error bars represent se received from five technical replicates performed on 16 plants.
With the Exception of the LHCII Trimers, Antenna Composition Is Not Affected by Removal of Lhcb1 or Lhcb2
amiLhcb1 plants contained residual amounts of Lhcb1 and had somewhat compensatory increases in the levels of Lhcb2 and Lhcb3, with Lhcb2 increasing by ∼50% and Lhcb3 by ∼30% (Figure 2A). The levels of Lhca and the Lhcb4-6 proteins were not significantly different from those of wild-type plants. Intriguingly, the level of STN7 was also increased in amiLhcb1 and the level of PPH1/TAP38 tended to be lower, but the latter difference was not statistically significant (Figure 2A). amiLhcb2 plants completely lacked Lhcb2 and had slightly (∼10%), but significantly, higher levels of Lhcb3 and Lhcb5 (Figure 2B). Lhcb1 tended also to be slightly upregulated, but this difference was not significant using the stringent criterion we applied (P < 0.01). The levels of all other proteins analyzed were unchanged (Figure 2B). It appears that the only compensations the plant made for the lack of one LHCII protein were within the LHCII trimers, leaving the other components of the light-harvesting antennas of PSI and PSII unchanged.
Figure 2.
LHCI, LHCII, and LHCII Kinase/Phosphatase Polypeptide Composition.
Protein content of amiLhcb1 (A) and amiLhcb2 (B). One microgram of total chlorophyll was loaded into each electrophoretic lane, except for quantification of STN7, STN8, PPH1/TAP38, and PBCP, to which 3 μg of chlorophyll was loaded. Immunoblotting of thylakoid protein preparations from the wild type, amiLhcb1 (-b1), and amiLhcb2 (-b2) was performed using monospecific antibodies. The numbers show signal strength in the amiRNA lines relative to that in the wild type, ±se obtained from analysis of thylakoid membranes isolated from four plants in three biological replicates. Asterisks denote significant difference (P < 0.01) from the wild type.
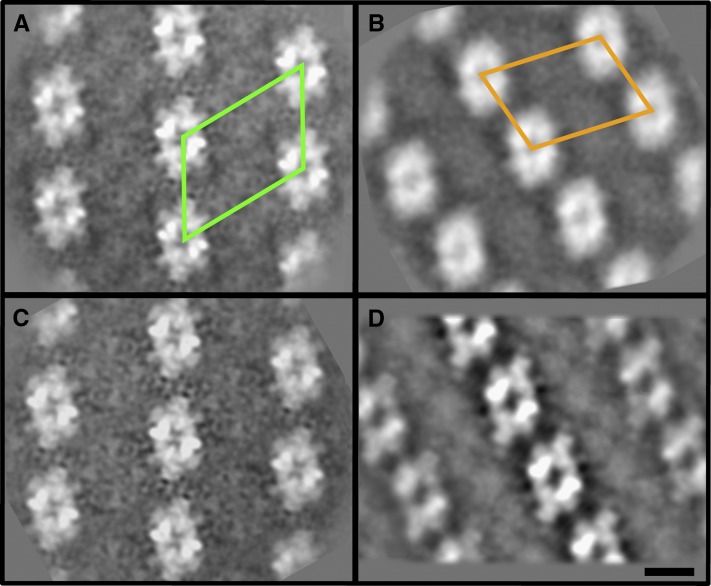
To get further information about the architecture of the PSII complex, we analyzed PSII supercomplexes in amiLhcb1 and amiLhcb2 using a combination of crystal and single-particle electron microscopy (EM) (Boekema et al., 2000). The shape and spacing of the PSII supercomplexes in the so-called “PSII membrane crystals” of amiLhcb2 showed that the supercomplexes of amiLhcb2 were indistinguishable from those of the wild type (Figures 3A and 3C). In amiLhcb1, the appearance of membrane crystals was very different from the wild type, showing change in the distances between the cores and as a result change in the surface of a unit cell (repeating motif of the crystal) from 534 nm2 in the wild type to 444 nm2 in amiLhcb1 (Figures 3A and 3B), and they reassembled C2S2M crystals previously only found in spinach (Spinacia oleracea) (481 nm2; Boekema et al., 2000) (Figure 3D). Two types of PSII particle were found in amiLhcb1 contributing to low resolution of the obtained arrays. The larger particle (Supplemental Figure 1A), which we named C2S2+, resembled a C2S2 particle with an extra mass at the position of the M-trimer. We believe that this additional mass corresponds to Lhcb3 and the antenna protein CP24, which are thought to be in direct contact with each other and located at this position (Kovács et al., 2006) (Supplemental Figure 1C), an interpretation consistent with the fact that CP24 and monomeric Lhcb3 were present in amiLhcb1. However, the most abundant complex in amiLhcb1 was C2S2, in which the S-trimers must be Lhcb2 homotrimers (Supplemental Figure 1B) as this is the only trimer present in amiLhcb1.
Figure 3.
Structure of Photosynthetic Complexes in amiLhcb1 and amiLhcb2.
Averaged projection map of PSII membrane crystals from wild-type Arabidopsis (A), amiLhcb1 (B), amiLhcb2 (C), and C2S2M crystals found so far only in spinach (D), from Boekema et al. (2000). Frames represent the unit cell: repeating motif of the crystal. Bar = 10 nm.
[See online article for color version of this figure.]
Lhcb1 Is Important for Thylakoid Structure Flexibility
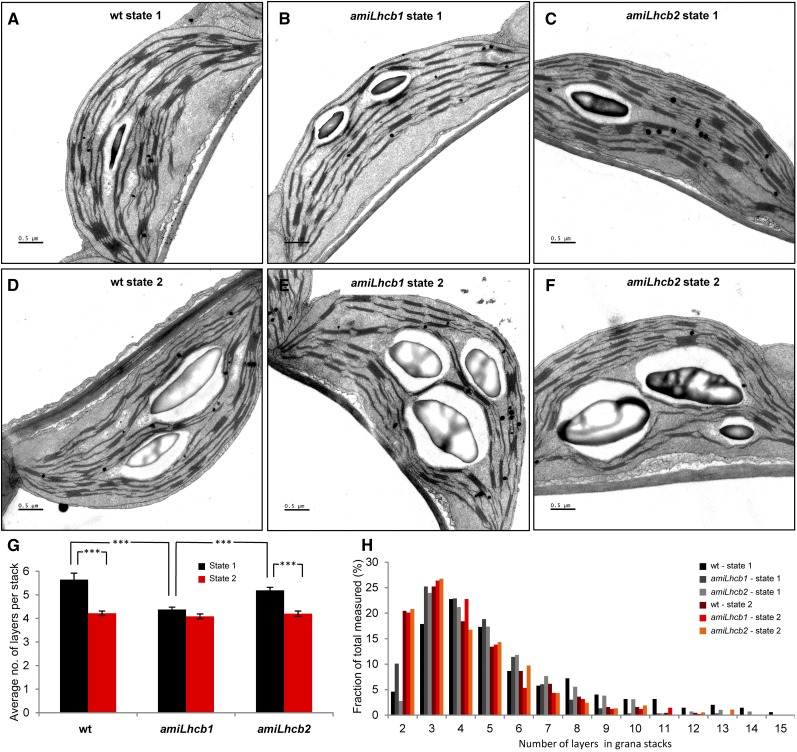
We used EM to investigate the changes in thylakoid ultrastructure between state 1 and 2 in wild-type, amiLhcb1, and amiLhcb2 plants. Plants were subjected to a mix of red and far-red light to induce state 1, or red light only to achieve state 2, followed by fixation in 2.5% glutaraldehyde under respective light. Ultrathin cross sections of leaves were cut and similar cells from palisade parenchyma were analyzed. As previously found for asLhcb2 plants, the thylakoid membranes were fully capable of stacking in the absence of LHCII proteins, and starch granules and other visible structures did not differ between the lines (Figure 4). When subjected to state 1 light, thylakoids form well-defined network of highly stacked grana with interconnecting lamellae (Figures 4A to 4C; Supplemental Figures 2A to 2C) in all analyzed samples; nevertheless, the number of grana and stacked layers was lower in amiLhcb1 (4.38 ± 0.10) than in the wild type (5.65 ± 0.28) or amiLhcb2 (5.19 ± 0.14). Red light results in partial destacking of grana and relaxation of thylakoid structure (Figures 3D to 3F; Supplemental Figures 2D to 2F) in the wild type (4.22 ± 0.10) and amiLhcb2 (4.20 ± 0.11), but not in amiLhcb1 (4.09 ± 0.10). This indicates that Lhcb1 is important for grana stacking and membrane reorganization during state transitions (Figures 4G and 4H).
Figure 4.
Electron Micrographs of State 1 and State 2 Adapted Thylakoid Membranes.
(A) to (C) Leaves of Arabidopsis wild type (A), amiLhcb1 (B), and amiLhcb2 (C) were subjected to 2 h of PSI light, and ultrathin cross sections were contrasted and analyzed. Bars = 1 μm.
(D) to (F) Cross sections of Arabidopsis wild-type (D), amiLhcb1 (E), and amiLhcb2 (F) thylakoid membranes subjected to PSII light and ultrathin cross sections were contrasted and analyzed. Bars = 1 μm.
(G) Average number of layers in grana stacks in state 1 and state 2. Error bars denote se obtained from 10 analyzed pictures (between 280 and 440 grana stacks for each condition). Asterisks refer to level of statistical significance (P > 0.05).
(H) Distribution of layers in grana stacks.
Moreover, when comparing the thylakoid membranes of the wild type and amiLhcb2 in state 1, we noticed that the amiLhcb2 grana were more evenly sized. We performed Levene’s test, examining the equality of variances, to confirm that there was a significant difference (P = 0.013) in variations in the number of layers in grana. This is most likely caused by the fact that amiLhcb2’s trimers are manly composed of Lhcb1 and as a consequence, the thylakoids are highly structured and packed more tightly in stacks of similar size.
Lhcb2 Mediates the Association of LHCII to PSI
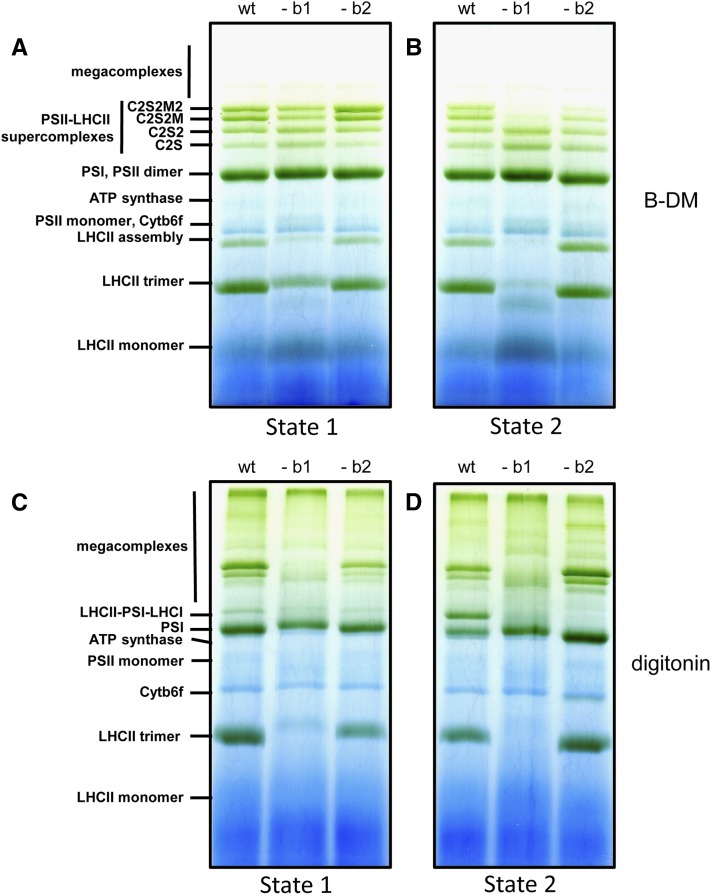
To investigate the pigment-protein complex composition of the thylakoids, we used two different kinds of native gel electrophoresis system, Blue Native (BN) PAGE and its variant, large-pore Blue Native (lpBN) PAGE (Järvi et al., 2011). Using standard BN PAGE, a large fraction of the thylakoid membrane pigment-protein complexes are solubilized with β-dodecyl maltoside (β-DM) and the complexes visualized represent the overall composition of the thylakoid relatively well. If a milder detergent, e.g., digitonin, which also preserves the larger megacomplexes, is used for thylakoid solubilization, only a fraction of the protein complexes (megacomplexes) can enter the BN gel. To avoid this problem, an lpBN system with large pores in both the stacking and the separation gel has been optimized for thylakoid protein complexes to allow the very large megacomplexes to enter the gel. Combining lpBN with digitonin solubilization of the thylakoid membrane, a method that makes it possible to preserve weak interactions between the protein complexes, allows separation and visualization of complexes containing different combinations of the individual protein complexes (PSI, PSII, and LHCII), formed particularly during state transitions (Pesaresi et al., 2009). It is worth noting that digitonin, due to its molecular structure, is not capable of solubilizing the protein complexes from dense grana stacks. Therefore, the thylakoid (mega)complexes that are solubilized by digitonin preferentially represent the complexes from nonstacked thylakoid regions, including grana margins and end membranes as well as the stroma thylakoids (Järvi et al., 2011).
When thylakoids from the wild type and amiLhcb1 were solubilized with β-DM and separated on BN PAGE, the pigment-protein complex composition of the plants was, as expected, different. No LHCII trimers were visualized in amiLhcb1, and megacomplexes were lacking (Supplemental Figure 3). BN combined with immunoblotting showed that the distribution of Lhcb2 was somewhat affected by the lack of Lhcb1, since less was found at the gel position corresponding to LHCII trimers and a little more in larger complexes (Supplemental Figure 3). The small residual amount of Lhcb1 that can sometimes be seen in the amiLhcb1 line (Supplemental Figure 3A) was not visible here. However, the distribution of Lhcb3 was drastically altered, as almost no Lhcb3 was found to be migrating as LHCII trimers but instead it was present as monomers, confirming that Lhcb3 is incapable of forming homotrimers (Jackowski and Pielucha, 2001; Caffarri et al., 2004). It appears that, in the absence of Lhcb1, the majority of trimers are Lhcb2 homotrimers and perhaps some (Lhcb2)2Lhcb3 heterotrimers, presumably since Lhcb3 forms stable trimers only in combination with Lhcb1. In amiLhcb2 thylakoids, there was no obvious effect on the distribution of either Lhcb1 or Lhcb3.
We then compared the thylakoid protein complexes of plants in state 1 and state 2 using lpBN gel electrophoresis of thylakoids solubilized with either β-DM or digitonin. In state 2, PSII complexes of amiLhcb1 solubilized with β-DM (supposed to mainly represent grana cores) had, like in growth light, very small antenna, as only C2S and C2S2 complexes were seen (Figure 5B). However, in state 1, the C2S2M and C2S2M2 complexes were also visualized from amiLhcb1, indicating that also M positions may be filled with LHC trimers even in the absence of Lhcb1 but they be confined to the grana cores, as little or no larger complexes are present in samples solubilized with digitonin (Figure 5D). Also, a weak band approximately at the position of the so-called LHCII assembly complex (Figures 5A and 5B), which likely represent the previously characterized “band 4” (Caffarri et al., 2009) appeared only under PSI light in amiLhcb1. In contrast, the PSI/PSII super- and megacomplex composition of amiLhcb2 plants mirrored that of the wild type (Figures 5A to 5D) with the exception of the so-called state transition-specific PSI-LHCII complex (Pesaresi et al., 2009; Järvi et al., 2011; Leoni et al., 2013), which was missing from the digitonin-solubilized amiLhcb2 samples (Figures 5C and 5D).
Figure 5.
lpBN Gel Electrophoresis of Thylakoid Membranes.
Thylakoid membranes subjected to either far-red ([A] and [C]) or red light ([B] and [D]). Thylakoid membrane complexes were solubilized with β-DM ([A] and [B]) or digitonin ([C] and [D]). The identities of the complexes resolved are indicated.
Taken together, these data show that in amiLhcb1, no LHCII trimers except Lhcb2 homotrimers accumulated to any great extent and that Lhcb2 and Lhcb3 could not compensate for the loss of Lhcb1 in maintaining the PSII supercomplex structure. This had a marked effect on the number of layers in grana and the pigment-protein complex composition of the thylakoid; under PSI light, the amiLhcb1 mutant retained some normal PSII-LHCII supercomplexes, while under PSII light, only some of the PSII centers had LHCII trimers attached at the S position, and some of them also had monomeric Lhcb3 and CP24 attached. Few or no free LHCII trimers that were not strongly attached to PSI or PSII appeared to be present upon state transition. In amiLhcb2, however, Lhcb1 was capable of compensating for Lhcb2 so that the sites that in wild type were occupied by Lhcb1/Lhcb2 heterotrimers were instead occupied by Lhcb1 homotrimers in amiLhcb2. The only exception was that the state transition-specific PSI-LHCII complex could not be formed in amiLhcb2, indicating the importance of Lhcb2 for binding of LHCII trimers to PSI.
State Transitions Occur Only When Both Lhcb1 and Lhcb2 Are Present
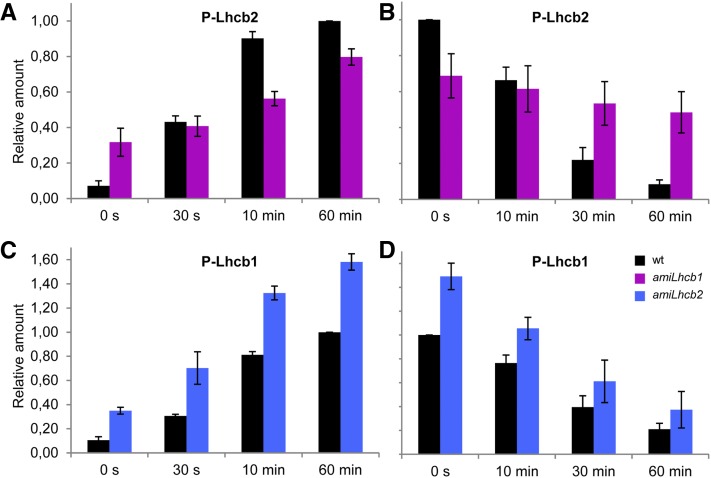
Since we have developed antibodies specific for the phosphorylated forms of Lhcb1 and Lhcb2 (Leoni et al., 2013), it was possible to quantify the phosphorylation and dephosphorylation kinetics of each of these proteins. The band intensities were measured and all of them were normalized to the phosphorylation level of the wild type after 60 min of red light treatment. We found that Lhcb2 phosphorylation kinetics were affected in amiLhcb1: The phosphorylation level at the beginning of the experiment was higher but the phosphorylation rate was much slower, and after 60 min of state 2 light treatment, the phosphorylation level was 20% lower than in the wild type (Figure 6A). The difference in Lhcb2 phosphorylation level in state 1 and state 2 conditions was therefore only 2-fold. Dephosphorylation in state 1 light was also slower than in the wild type (Figure 6B); therefore, the magnitude of the change in phosphorylation status of thylakoid membranes was much reduced, as confirmed by a total phosphoprotein stain using ProQ (Supplemental Figure 4). As the STN7 kinase was found at higher levels in amiLhcb1 than in the wild type, this suggests that the reduced kinetics were due to a decrease in the degree of activation of STN7 because the PQ pool was less reduced (see also Figure 7F below).
Figure 6.
LHCII Phosphorylation and Dephosphorylation Kinetics in amiLhcb1 and amiLhcb2 Arabidopsis Lines.
(A) and (B) Kinetics of wild-type and amiLhcb1 Lhcb2 phosphorylation after shifting from state 1 to state 2 light (A) and dephosphorylation after shifting from state 2 to state 1 light (B). Arabidopsis leaves were collected and snap frozen at different time points during the course of respective light treatment. One microgram of total chlorophyll was loaded and the phosphorylation was quantified and normalized to the maximum phosphorylation level of the wild type obtained after 60 min of red light treatment.
(C) and (D) Lhcb1 phosphorylation (C) and dephosphorylation (D) in the wild type and amiLhcb2. Error bars represent se obtained from three biological replicates.
[See online article for color version of this figure.]
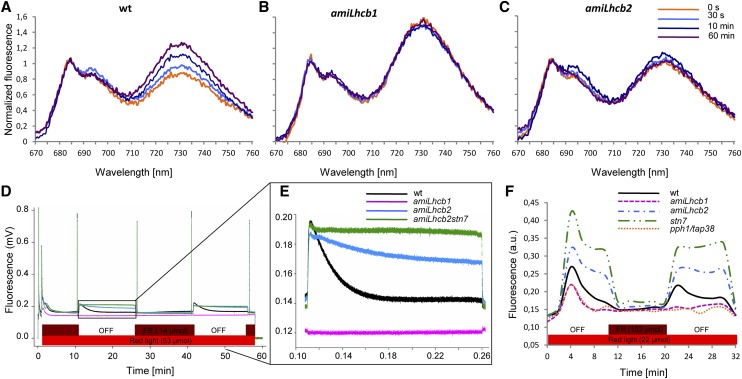
Figure 7.
Absence of State Transitions in amiLhcb1 and amiLhcb2.
(A) to (C) 77K chlorophyll fluorescence spectra before (0 s, orange) and 30 s (light blue), 10 min (dark blue), and 60 min (purple) after shift from state 1 to state 2 light for wild-type (A), amiLhcb1 (B), and amiLhcb2 (C) Arabidopsis lines.
(D) Pulse amplitude-modulated fluorescence traces after shifts from state 1 to state 2 light and back for wild-type (black), amiLhcb1 (magenta), amiLhcb2 (blue), and amiLhcb2stn7 (green) Arabidopsis lines. The bars at the bottom indicate illumination with red (shown in red) and far-red (dark red) light.
(E) Enlargement of a section of (D).
(F) PQ oxidation state (measured as F/Fm) of wild-type (black), amiLhcb1 (magenta), amiLhcb2 (blue), stn7 (green), and pph1/tap38 (orange) Arabidopsis lines after a shift from state 1 to state 2 light and back. Fluorescence is shown in arbitrary units.
In contrast, Lhcb1 phosphorylation was greatly increased in amiLhcb2 compared with the wild type, and it also started from a higher level; nevertheless, the kinetics, in absolute terms, were similar (Figures 6C and 6D). This finding was also confirmed using ProQ (Supplemental Figure 4). It appears that STN7 was activated by the reduced PQ pool and phosphorylated Lhcb1 over a longer period of time than in the wild type, while PPH1/TAP38 phosphatase activity seemed to be unaffected since the dephosphorylation kinetics of amiLhcb2 and the wild type in relative terms were similar.
It is noteworthy that neither amiLhcb1 nor amiLhcb2 plants were able to demonstrate light quality-dependent changes in low-temperature PSI and PSII fluorescence, which would be indicative of state transitions (Figures 7A to 7C). In wild-type plants, a state 1–state 2 transition brought about an increase in PSI fluorescence at around 730 nm, relative to PSII fluorescence at around 680 nm, as the functional attachment of a fraction of the LHCII trimers was transferred from PSII to PSI. This shift was absent in both amiLhcb1 and amiLhcb2. The relatively higher peak of PSI fluorescence in amiLhcb1 could be a consequence of differences in either emission or reabsorption of the emitted light due to the alteration in composition of the pigment-protein complexes in that line; the measurements were performed at equal chlorophyll concentration and data were normalized to the PSII peak. In any case, this is not relevant to the finding that no change occurred after the shift, nor did amiLhcb1 or amiLhcb2 show any typical state transitions as measured by a second, independent method, pulse amplitude-modulated chlorophyll fluorescence (Figure 7D). A detailed investigation of the fluorescence traces (Figure 7E) revealed that in amiLhcb1, changes in light quality failed to induce any significant changes in PSII fluorescence; apparently the balance of excitation between PSI and PSII was similar in both “state 1” and “state 2” light. In amiLhcb2, PSII light led to a greater increase in PSII fluorescence than in the wild type, but the exponential decline in fluorescence with a half-life (t1/2) of approximately 2 min due to energy redistribution to PSI was absent. No LHCII-PSI complexes are formed in amiLhcb2 (Figure 5D); hence, the very slow decrease in fluorescence (t1/2 >30 min) was obviously due to STN7-dependent Lhcb1 phosphorylation. This discovery was further confirmed by the observation that the amiLhcb2stn7 double mutant behaved like stn7 (Bellafiore et al., 2005).
We also measured the PQ oxidation state during a shift from state 1 light to state 2 light and back. When plants were shifted to state 2 light, the PQ pool in amiLhcb2 (as in stn7) became reduced and did not relax back in the same way as in the wild type (Figure 7F). In amiLhcb1, the oxidation state of the PQ pool was almost unaffected by fluctuations in light quality, a result consistent with the observed lack of changes in PSII fluorescence and state transitions and with the slow rate of phosphorylation. This resembled the situation in a pph1/tap38 mutant (Figure 7F). No single Lhcb1 or Lhcb2 T-DNA KO mutant differed significantly from the wild type in any of the aspects measured (for example, in the case of state transitions; Supplemental Figure 5). From this we conclude that both Lhcb1 and Lhcb2 are necessary, and neither is sufficient, for state transitions.
amiLhcb1 Plants Are Compromised in qE Quenching
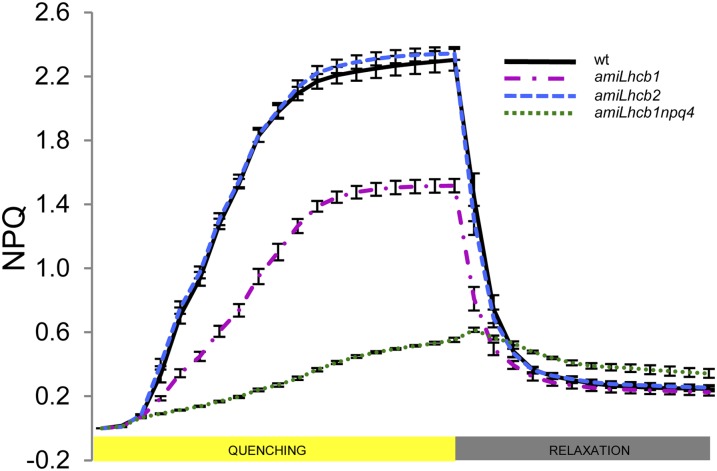
Acclimation of plants to varying light conditions is dependent on their capacity to keep the PQ pool oxidized despite large fluctuations in light intensity. Although the exact mechanism(s) of feedback deexcitation (the qE type of nonphotochemical quenching) are not yet fully understood, it is clear that LHCII trimers are involved in some way and that the PsbS protein together with the xanthophyll cycle pigments have the capacity to catalyze the transition of the antenna from a nonquenched to a quenched state. It has been demonstrated by studies of chlorophyll fluorescence quenching in liposomes composed of LHCII, PsbS, and zeaxanthin that carotenoid-dependent quenching take place via direct interactions of LHCII with PsbS (Wilk et al., 2013). amiLhcb2 plants displayed qE quenching like that of the wild type, while amiLhcb1 plants showed a decrease in qE (Figure 8). Again, no single Lhcb1 or Lhcb2 T-DNA KO mutant differed significantly from the wild type (Supplemental Figure 6). When exposed to varying intensities of white light, amiLhcb2 behaved much like the wild type, except at low-light intensities where state transitions (qT), not qE, govern the oxidation state of the PQ pool, while under high light, the inability of amiLhcb1 plants to induce qE resulted in the PQ pool becoming greatly reduced (Supplemental Figure 7). This would be consistent with the fact that Lhcb1 homotrimers would be fully competent to engage in qE quenching and that Lhcb1/Lhcb2 heterotrimers do not play a specific role in qE. However, if antenna size is reduced, as in amiLhcb1, qE is compromised. We believe that this is a direct effect due to a lack of quenching sites, rather than an indirect effect due to a reduction in the trigger, since the PQ pool was more oxidized, the amount of PsbS the same as in wild type, and the quenching in the amiLhcb1npq4 (Figure 8) double mutant was similar to that of npq4 (Li et al., 2000).
Figure 8.
Nonphotochemical Quenching in Examined Lines.
Feedback deexcitation (qE type of nonphotochemical quenching) induction and relaxation in wild-type (black), amiLhcb1 (magenta), amiLhcb2 (blue), and amiLhcb1npq4 (green) Arabidopsis lines after shifting to high light and back. Error bars are se.
DISCUSSION
Although state transitions are used as a classic textbook example of the regulation of photosynthesis, the exact mechanisms and the functional relevance of state transitions are still largely unresolved (for recent reviews, see Minagawa, 2011; Pesaresi et al., 2011; Pribil et al., 2014; Tikkanen and Aro, 2014). The causal link between LHCII phosphorylation and state transitions has been established for decades (Bennett et al., 1980; Horton and Black, 1980, 1981; Horton et al., 1981; Allen et al., 1981), but there are still several open questions concerning state transitions: (1) Which gene products undergo phosphorylation and participate in state transitions and (2) in which trimeric configurations? (3) At which positions in the PSII or PSI complexes these modifications take place? Furthermore, the exact thylakoid-wide location for LHCII phosphorylation and state transition is not known, i.e., does it take place in the grana, grana margins, or stroma-exposed regions? It is also unclear to what extent thylakoid protein complexes, and which ones, move physically and what large multiprotein complexes can be formed during state transitions. One of the limitations to these studies has been a lack of appropriate tools. With our specific antibodies against the phosphorylated forms of Lhcb1 and Lhcb2 and the Arabidopsis amiLhcb1 and amiLhcb2 lines described herein, is now possible to answer some of these questions.
In this article, we show that both Lhcb1 and Lhcb2 are necessary for state transitions and that neither alone is sufficient, although the reasons underlying this requirement are different. In the absence of Lhcb1, only a small number of Lhcb2 homotrimers are formed, and they are limited to grana cores. Additionally, negligible amount of Lhcb1/Lhcb2 heterotrimers incorporating the very small residual amount of Lhcb1 might possibly be formed, although we were unable to detect any Lhcb1 in LHCII trimers using BN (Supplemental Figure 3). This affects the grana structure and results in the absence of thylakoid membrane remodeling, which has been suggested to be typical for state transitions (Dietzel et al., 2011). The amount of LHCII trimers available for state transitions is clearly limited. Although Lhcb2 is phosphorylated in a light quality-dependent manner, this does not result in any change in the relative absorption profiles of PSI and PSII. Under these conditions, STN7 accumulates, perhaps as a compensatory response, although the effort is futile, as no LHCII will be functionally relocated to PSI. In the absence of Lhcb2, on the other hand, photosynthetic complexes of various combinations of PSI, PSII and LHCII, and LHCII trimers indistinguishable from those of the wild type are formed, with the exception of the state transition-specific PSI-LHCII complex, which is not formed. In these amiLhcb2 plants, the PQ pool is reduced after a shift to state 2 light, STN7 becomes activated, and Lhcb1 is hyperphosphorylated. However, this does not result in any change in the excitation of PSII or PSI caused by inability of Lhcb1 homotrimers to bind to PSI. Thus, excitation imbalance and PQ overreduction persist in amiLhcb2 despite Lhcb1 hyperphosphorylation. Interestingly, under these conditions, STN7 does not accumulate to levels greater than those in the wild type. The growth phenotypes of both amiLhcb1 and amiLhcb2 under fluctuating light confirms that state transitions are important under such conditions.
Understanding why both Lhcb1 and Lhcb2 are required for state transitions necessitates a more detailed consideration of the specific features and interactions of the two proteins. We have recently shown that Lhcb2 is more rapidly phosphorylated than Lhcb1 and that the state transition-specific PSI-LHCII complex contains only phospho-Lhcb2, not phospho-Lhcb1 (Leoni et al., 2013). Together with the findings presented in this contribution, that both Lhcb1 and Lhcb2 are necessary for state transitions but neither is sufficient, this provides strong evidence that the majority of trimers engaged in state transitions are (Lhcb1)2Lhcb2 heterotrimers. Although Lhcb1(Lhcb2)2 heterotrimers may exist in wild-type plants, they seem to be present in only minute amounts, as their presence has seldom been reported. Lhcb1 is by far the most abundant of the LHCII polypeptides and Lhcb1 homotrimers are the most abundant trimeric form; as Lhcb1, Lhcb2, and Lhcb3 are extremely similar in structure and can form heterotrimers, it is possible that the amount of each trimer formed is a simple consequence of the relative amounts of the proteins, with the exception of (Lhcb2)2Lhcb3 heterotrimers that, according to the data presented here, seem not to be stable. There is clearly a relationship between the Lhcb1/2/3 protein composition of a trimer and its location, but this relationship is not straightforward. It appears that in the wild type, only (Lhcb1)2Lhcb3 trimers can occupy the M-position in the PSII supercomplex (Galka et al., 2012); this specificity may be explained by the existence of an interface that promotes direct interaction between Lhcb3 and CP24 (Kovács et al., 2006). But why is it that (Lhcb2)2Lhcb3 and Lhcb1Lhcb2Lhcb3 heterotrimers do not readily form and attach to this position, and yet, in plants lacking Lhcb3, other trimers (presumably Lhcb1 homotrimers) occupy the position instead? One possibility is that the adjacent CP29 protein provides a surface with which only Lhcb1 can interact and that this interaction may be strong enough to bind an Lhcb1 homotrimer in the M-position in the absence of Lhcb3. The S-position may be more flexible, as it can obviously be occupied by Lhcb1/Lhcb2 heterotrimers (in wild-type plants) and Lhcb2 homotrimers (in amiLhcb1) and it is likely that for trimers attaching to the L-site, or not attaching to any photosystem, there may be little or no constraint acting on trimer composition other than factors influencing the stability of the trimer itself. We should also point out that under far-red light, the M-position in amiLhcb1 can also be filled with trimers, possibly Lhcb2 homotrimers (or heterotrimers, as a minute residual amount of Lhcb1 is present).
A definitive answer to these questions may have to await the availability of a high-resolution structure of the vascular plant PSII holocomplex. Matters are further complicated by the likelihood that the trimers that end up interacting with PSI in state 2 are recruited from the large pool of “extra” trimers (Wientjes et al., 2013a) that are not located in the S or M pockets but are nevertheless closely connected to PSII. The minute amount of Lhcb3 found in trimers attaching to PSI (Galka et al., 2012) may potentially originate either from M-trimers that functionally detach from PSII as a result of phosphorylation or from a small number of Lhcb3-containing trimers not located in the M-trimer position.
(Lhcb1)2Lhcb2 heterotrimers are believed to constitute a relatively minor component of LHCII, perhaps only 20 to 30% of all trimers. If these trimers are the only type that, after phosphorylation of Lhcb2, have the capacity to functionally connect to PSI, this explains why in seed plants only ∼25% of LHCII is engaged in state transitions (Allen, 1992; Vener, 2007) and thus settles an unresolved issue in our understanding of state transitions. Since it also seems that only one Lhcb2 monomer needs to be phosphorylated, a phosphorylation level of <10% of the Lhcb1/2/3 monomers is sufficient to bring about the maximum level of state transition. We should stress that the situation in green algae such as Chlamydomonas was previously believed to be different since there is no distinction between the functions of Lhcb1 and Lhcb2. Presumably all phospho-LHCII polypeptides can mediate functional attachment to PSI; hence, the magnitude of state transitions in Chlamydomonas was estimated to ∼80% (Delosme et al., 1996; Ferrante et al., 2012). However, a recent study suggested that also in Chlamydomonas only 10% of LHCII migrate to PSI, while the other detaching trimers are involved in nonphotochemical quenching (Ünlü et al., 2014). The situation in Chlamydomonas is different from that of seed plants as certain LHC subunits, not PsbS, provides most quenching capacity. In plants, on the other hand, LHCII trimers, presumably Lhcb1 homotrimers or (Lhcb1)2Lhcb2 heterotrimers, may dissociate from PSII and perform quenching by interaction with PsbS and zeaxanthin (Wilk et al., 2013).
Although almost identical in primary sequence, Lhcb1 and Lhcb2 have complementary functions: Lhcb1 makes up the bulk of the light-harvesting antenna and Lhcb2 enables rapid state transitions. It is clear that phosphorylated Lhcb2 provides the surface for interaction with PSI, which has been shown to occur on the side of PSI where the PsaH, PsaL, and PsaO polypeptides are located (Lunde et al., 2000; Jensen et al., 2004). The very subtle amino acid differences between Lhcb1 and Lhcb2 may be enough to determine whether each protein should bind to the (as yet not precisely defined) binding site on PSI; alternatively, amino acid substitutions may mean that Lhcb2 shifts to a different conformation from that of Lhcb1 after phosphorylation, thereby giving specificity to the process. In any case, the subtle sequence differences between the two proteins are so important that they have been conserved for ∼300 million years.
An intriguing conclusion from our study is that phosphorylation of Lhcb1, which has been assumed to drive state transitions, has a less obvious function. Grana structure is believed to be maintained by the attracting forces between opposite membranes (Standfuss et al., 2005), while the dimensions of grana stacks depend on curvature thylakoid (CURT1) proteins inducing membrane bending at grana margins (Armbruster et al., 2013). Absence of PSII core phosphorylation of stn8 increases the width of grana discs (Fristedt et al., 2009), which most likely is not the result of a change in PSII phosphorylation but rather due to a change in the activity of CURT1 proteins. The importance of LHCII for grana stacking has been previously shown, for example, in tobacco (Nicotiana tabacum), where constitutive expression of an Lhcb1 gene increased the height of grana (Labate et al., 2004). Although grana formation does not need Lhcb1 and Lhcb2, plants lacking both can form grana stacks (Andersson et al., 2003), our results demonstrate that the abundance of Lhcb1 modulates the height of the grana stacks (Figure 4). In contrast, the amiLhcb2 thylakoid membranes, mainly composed of Lhcb1 homotrimers, seem to be more structured in state 1 in comparison to the wild type, with more defined stacks and fewer stroma lamellae. Clearly, several factors contribute to grana stacking, allowing more flexibility to cope with changing light conditions.
In studies involving mutants of PSII subunits, Psb27 and PsbW have shown absence of PSII supercomplex formation and accelerated rates of state transitions, indicating that the release of PSII supercomplexes precedes state transition (Dietzel et al., 2011; García-Cerdán et al., 2011). Dietzel et al. (2011) proposed that prior to state transitions, CP43 has to be phosphorylated to allow for supercomplex rearrangement and D1, D2, and LHCII phosphorylation. This has been questioned as studies of organization of membranes by BN and EM during state 1 and state 2 suggested that the amount of supercomplexes does not change between states (Wientjes et al., 2013b); however, conditions inducing transitions were in this case different. Moreover, the majority of phosphorylated Lhcb1 and Lhcb2 in state 2 are found in super- and megacomplexes (Grieco et al., 2012; Leoni et al., 2013; Wientjes et al., 2013b) and S- and M-trimers probably do not move during state transitions. Instead, “extra” trimers are involved in state transitions, in state 2, mainly transferring energy to other trimers associated with PSII cores located at the grana margins (Wientjes et al., 2013a). Indeed, phosphorylation of these marginally located trimers might reduce interactions between the grana cores and margins, contributing to a decrease in stacking. The overaccumulation of STN7 in amiLhcb1 and the hypophosphorylation of Lhcb2 in PSI light suggest that Lhcb1 phosphorylation may facilitate flexibility at the terminal parts of grana structure, which enables better availability of Lhcb2 for STN7 (and/or other kinases). It is obvious that state transitions is a complex process that occur in several steps and that the measured changes in fluorescence is the integration of them.
Here, we suggest a two-step process for the transition from state 1 to 2. First, STN7 (and, to a small extent, also STN8) phosphorylates LHCII, which, however, has different consequences for its subunits. Phosphorylation of Lhcb1 causes loosening of the grana structure, maybe by charge repulsion, and release of the “extra” LHCII trimers, while Lhcb2 phosphorylation may enhance the process. Second, (Lhcb1)2Lhcb2 heterotrimers with phosphorylated Lhcb2 mediate the interaction and energy transfer to PSI. If the second step is blocked, like in amiLhcb2 or mutants lacking, e.g., PsaL, the observed fluorescence decline may be caused only by detachment of extra antennae from PSII. This detachment may be what we observe as a slower “state transition-like” process by monitoring fluorescence changes in amiLhcb2 after a change in light quality. This process, also dependent on the presence of STN7, may be regarded as a complementary, Lhcb1-dependent mechanism (which we suggest could be named “qT2”) acting over a longer time scale and only apparent in plants lacking Lhcb2, in wild-type plants obscured by the “typical state transition” process. This split of the mechanism for functional displacement of LHCII into two components with different kinetics could, from a theoretical point of view, be highly adaptive, allowing plants to ensure proper regulation under a wide range of conditions.
In this context, it is also important to note that the phosphorylation site of Lhcb1 is, in contrast to that of Lhcb2, not strictly conserved, as in many species there are Lhcb1 gene products that lack the phosphorylatable Thr residue (Leoni et al., 2013). Hence, there is apparently no selective pressure acting to maintain all Lhcb1 polypeptides as substrates for STN7, which in any case has a higher affinity for phosphorylating Lhcb2 (Leoni et al., 2013).
In conclusion, our data allow us to place the regulation of light harvesting in land plants more accurately into an evolutionary perspective. Green algae regulate light harvesting by both qE and qT. The “land plant type” of qE (which is dependent on the PsbS protein) evolved around the time when plants colonized land (∼400 MYA); bryophytes gained the PsbS-dependent qE but they also retain LhcSR-dependent (green alga-type) qE (Alboresi et al., 2008, 2010), whereas vascular plants have lost LhcSR. Our data indicate that the vascular plant type of state transitions evolved relatively shortly after land colonization (∼300 to 400 MYA), after the split of the bryophyte and spermatophyte (gymnosperms and angiosperms) lineages but before the split of the conifer and angiosperm lineages. The optimization of these two processes may have contributed to the enormous evolutionary success of the spermatophytes.
METHODS
Growth Conditions
Arabidopsis thaliana plants were grown under an 8/16-h-light/dark photoperiod at either 120 or 250 μmol photons m−2 s−1 irradiance and 75% humidity. The fluctuating light conditions were achieved by repeated cycles of low light (50 μmol photons m−2 s−1 for 5 min) and high light (500 μmol photons m−2 s−1 for 1 min).
Construction of amiRNA Lines
amiRNA sequences were designed to silence either the five Lhcb1 (Lhcb1.1-.5) or the three Lhcb2 (Lhcb2.1-3) Arabidopsis genes using WMD3 (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi?page=Home;project=stdwmd) and checked as described by Schwab et al. (2006) and Ossowski et al. (2008). Two separate sequences were generated for Lhcb1, amiLhcb1v1 (5′-TTCCGTACCATGGGGTGCCTG-3′) and amiLhcb1v2 (5′-TCAGCCTAGGGCTGCGACCAT-3′), and two for Lhcb2, amiLhcb2v1 (5′-TTAGACTTGACGGTACGGCAC-3′), and amiLhcb2v2 (5′-TTAGACTTGACGGTATGACCC-3′). The sequences were used to generate four oligonucleotides for overlapping PCR on a pRS300 matrix. Two oligo A and oligo B sequences were used to obtain the final amiRNA product. The oligo A sequence contained CACC nucleotides at the 5′ end, allowing incorporation of amiRNA into a pENTR/SD/D-TOPO vector (Invitrogen) for directional cloning into the binary vector pB7WG2D,1, which contains R1 and R2 attachment sites and the BAR gene (conferring resistance to the herbicide Basta). Arabidopsis Columbia-0 plants (or stn7 [Bellafiore et al., 2005] or npq4 [Li et al., 2000]) were transformed using the floral dip method (Clough and Bent, 1998). T0 plants were selected on Basta and screened for the presence of amiLhcb1 and amilhcb2 constructs using oligo A and oligo B primers and then for depletion of Lhcb1 and Lhcb2 using immunoblotting. Only amiLhcb1v1 was efficient in repressing Lhcb1 gene expression; plants originating from 30 individual transformation events were screened and the lines amiLhcb1v1.14 and amiLhcb1v1.3, which had the lowest Lhcb1 levels, were used for further analysis. Both amiLhcb2v1 and amiLhcb2v2 efficiently silenced Lhcb2 gene expression; line amiLhcb2v2.4 was used for further analysis.
Induction of State Transitions
Arabidopsis plants were subjected to red/far-red light treatment using a customized LED light source (SL 3500-R-D) from Photon System Instruments as previously described by Leoni et al. (2013). To investigate the kinetics of phosphorylation, the plants were first dephosphorylated for 60 min by applying a mix of red and far-red light, and “0 s” sample was collected and snap frozen in liquid nitrogen at the end of dephosphorylation period. Then, the leaves were collected after 30 s, 10 min, and 60 min of red light treatment. Dephosphorylation kinetics was studied by the opposite treatment: First, the samples were fully phosphorylated for 60 min by applying red light, and 0 s sample was collected at the end. Then, far-red light was added to induce dephosphorylation. Samples were collected after 10, 30, and 60 min.
Pigment and Protein Analyses
Chlorophyll content was measured according to Porra et al. (1989), and thylakoid membranes were isolated according to Järvi et al. (2011) from fresh or frozen (in liquid nitrogen) leaves. All of the extraction buffers contained 10 mM NaF to inhibit the phosphatase activity. Immunoblot analysis was performed on thylakoid membranes according to Leoni et al. (2013). The monospecific antibodies used were Lhca1 (Ganeteg et al., 2001; Agrisera AS01 005; 1:5000), Lhca2 (Ganeteg et al., 2001; Agrisera AS01 006; 1:5000), Lhca3 (Król et al., 1995; 1:2000; Agrisera AS01 007), Lhca4 (Ganeteg et al., 2001; 1:5000; Agrisera AS01 008; 1:1000), Lhca5 (Ganeteg et al., 2001; Agrisera AS05 082; 1:5000), PsaC (Agrisera AS10 939; 1:10,000), Lhcb1 (Jansson et al., 1997; 1:5000; Agrisera AS09 522), P-Lhcb1 (Leoni et al., 2013; 1:5000; AS13 2704), Lhcb2 (Jansson et al., 1997; 1:5000; Agrisera AS01 003), P-Lhcb2 (Leoni et al., 2013; 1:5000; AS13 2705), Lhcb3 (Damkjaer et al., 2009; 1:1000; Agrisera AS01 002), Lhcb4 (Jansson et al., 1997; 1:7000; Agrisera AS04 045), Lhcb5 (Jansson et al., 1997; 1:1000; Agrisera AS01 009), Lhcb6 (Andersson et al., 2003; Agrisera AS01 010; 1:1000), PsbD (Agrisera AS06 146; 1:10,000), PsbS (Agrisera AS09 533; 1:2000), STN7 (Agrisera AS10 1611; 1:1000), STN8 (Agrisera AS10 1601; 1:1000), PPH1/TAP38 (provided by Roberto Barbato; 1:1000), and PBCP (provided by Michel Goldschmidt-Clermont; 1:1000). The GE Healthcare ECL reagents were applied for detection of the immunoreaction. The chemiluminescence was recorded on blots where the antibody signal was in the linear range using a FUJI LAS-3000 cooled CCD camera and detected bands were quantified using the Multi Gauge application. ProQ Diamond (Molecular Probes) staining of SDS-PAGE was performed according to the manufacturer's instructions. Native PAGE analysis was performed according to Sirpiö et al. (2011).
Electron Microscopy
The plants were subjected to 2 h of red light or 2 h of red and far-red light to induce state 1 or state 2, respectively. Thylakoid ultrastructure was analyzed according to Brouwer et al. (2012). All grana, on 10 pictures of chloroplasts for each treatment, were quantified to establish the mean number of layers forming grana of the wild type, amiLhcb1, and amiLhcb2 in state 1 and state 2. Two-way ANOVA was performed to assess the significance in the changes in grana structure between the treatment and lines, while the differences in variances in grana size were examined using Levene’s test (Levene, 1960). PSII supercomplexes were analyzed by crystal and single particle EM according to Ruban et al. (2003).
Chlorophyll Fluorescence Analysis
State transitions were induced according to Leoni et al. (2013) as described above. 77K fluorescence spectra of thylakoid samples (5 μg chlorophyll/mL in 50% glycerol, 50 mM HEPES-KOH, pH 7.5, 10 mM MgCl2, 100 mM sorbitol, and 10 mM NaF) were measured on a Fluoromax-2 fluorometer. Excitation wavelength was 435 nm, increment 0.5 nm and integration time 2 s. Spectra were normalized with respect to the 685-nm peak. A WALTZ Dual PAM-100 fluorometer was used to measure qE and state transitions according to Damkjaer et al. (2009). Plants were dark-adapted for 1 h prior to the measurements. The PQ pool oxidation state was measured according to Grieco et al. (2012).
Analysis of Single T-DNA KO Mutants
Single Lhcb1 and Lhcb2 T-DNA KO mutants (N662639/SALK_059893C, Lhcb1.1; N597368/SALK_097368, Lhcb1.2; N846666/SAIL_1261_C06, Lhcb1.3; N549351/SALK_049351, Lhcb1.4; N657479/SALK_005774C, Lhcb2.1; N651088/SALK_151088, Lhcb2.2) were selfed to produce homozygous lines, grown, and analyzed as described above for polypeptide composition, Lhcb1 and Lhcb2 phosphorylation, state transitions, and qE.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: Lhcb1.1 (At1g29920), Lhcb1.2 (At1g29910), Lhcb1.3 (At1g29930), Lhcb1.4 (At2g34430), Lhcb1.5 (At2g34420), Lhcb2.1 (At2g05100), Lhcb2.2 (At2g05070), and Lhcb2.3 (At3g27690).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PSII Complex Topography in amiLhcb1 Derived from Single-Particle Analysis of Membrane Crystals.
Supplemental Figure 2. Micrographs of Leaf Cross-Section Demonstrating Changes in Thylakoid Structure during State Transitions.
Supplemental Figure 3. BN Gel Electrophoresis of Thylakoid Complexes and Their Lhcb1-3 Protein Content.
Supplemental Figure 4. Visualization of Phosphorylated Thylakoid Proteins with ProQ Diamond Staining.
Supplemental Figure 5. Pulse-Amplitude Modulated Fluorescence Traces.
Supplemental Figure 6. Nonphotochemical Quenching in Examined Lines.
Supplemental Figure 7. PQ Reduction State (Measured as F/Fm).
Supplementary Material
Acknowledgments
We thank Virpi Paakkarinen, Kathryn Robinson, and Lenore Johansson for technical assistance and Anett Z. Kiss, Gergely Molnar, and Rishikesh Bhalerao for technical advice. This work was supported by the European Union project FP7-PEOPLE-ITN-2008 “HARVEST: Control of Light Use Efficiency in Plants and Algae From Light to Harvest,” the Swedish Research Council (VR), the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (Formas), and the Academy of Finland (Grants 273870, 271832, 138703, and 260094).
AUTHOR CONTRIBUTIONS
M.P. constructed the amiRNA lines, performed the growth, pigment, and thylakoid ultrastructure and most of the immunoblot and fluorescence analyses, and contributed to the native PAGE and EM analysis. M.S. performed most of the native PAGE and some of the immunoblot analysis. D.A.S. performed the crystal and single-particle EM analysis. M.T. carried out the PQ oxidation analysis. E.J.B. supervised the crystal and single-particle EM analysis. E.-M.A. supervised the native PAGE and participated in the design of the study. S.J. coordinated and designed the study and wrote most of the article.
Glossary
- PSII
photosystem II
- PSI
photosystem I
- PQ
plastoquinone
- KO
knockout
- MYA
million years ago
- amiRNA
artificial microRNA
- EM
electron microscopy
- BN
Blue Native
- lpBN
large-pore Blue Native
- β-DM
β-dodecyl maltoside
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alboresi, A., Caffarri, S., Nogue, F., Bassi, R., Morosinotto, T. (2008). In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: identification of subunits which evolved upon land adaptation. PLoS ONE 3: e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi, A., Gerotto, C., Giacometti, G.M., Bassi, R., Morosinotto, T. (2010). Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc. Natl. Acad. Sci. USA 107: 11128–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, J.F. (1992). Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta 1098: 275–335 [DOI] [PubMed] [Google Scholar]
- Allen, J.F., Bennett, J., Steinback, K.E., Arntzen, C.J. (1981). Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291: 25–29 [Google Scholar]
- Andersson, J., Wentworth, M., Walters, R.G., Howard, C.A., Ruban, A.V., Horton, P., Jansson, S. (2003). Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II - effects on photosynthesis, grana stacking and fitness. Plant J. 35: 350–361 [DOI] [PubMed] [Google Scholar]
- Armbruster, U., et al. (2013). Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell 25: 2661–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook, J.R., Smith, S.M., Ellis, R.J. (1980). Molecular cloning and sequencing of cDNA encoding the precursor to the small subunit of chloroplast ribulose-1,5-bisphosphate carboxylase. Nature 287: 692–697 [Google Scholar]
- Bellafiore, S., Barneche, F., Peltier, G., Rochaix, J.-D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Bennett, J. (1977). Phosphorylation of chloroplast membrane polypeptides. Nature 269: 344–346 [Google Scholar]
- Bennett, J., Steinback, K.E., Arntzen, C.J. (1980). Chloroplast phosphoproteins: regulation of excitation energy transfer by phosphorylation of thylakoid membrane polypeptides. Proc. Natl. Acad. Sci. USA 77: 5253–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema, E.J., Van Roon, H., Van Breemen, J.F.L., Dekker, J.P. (1999). Supramolecular organization of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. Eur. J. Biochem. 266: 444–452 [DOI] [PubMed] [Google Scholar]
- Boekema, E.J., van Breemen, J.F.L., van Roon, H., Dekker, J.P. (2000). Arrangement of PSII supercomplexes in crystalline macrodomains within the thylakoid membrane of green plants. J. Mol. Biol. 301: 1123–1133 [DOI] [PubMed] [Google Scholar]
- Bonardi, V., Pesaresi, P., Becker, T., Schleiff, E., Wagner, R., Pfannschmidt, T., Jahns, P., Leister, D. (2005). Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Brouwer, B., Ziolkowska, A., Bagard, M., Keech, O., Gardeström, P. (2012). The impact of light intensity on shade-induced leaf senescence. Plant Cell Environ. 35: 1084–1098 [DOI] [PubMed] [Google Scholar]
- Caffarri, S., Croce, R., Cattivelli, L., Bassi, R. (2004). A look within LHCII: differential analysis of the Lhcb1-3 complexes building the major trimeric antenna complex of higher-plant photosynthesis. Biochemistry 43: 9467–9476 [DOI] [PubMed] [Google Scholar]
- Caffarri, S., Kouřil, R., Kereïche, S., Boekema, E.J., Croce, R. (2009). Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28: 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., Bent, A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Damkjaer, J.T., Kereïche, S., Johnson, M.P., Kovacs, L., Kiss, A.Z., Boekema, E.J., Ruban, A.V., Horton, P., Jansson, S. (2009). The photosystem II light-harvesting protein Lhcb3 affects the macrostructure of photosystem II and the rate of state transitions in Arabidopsis. Plant Cell 21: 3245–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delosme, R., Olive, J., Wollman, F.-A. (1996). Change in light energy distribution upon state transitions: an in vivo photoacoustic study of the wild-type and photosynthesis mutants from Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1273: 150–158 [Google Scholar]
- Dietzel, L., Bräutigam, K., Steiner, S., Schüffler, K., Lepetit, B., Grimm, B., Schöttler, M.A., Pfannschmidt, T. (2011). Photosystem II supercomplex remodeling serves as an entry mechanism for state transitions in Arabidopsis. Plant Cell 23: 2964–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrad, D., Grossman, A.R. (2004). A genome’s-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr. Genet. 45: 61–75 [DOI] [PubMed] [Google Scholar]
- Ferrante, P., Ballottari, M., Bonente, G., Giuliano, G., Bassi, R. (2012). LHCBM1 and LHCBM2/7 polypeptides, components of major LHCII complex, have distinct functional roles in photosynthetic antenna system of Chlamydomonas reinhardtii. J. Biol. Chem. 287: 16276–16288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt, R., Willig, A., Granath, P., Crèvecoeur, M., Rochaix, J.-D., Vener, A.V. (2009). Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 21: 3950–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galka, P., Santabarbara, S., Khuong, T.T., Degand, H., Morsomme, P., Jennings, R.C., Boekema, E.J., Caffarri, S. (2012). Functional analyses of the plant photosystem I-light-harvesting complex II supercomplex reveal that light-harvesting complex II loosely bound to photosystem II is a very efficient antenna for photosystem I in state II. Plant Cell 24: 2963–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeteg, U., Strand A, Gustafsson, P., Jansson, S. (2001). The properties of the chlorophyll a/b-binding proteins Lhca2 and Lhca3 studied in vivo using antisense inhibition. Plant Physiol. 127: 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cerdán, J.G., Kovács, L., Tóth, T., Kereïche, S., Aseeva, E., Boekema, E.J., Mamedov, F., Funk, C., Schröder, W.P. (2011). The PsbW protein stabilizes the supramolecular organization of photosystem II in higher plants. Plant J. 65: 368–381 [DOI] [PubMed] [Google Scholar]
- Grieco, M., Tikkanen, M., Paakkarinen, V., Kangasjärvi, S., Aro, E.-M. (2012). Steady-state phosphorylation of light-harvesting complex II proteins preserves photosystem I under fluctuating white light. Plant Physiol. 160: 1896–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankamer, B., Nield, J., Zheleva, D., Boekema, E., Jansson, S., Barber, J. (1997). Isolation and biochemical characterisation of monomeric and dimeric photosystem II complexes from spinach and their relevance to the organisation of photosystem II in vivo. Eur. J. Biochem. 243: 422–429 [DOI] [PubMed] [Google Scholar]
- Horton, P., Black, M.T. (1980). Activation of adenosine-′-triphosphate-induced quenching of chlorophyll fluorescence by reduced plastoquinone. The basis of state I-state II transitions in chloroplasts. FEBS Lett. 119: 141–144 [Google Scholar]
- Horton, P., Black, M.T. (1981). Light-dependent quenching of chlorophyll fluorescence in pea chloroplasts induced by adenosine 5′-triphosphate. Biochim. Biophys. Acta 635: 53–62 [DOI] [PubMed] [Google Scholar]
- Horton, P., Allen, J.F., Black, M.T., Bennett, J. (1981). Regulation of phosphorylation of chloroplast membrane polypeptides by the redox state of plastoquinone. FEBS Lett. 125: 193–196 [Google Scholar]
- Jackowski, G., Pielucha, K. (2001). Heterogeneity of the main light-harvesting chlorophyll a/b-protein complex of photosystem II (LHCII) at the level of trimeric subunits. J. Photochem. Photobiol. B 64: 45–54 [DOI] [PubMed] [Google Scholar]
- Jansson, S. (1994). The light-harvesting chlorophyll a/b-binding proteins. Biochim. Biophys. Acta 1184: 1–19 [DOI] [PubMed] [Google Scholar]
- Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4: 236–240 [DOI] [PubMed] [Google Scholar]
- Jansson, S., Gustafsson, P. (1990). Type I and type II genes for the chlorophyll a/b-binding protein in the gymnosperm Pinus sylvestris (Scots pine): cDNA cloning and sequence analysis. Plant Mol. Biol. 14: 287–296 [DOI] [PubMed] [Google Scholar]
- Jansson, S., Virgin, I., Gustafsson, P., Andersson, B., Oquist, G. (1992). A nomenclature for the genes encoding the chlorophyll a/b-binding proteins of higher plants. Plant Mol. Biol. Rep. 10: 242–253 [Google Scholar]
- Jansson, S., Stefansson, H., Nystrom, U., Gustafsson, P., Albertsson, P.-Å. (1997). Antenna protein composition of PS I and PS II in thylakoid subdomains. Biochem. Biophys. Acta Bioenergetics 1320: 297–309 [Google Scholar]
- Jensen, P.E., Haldrup, A., Zhang, S., Scheller, H.V. (2004). The PSI-O subunit of plant photosystem I is involved in balancing the excitation pressure between the two photosystems. J. Biol. Chem. 279: 24212–24217 [DOI] [PubMed] [Google Scholar]
- Järvi, S., Suorsa, M., Paakkarinen, V., Aro, E.-M. (2011). Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem. J. 439: 207–214 [DOI] [PubMed] [Google Scholar]
- Kovács, L., Damkjaer, J., Kereïche, S., Ilioaia, C., Ruban, A.V., Boekema, E.J., Jansson, S.Horton, P. (2006). Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. Plant Cell 18: 3106–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Król, M., Spangfort, M.D., Huner, N.P., Oquist, G., Gustafsson, P., Jansson, S. (1995). Chlorophyll a/b-binding proteins, pigment conversions, and early light-induced proteins in a chlorophyll b-less barley mutant. Plant Physiol. 107: 873–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni, C., Pietrzykowska, M., Kiss, A.Z., Suorsa, M., Ceci, L.R., Aro, E.-M., Jansson, S. (2013). Very rapid phosphorylation kinetics suggest a unique role for Lhcb2 during state transitions in Arabidopsis. Plant J. 76: 236–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labate, M.T., Ko, K., Ko, Z.W., Pinto, L.S., Real, M.J., Romano, M.R., Barja, P.R., Granell, A., Friso, G., van Wijk, K.J., Brugnoli, E., Labate, C.A. (2004). Constitutive expression of pea Lhcb 1-2 in tobacco affects plant development, morphology and photosynthetic capacity. Plant Mol. Biol. 55: 701–714 [DOI] [PubMed] [Google Scholar]
- Levene, H. (1960). Robust tests for equality of variances. In Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling, I. Olkin, S.G. Ghurye, W. Hoeffding, W.G. Madow, and H.B. Mann, eds (Stanford, CA: Stanford University Press), pp. 278–292 [Google Scholar]
- Li, X.-P., Björkman, O., Shih, C., Grossman, A.R., Rosenquist, M., Jansson, S., Niyogi, K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Lunde, C., Jensen, P.E., Haldrup, A., Knoetzel, J., Scheller, H.V. (2000). The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408: 613–615 [DOI] [PubMed] [Google Scholar]
- Michel, H., Griffin, P.R., Shabanowitz, J., Hunt, D.F., Bennett, J. (1991). Tandem mass spectrometry identifies sites of three post-translational modifications of spinach light-harvesting chlorophyll protein II. Proteolytic cleavage, acetylation, and phosphorylation. J. Biol. Chem. 266: 17584–17591 [PubMed] [Google Scholar]
- Minagawa, J. (2011). State transitions—the molecular remodeling of photosynthetic supercomplexes that controls energy flow in the chloroplast. Biochim. Biophys. Acta 1807: 897–905 [DOI] [PubMed] [Google Scholar]
- Ossowski, S., Schwab, R., Weigel, D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Pesaresi, P., Hertle, A., Pribil, M., Kleine, T., Wagner, R., Strissel, H., Ihnatowicz, A., Bonardi, V., Scharfenberg, M., Schneider, A., Pfannschmidt, T., Leister, D. (2009). Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21: 2402–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi, P., Pribil, M., Wunder, T., Leister, D. (2011). Dynamics of reversible protein phosphorylation in thylakoids of flowering plants: the roles of STN7, STN8 and TAP38. Biochim. Biophys. Acta 1807: 887–896 [DOI] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., Kriedmann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Pribil, M., Pesaresi, P., Hertle, A., Barbato, R., Leister, D. (2010). Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol. 8: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribil, M., Labs, M., Leister, D. (2014). Structure and dynamics of thylakoids in land plants. J. Exp. Bot. 65: 1955–1972 [DOI] [PubMed] [Google Scholar]
- Rinalducci, S., Larsen, M.R., Mohammed, S., Zolla, L. (2006). Novel protein phosphorylation site identification in spinach stroma membranes by titanium dioxide microcolumns and tandem mass spectrometry. J. Proteome Res. 5: 973–982 [DOI] [PubMed] [Google Scholar]
- Rintamäki, E., Martinsuo, P., Pursiheimo, S., Aro, E.-M. (2000). Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl. Acad. Sci. USA 97: 11644–11649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban, A.V., Wentworth, M., Yakushevska, A.E., Andersson, J., Lee, P.J., Keegstra, W., Dekker, J.P., Boekema, E.J., Jansson, S., Horton, P. (2003). Plants lacking the main light-harvesting complex retain photosystem II macro-organization. Nature 421: 648–652 [DOI] [PubMed] [Google Scholar]
- Schwab, R., Ossowski, S., Riester, M., Warthmann, N., Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov, A., Ingelsson, B., Samol, I., Andres, C., Kessler, F., Rochaix, J.-D., Vener, A.V., Goldschmidt-Clermont, M. (2010). The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö, S., Suorsa, M., Aro, E.-M. (2011). Analysis of thylakoid protein complexes by two-dimensional electrophoretic systems. Methods Mol. Biol. 775: 19–30 [DOI] [PubMed] [Google Scholar]
- Standfuss, J., Terwisscha van Scheltinga, A.C., Lamborghini, M., Kühlbrandt, W. (2005). Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. EMBO J. 24: 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen, M., Aro, E.-M. (2014). Integrative regulatory network of plant thylakoid energy transduction. Trends Plant Sci. 19: 10–17 [DOI] [PubMed] [Google Scholar]
- Vainonen, J.P., Hansson, M., Vener, A.V. (2005). STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of photosystem II core proteins. J. Biol. Chem. 280: 33679–33686 [DOI] [PubMed] [Google Scholar]
- Vener, A.V. (2007). Environmentally modulated phosphorylation and dynamics of proteins in photosynthetic membranes. Biochim. Biophys. Acta 1767: 449–457 [DOI] [PubMed] [Google Scholar]
- Vener, A.V., Harms, A., Sussman, M.R., Vierstra, R.D. (2001). Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes of Arabidopsis thaliana. J. Biol. Chem. 276: 6959–6966 [DOI] [PubMed] [Google Scholar]
- Wientjes, E., van Amerongen, H., Croce, R. (2013a). LHCII is an antenna of both photosystems after long-term acclimation. Biochim. Biophys. Acta 1827: 420–426 [DOI] [PubMed] [Google Scholar]
- Wientjes, E., Drop, B., Kouřil, R., Boekema, E.J., Croce, R. (2013b). During state 1 to state 2 transition in Arabidopsis thaliana, the photosystem II supercomplex gets phosphorylated but does not disassemble. J. Biol. Chem. 288: 32821–32826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk, L., Grunwald, M., Liao, P.-N., Walla, P.J., Kühlbrandt, W. (2013). Direct interaction of the major light-harvesting complex II and PsbS in nonphotochemical quenching. Proc. Natl. Acad. Sci. USA 110: 5452–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ünlü, C., Drop, B., Croce, R., van Amerongen, H. (2014). State transitions in Chlamydomonas reinhardtii strongly modulate the functional size of photosystem II but not of photosystem I. Proc. Natl. Acad. Sci. USA 111: 3460–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.