Pollen tubes are tubular structures that extend by tip growth. Upon overexpression of merely a single endogenous molecule, LePRK1, or a portion of it lacking its extracellular domain, pollen tube cells switched to an alternative mechanism for extension of the leading edge that is analogous to the blebbing growth mode reported for Dictyostelium and for Drosophila stem cells.
Abstract
The tubular growth of a pollen tube cell is crucial for the sexual reproduction of flowering plants. LePRK1 is a pollen-specific and plasma membrane–localized receptor-like kinase from tomato (Solanum lycopersicum). LePRK1 interacts with another receptor, LePRK2, and with KINASE PARTNER PROTEIN (KPP), a Rop guanine nucleotide exchange factor. Here, we show that pollen tubes overexpressing LePRK1 or a truncated LePRK1 lacking its extracellular domain (LePRK1ΔECD) have enlarged tips but also extend their leading edges by producing “blebs.” Coexpression of LePRK1 and tomato PLIM2a, an actin bundling protein that interacts with KPP in a Ca2+-responsive manner, suppressed these LePRK1 overexpression phenotypes, whereas pollen tubes coexpressing KPP, LePRK1, and PLIM2a resumed the blebbing growth mode. We conclude that overexpression of LePRK1 or LePRK1ΔECD rewires pollen tube growth to a blebbing mode, through KPP- and PLIM2a-mediated bundling of actin filaments from tip plasma membranes. Arabidopsis thaliana pollen tubes expressing LePRK1ΔECD also grew by blebbing. Our results exposed a hidden capability of the pollen tube cell: upon overexpression of a single membrane-localized molecule, LePRK1 or LePRK1ΔECD, it can switch to an alternative mechanism for extension of the leading edge that is analogous to the blebbing growth mode reported for Dictyostelium and for Drosophila melanogaster stem cells.
INTRODUCTION
A pollen tube is a fast-growing plant cell that extends at its cellular leading edge (the tip) over macroscopic distances (centimeters) after it protrudes from a germination pore of a hydrated pollen grain (a process called pollen germination). Using pollen tubes to deliver sperm cells for fertilization (siphonogamy) is an evolutionary innovation of seed plants (Rounds and Bezanilla, 2013). From the haustorial pollen tubes of cycads and ginkgo (Ginkgo biloba), to the sometimes branched pollen tubes of conifers (deWin et al., 1996; Fernando et al., 2005), to the nonbranched pollen tubes of flowering plants, the shape of the growing pollen tube became more and more restricted to a hemispherical tip capping a cylinder with a uniform diameter (Kroeger and Geitmann, 2012). An alternative method of extending a cellular leading edge, by continuously producing blebs (i.e., rounded protrusions), has been reported for Dictyostelium (Zatulovskiy et al., 2014) and Drosophila melanogaster stem cells (Charras and Paluch, 2008) but not in pollen tubes.
Pollen tubes extend exclusively at the tip and maintain a tubular shape with an intact cell wall and plasma membrane via the joint efforts of turgor and the structure of the cell wall (Hepler et al., 2013). Turgor provides the force that drives cell expansion. The cell wall is more expandable in the hemispherical tip and more rigid in the cylinder, and tip-targeted exocytosis of new cell wall materials and membranes increases the expandability of the tip surface. The internal machinery that supports pollen tube growth is the dynamic actin cytoskeleton, which is composed of thick, long F-actin bundles aligned to the long axis in the cylinder tube, and a fine, short F-actin–formed fringe located in the subapical region (i.e., the hemispherical tip region excluding the extreme apical part), which rapidly turns over and keeps pace with growth (Cheung and Wu, 2008; Mollet et al., 2013). F-actin bundles serve as “tracks” for transporting subcellular organelles and also participate in cytoplasmic streaming. “Reverse fountain” cytoplasmic streaming is a hallmark of pollen tube cell viability (Chebli et al., 2013). A tip-focused cytosolic [Ca2+] gradient, 3 to 10 μM at the hemispherical tip and 100 to 200 nM in the tube cylinder, spatially regulates the activity of different actin binding proteins, contributing to shaping the dynamic actin structure (Hepler et al., 2013). Perturbations affecting cell wall properties, actin dynamics, or cytosolic [Ca2+] often lead to morphological changes such as tip swelling (Cárdenas et al., 2005; Mollet et al., 2013), probably because the cell wall at the tip is more flexible than the cell wall of the tube shank.
The mechanism of pollen tube growth is typical of polar growth. As in yeast, which also exhibit polar growth, the requirement for Rho family small guanine nucleotide binding proteins (namely, ROP or RAC) in polarity establishment has been demonstrated in pollen tubes (reviewed in Wedlich-Soldner and Li, 2008; Yang, 2008; Slaughter et al., 2009). GTP-bound active ROP localizes at the plasma membrane of the hemispherical tip, and the amount of tip-localized active ROP correlates with the speed of pollen tube extension (Hwang et al., 2010). Overexpressing GTP-bound ROP caused significant tip swelling, so that the pollen tubes ballooned and elongation was arrested, while overexpressing the GDP-bound ROP arrested tube growth without significant changes to the tubular shape (Lin and Yang, 1997). In pollen tubes, the ROP molecular switches are activated by RopGEFs containing a plant-specific ROP nucleotide exchanger domain (Berken et al., 2005; Thomas et al., 2007), and cytoplasmic RopGEFs interact with cell surface–located receptor-like kinases (RLKs) (Kaothien et al., 2005; Zhang and McCormick, 2007). Although the morphological changes caused by the manipulation of RopGEFs or RLKs have been reported to be tip enlargement (Lin and Yang, 1997; Kaothien et al., 2005; Chang et al., 2013), the specific linkage between the ROP GTPase module (including the ROP core switch and its regulatory links such as RopGEF) and the pollen tube growth machinery (i.e., actin and actin binding proteins) has not been elucidated.
In tomato (Solanum lycopersicum), LePRK1 and LePRK2 are two pollen-specific receptor kinases localized at the pollen tube plasma membrane (Muschietti et al., 1998; Kim et al., 2002). These kinases interact with KINASE PARTNER PROTEIN (KPP) (Kaothien et al., 2005), a RopGEF that activates a tomato ROP in vitro (Löcke et al., 2010). LePRK2 is a positive regulator of pollen tube growth (Zhang et al., 2008), as antisense LePRK2 pollen tubes grow slower and exhibit an attenuated response to the growth-promoting signals STYLE INTERACTOR FOR LePRKs (STIL) (Wengier et al., 2010) and STIGMA-SPECIFIC PROTEIN1 (Huang et al., 2014). Furthermore, the phosphorylation status of particular motifs in the juxtamembrane domain of LePRK2 reciprocally affected pollen tube growth rate (Salem et al., 2011). STIL triggers specific dephosphorylation of LePRK2, followed by dissociation of the LePRK1/LePRK2 complex (Wengier et al., 2003, 2010). However, LePRK1 has been less studied, because unlike LePRK2, its expression does not increase upon pollen germination (Muschietti et al., 1998) and its kinase activity is not required to form the LePRK1/LePRK2 complex, whereas LePRK2 kinase activity is required (Wengier et al., 2003). Here, we report that pollen tubes in which LePRK1 expression was reduced by RNA interference (RNAi) grew slower, similar to antisense LePRK2 pollen tubes (Zhang et al., 2008). However, the overexpression phenotypes for LePRK1 and LePRK2 were very different; LePRK2 overexpression resulted in slightly enlarged tips (Zhang et al., 2008), whereas LePRK1 overexpression caused drastic morphological changes in growing pollen tubes, which extended by continuously forming blebs at the leading edge. Coexpression assays showed that accumulation of LePRK1 at the tip plasma membrane can rewire cellular machineries to attain this blebbing growth mode. Furthermore, we showed that PLIM2a, which binds and bundles actin filaments, can form a complex with KPP that interacts with LePRK1 at the plasma membrane of the hemispherical tip. Overexpressing LePRK1 increased membrane-anchored actin bundling at the tip and resulted in the switch to blebbing growth, whereas coexpressing PLIM2a increased global actin bundling in pollen tubes, canceling the effect of LePRK1 overexpression and thus switching back to a tubular growth mode, albeit with a slower growth rate.
RESULTS
Transient Overexpression of LePRK1 Alters the Morphology of the Pollen Tube Tip
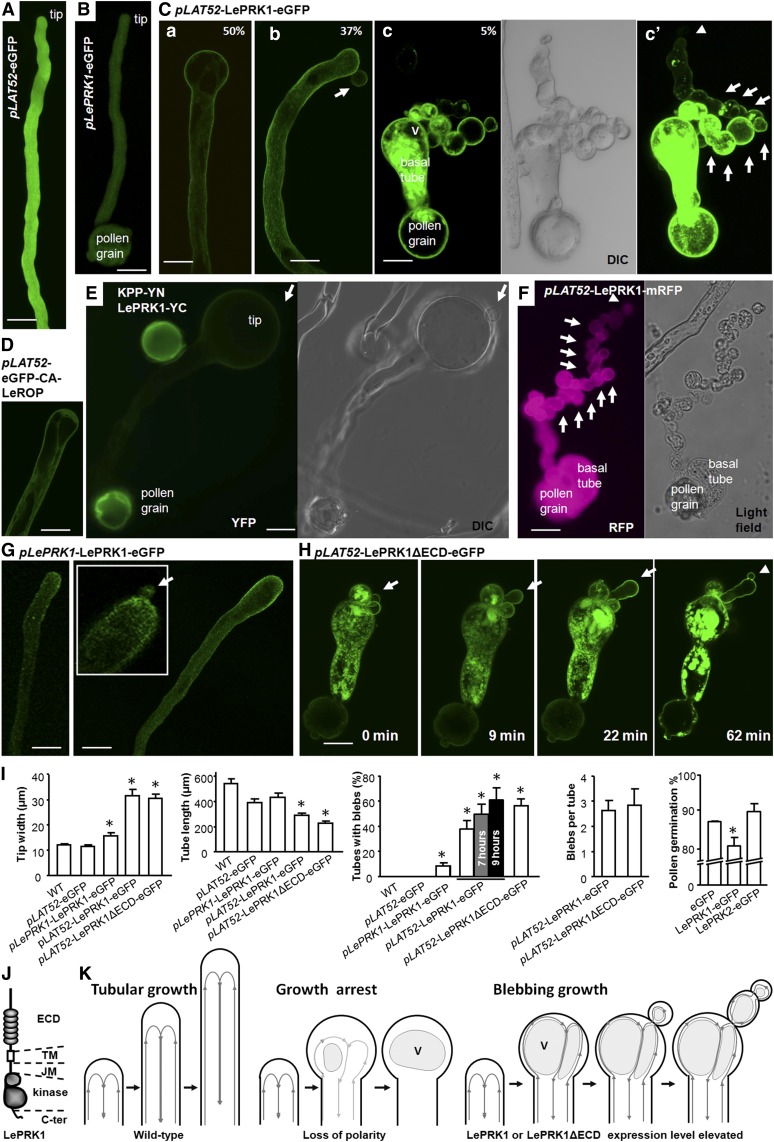
To elucidate the biological function of LePRK1, we transiently overexpressed LePRK1-eGFP (for enhanced green fluorescent protein) in tomato and tobacco (Nicotiana tabacum) pollen under the control of the pollen-specific LAT52 promoter (pLAT52) (Twell et al., 1990). After particle bombardment, pollen grains cultured in germination medium germinate and the resulting pollen tubes grow in vitro for up to 30 h, allowing live cell imaging of pollen morphologies and subcellular localization of fluorescently tagged proteins. Both nontransformed pollen and pollen expressing eGFP grew cylindrical tubes with hemispherical tips (Figures 1A and 1B; Supplemental Figure 1A). However, pollen expressing LePRK1-eGFP had a lower germination percentage than wild-type pollen (Figure 1I; Supplemental Figure 1B), and those that did germinate grew shorter tubes with enlarged (balloon-like) tips, from which additional spherical blebs often continued to form (Figure 1C, a to c; Supplemental Figure 1B and Supplemental Movies 1 and 2). Occasionally (∼12% of the tubes with blebs), more blebs developed from a previous bleb, one by one (Figure 1C, c and c′). The overexpression phenotypes in tobacco (Figure 1) and tomato (Supplemental Figure 1) were similar; therefore, we used tobacco pollen in subsequent bombardment assays, as it is easier to collect large amounts of tobacco pollen.
Figure 1.
Transient Overexpression of LePRK1 Resulted in Pollen Tube Tip Swelling and Blebbing.
(A) to (H) Representative tobacco pollen tubes expressing various constructs as indicated. In cases where pollen tubes were very long, only the region near the tip is shown. All photographs were taken under the GFP channel unless indicated otherwise. Arrows point to blebs. Arrowheads point to the tip of a series of contiguous blebs. pLAT52, LAT52 promoter; pLePRK1, LePRK1 native promoter. In (C), (a) to (c) show the variation in phenotypes; the frequency of the phenotype is indicated by percentage. (c′) is an overexposure of (c). eGFP-CA-LeROP indicates constitutively active LeROP with eGFP fused at the N terminus. KPP-YN and LePRK1-YC are BiFC constructs. The arrows in (E) point to a bleb visible in the differential interference contrast image but not in the YFP channel. The inset in (G) shows an enlarged image of the tip with a small bleb (arrow). (H) shows four images from a time-lapse movie (Supplemental Movie 4) of a representative tobacco pollen tube expressing LePRK1 lacking the extracellular Leu-rich repeat domain (LePRK1ΔECD-eGFP). Note that one of the blebs continued to grow and formed another bleb at the tip of a previously formed bleb. Bars = 20 μm.
(I) Measurements of pollen tubes in transient assays. White bars represent measurements after 4 to 6 h of cultivation; the gray and black bars are after 7 and 9 h, respectively. Asterisks indicate significant differences from the wild type or the eGFP control (P < 0.01, Student’s t test). n = 3 independent experiments. Error bars indicate se.
(J) Diagram of the LePRK1 structure (detail in Supplemental Figure 3A). JM, juxtamembrane domain; TM, transmembrane domain.
(K) Model illustrating pollen tube growth modes: normal tubular growth (left), growth arrest (middle), and blebbing mode (right). V, large vacuole. Gray lines with arrows indicate the cytostream.
Tip swelling (Figure 1K) can be due to a loss of polarity (Yang, 1998), to increased actin polymerization (Cárdenas et al., 2005), or to reduced cell wall stiffness (Mollet et al., 2013). The cytoplasmic portion of LePRK1 can interact with KPP (Kaothien et al., 2005). Although LePRK1 localized at the plasma membrane of the entire pollen tube (Kim et al., 2002), LePRK1 preferentially recruited KPP at the apical membrane (Figure 1E). Because KPP is a RopGEF that activates LeROP (Berken et al., 2005; Löcke et al., 2010) and because overexpressing constitutively active LeROP also resulted in tip swelling (Figure 1D), it was not surprising that overexpressing LePRK1 led to enlarged tips. However, the phenotype of pollen tube blebbing (Figure 1K) is more intriguing. Presumably, growing a bleb, at least at the beginning, would require exocytosis to be focused in a more restricted area (i.e., at a point instead of a dome), which suggests greater polarity, whereas an enlarged exocytosis area would suggest less polarity. Furthermore, a blebbing phenotype has not been reported in pollen tubes overexpressing ROP or KPP.
We performed more analyses to clarify the relationship between LePRK1 overexpression and blebbing. First, the overall percentage of pollen tubes with blebs increased from 42 to 61% as the culturing time extended from 4 to 6 h to 9 h (Figure 1I). Considering that the time needed for gene expression, protein production, and downstream events might vary among individual pollen tubes, it is reasonable that not every pollen tube expressing LePRK1-eGFP formed blebs. Second, when measured at 4 to 6 h of culturing, ∼50% of the pollen tubes with moderate or high expression levels of LePRK1-eGFP formed one or more spherical blebs, as did ∼20% of the pollen tubes with low expression (Supplemental Figures 2C and 2D). This result indicates that the phenotype of forming additional blebs was not restricted to pollen tubes with very high expression levels of LePRK1-eGFP (as judged by fluorescence intensity). Third, the blebbing phenotype also occurred (6% frequency) (Figure 1G; Supplemental Movie 3) in pollen tubes in which LePRK1 was expressed from its own promoter. This result indicates that the blebbing phenotype does not require a strong promoter. Fourth, we used a different fluorescent protein, mRFP, which is a monomeric red fluorescent protein derived from DsRed (Campbell et al., 2002). Pollen tubes expressing mRFP were the same as wild-type pollen tubes (Figure 2A), while pollen tubes transiently expressing LePRK1-mRFP exhibited the same morphological changes as LePRK1-eGFP (Supplemental Figure 2A). Figure 1F shows one pollen tube that formed ∼20 tandem blebs. Furthermore, pollen tubes coexpressing LePRK1 and eGFP on separate constructs also formed blebs (Supplemental Figure 2B), indicating that LePRK1 overexpression per se, and not the fluorescent protein fusion, was the causal factor for blebbing growth.
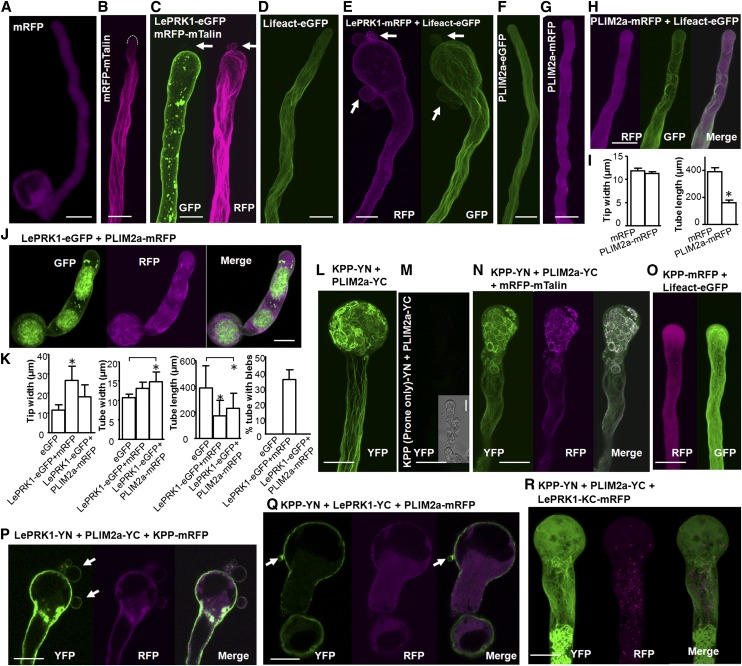
Figure 2.
Coexpression of PLIM2a Suppressed Pollen Tube Blebbing Caused by LePRK1 Overexpression, and Coexpression of KPP Restored Pollen Tube Blebbing.
(A) to (H), (J), and (L) to (R) Representative tobacco pollen tubes transiently expressing the denoted constructs or construct combinations. All genes were driven by the LAT52 promoter. Arrows point to blebs. The dotted line in (B) outlines the margin of the pollen tube tip. Bars = 20 μm.
(I) and (K) Measurements of pollen tubes in transient assays after 4 to 6 h of cultivation. Asterisks indicate a significant difference from the mRFP or eGFP control (P < 0.01, Student’s t test). n = 3 independent experiments. Error bars indicate se.
The C-Terminal Tail and Plasma Membrane Localization Are Required for LePRK1 to Cause Pollen Tube Blebbing
LePRK1 has an extracellular domain (ECD) containing six Leu-rich repeats, a single transmembrane domain, and a cytoplasmic portion composed of a juxtamembrane domain, a protein kinase domain, and a 29-amino acid tail at the C terminus (Figure 1J; Supplemental Figure 3A). To determine which portions of LePRK1 were required for the blebbing phenotype, we tested various truncated or point-mutated versions of LePRK1 by transiently expressing them in pollen tubes (Supplemental Figures 3A and 3B). Pollen tubes expressing a construct that lacked the extracellular domain (LePRK1ΔECD) also had enlarged tips and grew with blebs (Supplemental Figures 3C and 3D). The percentage of tubes with blebs was even higher than in pollen overexpressing full-length LePRK1 (Figure 1I). Tracking the growth of one pollen tube overexpressing LePRK1ΔECD clearly showed that a new bleb formed from a previously formed bleb (Figure 1H; Supplemental Movie 4), demonstrating that blebbing pollen tubes are still growing, extending their leading edges. Thus, the region of the protein responsible for blebbing was narrowed to the transmembrane and/or the cytoplasmic regions of LePRK1.
Overexpressing a construct lacking the kinase domain and the C-terminal tail (LePRK1ΔKC) yielded tubes with enlarged tips, but they lacked blebs (Supplemental Figure 3F), indicating that the kinase domain and/or C-terminal tail are required for blebbing. mLePRK1 is a kinase-dead version of LePRK1, in which the Lys residue in subdomain II of the conserved kinase domain was changed to Arg (K396R) (Muschietti et al., 1998). Overexpressing mLePRK1 resulted in enlarged tips with blebs (Supplemental Figure 3J), indicating that kinase activity was not required. Overexpressing a construct lacking the C-terminal tail (LePRK1ΔC) yielded significantly shorter tubes, which usually stopped growing and had no blebs (Supplemental Figure 3E). These results indicate that the C-terminal tail is required for blebbing. Note that the C-terminal tail is also required for the LePRK1–KPP interaction, as LePRK1ΔC did not interact with KPP in a bimolecular fluorescence complementation (BiFC) assay (Supplemental Figure 3K), whereas LePRK1 did (Figure 1E).
Expression of any of three truncated constructs lacking the extracellular domain and the transmembrane domain (LePRK1-JKC, LePRK1-JK, and LePRK1-KC) had the expected loss of plasma membrane localization and yielded tubular pollen tubes and no blebs (Supplemental Figures 3G and 3I). Therefore, we concluded that plasma membrane localization was required for blebbing. In line with this, we reevaluated pollen tubes expressing LePRK1-eGFP, LePRK1-mRFP, or LePRK1ΔECD-eGFP and noticed that, in blebbing pollen tubes, these proteins at least partially accumulated at the plasma membrane (Figures 1Cb and 1G), but blebbing was never observed in the absence of membrane-localized LePRK1 or LePRK1ΔECD.
Further examination of blebbing pollen tubes provided more information in understanding this phenotype. Judged by the plasma membrane–localized LePRK1-eGFP or LePRK1-mRFP (Figure 1; Supplemental Figures 1 to 3), these blebbing pollen tubes still have an intact plasma membrane. Furthermore, the blebs have cytoplasmic streams that are linked with the primary tube (Supplemental Movies 1 to 4). The large vacuoles, which are located distally (i.e., well away from the tip) in wild-type pollen, now were moving toward the proximal (near the tip) part of the tube (Figure 1C; Supplemental Movie 2). The images of pollen tubes overexpressing LePRK1 (e.g., the tube in Figures 1Cc and 1F) show that a number of tandem blebs (which presumably formed sequentially one by one from previously formed blebs) extended from the tube. Measured along/parallel to the longest backbone of the connecting cytoplasm/axis of growth, the most distal bleb was more than 150 μm away from the primary tube (which was ∼60 μm long). Thus, such pollen tubes also extend their leading edge by continuously producing blebs, a mode of growth quite different from that of wild-type tubes, which extend a tubular shape. Therefore, we consider this phenotype as a change in growth mode (from a normal tubular growth mode to an abnormal blebbing mode).
Tomato has six LePRKs (Tomato Genome Consortium, 2012); all except LePRK6 are expressed in pollen (Supplemental Figure 4A). LePRK1 and LePRK2 share 54% identity and 80% similarity at the amino acid level (Muschietti et al., 1998); LePRK3 is less similar (Kim et al., 2002). LePRK2 is highly phosphorylated in pollen membranes (Salem et al., 2011), and LePRK1 is also possibly phosphorylated (Supplemental Figure 5). Pollen tubes expressing LePRK3-eGFP under the control of the pollen-specific LAT52 promoter were similar to wild-type pollen tubes, while those expressing LePRK4-eGFP or LePRK5-eGFP had slightly swollen tips (Supplemental Figure 4B), similar to those of tubes overexpressing LePRK2. None of these pollen tubes formed blebs. Thus, the overexpression phenotype of LePRK1 is unique within this clade.
The Actin Bundling Protein PLIM2a Suppressed the Blebbing Phenotype Caused by LePRK1 Overexpression
To understand the mechanisms underlying the cellular morphological changes that result in bleb formation, we assessed downstream cellular events in pollen tubes transiently expressing LePRK1. Actin cytoskeleton dynamics underlie many pollen tube growth behaviors (Cheung and Wu, 2008; Staiger et al., 2010; Thomas, 2012); thus, components of the actin cytoskeleton are candidates for downstream effectors of LePRK1. Therefore, we used mTalin (Kost et al., 1998) or Lifeact (Vidali et al., 2009) to visualize F-actin structures in pollen tubes overexpressing LePRK1. Control tubes showed longitudinally oriented long actin bundles along the shank of the tube but not in the hemispherical tip region, with fine filaments close to the hemispherical tip region (Figures 2B and 2D). However, when LePRK1-eGFP or LePRK1-mRFP was expressed together with mTalin or Lifeact, thicker long actin bundles were visible in the tube shank, and they also extended and often bent into the ballooned tip area and even into the blebs (Figures 2C and 2E). We concluded that the accumulation of LePRK1 in growing pollen tubes promotes actin bundle formation.
LePRK1 lacks apparent actin binding motifs, so it was unclear how its overexpression might affect actin cytoskeleton dynamics. Therefore, we assessed several putative actin binding proteins from tomato that had been identified in yeast two-hybrid screens (Kaothien et al., 2005) as potential interactors of the LePRK1-LePRK2-KPP complex. Tomato ADF, a putative actin depolymerizing factor/cofilin protein (Solyc03g025750.2.1), was obtained with the LePRK1 cytoplasmic domain. Tomato δLIM2 (Solyc08g007940.2.1) and PLIM2a (Solyc01g094320.1.1; Supplemental Figure 6A) contain two LIM domains (Arnaud et al., 2007) and were obtained with the LePRK2 cytoplasmic domain and KPP, respectively. Both ADF and δLIM2 can interact with LePRK1 when transiently expressed in pollen tubes (Supplemental Figures 7A and 7B). Coexpression of ADF or δLIM2 with LePRK1-eGFP did not reduce the frequency of blebbing pollen tubes (Supplemental Figures 7F and 7G), but coexpression of PLIM2a-mRFP with LePRK1-eGFP completely suppressed the blebbing phenotype (Figure 2J); the pollen tubes appeared short and fat, and no blebs formed (Figures 2J and 2K).
The subcellular localization of LePRK1-eGFP also changed in tubes expressing both LePRK1-eGFP and PLIM2a-mRFP. In wild-type pollen (Muschietti et al., 1998) or when overexpressed, LePRK1 was mostly plasma membrane localized (Figures 1C, 1F, and 1G). By contrast, when coexpressed with PLIM2a, only a small amount of LePRK1-eGFP was plasma membrane localized and most was cytoplasmic, discontinuously constrained within capsule-like structures formed by short band-like PLIM2a (Figure 2J). This localization of PLIM2a was different from when it was expressed alone or with an F-actin marker (Figures 2F to 2H). There was little overlap between the subcellular localization positions of PLIM2a-mRFP and LePRK1-eGFP (Figure 2J). Consistent with this, coexpressing LePRK1 and PLIM2a in BiFC constructs yielded only a very weak signal, and these pollen tubes were also fat and short, with no blebs (Supplemental Figure 7C), like pollen tubes coexpressing PLIM2a-mRFP with LePRK1-eGFP.
Tomato PLIM2a belongs to the pollen-expressed PLIM2 clade of the LIM protein family (Supplemental Figure 6A). Tomato has three members in this clade, as does Arabidopsis thaliana (Papuga et al., 2010). Transient coexpression of PLIM2b-mRFP (Solyc10g017520.2.1) or PLIM2c-mRFP (Solyc05g049870.2.1) with LePRK1-eGFP in tobacco did not abolish pollen tube blebbing (Supplemental Figure 8), suggesting that the bleb suppression activity of PLIM2a was specific.
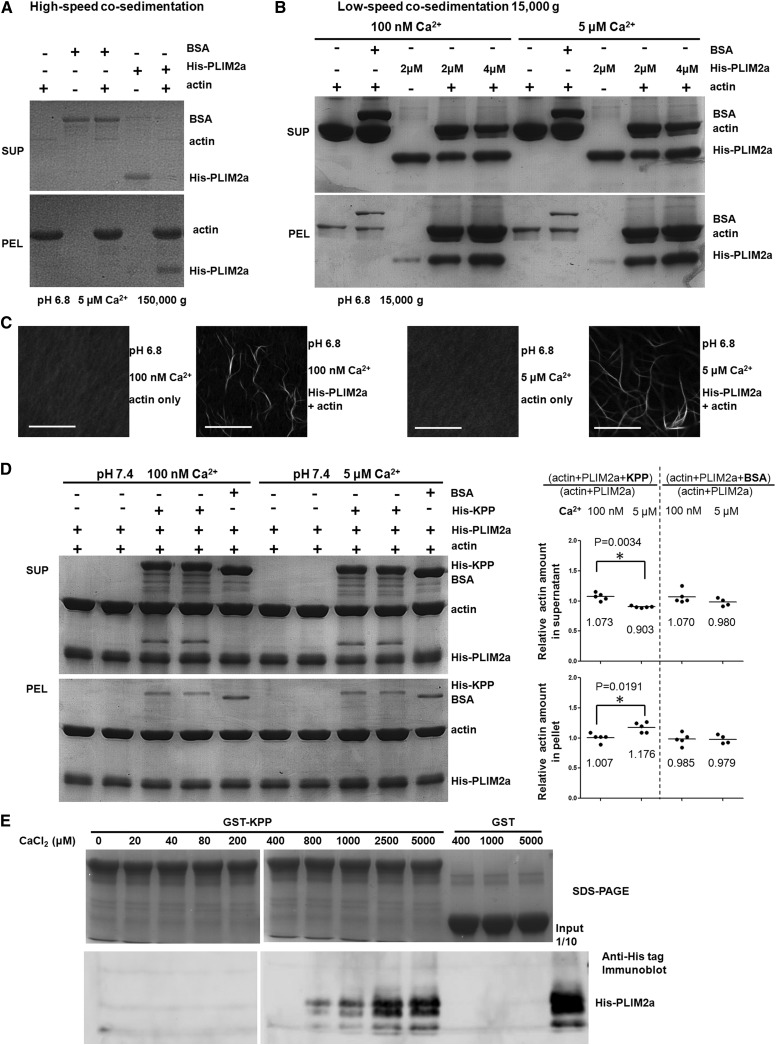
To determine whether tomato PLIM2a promotes actin bundling, as shown for its homologs in Arabidopsis (Papuga et al., 2010), we tested its actin binding and bundling activities in vitro. PLIM2a showed actin binding activity in a high-speed cosedimentation assay (Figure 3A). In a low-speed cosedimentation assay, most actin was detected in the supernatant fraction, but in the presence of PLIM2a, actin massively sedimented, indicating the presence of higher order actin structures (Figure 3B). The promotive effect on actin bundling of PLIM2a was verified by fluorescence microscopy after Alexa Fluor 488 phalloidin labeling (Figure 3C). The promotive effect of Arabidopsis PLIM2a on actin bundling was reported to be dependent on Ca2+ concentration and pH (Papuga et al., 2010). By contrast, tomato PLIM2a promoted actin bundling at both low and high Ca2+ concentrations, within a pH range of 6.2 to 7.4 (Supplemental Figure 9). Tomato PLIM2a belongs to the same clade as the tobacco pollen-expressed LIMs, Nt-PLIM2 (Eliasson et al., 2000) and Nt-PLIM2b (Cheung et al., 2008), and is slightly more similar to Nt-PLIM2. Transiently overexpressing PLIM2a-mRFP caused tobacco pollen tubes to grow slower (Figure 2I), similar to pollen tubes highly expressing Nt-PLIM2b (Cheung et al., 2008). These results show that tomato PLIM2a is an actin bundling protein.
Figure 3.
Tomato PLIM2a Promotes F-Actin Bundling in Vitro and Interacts with KPP.
(A) and (B) High-speed (A) and low-speed (B) cosedimentation assays for in vitro actin bundling. PEL, pellet fraction; SUP, supernatant fraction.
(C) Direct visualization of actin bundles induced by PLIM2a at pH 6.8. Bars = 20 μm.
(D) Low-speed cosedimentation assay for in vitro actin bundling of the KPP and PLIM2a combination at two different Ca2+ concentrations at pH 7.4. The left panel shows a representative experiment; the chart at right shows a statistical analysis of the relative amount of actin from five independent experiments.
(E) In vitro pull-down assays. His-PLIM2a can be coprecipitated with GST-KPP under high-CaCl2 concentrations.
KPP Links LePRK1 and PLIM2a and Restored Pollen Tube Blebbing
We then asked how PLIM2a was connected to LePRK1. The LePRK1–PLIM2a interaction, as visualized by BiFC, was very weak (Supplemental Figure 7C); furthermore, the BiFC signal was distributed only in the annular region of the shank membrane and in the cytoplasm around the tip (a region corresponding to the endocytosis zone; Supplemental Figure 7D). This region is also known as the alkaline band, where the pH is around 7.4, higher than other regions of the pollen tube, where the pH is around 6.8 (Hepler et al., 2013). Because LePRK1 can recruit KPP to the tip hemispherical plasma membrane (Figure 1E) and the KPP–PLIM2a interaction is obvious, as visualized by BiFC (Figure 2L; Supplemental Figure 6B) and in the yeast two-hybrid system (Supplemental Figure 6C), we hypothesized that PLIM2a might interact with LePRK1 indirectly through KPP. Consistent with this, when KPP-mRFP was coexpressed with LePRK1-YN and PLIM2a-YC, the BiFC signal of LePRK1 and PLIM2a was easily detectable at the apical plasma membrane (Figure 2P), and these tubes resumed blebbing (Figure 2P). This suggests that KPP and PLIM2a are functionally connected with LePRK1 in regulating blebbing.
The BiFC assay not only demonstrated an interaction between PLIM2a and KPP in transiently transformed tobacco pollen tubes but also showed an interesting localization pattern. Notably, the BiFC signals mimicked the distribution of longitudinal long actin bundles in the shank and the ring-like actin bundles at the ballooned tips (Figure 2L; Supplemental Figure 6B). As a control, we used the plant-specific ROP nucleotide exchanger domain of KPP, which did not interact with PLIM2a in BiFC (Figure 2M). mRFP-mTalin labels F-actin in tobacco pollen tubes (Figure 2B). The PLIM2a-KPP complex colocalized with F-actin (Figure 2N), which formed rings in the tip region and formed bundles in the tube shank, while the overall distribution of PLIM2a or KPP was diffuse in the cytoplasm when either coexpressed (Supplemental Figure 7I) or expressed individually (Figures 2F and 2G; Supplemental Figure 7H). In tubes overexpressing PLIM2a alone, actin was in only a few small half rings in the shank (Figure 2H), while when KPP was overexpressed alone, fine actin filaments formed all along the tube, with some aggregates in the tip region (Figure 2O). Therefore, the tip-localized actin rings (Figure 2L) required both PLIM2a and KPP.
The difference in the KPP-PLIM2a BiFC signal pattern between the tip and shank (rings versus long cables) led us to explore the influence of Ca2+ concentration on KPP-PLIM2a. Although PLIM2a promoted actin bundling at both low and high Ca2+ concentrations (Figures 3B to 3D), adding KPP to the in vitro bundling assay at pH 7.4 at high Ca2+ concentrations (5 μM, similar to the concentration of Ca2+ at the pollen tube tip; Hepler et al., 2013) slightly reduced the amount of actin in the supernatant (by 10%) and slightly increased the amount of actin in the pellet (by 18%), indicating a limited enhancement of PLIM2a-mediated actin bundling. At low Ca2+ concentrations (100 nM, similar to the concentration of Ca2+ in the basal pollen tube; Hepler et al., 2013), the addition of KPP had no effect on PLIM2a-mediated bundling (Figure 3D; Supplemental Figure 10). Moreover, in vitro binding assays showed that His-PLIM2a coprecipitated with GST-KPP when the Ca2+ concentration was 800 μM or higher (Figure 3E), indicating that the interaction between PLIM2a and KPP is Ca2+ dependent. Note that 800 μM is much higher than the normal cytosolic Ca2+ concentration in pollen tubes, although it is close to the extracellular Ca2+ concentration; therefore, it is not clear whether this PLIM2a-KPP in vitro binding requirement reflects the in vivo situation.
Interactions between LePRK1 and Its Various Binding Partners Affect the Pollen Tube Blebbing Phenotype
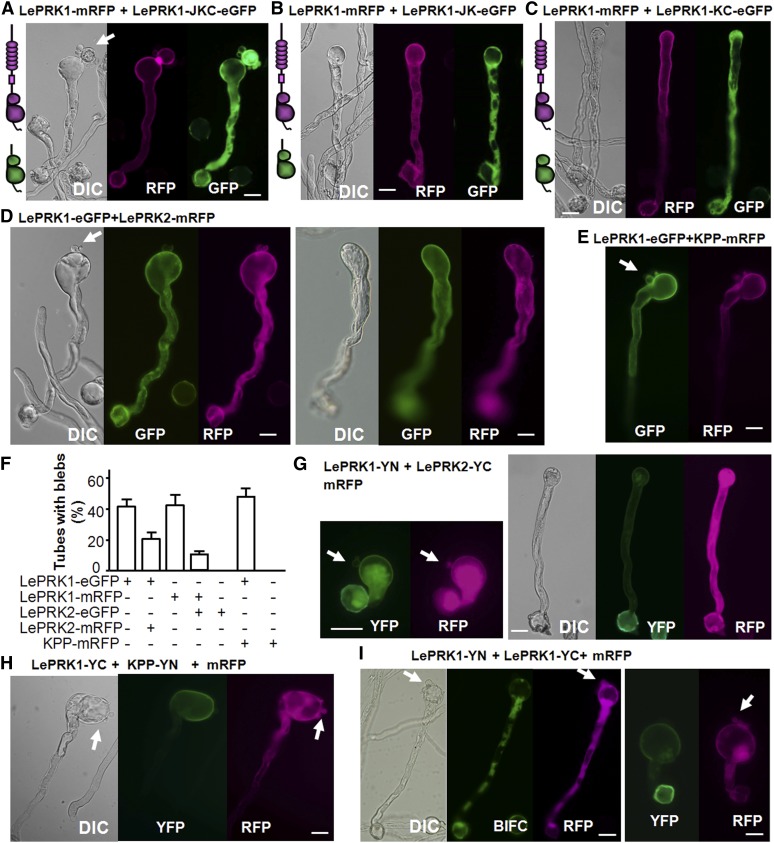
We further tested our hypothesis that overexpressing LePRK1 switched the pollen tube growth mode via KPP and PLIM2a action by observing the growth mode after perturbing interactions between LePRK1, KPP, or PLIM2a. A truncated version of LePRK1, LePRK1-JKC (LePRK1 lacking its ECD and transmembrane domain), interacted with LePRK1 (Supplemental Figure 11B) and KPP (Supplemental Figure 11J) and so should not be able to sequester PLIM2a from interacting with the apical membrane–localized LePRK1, and indeed the blebbing phenotype was retained (Figure 4A; Supplemental Figure 11B). By contrast, another truncated version of LePRK1, LePRK1-JK (LePRK1 lacking its ECD, transmembrane domain, and C-terminal domain; Supplemental Figure 3A), was not able to interact with full-length LePRK1 or KPP (Supplemental Figures 11C and 11K) but retained the ability to interact with PLIM2a (Supplemental Figure 11O). Yet another truncated version of LePRK1, LePRK1-KC (LePRK1 lacking its ECD, transmembrane domain, and juxtamembrane domain; Supplemental Figure 3A), also was not able to interact with full-length LePRK1 (Supplemental Figure 11D) but retained the ability to interact with KPP and PLIM2a (Supplemental Figures 11L and 11P). Consistent with the notion that cytoplasmic molecules would sequester KPP or PLIM2a, thereby reducing their interactions with apical membrane–localized LePRK1 and thus abolishing blebbing, when LePRK1-KC or LePRK1-JK was coexpressed with full-length LePRK1 (Figures 4B and 4C; Supplemental Figures 11C and 11D) the blebbing phenotype was abolished. We explored a bit further to understand how LePRK1-KC might modulate blebbing through KPP and PLIM2a. Figure 2R shows that, when coexpressed with LePRK1-KC-mRFP, the KPP-PLIM2a complex (as visualized by the BiFC signal) mostly localized in the cytoplasm of the pollen tube shank, not in the tip region. In comparison with Figure 2L and Supplemental Figure 6B, this result indicates that cytoplasmic LePRK1-KC caused the KPP-PLIM2a complex to retreat from the pollen tube tip region, inhibiting the blebbing.
Figure 4.
Coexpressing LePRK1-JK, LePRK1-KC, or LePRK2 with LePRK1 Alleviated the LePRK1 Overexpression Phenotype.
(A) to (E) Representative pollen tubes coexpressing the denoted eGFP and mRFP fusion combinations.
(F) Measurements of pollen tubes with blebs.
(G) to (I) Representative pollen tubes coexpressing BiFC constructs with mRFP.
Arrows indicate blebs at the tip. All genes were expressed transiently in tobacco pollen tubes and driven by the LAT52 promoter. DIC, differential interference contrast. Bars = 30 μm.
LePRK2 and KPP are both binding partners of LePRK1 at the pollen tube plasma membrane (Wengier et al., 2003; Kaothien et al., 2005), but their influences on the percentage of pollen tube blebbing caused by LePRK1 overexpression are different. In transient expression experiments, coexpressing LePRK2 with LePRK1 significantly reduced the percentage of pollen tube blebbing (Figures 4D and 4F), while coexpression of KPP with LePRK1 did not change the percentage of pollen tube blebbing (Figures 4E and 4F).
Interactions among LePRK1 and some of its binding partners not only affected the blebbing phenotype but also the localization preference of LePRK1 between basal tubes and newly formed blebs. It is noteworthy that LePRK1 and PLIM2a, when coexpressed with KPP and visualized by BiFC, localized in the plasma membrane of the enlarged tip, particularly in “dents” (i.e., the deformed plasma membrane) (Figure 2P). In transient expression experiments, LePRK1-eGFP or LePRK1-mRFP, when expressed alone, was plasma membrane localized in both the basal tube tip and blebs (Figures 1C, 1F, and 1G), while the LePRK1-KPP BiFC signal was very strong in the basal tube tip but very weak in blebs (Figures 1C and 4H), suggesting a preference of the LePRK1-KPP complex for the basal tube plasma membrane. Similarly, the BiFC signal of LePRK1-YN and LePRK1-YC was strong in the basal tube tip but not visible in blebs (Figure 4I), suggesting that LePRK1 homodimers are depleted from blebs and perhaps that LePRK1 monomers are present in blebs. By contrast, the similar intensities of the LePRK1-LePRK2 BiFC signal (Figure 4G) suggest that the LePRK1-LePRK2 complex is present on plasma membranes of both the basal tube and newly formed bleb. Mechanically, extension by blebbing should require that the plasma membranes of the basal tube and forming bleb behave differently (i.e., the basal tube membrane does not expand whereas the bleb membrane does). The different localization preferences among different LePRK1 complexes might be related to this difference in membrane behavior.
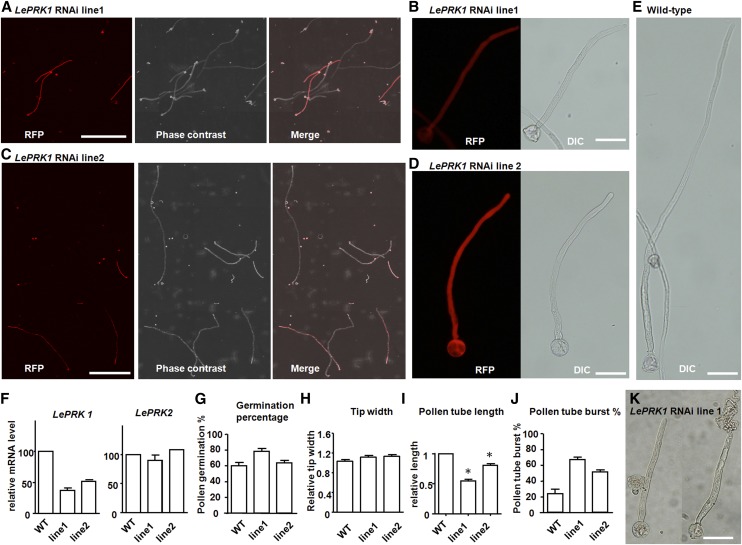
In Transgenic Tomato Plants, LePRK1 RNAi Pollen Tubes Grew Slower and LePRK1 Overexpressing Pollen Arrested Early in Tube Formation
In addition to addressing LePRK1 function by transient assays, we generated transgenic tomato plants carrying an inverted repeat sequence against LePRK1 driven by the pollen-specific LAT52 promoter and a separate mRFP gene also driven by pLAT52. Two independent transgenic lines of LePRK1 RNAi had ∼50% RFP-expressing pollen (Figures 5A and 5C). Figure 5F shows that the expression level of LePRK1 was reduced in both lines, while the expression of LePRK2 was not. Mature pollen of these LePRK1 RNAi lines appeared normal, and when cultured in vitro, they had a germination percentage similar to that of the wild type (Figure 5G). The morphology of the pollen tubes (Figures 5B, 5D, and 5H) was similar to that of the wild type (Figure 5E). However, compared with nonfluorescent wild-type tubes in the same culture, the LePRK1 RNAi pollen tubes (mRFP expressing) were significantly shorter (Figures 5A, 5C, and 5I). Time-lapse microscopy showed that LePRK1 RNAi pollen tubes grew ∼3-fold slower than wild-type pollen tubes (Supplemental Movies 5 and 6). In addition, LePRK1 RNAi pollen tubes burst more often (Figure 5J; Supplemental Movie 7). We concluded that LePRK1 plays a role in normal pollen tube growth to support high-speed growth and to maintain tube integrity.
Figure 5.
Transgenic LePRK1 RNAi Pollen Germinates Normally but Grows Shorter Tubes Than Wild-Type Pollen.
(A) to (D) Representative images of pollen tubes from transgenic tomato expressing LePRK1 RNAi and RFP constructs.
(E) A representative wild-type tomato pollen tube.
Photographs in (A) and (C) were taken after 4 h of in vitro germination. Note that the plants are heterozygotes, containing around 50% LePRK1 RNAi pollen tubes expressing RFP and 50% wild-type pollen tubes not expressing RFP. DIC, differential interference contrast. White bars = 500 μm; black bars = 50 μm.
(F) Quantitative RT-PCR of LePRK1 and LePRK2 mRNA levels using total RNA of mature pollen as template. n = 2 independent experiments.
(G) to (J) Measurements of LePRK1 RNAi pollen tubes and wild-type pollen tubes after 4 h of in vitro germination. Asterisks indicate significant differences from the wild-type control (P < 0.01, Student’s t test). n = 3 independent experiments. Error bars indicate se.
(K) A live pollen tube (left) and a burst pollen tube (right).
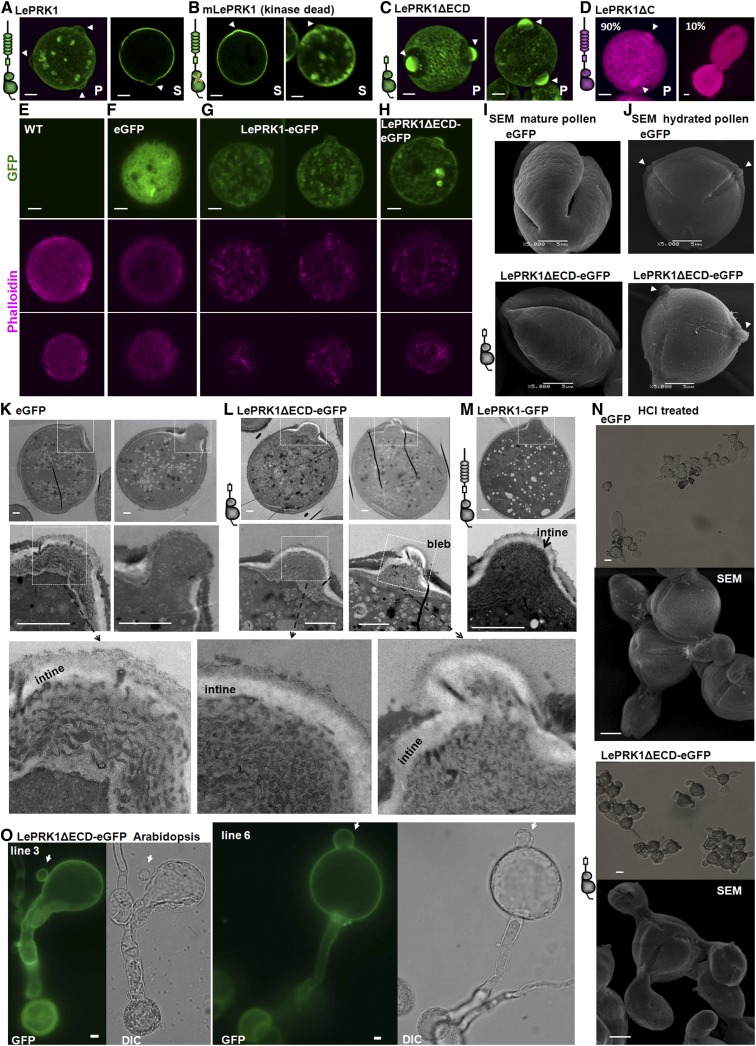
We also generated transgenic tomato plants expressing pLAT52-LePRK1-eGFP. Two independent transgenic lines overexpressing LePRK1 had mature pollen that appeared normal, of which 50% expressed LePRK1-GFP. Unlike the pollen expressing GFP only, which could grow pollen tubes normally in vitro (Supplemental Figure 12A), the LePRK1-GFP–expressing pollen hydrated but at most had hemispherical protrusions at the potential germination pores and never extended a tube (Figure 6A; Supplemental Figure 12B); the protrusions were no longer than 3 μm (about one-tenth of the diameter of a normal hydrated tomato pollen grain). Supplemental Movie 8 shows that the LePRK1-GFP–expressing pollen also exhibited cytosolic vesicle movement, indicating viability. In the protrusions, LePRK1-eGFP was localized mainly in the plasma membrane. Consistent with the germination-failure phenotype, the ratio of kanamycin resistance to sensitivity in the T1 progeny was ∼1:1, and when heterozygous plants were crossed as males with the wild type, none of the resulting seeds were kanamycin resistant, indicating a block in male transmission.
Figure 6.
Transgenic Pollen Overexpressing Wild-Type or Mutated Versions of LePRK1 Have Defects in Extending a Pollen Tube.
(A) to (N) show tomato pollen tubes, and (O) shows Arabidopsis pollen tubes.
(A) to (D) Representative images of transgenic pollen expressing LePRK1-eGFP (A), kinase-dead mLePRK1-eGFP (B), LePRK1ΔECD-eGFP (C), or LePRK1ΔC-mRFP (D). P, projection of a series of confocal slices, S, single confocal section.
(E) to (H) Actin structures, visualized by Alexa Fluor 568 (red) phalloidin, in hydrated tomato pollen of the wild type (E) or that stably expressing eGFP (F), LePRK1-eGFP (G), or LePRK1ΔECD-eGFP (H). GFP is shown in the top panels and phalloidin staining in the middle and bottom panels. The middle panels show single confocal slices at the equatorial plane, and the bottom panels show single confocal slices at a plane near the periphery.
(I) and (J) Representative scanning electron microscopy images of mature pollen (I) or hydrated pollen (J) expressing eGFP alone or LePRK1ΔECD-eGFP as indicated.
(K) to (M) Representative transmission electron microscopy images of pollen expressing eGFP (K), LePRK1ΔECD-eGFP (L), and LePRK1-eGFP (M). LePRK1-eGFP pollen grains were isolated by fluorescence-activated cell sorting.
(N) HCl treatment caused protrusion from all three germination pores in transgenic pollen expressing eGFP or LePRK1ΔECD-eGFP.
(O) Representative pollen tubes from Arabidopsis transgenic lines expressing LePRK1ΔECD-eGFP. DIC, differential interference contrast.
Arrowheads point to germination pores, and arrows point to blebs. Diagrams of LePRK1 and its mutated versions are included at the left of relevant panels to facilitate understanding. Bars in (A) to (J), (N), and (O) = 5 μm; bars in (K) to (M) = 2 μm.
To further dissect which region was responsible for the pollen germination defect, we also generated transgenic tomato plants with the pLAT52-mLePRK1-eGFP, pLAT52-LePRK1ΔECD-eGFP, or pLAT52-LePRK1ΔC-mRFP construct. All these plants had mature pollen that appeared normal, of which 50% expressed GFP or RFP. Transgenic pollen expressing mLePRK1-eGFP (from two independent transgenic events) also did not germinate (Figure 6B). Approximately 10% of the transgenic pollen expressing LePRK1ΔC-mRFP (from four independent transgenic events) germinated, and these grew very short and fat tubes (Figure 6D). Because the transgenic pollen did not form normal tubes, it was not surprising that homozygotes were not obtained from these transgenic plants.
Transgenic pollen expressing LePRK1ΔECD-eGFP (from five independent transgenic events) all appeared to hydrate normally with the three apertures (i.e., germination pores) exposed (Figure 6J). The GFP-expressing pollen from four of these lines never germinated (Figure 6C; Supplemental Figure 12C). Homozygotes were not obtained from these four lines. In the fifth line, although the expression level (as judged by GFP) in almost all of the pollen was similar to that in the four lines whose pollen did not germinate, ∼1% of the pollen grains with very weak GFP did germinate (Supplemental Figure 12C). These tubes had membrane fluorescence only at the shank, near the grain (Supplemental Figure 12C). A homozygote was obtained from this line. In both the T1 and T2 generations from this homozygote, >95% of the pollen expressing LePRK1ΔECD-eGFP at similar levels did not germinate, but 2.6% of the pollen (weakly expressing LePRK1ΔECD-eGFP) did germinate. Except for this rare LePRK1ΔECD-eGFP progeny from line 5, the constructs that caused germination failure in stably transformed pollen are those that caused pollen tube blebbing in transient assays (LePRK1, mLePRK1, and LePRK1ΔECD); the LePRK1ΔC construct that allowed some pollen germination in stably transformed pollen did not cause pollen tube blebbing in the transient assay. Nevertheless, this rare LePRK1ΔECD-eGFP homozygote provided an opportunity to examine the pollen by electron microscopy, without worrying about the presence of 50% wild-type pollen in heterozygous plants.
LePRK1ΔECD-eGFP in transgenic pollen preferentially localized at the plasma membrane region of the germination pore, forming a “cap” (Figure 6C). Scanning electron microscopy showed that the surface of the hydrated LePRK1ΔECD-eGFP pollen was similar to that of wild-type pollen (Figures 6I and 6J). Transmission electron microscopy showed that the intine, which covers the germination pore, was thicker in homozygous transgenic LePRK1ΔECD-eGFP pollen than in wild-type pollen (Figures 6K and 6L) and was occasionally deformed (formed a tiny bleb; Figure 6L). Pollen expressing LePRK1-eGFP, which were harvested by fluorescence-activated cell sorting from heterozygous LePRK1-eGFP transgenic plants, also had a thickened intine (Figure 6M). The intine is a pectin-enriched cell wall layer (Suarez-Cervera et al., 2002; Castro et al., 2013). Treating mature tomato pollen with 1 mol/L hydrochloric acid, a treatment often used in extracting pectin, caused tube-like protrusions (up to 20 μm in length; Figure 6N). Treating transgenic LePRK1ΔECD-eGFP pollen with 1 mol/L hydrochloric acid caused similar protrusions (Figure 6N [note that these protrusions are far longer than the protrusions that transgenic LePRK1ΔECD-eGFP pollen or LePRK1-eGFP pollen make in pollen germination medium]; compare with Figures 6A to 6C and 6J). These results suggest that LePRK1ΔECD-overexpressing pollen grains hold the potential to protrude short tubes. We think it most likely that the germination pore–localized LePRK1ΔECD locked (i.e., prevented) pollen tube protrusion.
Transient overexpression of LePRK1 promoted actin bundle formation (Figures 2C and 2E), so we also observed F-actin decoration (labeled by phalloidin) in hydrated tomato pollen grains. Figures 6E and 6F show that fine actin filaments were distributed throughout wild-type pollen grains, with preferential accumulation at the periphery beneath the plasma membrane (cortical region) and slight depletion in the center. By contrast, actin filaments in both LePRK1 and LePRK1ΔECD transgenic pollen were severely thickened and formed many short bundles (Figures 6G and 6H). As these distribution patterns differed from those in the wild type, we concluded that LePRK1 also promotes actin bundle formation in pollen grains. Similar actin behaviors were found in the Arabidopsis fimbrin5 mutant; FIMBRIN5 encodes a pollen-expressed actin bundling factor (Wu et al., 2010).
Although the promotion of actin bundles was similar in both transient and stable transformed pollen tubes, we still did not know whether the blebbing phenotype was only produced in transient assays. Therefore, we transformed LePRK1ΔECD-eGFP, which caused a high percentage of pollen tube blebbing, into another plant species, Arabidopsis, using the LAT52 promoter, which also functions in Arabidopsis (Twell et al., 1990). We obtained six independent Arabidopsis transgenic lines (Supplemental Figures 13A and 13E) expressing LePRK1ΔECD-eGFP in pollen, with differing levels of GFP expression. In each line, ∼50% of the pollen expressed GFP, so they were likely heterozygotes. In lines 1 and 2, the GFP signal was weak and the pollen tubes appeared normal (Supplemental Figure 13C), similar to wild-type Arabidopsis pollen tubes growing in vitro (Supplemental Figures 13B and 13F). In line 3, the GFP signal was stronger and the germination percentage of the GFP-positive pollen grains was reduced to 25% (Supplemental Figure 13D). Of those that did germinate, ∼50% had wider tips, and of these, ∼10% formed blebs (Figure 6O; Supplemental Figure 13C). In line 4, with the strongest GFP expression, the GFP-positive grains did not germinate (Supplemental Figure 13C). In lines 5 and 6, 20 to 30% of the GFP-positive pollen tubes formed blebs (Supplemental Figures 13G and 13H). This result indicates that LePRK1ΔECD also confers a pollen tube blebbing phenotype to Arabidopsis.
DISCUSSION
Pollen tubes are fast-growing cells that continuously extend at their leading edge; the tubular cell shape is conserved throughout angiosperms (Rounds and Bezanilla, 2013). Pollen tubes of LePRK1 RNAi tomato lines grew slower and burst more often (Figure 5), indicating that LePRK1 is a component of the normal pollen tube growth machinery and probably functions in pollen tube integrity. The tomato pollen stably overexpressing LePRK1 arrested at a very early stage in tube extension, likely due to the globular shape of pollen being maintained too strongly. Notably, mere transient overexpression of LePRK1, or a portion of it, namely LePRK1ΔECD, enables a pollen tube to switch to a different method of advancing its leading edge, by forming blebs (Figures 1C and 1F). This is a distinct growth mode (Figure 1K) not previously reported in pollen tubes. Given that the natural variation of individual gene expression can be large among pollen grains with the same genotype (as we estimated by measuring LePRK1ΔECD-GFP fluorescence variation among individual pollen grains from the same flower; Supplemental Figure 14), it is possible that in some pollen grains the native LePRK1 expression is much higher than the average level and blebbing could occur. Our work might have exposed a hidden potential of pollen tube cells. However, blebbing growth occurs in other organisms, such as the primordial germ cells of zebrafish and Drosophila, which use blebs to migrate (Jaglarz and Howard, 1995; Blaser et al. 2006), and Dictyostelium cells, which produce blebs to migrate when exposed to a chemoattractant (Zatulovskiy et al., 2014). Recent studies of these blebbing cells indicate a nonsteady actin behavior, called an actin traveling wave, that is responsible for cell motility (Allard and Mogilner, 2013). The mechanism underlying blebbing growth among these evolutionarily diverged cells converges in some aspects, including bundling actin from plasma membranes of the leading edge, which might be an important principle for cell blebbing.
Using in vitro pull-down, yeast two-hybrid, in vivo BiFC, and pollen tube coexpression assays, this work provides a solid linkage between a plasma membrane–localized RLK and actin filaments (LePRK1-KPP-PLIM2a-actin bundle), and because KPP is a RopGEF, it also provides a specific link between the Rop small GTPase molecular switch and actin filaments. Actin bundles often play important roles in cell morphogenesis (Smith, 2003; Thomas, 2012). PLIM2a belongs to the actin bundling LIM protein family; several members of this family, including Arabidopsis PLIMs and Lilium longiflorum LIM1, have been reported to play roles in supporting pollen tube elongation (Wang et al., 2008; Papuga et al., 2010; Staiger et al., 2010; Ye et al., 2013). In addition, a tobacco PLIM2 can form ring-like structures when highly overexpressed in pollen tubes (Cheung et al., 2008). Because the LePRK1-KPP complex preferentially localized at the tip plasma membrane, and the tip cell wall is less rigid, anchoring actin bundles to the tip provides a possibility for local membrane deformation and, therefore, a way to change the morphology of this tip-growing cell. Actin bundles have been reported to link with vacuoles through vacuolar H+-ATPase subunits (Ma et al., 2012), which might account for vacuole movement toward the tip in blebbing pollen tubes.
Blebbing growth is still polar growth, and the ROP module, the conserved core in regulating cell polarity, is still functioning. LePRK1 might be a regulatory link that modulates pollen tube morphology by affecting the ROP module through the RopGEF KPP (Figure 2). In line with the idea that biological systems are built from adaptable core modules, which allow changes in regulatory linkages to functionally vary morphological outputs (Wedlich-Soldner and Li, 2008), it is feasible that manipulation of the LePRK1 expression level, as a regulatory linkage, might cause a growing pollen tube to produce specific morphogenetic changes, whereas changes to ROP, at the core of cell polarity, often leads to growth arrest (Figure 1K) (Lin and Yang, 1997; Hwang et al., 2010).
Overexpressing a single endogenous molecule, LePRK1, changed the growth mode from tubular to blebbing, indicating that LePRK1’s function in the complex machinery of pollen tube growth is special. The blebbing growth phenotype was not seen when other members of the LePRK1 clade were overexpressed. For example, overexpressing LePRK2 caused pollen tube tip swelling and sometimes hockey stick–like tubes (Salem et al., 2011), and overexpressing LePRK3, LePRK4, or LePRK5 caused only slight swelling of the tip (Supplemental Figure 5). Among the five homologs in Arabidopsis, PRK2 and PRK3 overexpression in tobacco pollen tubes caused swollen tips, while PRK1 overexpression did not cause tip swelling but promoted pollen tube growth (Chang et al., 2013). Overexpression of tobacco PRK2, which is the closest tobacco homolog of LePRK1, caused bifurcated pollen tubes (Zou et al., 2011). Although overexpressing At-PRKs did not cause pollen tube blebbing, expressing a portion of LePRK1 (i.e., LePRK1ΔECD) in Arabidopsis pollen caused pollen tube blebbing (Figure 6O; Supplemental Figure 11). These results indicate that LePRK1ΔECD is capable of enabling a pollen tube growth mode transition from tubular to blebbing in different plant species, including tomato, tobacco, and Arabidopsis.
METHODS
Plant Material and in Vitro Pollen Germination
Transgenic tomato (Solanum lycopersicum cv VF36) lines were generated as described (McCormick, 1991). For overexpression of LePRK1 and its variants, target fragments fused with eGFP (from pPK100; Blanvillain et al., 2011) or mRFP (from pMT-mRFP1; Toews et al., 2004) and driven by the LAT52 promoter were inserted into pCAMBIA2300, then Agrobacterium tumefaciens LBA4404 (Hoekema et al., 1983) carrying these plasmids was used to transform tomato. For RNAi transgenic lines, an intron-spliced hairpin RNA construct against LePRK1 cDNA was generated according to Wesley et al. (2001). The RNAi construct of LePRK1 was driven by the LAT52 promoter and terminated by the cauliflower mosaic virus 35S terminator. The inverted repeat sequences against the first 500 bp of the LePRK1 cDNA was spaced by the LAT52 intron. All the primers and cloning sites are provided in Supplemental Tables 1 and 2.
Tomato and tobacco (Nicotiana tabacum cv Gexin No. 1) were grown under standard greenhouse conditions. Mature pollen grains were collected and germinated as described (Zhang et al., 2008) with minor modifications. Freshly collected mature pollen grains was obtained by vibrating anthers of open flowers with a biovortexer (BioSpec Products) and then germinated in vitro in pollen germination medium [20 mM MES, pH 6.0, 3 mM Ca(NO3)2, 1 mM KCl, 0.8 mM MgSO4, 1.6 mM boric acid, 2.5% (w/v) Suc, and 24% (w/v) polyethylene glycol, molecular weight 3350] at 25°C on 6- or 24-well plates rotated horizontally at 60 rpm (tomato) or 150 rpm (tobacco).
Transgenic Arabidopsis thaliana Columbia-0 lines were generated as described (Clough and Bent, 1998). The same construct expressing LePRK1ΔECD-eGFP driven by the LAT52 promoter used in tomato transformation was used to transform Arabidopsis using Agrobacterium strain GV3101. Transgenic lines were selected based on kanamycin resistance and mature pollen fluorescence. For in vitro pollen germination, fully opened Arabidopsis flowers of the T2 generation were collected in the morning and dipped into liquid pollen germination medium, pH 7.1, to release pollen, which was then cultured for germination for 4 to 7 h at 22°C.
Pollen Bombardment Assay and Imaging Analysis
The pollen-specific LAT52 promoter (Twell et al., 1990) was used in all bombardment assays, unless specified otherwise. Pollen bombardment was performed as described (Zhang et al., 2008) using 10 μg of plasmid for each bombardment. For cobombardment, 5 μg of each construct was used. After 4 h of incubation, bombarded pollen tubes were observed and images were captured using an Olympus BX51 microscope fitted with an Olympus DP71 digital camera or with a confocal microscope (Olympus Fluoview FV1000). The bombardment experiments for each construct or combination of constructs were performed at least three independent times, and each time at least 20 fluorescent pollen tubes were observed; representative images for each phenotype are shown. For each experiment, pollen tube lengths, pollen tube tip widths, and the fluorescence intensities of at least 20 pollen tubes in each category were measured using ImageJ (Schneider et al., 2012) and Image pro plus 7.0 software.
The BiFC constructs were generated according to Bracha-Drori et al. (2004) with minor modifications. Briefly, the yellow fluorescent protein (YFP) was split between residues 172 and 173 into two nonoverlapping N-terminal (YN) and C-terminal (YC) fragments. Genes of interest were cloned in frame either upstream or downstream of YC or YN. The sequence encoding LeROP was amplified by PCR from tomato pollen cDNA, and QuikChange (Stratagene) PCR-based mutagenesis was used to create mutant sequences encoding CA-LeROP (G15V) as described by Löcke et al. (2010). After sequence confirmation, mutated LeROP cDNAs were inserted in frame at the 3′ end of the eGFP coding sequence.
RNA Extraction and Quantitative RT-PCR
Total RNA from mature pollen was extracted using RNAiso plus (TaKaRa) according to the manufacturer’s protocol. cDNA was generated by M-MLV (TaKaRa). Quantitative PCR was performed using SYBR Green on an iCycler (Bio-Rad). The primers used to amplify fragments of LePRK1, LePRK2, and a tomato ACTIN gene were synthesized as described (Zhang et al., 2008).
F-Actin Staining
The actin cytoskeleton was visualized as described (Wu et al., 2010) with slight modifications. Briefly, pollen grains were fixed with 300 μM 3-maleimidobenzoic acid N-hydroxysuccinimide ester in pollen germination medium. The pollen grains were subsequently extracted with 0.05% Nonidet P-40 in germination medium for 10 min. Fixed pollen grains were then rinsed three times in 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 400 mM Suc containing 0.05% Nonidet P-40 for 10 min each and then stained overnight at 4°C with 200 nM Alexa Fluor 568 phalloidin (Molecular Probes) in 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 400 mM Suc containing 0.05% Nonidet P-40.
Scanning Electron Microscopy
Pollen was fixed in 50% ethanol, 5% acetic acid, and 3.7% formaldehyde for more than 2 h at 4°C. Subsequently, the samples were dehydrated in a graded ethanol series (60, 70, 80, 90, 95, and 100% [twice]) for at least 5 min and not more than 10 min each. Dehydrated samples were desiccated with a critical point drying machine (JCPD-5; JEOL), fixed in stages for conductive coating, and viewed with a scanning electron microscope (JSM-6360LV; JEOL).
In the HCl treatment, pollen was treated with 1 M HCl for 5 s, harvested by centrifugation (100g, 3 min), and then fixed with 50% ethanol, 5% acetic acid, and 3.7% formaldehyde for observation by differential interference contrast or scanning electron microscopy.
Transmission Electron Microscopy
Mature pollen or pollen cultured for different times was fixed in 2.5% (v/v) glutaraldehyde in 50 mM phosphate buffer, pH 7.2, for no more than 4 weeks at 4°C. Thereafter, the samples were rinsed thoroughly with 50 mM phosphate buffer and later postfixed with 1% (w/v) osmium tetroxide in 50 mM phosphate buffer, pH 7.2, for 2 h at 4°C. Subsequently, all samples were dehydrated in a graded ethanol series and then embedded in Epon 812 resin. Ultrathin sections of the samples were cut with a diamond knife, collected on copper grids, and viewed with a transmission electron microscope (Hitachi H-7650).
Expression and Purification of Recombinant PLIM2a
GST-tagged and His-tagged tomato PLIM2a and KPP were expressed in Rosetta bacteria and purified using Glutathione Sepharose 4B (GE Healthcare) and Ni-NTA (Qiagen) following procedures described by the respective manufacturers. Purified proteins were concentrated with a centrifugal filter (Amicon), buffer-exchanged into actin bundling buffer or in vitro pull-down buffer using a 10,000 molecular weight cutoff dialysis cassette (Amicon), and stored on ice. Proteins were preclarified at 150,000g, their concentration were determined with UV spectrophotometry, and they were checked for molecular weight by SDS-PAGE analysis.
Yeast Two-Hybrid Assay
The Matchmaker GAL4 Two-Hybrid System (Clontech) was used. Sl-PLIM2a was inserted into pGADT7 and KPP was inserted into pGBKT7. Saccharomyces cerevisiae AH109 was used as the host strain. Cotransformation was confirmed on minimal medium lacking Leu and Trp, and interactions were assayed on minimal medium lacking Leu, Trp, adenine, and His. Colonies were photographed after 2 weeks.
In Vitro Binding Assays
In vitro binding assays were performed as described (Tang et al., 2004) except that indicated concentrations of CaCl2 were added to the coimmunoprecipitation buffer, pH 7.5. GST or GST-KPP (each ∼100 pmol) were inputs; His-PLIM2a (∼100 pmol) was added to the coimmunoprecipitation buffer. The pulled down His-PLIM2a was detected by anti-His tag antibody (CWBiotech).
In Vitro Actin Binding and Bundling Assays
The actin binding and actin bundling activities of His-PLIM2a were assessed according to Papuga et al. (2010), with modifications, following procedures described by the manufacturer. In both cases, nonmuscle actin (Cytoskeleton) was polymerized for 0.5 h in 50 mM KCl, 2 mM MgCl2, 1 mM ATP, and 0.5 mM DTT and then incubated with different protein combinations for 30 min. The reaction medium was buffered with 7 mM MES and 10 mM PIPES, pH 6.2, or 7 mM PIPES and 10 mM Tris, pH 6.8 and 7.4, and was supplemented with either EGTA (low-[Ca2+] conditions) or CaCl2 (high-[Ca2+] conditions).
In actin binding experiments, samples were centrifuged at 150,000g for 1.5 h at 24°C to pellet actin filaments. The presence of the corresponding proteins in the resulting supernatants (F-actin–unbound fraction) and pellets (F-actin–bound fraction) was analyzed by SDS-PAGE and Coomassie Brilliant Blue R 250 staining.
In actin bundling experiments, samples were centrifuged at 15,000g for 1.5 h at 24°C to pellet higher order F-actin structures. The presence of actin in the resulting supernatants (non–cross-linked AFs) and pellets (cross-linked AFs) was analyzed by SDS-PAGE and Coomassie Brilliant Blue R 250 staining. The presence of actin bundles in samples was also checked by direct visualization using fluorescence microscopy. An aliquot of the copolymerized actin samples was labeled with 4 μM Alexa Fluor 488 phalloidin (Invitrogen).
Two-Dimensional Gel Electrophoresis
Proteins and membrane fractions were isolated from mature tomato pollen as described (Muschietti et al., 1998). Two-dimensional gel electrophoresis and immunoblotting with an anti-LePRK1 antibody was as described (Salem et al., 2011).
Phylogenetic Analysis
LePRK protein sequences were used to identify Arabidopsis, maize (Zea mays), rice (Oryza sativa), and tomato orthologs by BLAST against the National Center for Biotechnology Information database and the Solanaceae Genomics Network (http://solgenomics.net/). Protein sequences were initially aligned using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (Chenna et al., 2003). Alignment documents (Supplemental Data Sets 1 and 2) downloaded from the website were loaded into MEGA5 (Tamura et al., 2011) to construct a neighbor-joining tree using the Jones-Taylor-Thornton model (MEGA5; Tamura et al., 2011), calculated by a bootstrap method (1000 replicates). The PLIM2a phylogenetic tree was generated using the same method (Supplemental Data Set 3).
Accession Numbers
GenBank accession numbers of the genes used in this article are as follows: U58474 for LePRK1 (Solyc05g047570.2.1), U58473 for LePRK2 (Solyc07g017230.2.1), NM_001247554.1 for LePRK3 (Solyc05g025780.2.1), XM_004251647.1 for LePRK4 (Solyc12g009190.1.1), XM_004236321.1 for LePRK5 (Solyc03g124050.2.1), XM_004245387.1 for LePRK6 (Solyc08g069170.1.1), AY730762 for KPP (Solyc03g120650.1.1), XM_004233517.1 for Sl-ROP (Solyc02g062020.1.1), KF387649 for PLIM2a (Solyc01g094320.1.1), XM_004244558.1 for Sl-δLIM2 (Solyc08g007940.1.1), XM_004234403.1 for Sl-ADF (Solyc03g025750.2.1), and XM_004236699.1 for Sl-ACTIN4 (Solyc04g011500.2.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transient Overexpression of LePRK1 in Tomato Pollen Caused Pollen Tube Blebbing.
Supplemental Figure 2. Analysis of Tobacco Pollen Tubes Overexpressing LePRK1.
Supplemental Figure 3. The C-Terminal Tail and Membrane Localization of LePRK1, but Not Its Kinase Activity, Are Required for Blebbing Pollen Tubes.
Supplemental Figure 4. Mild Phenotypes When LePRK3, 4, or 5 Is Transiently Overexpressed in Tobacco Pollen.
Supplemental Figure 5. LePRK1 Is Phosphorylated in Tomato Pollen.
Supplemental Figure 6. Phylogenetic Tree of Plant LIM Domain Proteins and Interaction between PLIM2a and KPP.
Supplemental Figure 7. Sl-ADF and Sl-δLIM2 Form a Complex with LePRK1, but Neither Interferes with the Pollen Tube Blebbing Caused by LePRK1 Overexpression.
Supplemental Figure 8. PLIM2b and PLIM2c Did Not Abolish Pollen Tube Blebbing When Transiently Coexpressed with LePRK1.
Supplemental Figure 9. Low-Speed Sedimentation Assay.
Supplemental Figure 10. Low-Speed Sedimentation Assay for Actin Bundling Promotion Effects.
Supplemental Figure 11. LePRK1 Lacking the Juxtamembrane Domain Region or the C-Terminal Region Prevents LePRK1 from Forming a Complex with Itself but Facilitates LePRK1 Forming a Complex with PLIM2a and the Formation of Ring-Like Structures.
Supplemental Figure 12. Pollen of Transgenic Tomato Plants Overexpressing LePRK1-eGFP or LePRK1ΔECD-eGFP Did Not Geminate.
Supplemental Figure 13. Analysis of Arabidopsis Lines Expressing pLAT52-LePRK1ΔECD-eGFP.
Supplemental Figure 14. Fluorescence Variation among Individual Pollen Grains from a Single Flower of a Homozygous LePRK1ΔECD-eGFP Transgenic Tomato Plant.
Supplemental Table 1. Detailed Information for Plasmid Construction.
Supplemental Table 2. Primer Sequences.
Supplemental Movie 1. A Representative Tobacco Pollen Tube Transiently Overexpressing LePRK1-eGFP Driven by the LAT52 Promoter Forms a Bleb.
Supplemental Movie 2. A Representative Tobacco Pollen Tube Transiently Overexpressing LePRK1-eGFP Driven by the LAT52 Promoter, with Growing Blebs.
Supplemental Movie 3. A Representative Tobacco Pollen Tube Transiently Expressing LePRK1-eGFP Driven by the LePRK1 Promoter Forms a Small Bleb.
Supplemental Movie 4. A Representative Tobacco Pollen Tube Transiently Overexpressing LePRK1ΔECD-eGFP Driven by the LAT52 Promoter Forms a Bleb from an Existing Bleb.
Supplemental Movie 5. A Representative Wild-Type Tomato Pollen Tube Grew in Vitro.
Supplemental Movie 6. A Representative Transgenic Tomato Pollen Tube Expressing a LePRK1 RNAi Construct Grew in Vitro.
Supplemental Movie 7. A Representative Transgenic Tomato Pollen Tube Expressing a LePRK1 RNAi Construct Grew in Vitro and Burst.
Supplemental Movie 8. A Representative Transgenic Tomato Pollen Grain Overexpressing LePRK1-eGFP Driven by the LAT52 Promoter Has Moving Vesicles in the Cytoplasm.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analyses of PRK Proteins Shown in Supplemental Figure 4C.
Supplemental Data Set 2. Text File of the Alignment Used for the Phylogenetic Analyses of Tomato and Tobacco PRK Proteins Shown in Supplemental Figure 4D.
Supplemental Data Set 3. Text File of the Alignment Used for the Phylogenetic Analyses of Plant LIM Domain Proteins Shown in Supplemental Figure 6A.
Supplementary Material
Acknowledgments
We thank Wei Zhang and Ying-Yu Si for help with Arabidopsis transformation, Ji-Rong Huang, Li-Jia Qu, and Peng Zhang for insightful discussions, rotating graduate student Fang-Yuan He for assistance with PLIM2a expression, and Ting-Lu Yuan for assistance with manuscript preparation. This work was supported by the Natural Science Foundation of China (Grant 31170291 to W.-H.T.), the Ministry of Science and Technology of China (Grant 2011CB944604 to D.Z. and Grants 2011CB100702 and 2012CB944801 to W.-H.T.), the Chinese Academy of Sciences (Grant XDB11020500 to W.-H.T.), and the USDA (Grant CRIS-5335-21000-020-00D to S.M.).
AUTHOR CONTRIBUTIONS
W.-H.T. and D.Z. designed experiments. C.-P.G., X.D., H.-K.L., W.-J.H., S.-J.W. X.-Y.G., and M.L.B. performed experiments. W.-H.T., H.-K.L., D.Z., J.M., and S.M. analyzed data. W.-H.T. and S.M. wrote the article.
Glossary
- RLK
receptor-like kinase
- ECD
extracellular domain
- BiFC
bimolecular fluorescence complementation
- RNAi
RNA interference
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Allard, J., Mogilner, A. (2013). Traveling waves in actin dynamics and cell motility. Curr. Opin. Cell Biol. 25: 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud, D., Déjardin, A., Leplé, J.C., Lesage-Descauses, M.C., Pilate, G. (2007). Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res. 14: 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berken, A., Thomas, C., Wittinghofer, A. (2005). A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436: 1176–1180 [DOI] [PubMed] [Google Scholar]
- Blanvillain, R., Young, B., Cai, Y.M., Hecht, V., Varoquaux, F., Delorme, V., Lancelin, J.M., Delseny, M., Gallois, P. (2011). The Arabidopsis peptide kiss of death is an inducer of programmed cell death. EMBO J. 30: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser, H., Reichman-Fried, M., Castanon, I., Dumstrei, K., Marlow, F.L., Kawakami, K., Solnica-Krezel, L., Heisenberg, C.P., Raz, E. (2006). Migration of zebrafish primordial germ cells: A role for myosin contraction and cytoplasmic flow. Dev. Cell 11: 613–627 [DOI] [PubMed] [Google Scholar]
- Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., Ohad, N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas, L., Lovy-Wheeler, A., Wilsen, K.L., Hepler, P.K. (2005). Actin polymerization promotes the reversal of streaming in the apex of pollen tubes. Cell Motil. Cytoskeleton 61: 112–127 [DOI] [PubMed] [Google Scholar]
- Castro, A.J., Suárez, C., Zienkiewicz, K., Alché, J.d.D., Zienkiewicz, A., Rodríguez-García, M.I. (2013). Electrophoretic profiling and immunocytochemical detection of pectins and arabinogalactan proteins in olive pollen during germination and pollen tube growth. Ann. Bot. (Lond.) 112: 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F., Gu, Y., Ma, H., Yang, Z. (2013). AtPRK2 promotes ROP1 activation via RopGEFs in the control of polarized pollen tube growth. Mol. Plant 6: 1187–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras, G., Paluch, E. (2008). Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol. 9: 730–736 [DOI] [PubMed] [Google Scholar]
- Chebli, Y., Kroeger, J., Geitmann, A. (2013). Transport logistics in pollen tubes. Mol. Plant 6: 1037–1052 [DOI] [PubMed] [Google Scholar]
- Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T.J., Higgins, D.G., Thompson, J.D. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., Wu, H.M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59: 547–572 [DOI] [PubMed] [Google Scholar]
- Cheung, A.Y., Duan, Q.H., Costa, S.S., de Graaf, B.H., Di Stilio, V.S., Feijo, J., Wu, H.M. (2008). The dynamic pollen tube cytoskeleton: Live cell studies using actin-binding and microtubule-binding reporter proteins. Mol. Plant 1: 686–702 [DOI] [PubMed] [Google Scholar]
- Clough, S.J., Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- deWin, A.H.N., Knuiman, B., Pierson, E.S., Geurts, H., Kengen, H.M.P., Derksen, J. (1996). Development and cellular organization of Pinus sylvestris pollen tubes. Sex. Plant Reprod. 9: 93–101 [Google Scholar]
- Eliasson, A., Gass, N., Mundel, C., Baltz, R., Kräuter, R., Evrard, J.-L., Steinmetz, A. (2000). Molecular and expression analysis of a LIM protein gene family from flowering plants. Mol. Gen. Genet. 264: 257–267 [DOI] [PubMed] [Google Scholar]
- Fernando, D.D., Lazzaro, M.D., Owens, J.N. (2005). Growth and development of conifer pollen tubes. Sex. Plant Reprod. 18: 149–162 [Google Scholar]
- Hepler, P.K., Rounds, C.M., Winship, L.J. (2013). Control of cell wall extensibility during pollen tube growth. Mol. Plant 6: 998–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema, A., Hirsch, P.R., Hooykas, P.J.J., Schilperoort, R.A. (1983). A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303: 179–180 [Google Scholar]
- Huang, W.J., Liu, H.K., McCormick, S., Tang, W.H. (2014). Tomato pistil factor STIG1 promotes in vivo pollen tube growth by binding to phosphatidylinositol 3-phosphate and the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 26: 2505–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, J.U., Wu, G., Yan, A., Lee, Y.J., Grierson, C.S., Yang, Z. (2010). Pollen-tube tip growth requires a balance of lateral propagation and global inhibition of Rho-family GTPase activity. J. Cell Sci. 123: 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglarz, M.K., Howard, K.R. (1995). The active migration of Drosophila primordial germ cells. Development 121: 3495–3503 [DOI] [PubMed] [Google Scholar]
- Kaothien, P., Ok, S.H., Shuai, B., Wengier, D., Cotter, R., Kelley, D., Kiriakopolos, S., Muschietti, J., McCormick, S. (2005). Kinase partner protein interacts with the LePRK1 and LePRK2 receptor kinases and plays a role in polarized pollen tube growth. Plant J. 42: 492–503 [DOI] [PubMed] [Google Scholar]
- Kim, H.U., Cotter, R., Johnson, S., Senda, M., Dodds, P., Kulikauska, R., Tang, W., Ezcura, I., Herzmark, P., McCormick, S. (2002). New pollen-specific receptor kinases identified in tomato, maize and Arabidopsis: The tomato kinases show overlapping but distinct localization patterns on pollen tubes. Plant Mol. Biol. 50: 1–16 [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Kroeger, J., Geitmann, A. (2012). The pollen tube paradigm revisited. Curr. Opin. Plant Biol. 15: 618–624 [DOI] [PubMed] [Google Scholar]
- Lin, Y., Yang, Z. (1997). Inhibition of pollen tube elongation by microinjected anti-Rop1Ps antibodies suggests a crucial role for Rho-type GTPases in the control of tip growth. Plant Cell 9: 1647–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löcke, S., Fricke, I., Mucha, E., Humpert, M.L., Berken, A. (2010). Interactions in the pollen-specific receptor-like kinases-containing signaling network. Eur. J. Cell Biol. 89: 917–923 [DOI] [PubMed] [Google Scholar]
- Ma, B., Qian, D., Nan, Q., Tan, C., An, L., Xiang, Y. (2012). Arabidopsis vacuolar H+-ATPase (V-ATPase) B subunits are involved in actin cytoskeleton remodeling via binding to, bundling, and stabilizing F-actin. J. Biol. Chem. 287: 19008–19017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, S. (1991). Transformation of tomato with Agrobacterium tumefaciens. In Plant Tissue Culture Manual: Fundamentals and Applications, Vol. B6, K. Lindsey, ed (Amsterdam: Kluwer), pp. 1–9. [Google Scholar]
- Mollet, J.C., Leroux, C., Dardelle, F., Lehner, A. (2013). Cell wall composition, biosynthesis and remodeling during pollen tube growth. Plants 2: 107–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschietti, J., Eyal, Y., McCormick, S. (1998). Pollen tube localization implies a role in pollen-pistil interactions for the tomato receptor-like protein kinases LePRK1 and LePRK2. Plant Cell 10: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papuga, J., Hoffmann, C., Dieterle, M., Moes, D., Moreau, F., Tholl, S., Steinmetz, A., Thomas, C. (2010). Arabidopsis LIM proteins: A family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 22: 3034–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounds, C.M., Bezanilla, M. (2013). Growth mechanisms in tip-growing plant cells. Annu. Rev. Plant Biol. 64: 243–265 [DOI] [PubMed] [Google Scholar]
- Salem, T., Mazzella, A., Barberini, M.L., Wengier, D., Motillo, V., Parisi, G., Muschietti, J. (2011). Mutations in two putative phosphorylation motifs in the tomato pollen receptor kinase LePRK2 show antagonistic effects on pollen tube length. J. Biol. Chem. 286: 4882–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A., Rasband, W.S., Eliceiri, K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter, B.D., Smith, S.E., Li, R. (2009). Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb. Perspect. Biol. 1: a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L.G. (2003). Cytoskeletal control of plant cell shape: Getting the fine points. Curr. Opin. Plant Biol. 6: 63–73 [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., Poulter, N.S., Henty, J.L., Franklin-Tong, V.E., Blanchoin, L. (2010). Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 61: 1969–1986 [DOI] [PubMed] [Google Scholar]
- Suarez-Cervera, M., Arcalis, E., Le Thomas, A., Seoane-Camba, J.A. (2002). Pectin distribution pattern in the apertural intine of Euphorbia peplus L. (Euphorbiaceae) pollen. Sex. Plant Reprod. 14: 291–298 [Google Scholar]
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W., Kelley, D., Ezcurra, I., Cotter, R., McCormick, S. (2004). LeSTIG1, an extracellular binding partner for the pollen receptor kinases LePRK1 and LePRK2, promotes pollen tube growth in vitro. Plant J. 39: 343–353 [DOI] [PubMed] [Google Scholar]
- Thomas, C. (2012). Bundling actin filaments from membranes: Some novel players. Front. Plant Sci. 3: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C., Fricke, I., Scrima, A., Berken, A., Wittinghofer, A. (2007). Structural evidence for a common intermediate in small G protein-GEF reactions. Mol. Cell 25: 141–149 [DOI] [PubMed] [Google Scholar]
- Toews, M.W., Warmbold, J., Konzack, S., Rischitor, P., Veith, D., Vienken, K., Vinuesa, C., Wei, H., Fischer, R. (2004). Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45: 383–389 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium . (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell, D., Yamaguchi, J., McCormick, S. (1990). Pollen-specific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109: 705–713 [DOI] [PubMed] [Google Scholar]
- Vidali, L., Rounds, C.M., Hepler, P.K., Bezanilla, M. (2009). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H.J., Wan, A.R., Jauh, G.Y. (2008). An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 147: 1619–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner, R., Li, R. (2008). Yeast and fungal morphogenesis from an evolutionary perspective. Semin. Cell Dev. Biol. 19: 224–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengier, D., Valsecchi, I., Cabanas, M.L., Tang, W.H., McCormick, S., Muschietti, J. (2003). The receptor kinases LePRK1 and LePRK2 associate in pollen and when expressed in yeast, but dissociate in the presence of style extract. Proc. Natl. Acad. Sci. USA 100: 6860–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengier, D.L., Mazzella, M.A., Salem, T.M., McCormick, S., Muschietti, J.P. (2010). STIL, a peculiar molecule from styles, specifically dephosphorylates the pollen receptor kinase LePRK2 and stimulates pollen tube growth in vitro. BMC Plant Biol. 10: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wu, Y., Yan, J., Zhang, R., Qu, X., Ren, S., Chen, N., Huang, S. (2010). Arabidopsis FIMBRIN5, an actin bundling factor, is required for pollen germination and pollen tube growth. Plant Cell 22: 3745–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (1998). Signaling tip growth in plants. Curr. Opin. Plant Biol. 1: 525–530 [DOI] [PubMed] [Google Scholar]
- Yang, Z. (2008). Cell polarity signaling in Arabidopsis. Annu. Rev. Cell Dev. Biol. 24: 551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J.R., Zhou, L.M., Xu, M.L. (2013). Arabidopsis LIM proteins PLIM2a and PLIM2b regulate actin configuration during pollen tube growth. Biol. Plant. 57: 433–441 [Google Scholar]
- Zatulovskiy, E., Tyson, R., Bretschneider, T., Kay, R.R. (2014). Bleb-driven chemotaxis of Dictyostelium cells. J. Cell Biol. 204: 1027–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., Wengier, D., Shuai, B., Gui, C.P., Muschietti, J., McCormick, S., Tang, W.H. (2008). The pollen receptor kinase LePRK2 mediates growth-promoting signals and positively regulates pollen germination and tube growth. Plant Physiol. 148: 1368–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., McCormick, S. (2007). A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Y., Aggarwal, M., Zheng, W.G., Wu, H.M., Cheung, A.Y. (2011). Receptor-like kinases as surface regulators for RAC/ROP-mediated pollen tube growth and interaction with the pistil. AoB Plants 2011: plr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.