Gain- and loss-of-function studies revealed that the rice KNOX transcription factor OSH1 negatively regulates the brassinosteroid (BR) phytohormone pathway. Genomics approaches identified BR catabolism genes as direct targets of OSH1 in the shoot meristems. RNAi knockdown studies suggested that BR catabolism is important for shoot meristem function and for regulation of panicle branch angle.
Abstract
In flowering plants, knotted1-like homeobox (KNOX) transcription factors play crucial roles in establishment and maintenance of the shoot apical meristem (SAM), from which aerial organs such as leaves, stems, and flowers initiate. We report that a rice (Oryza sativa) KNOX gene Oryza sativa homeobox1 (OSH1) represses the brassinosteroid (BR) phytohormone pathway through activation of BR catabolism genes. Inducible overexpression of OSH1 caused BR insensitivity, whereas loss of function showed a BR-overproduction phenotype. Genome-wide identification of loci bound and regulated by OSH1 revealed hormonal and transcriptional regulation as the major function of OSH1. Among these targets, BR catabolism genes CYP734A2, CYP734A4, and CYP734A6 were rapidly upregulated by OSH1 induction. Furthermore, RNA interference knockdown plants of CYP734A genes arrested growth of the SAM and mimicked some osh1 phenotypes. Thus, we suggest that local control of BR levels by KNOX genes is a key regulatory step in SAM function.
INTRODUCTION
In contrast to animals that complete organ formation during embryogenesis, plants continue to produce organs throughout their life cycle. In flowering plants, formation of aboveground organs relies on the shoot apical meristem (SAM). The SAM is an indeterminate structure comprised of self-renewing stem cells in its center and daughter cells at its periphery. Lateral organs such as leaves and flowers initiate from the flank of the SAM at the expense of stem cells. Cells in the SAM are nucleocytoplasmic, small, and divide slowly, whereas those within lateral organs are vacuolated, elongate, and have increased rates of cell division (Steeves and Sussex, 1989). Thus, the transition from the indeterminate SAM to determinate lateral organs is likely to include various cytological and physiological changes.
Indeterminancy of the SAM is maintained by Class I knotted1-like homeobox (KNOX) transcription factors. KNOX genes are specifically expressed in the SAM and are excluded from lateral organ primordia. Genetic studies have shown that ectopic expression of KNOX genes in leaves causes proximal distal patterning defects, whereas loss of KNOX function results in failure of SAM formation and maintenance (Freeling and Hake, 1985; Vollbrecht et al., 2000; Tsuda et al., 2011). KNOX genes have been shown to activate cytokinin (CK) biosynthesis and repress gibberellin (GA) and lignin production (Sakamoto et al., 2001; Mele et al., 2003; Jasinski et al., 2005; Yanai et al., 2005; Bolduc and Hake, 2009). Recently, a study using chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) revealed that maize (Zea mays) KNOTTED1 (KN1) targets more than 5000 loci (Bolduc et al., 2012). Transcription factors and hormonal pathway genes are overrepresented as targets. Nevertheless, functional and regulatory relationships between KNOX proteins and the majority of their targets remain unknown.
Brassinosteroids (BRs) are growth-promoting phytohormones involved in diverse aspects of plant growth and development (Clouse and Sasse, 1998). BR promotes differentiation through activation of a large number of genes related to cell elongation and cell wall modification (Sun et al., 2010). It was recently reported that BR also plays important roles in organ boundaries in Arabidopsis thaliana. LATERAL ORGAN BOUNDARIES proteins directly activate phyB activation-tagged suppressor1 (BAS1), which encodes a BR catabolism enzyme and represses BR accumulation and overgrowth of the boundary between axillary branches and subtending leaves (Neff et al., 1999; Bell et al., 2012). In addition, BRASSINAZOLE RESISTANT1 (BZR1), which encodes a transcription factor in the BR signaling pathway, is excluded from boundaries between the SAM and leaf primordia, allowing expression of CUP SHAPED COTYLEDON (CUC) genes to repress growth in the boundary (Gendron et al., 2012). Thus, these studies indicate the importance of BR regulation during boundary development in Arabidopsis.
We determined that OSH1 negatively regulates the BR pathway from observations of osh1 loss-of-function mutants and inducible overexpression. To gain further understanding of KNOX gene function, we performed a genome-wide analysis of the OSH1 downstream pathway using ChIP-seq combined with transcriptome analyses. Among the bound and modulated targets, three BR catabolism genes encoding CYP734As were rapidly upregulated upon OSH1 induction. The expression domain of CYP734A6 overlapped with that of OSH1, indicating that the genes function in the same area within the shoot apex. Comparison of ChIP-seq data between rice (Oryza sativa) and maize suggest that this regulation is evolutionarily conserved. Furthermore, RNA interference (RNAi) knockdown of these BR catabolism genes resulted in an arrested shoot and mimicked some aspects of the osh1 mutant, indicating the importance of BR catabolism in SAM maintenance.
RESULTS
Induced Overexpression of OSH1 Results in a BR-Insensitive Phenotype
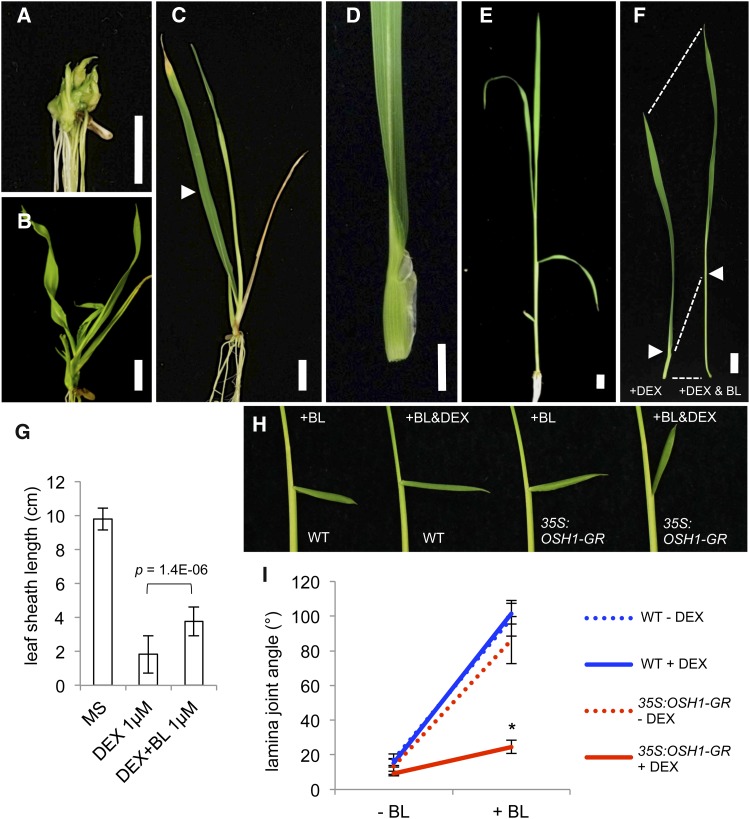
To gain a better understanding of networks downstream of KNOX transcription factors, we made a dexamethasone (DEX)-inducible OSH1 overexpressor using a rat glucocorticoid receptor (GR) domain fusion protein (35S:OSH1-GR) (Aoyama and Chua, 1997). At the T2 generation, we selected lines homozygous for the transgene and examined their phenotypes with and without DEX treatment. 35S:OSH1-GR plants grown on DEX-containing media for 3 weeks showed DEX-dose-dependent phenotypes. When grown with 10 µM DEX, 35S:OSH1-GR plants had multiple shoots, a morphology typical of severe KNOX overexpressors in rice (Figure 1A) (Sentoku et al., 2000). Treatment with 1 µM DEX led to twisted leaf blades and extremely shortened leaf sheaths (Figure 1B). At 0.1 μM DEX, the short sheath phenotype was still observed and leaf blades were erect (Figures 1C and 1D). These transgenic plants showed no abnormality without DEX (Figure 1E), and control nontransgenic plants grown with DEX were also normal, confirming these leaf phenotypes were caused by induced overexpression of OSH1.
Figure 1.
Induction of OSH1 Overexpression Leads to BR Insensitivity.
(A) to (D) 35S:OSH1-GR plants grown on 10 μM (A), 1 μM (B), and 0.1 μM ([C] and [D]) DEX.
(D) The fourth leaf labeled with arrowhead in (C) has an erect leaf blade and extremely shortened leaf sheath.
(E) A 3-week-old 35S:OSH1-GR plant grown without DEX.
(F) 35S:OSH1-GR plants grown for 3 weeks on 1 μM DEX (left) and 1 μM DEX plus 1 μM BL (right). Arrowheads and dashed lines indicate the position of the lamina joint.
(G) Quantification of fourth leaf sheath length (n = 15).
(H) Lamina joint assay of wild-type and 35S:OSH1-GR plants in the presence of 500 ng BL with/without 100 ng DEX.
(I) Quantification of lamina joint angle of second leaves (n = 10). An asterisk represents the statistically significant difference at P = 1.7E-11 compared with the angle of 35S:OSH1-GR without DEX.
Bars = 1 cm in (A) to (F). Error bars represent sd in (G) and (I).
Some 35S:OSH1-GR DEX-induced phenotypes such as twisted, erect leaf blades and extremely shortened leaf sheaths are characteristic of BR-deficient or -insensitive mutants of rice (Yamamuro et al., 2000; Hong et al., 2002). To test if the OSH1 overexpressor phenotype was caused by deficiency in the BR pathway, we examined the effect of brassinolide (BL) treatment, a biologically active BR. Although the phenotype of 35S:OSH1-GR with 10 μM DEX was not changed by addition of BL, this treatment attenuated the shortened leaf sheath phenotype obtained at lower concentrations of DEX (Figures 1F and 1G). This result supports the idea that a subset of OSH1 overexpression phenotypes was caused by repression of the BR pathway.
The fact that BL treatment did not affect the 35S:OSH1-GR phenotype at high concentrations of DEX suggested that overexpression of OSH1 reduced the sensitivity to BR. To test this hypothesis, we performed a lamina joint test for BR sensitivity (Figures 1H and 1I). In rice, BR enhances lamina joint bending by promoting adaxial cell elongation in auricles (Wada et al., 1981; Cao and Chen, 1993), while BR-deficient and -insensitive mutants show a decreased leaf angle (Yamamuro et al., 2000; Hong et al., 2002). If OSH1 overexpression causes repression of BR biosynthesis, application of BR onto developing leaves should increase the lamina joint angle, whereas if it causes BR insensitivity, BR application would have no effect. When wild-type seedlings were treated with BL, lamina joint angle greatly increased with or without DEX. In contrast, although 35S:OSH1-GR plants responded to BL similarly to the wild type in the absence of DEX, the angle barely increased and leaf blades remained upright when they were treated with both BL and DEX (Figures 1H and 1I). Therefore, overexpression of OSH1 caused BR insensitivity, which suggests that OSH1 represses the BR pathway by inhibiting BR signaling and/or by accelerating its inactivation, but not by inhibiting BR biosynthesis.
osh1 Mutants Showed BR Overproduction/Hypersensitive Phenotype
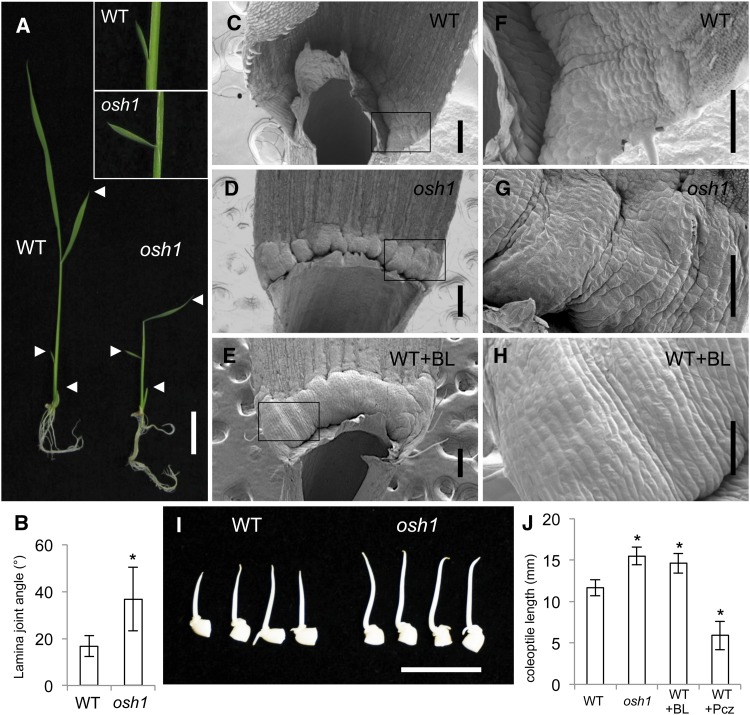
We then asked if a loss-of-function mutant of OSH1 (osh1) shows BR-related phenotypes. The lamina joint angle of the second leaf was increased in osh1 compared with that of the wild type (Figures 2A and 2B). Scanning electron microscopy revealed that auricle tissue in osh1 protruded outward (Figures 2C, 2D, 2F, and 2G). In the wild type, elongated cells were observed only in the proximal part of the auricle. In contrast, osh1 mutants had elongated cells not only in the proximal region but also in the distal part of the protruding auricle. This phenotype was also present in BL-treated wild-type plants (Figures 2E and 2H). Stimulation of coleoptile elongation is known as another BR response in monocots. Exogenous BL treatment increases coleoptile elongation (Yamamuro et al., 2000) and the inhibitor of BR synthesis, propiconazole decreases coleoptile elongation (Hartwig et al., 2012). osh1 mutants had longer coleoptiles than the wild type when seeds were germinated under submerged conditions (Figures 2I and 2J). The increase in coleoptile length was similar to that obtained by treating the wild type with BL (Figure 2J). The BR overproduction and/or hypersensitive phenotype of osh1 support our hypothesis that OSH1 negatively regulates the BR pathway.
Figure 2.
Loss-of-Function Mutant of OSH1 Shows BR Overproduction/Hypersensitive Phenotype.
(A) Two-week-old seedlings of the wild type (left) and osh1 (right). Enlarged pictures of the second leaf lamina joint from the wild type and osh1 are shown in insets. Arrowheads represent the position of the first three leaves.
(B) Quantification of second lamina joint angles of the wild type and osh1 (n = 30).
(C) to (E) Scanning electron microscopy of second leaf lamina joint in the wild type (C), osh1 (D), and the wild type treated with 100 ng of BL (E).
(F) to (H) Enlarged pictures of auricle region. Boxed regions in (C) to (E) were enlarged in (F) to (H), respectively.
(I) osh1 has longer coleoptiles than the wild type. Seeds were imbibed in water for 2 d at 30°C.
(J) Quantification of coleoptile length. The 10 longest coleoptiles from twenty seeds were measured at 2 d after imbibition.
Bars = 2 cm in (A), 200 μm in (C) to (E), 100 μm in (F) to (H), and 1 cm in (I). Error bars represent sd in (B) and (J). Asterisks indicate statistically significant differences at P < 1.00E-5 compared with wild-type control.
osh1 Mutants Showed Boundary Defects in Leaves and in the SAM
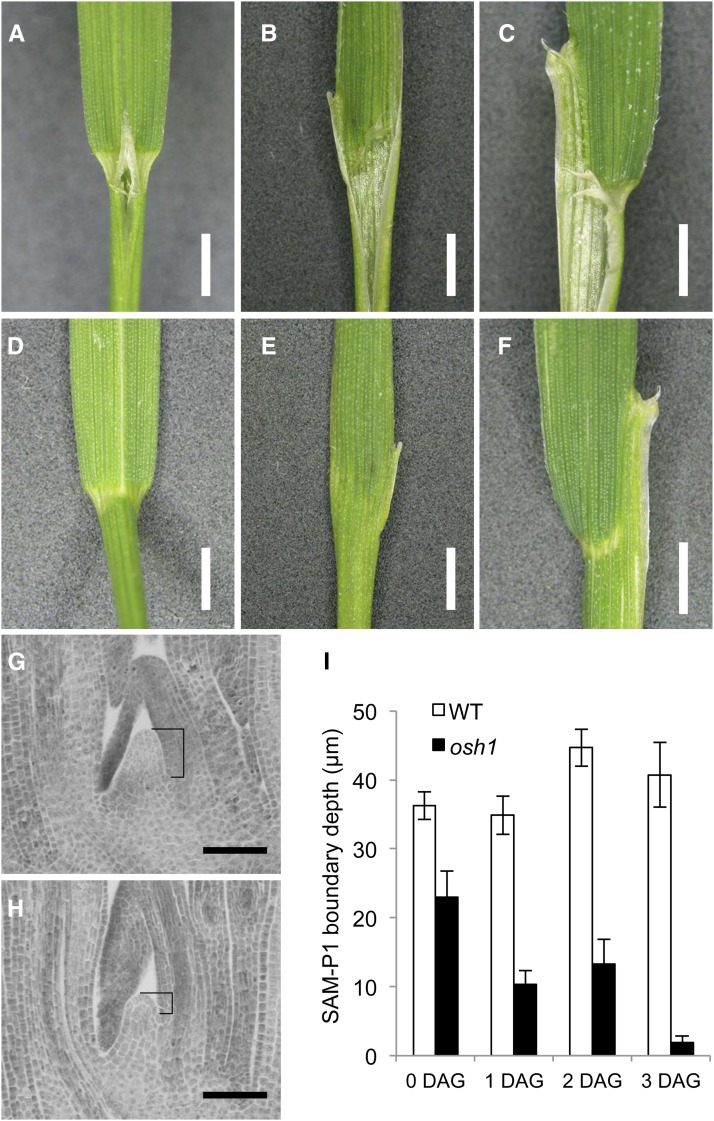
Wild-type leaf blade and sheath are separated by a distinct band of auricle tissue, visible from both adaxial and abaxial sides (Figures 3A and 3D). At low frequency (∼2%), osh1 mutants showed abnormal blade/sheath boundaries. In these mutants, the ligule was discontinuous, abnormal flaps formed along the leaf margin, and auricle tissue was partially (Figures 3C and 3F) or completely lost (Figures 3B and 3E). osh1 mutants also showed a boundary defect in the SAM. As previously reported (Tsuda et al., 2011), the boundary between the SAM and P1 leaf primordium was shallower in osh1 mutants than in the wild type (Figures 3G to 3I). The boundary depth decreased until osh1 mutants lost the SAM at 3 to 4 d after germination, whereas it was maintained at constant level in the wild type. Thus, osh1 mutants show boundary defects in the SAM and leaves.
Figure 3.
osh1 Mutant Shows Boundary Defects in Leaves and SAM.
(A) to (C) Adaxial view of lamina joint of the wild type in (A) and osh1 in (B) and (C).
(D) to (F) Abaxial view of (A) to (C) is shown in (D) to (F), respectively.
(G) and (H) Shoot apex sections of the wild type and osh1 at 2 d after imbibition. Brackets represent the depth of boundary between the SAM and P1 leaf primordia.
(I) Quantification of SAM-P1 boundary depth during germination (n = 10).
Error bars indicate sd. Bars = 2 mm in (A) to (F) and 50 μm in (G) and (H).
Genome-Wide Identification of OSH1 Direct Targets by ChIP-seq
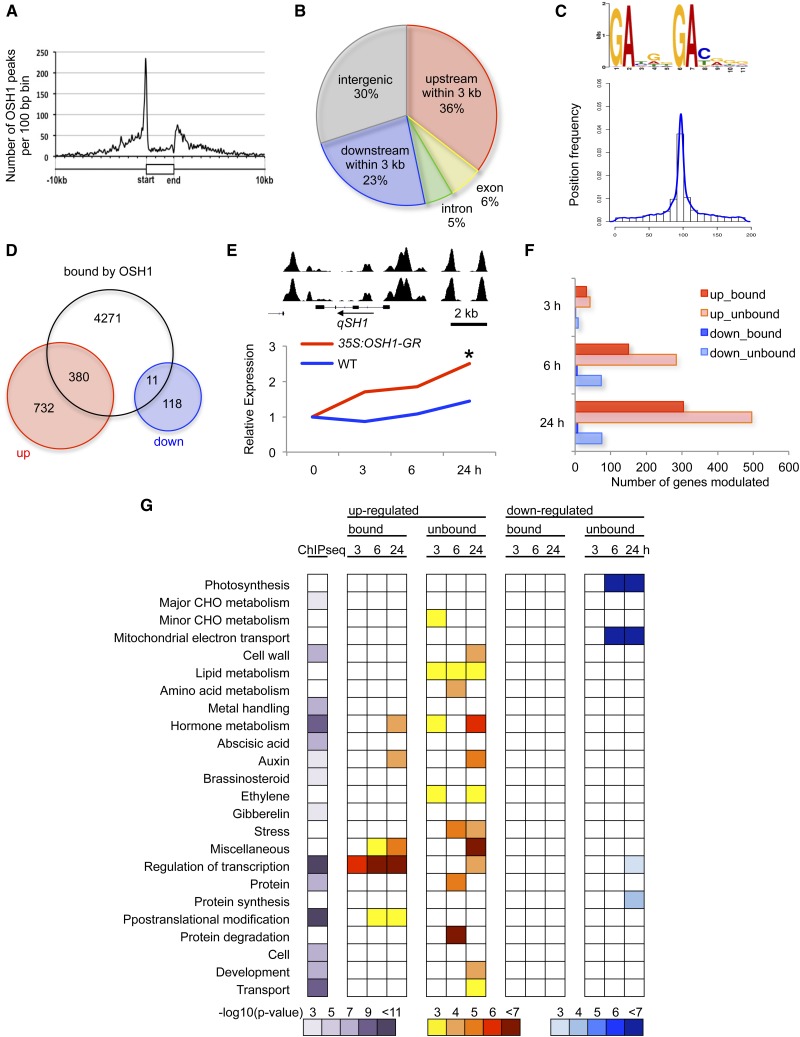
To determine the potential OSH1 direct targets, we performed ChIP-seq. Young panicles at the 5-mm stage were collected and OSH1-bound chromatin was immunoprecipitated using an α-OSH1 specific antibody. As negative controls, we used control IgG. By comparing ChIP-seq data (from two biological replicates) between α-OSH1 and control IgG, 5547 OSH1-bound peaks were reproducibly detected. These peaks were assigned to the closest genes, thereby deriving 4662 genes as potential OSH1 direct targets (Supplemental Data Set 1). We previously reported that five class I KNOX genes, including OSH1 itself, were directly regulated by OSH1 (Tsuda et al., 2011). All of these KNOX loci were successfully identified as potential direct targets (Supplemental Figure 1). We also validated 18 newly identified OSH1-bound loci by ChIP followed by quantitative PCR (qPCR) (Supplemental Figure 2), confirming the reliability of our ChIP-seq data.
OSH1 binds preferentially to the vicinity of genic regions, especially 5′ upstream regions (Figure 4A). Indeed, 59% of the peaks were located within 3 kb of coding regions (Figure 4B). Significant numbers of peaks (30%), however, were located greater than 3 kb from a gene, suggesting that cis-acting elements for OSH1 binding also exist as distal regulatory regions.
Figure 4.
Genome-Wide Analyses of OSH1 Downstream Regulatory Gene Network.
(A) Distribution of OSH1 binding peaks relative to genic region. The gene model normalized to the average gene length in rice is shown as a white box.
(B) Distribution of OSH1 binding peaks across the genomic features. Peaks found within 3 kb upstream from the transcription start sites or downstream from the last exon were assigned into upstream and downstream, respectively. Peaks found further upstream or downstream were assigned into intergenic region.
(C) The most enriched motif in OSH1 binding peaks (top) and its distribution in 200-bp region of peaks (bottom). The peak summit was positioned in the center. Vertical axis shows the position frequency of OSH1 bound peak summits.
(D) Venn diagram of OSH1-bound and differentially expressed genes in 35S:OSH1-GR after DEX treatment.
(E) OSH1 directly targets and upregulates qSH1. ChIP-seq and RNA-seq data are shown in top and bottom, respectively. Red is 35S:OSH1-GR plus DEX and blue is the wild type with DEX. An asterisk represents significant upregulation at P < 0.05 compared with the expression level before DEX treatment.
(F) Number of genes differentially expressed in 35S:OSH1-GR. Horizontal axis represents the number of genes modulated after DEX treatment.
(G) Functional categories enriched in OSH1-bound genes and in differentially expressed genes in 35S:OSH1-GR.
We searched DNA motifs enriched among OSH1-bound regions. The most frequently found motif (Motif 1) consists of two GA cores separated by 3 bp with G enriched in between (Figure 4C; Supplemental Figure 3). The second GA was frequently followed by C/T. Although other nucleotide positions were highly degenerated, this motif was preferentially found in the center of OSH1 binding peaks, supporting the idea that OSH1 in fact binds this motif in vivo. Motif 3, which contains multiple (G/T)GATs, was also found in the center of peak region (Supplemental Figure 3). Several other motifs were distributed in peripheral regions of OSH1 binding peaks; hence, they could be binding sites for other transcription factors functioning in the same genomic context as OSH1.
Gene Expression Profiling by RNA-seq during OSH1 Induction
To determine which targets were modulated, we investigated the time-course transcriptional profile of inducible overexpression of OSH1 by RNA-seq. To capture transcriptional response upon OSH1 induction, we chose young leaf tissue that does not normally express OSH1. Young leaves from 3-week-old 35S:OSH1-GR and wild-type plants were harvested before (0 h) and after (3, 6, and 24 h) DEX treatment. In total, 1241 genes were modulated only in 35S:OSH1-GR (P < 0.05; Figure 4D; Supplemental Data Set 2). Among them, 391 genes (31.5%) were bound by OSH1, and most of them (380 genes; 97%) were upregulated. Only 11 direct targets were downregulated. These data suggest that OSH1 acts as a transcriptional activator.
To understand the pattern of differential gene expression over time, we followed the activity of qSH1, which encodes an putative ortholog of Arabidopsis BELL-type homeodomain (BLH) protein PENNYWISE (Konishi et al., 2006). BLH and KNOX proteins form heterodimers that are thought to be required for functional binding (Smith et al., 2002). The qSH1 locus was bound by OSH1 both 5′ and 3′ of the coding region and transcription increased during induction (Figure 4E). The small number of modulated targets at 3 h and the increase in number of upregulated genes over time (Figure 4F) may be explained by a requirement to first activate qSH1.
Functional classification of modulated OSH1 targets revealed that transcription regulators were first upregulated at 3 h (Figure 4G). This category was not overrepresented in the unbound gene list, suggesting that one of the major functions of KNOX proteins is a direct activation of other groups of transcriptional regulators. At later time points (6 and 24 h), genes for posttranslational protein modification were also upregulated. Because this upregulation was also specific to OSH1-bound genes, this result indicates that direct activation of genes involved in protein posttranslational modification is another characteristic function of OSH1. In unbound categories, genes for metabolism of CHO, lipid, and ethylene were upregulated within 3 h of induction. These genes could be the targets of transcriptional regulators activated by OSH1. Categories involved in photosynthesis and mitochondrial electron transport were enriched in OSH1-unbound downregulated genes. Since OSH1 localizes to the nucleus and not to organelles, these data suggest OSH1 indirectly represses these genes.
BR Catabolism Genes Targeted by OSH1
Observations of OSH1 gain- and loss-of-function phenotypes suggest that OSH1 represses the BR pathway by inhibiting its signaling or accelerating its inactivation. Indeed, among seven OSH1 direct target genes that were upregulated at all time points after OSH1 induction (Supplemental Figure 4), one encoded CYP734A4, a rice homolog of Arabidopsis BAS1. BAS1 encodes a cytochrome P450 and inactivates BR by C26 hydroxylation (Neff et al., 1999). Because a previous study showed that three rice BAS1 homologs, CYP734A2, CYP734A4, and CYP734A6 also inactivate BR and its precursors (Sakamoto et al., 2011), we performed further analyses on this gene family.
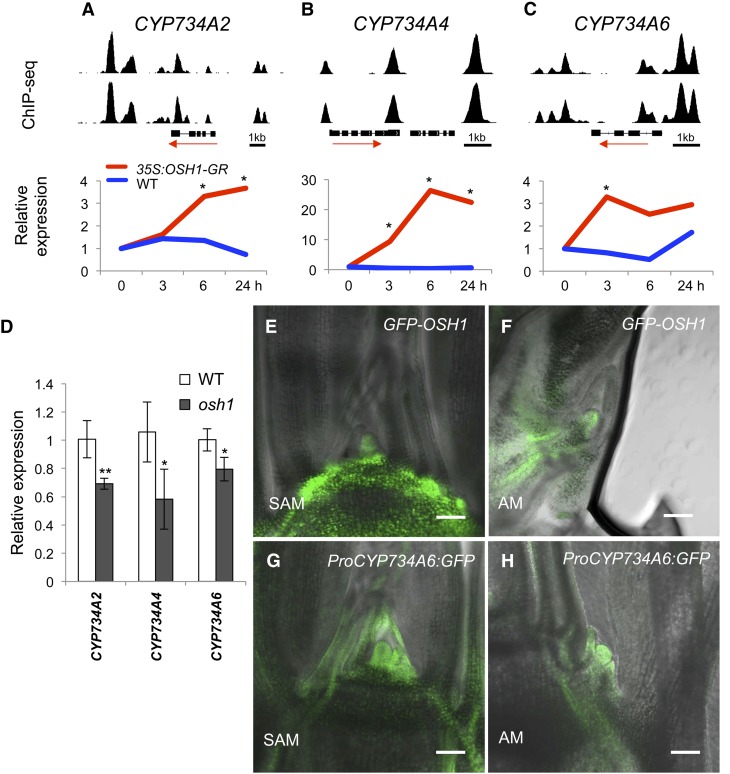
The rice genome has four BAS1 homologs (CYP734A2, CYP734A4, CYP734A5, and CYP734A6), and three of them (CYP734A2, CYP734A4, and CYP734A6) are expressed in shoot tissue (Sakamoto et al., 2011). In our ChIP-seq data, these three genes were found to be OSH1 direct targets. Large OSH1 binding peaks were found in the 5′ or 3′ flanking region, and moderate peaks were also found in the 3′ untranslated region or intron (Figures 5A to 5C). Several Motif 1 sequences as well as one or two TGAC/GTCA sequence, which was previously shown to serve as an in vitro KNOX binding site, were found in OSH1 binding regions on each loci (Supplemental Figure 5) (Smith et al., 2002). By chromatin immunoprecipitation followed by qPCR, OSH1 binding in vivo was detected in shoot apex tissue (Supplemental Figure 6), indicating OSH1 also directly binds these loci in vegetative shoot meristems. CYP734A2 was upregulated at 6 and 24 h after DEX treatment (Figure 5A), CYP734A4 was rapidly upregulated after OSH1 induction (Figure 5B), and CYP734A6 was significantly upregulated at 3 h (Figure 5C). Rapid upregulation was not observed with other BR-related genes bound by OSH1 (Supplemental Figure 7). We also determined that expression levels of CYP734A genes were reduced in osh1 mutants compared with the wild type (Figure 5D), consistent with activation of these genes by OSH1. Overexpression of the CYP734A genes caused severe BR-deficient phenotype such as twisted leaf blade and shortened leaf sheath (Sakamoto et al., 2011), similar to the phenotypes seen with overexpression of OSH1 (Figure 1). These results are consistent with our observation that overexpression of OSH1 causes BR insensitivity.
Figure 5.
BR Catabolism Enzyme Genes Are Directly Activated by OSH1.
(A) to (C) ChIP-seq (top) and RNA-seq (bottom) data for CYP734A2, CYP734A4, and CYP734A6 in (A) to (C), respectively. Relative expression level calculated from RNA-seq data is shown in bottom of each panel. Asterisks represent significant upregulation at P < 0.05 (based on cuffdiff analysis) compared with the expression level before DEX treatment.
(D) Expression levels of CYP734A genes relative to the wild type are reduced in osh1 mutants at 1 d after imbibition. The data represent the average of three biological replicates. Error bars represent sd. Asterisks indicate significant reduction at *P < 0.05 and at **P < 0.01 compared with the expression level in the wild type in a Student’s t test.
(E) to (H) Confocal images of GFP reporter expression of OSH1 in (E) and (F) and CYP734A6 in (G) and (H). GFP images were merged with transmitted light images. AM, axillary meristem. Bars =100 μm.
To examine the expression pattern of CYP734A6, which has strong OSH1 binding peaks in its promoter, we made a green fluorescent protein (GFP) reporter with 1.6 kb upstream, the GFP coding region, and 1 kb downstream. The GFP expression of this reporter line largely overlapped with expression of OSH1 (Figures 5E and 5F) in the shoot apex. ProCYP734A6:GFP was strongly expressed in the SAM, stem, and young leaf primordia (Figure 5G). Expression was also detected in the meristem and young leaves of axillary buds (Figure 5H). Taken together, our genome-wide and expression analyses revealed that OSH1 directly activates the expression of BR inactivating enzyme genes in shoot meristems.
BR Catabolism Is Important for SAM Function and Boundary Formation in Leaves
To examine the biological importance of BR inactivation, we made RNAi knockdown lines of CYP734A genes and observed their phenotype at the T0 generation. Using a part of the CYP734A2 coding region that shows high degrees of sequence identity among the three CYP734A genes, the expression level of all three genes was reduced in these lines (Figure 6A). In the juvenile seedling stage, CYP734A RNAi plants showed some knox loss-of-function phenotypes. At low frequency (3.4%), plants showed the discontinuous leaf blade/sheath boundary similar to boundary defects seen in osh1 mutants (Figure 6B). In addition, these lines occasionally (5.4%) produced very rigid shoots from which new leaves did not emerge (Figures 6C and 6D). Tissue sections revealed that the cells in the stem were abnormally elongated (compared with Figures 6E and 6F). The SAM was still present; however, tissue was hardened and cells in the SAM and young leaves were more vacuolated than the wild type, suggesting that the cells prematurely differentiated and growth of these shoots was arrested. Importantly, the boundary between the SAM and P1 leaf primordium was shallower in these shoots, similar to osh1 (Figures 6G and 6H). Thus, CYP734A knockdown plants mimicked numerous aspects of the osh1 phenotype.
Figure 6.
CYP734A2 RNAi Knockdown Lines Mimic Some Aspects of osh1 Mutants.
(A) Expression level of CYP734A genes in CYP734A2 RNAi knockdown calli calculated as the ratio to the expression level in wild-type callus. The data represent the average of two biological replicates with three technical replicates. Error bars represent sd. Asterisks indicate significant reduction at P < 0.01 compared with the expression in wild-type callus in a Student’s t test.
(B) Discontinuous leaf sheath/blade boundary phenotype observed in CYP734A2 RNAi plants. An arrowhead indicates a discontinuous ligule.
(C) and (D) CYP734A2 RNAi plants occasionally show an arrested shoot phenotype. The arrested shoots are indicated with arrowheads in (C).
(E) to (H) Tissue sections of shoot apices in the wild type in (E) and in arrested shoot of CYP734A2 RNAi in (F). (G) and (H) are enlarged images of the SAM in (E) and in (F), respectively. Arrowhead indicates boundary between the SAM and P1 leaf primordium.
(I) and (J) Panicles of the wild type in (I) and CYP734A2 RNAi line in (J). RNAi lines had larger branch angles than wild type.
(K) and (L) Images of base of the primary branches in the wild type in (K) and in CYP734A2 RNAi in (L). RNAi lines have enlarged pulvinus (arrowheads).
Bars = 2 mm in (B), (K), and (L), 2 cm in (D), 200 μm in (E) and (F), and 50 μm in (G) and (H).
In the reproductive stage, these knockdown lines produced panicles with larger branch angles (Figures 6I and 6J). At the base of a primary branch, they produced a well-developed pulvinus that was not observed in wild-type plants (Figures 6K and 6L). Taken together, these results suggest that BR inactivation plays important roles not only in the maintenance of proper shoot meristem functionality, but also in repression of pulvinus growth to regulate panicle branch angles.
The Regulation of CYP734A6 by KNOX Protein Is Conserved in Maize
To examine the importance of BR regulation by KNOX genes across species, we asked if KN1 also directly regulates BR inactivating enzyme expression in maize (Bolduc et al., 2012). GRMZM2G138750, a gene closely related to rice CYP734A6 (Sakamoto et al., 2011), was found to be a direct target of KN1 (Supplemental Figure 8). This gene was upregulated in leaves of Kn1-N gain-of-function mutants (Supplemental Figure 8). Although only a weak KN1 binding region was assigned, GRMZM2G045319, putatively orthologous to CYP734A2 in rice (Sakamoto et al., 2011), was also upregulated in Kn1-N leaves. Both genes were strongly expressed in the SAM compared with leaves. Thus, these data suggest that the regulation of BR-inactivating genes by KNOX proteins is a distinct regulatory module that has been evolutionarily conserved in grasses.
DISCUSSION
We provide evidence that the rice KNOX protein OSH1 promotes local inactivation of a class of phytohormone, BRs. Gain and loss of function of OSH1 caused BR-insensitive and overproduction/hypersensitive phenotypes, respectively. Genome-wide study of OSH1 binding regions and time-course transcriptome profiling revealed that three genes encoding BR inactivation enzymes were directly activated by OSH1. RNAi knockdown studies revealed that BR inactivation was important for maintenance of SAM activity. CYP734A2 RNAi shoots occasionally displayed signs of differentiation such as abnormally elongated stem and vacuolated cells in the SAM, with eventual arrest of leaf initiation. Furthermore, BR inactivation also affected the regulation of inflorescence branch angles, an important agronomic trait to reduce seed shattering (Ishii et al., 2013). Taken together, we propose that the local inactivation of BR is a key regulatory step for SAM function and inflorescence architecture.
Following leaf initiation from the SAM, various cellular events are activated, such as cell division, cell elongation, and cell wall differentiation. Throughout differentiation, cells are likely to undergo a range of cytological and physiological transitions with phytohormones playing essential roles. KNOX proteins regulate a wide range of phytohormone pathways to maintain SAM indeterminancy. For example, they promote CK production by activation of CK biosynthesis enzyme genes (Jasinski et al., 2005; Yanai et al., 2005) and lower GA levels by repressing GA biosynthesis and activating GA catabolism (Sakamoto et al., 2001; Bolduc and Hake, 2009). The combination of increased CK and reduced GA is critical given that constitutive GA signaling combined with a reduced CK level impairs shoot meristem functions (Jasinski et al., 2005). CK is required for maintenance of the SAM (Kurakawa et al., 2007), and GA contributes to cell differentiation of lateral organs and stems by promoting cell elongation and cell wall formation (Claeys et al., 2014). The auxin pathway is also regulated by KNOX proteins (Bolduc et al., 2012). Auxin triggers initiation of lateral primordia and accumulates distally in developing leaves (Reinhardt et al., 2003; O'Connor et al., 2014).
We show that a fourth hormone, BR, is a critical component of KNOX function in the SAM. BR is perceived by the receptor BRASSINOSTEROID INSENSITIVE1 and activates a signaling cascade (Wang et al., 2012). This activates BZR transcription factors that regulate BR-responsive genes as outputs of this phytohormone activity. Genome-wide analysis of BR-regulated BZR1 target genes revealed that activation of genes involved in cell elongation and cell wall modification was one of the most prominent functions of BR activity (Sun et al., 2010). Our findings suggest that KNOX proteins repress these cellular processes by activating BR catabolism to keep meristematic cells slowly dividing, nucleocytoplasmic enriched, and isodiametric. The notion that repression of cell elongation and cell wall modification is important for the function of the SAM is supported by the RNAi knockdown study of CYP734A genes. CYP734A2 knockdown plants showed abnormally elongated stems, vacuolated cells in the SAM that are usually seen in more differentiated tissues, and occasional arrest of shoot growth. Therefore, BR inactivation by KNOX proteins is an indispensable mechanism for repression of the onset of precocious cell differentiation in meristems.
osh1 mutants occasionally show blade/sheath boundary defects in leaves. This boundary is thought to be established at an early stage of leaf development, such as P2-3 (Sylvester et al., 1990). Although KNOX transcript does not accumulate in developing leaves (Jackson et al., 1994; Sentoku et al., 1999), KNOX proteins accumulate at the base of leaves due to intercellular trafficking from the meristem (Jackson, 2002). One possibility is that KNOX proteins at the base of leaves participate in establishment of the blade/sheath boundary. From analysis of gain-of-function mutants, KNOX proteins appear to promote proximal cell identity. Ectopic expression of maize Knotted1 in leaves results in the formation of more proximal tissues, i.e., sheath and ligule, in the distal blade (Freeling and Hake, 1985). We propose that KNOX proteins function in leaf development to keep BR levels low in the SAM and base of lateral organs such as leaves and coleoptiles via activation of CYP734As. Low levels of BR may help establish the sheath portion of the leaf as well as repress coleoptile overgrowth. A role for regulation of the BR pathway in boundary establishment is also seen in Arabidopsis (Gendron et al., 2012). BZR1 is excluded from the boundary between the SAM and leaf primordia, allowing CUC expression in the boundary. A low level of BR might be required for expression of other regulators that define blade/sheath boundary, although they have not yet been identified.
Local inactivation of BR is likely to be important for various aspects of plant development. For example, RNAi knockdown plants of CYP734As showed derepression of pulvinus growth leading to a large panicle branch angle. osh1 mutants reach maturity when they are regenerated from callus; however, they show no abnormality in panicle branch angles (Tsuda et al., 2011). Thus, other transcription factors must regulate BR inactivation genes in the pulvinus. Recently, it was reported that LIGULELESS1 (LG1) promotes pulvinus growth in wild rice (Oryza rufipogon) (Ishii et al., 2013). In maize, loss of function of lg1 results in a reduced tassel branch angle (M. Lewis and S. Hake, unpublished data). It is an intriguing question whether LG1 and BR interact to regulate pulvinus growth. LG1 may repress CYP734As to allow growth of the pulvinus, or the BR pathway might activate LG1 expression. Wavy auricle in blade1 (WAB1) encodes a maize TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) transcription factor that leads to LG1 accumulation (M. Lewis and S. Hake, unpublished data). In Arabidopsis, it is known that BR biosynthesis is activated through direct regulation of the BR biosynthetic gene DWARF4 by TCP1 and that, in turn, BR activates TCP1 expression (Guo et al., 2010). Based on these facts, lg1 may be activated by BR, and this regulation could be mediated by the TCP transcription factor WAB1. Alternatively, it is also possible that LG1 activates targets independent of BR.
Whether KNOX proteins function primarily as transcriptional activators remains an open question. The majority (97%) of direct targets modulated upon OSH1 induction were upregulated. Although this finding highlights the functional aspect of OSH1 as a transcriptional activator, more than 90% of OSH1 direct targets were not differentially expressed. It is possible that KNOX proteins require a longer time period than the 24-h incubation we used to modulate their target expression. For example, Arabidopsis floral homeotic regulator AGAMOUS (AG) requires about 2 d to induce one of its direct targets, KNUCKLES (KNU), to terminate stem cell activity in the floral meristems (Sun et al., 2009). AG reduces trimethylation at Histone H3 Lysine 27 residue in the KNU locus by the cell-division-dependent eviction of Polycomb proteins (Sun et al., 2014). Therefore, it will be intriguing to monitor the status of chromatin modification and transcriptome profiles in the OSH1 induction system over longer time periods.
METHODS
Plant Material and Growth Conditions
Japonica rice (Oryza sativa) variety Nipponbare or T65 was used as the wild type. T65 was used for transformation. The GFP-OSH1 reporter line was described previously (Tsuda et al., 2011). Plants were grown on Murashige and Skoog (MS) medium for 3 weeks at 26°C under 14-h-light and 10-h-dark condition. For feeding experiments, BL and DEX were added into MS medium at the concentration indicated in figure legends.
Lamina Joint Test
The lamina joint tests were performed as described by Hong et al. (2003). Rice seeds were surface sterilized in 1.5% sodium hypochlorite containing a drop of Tween 20 for 15 min and washed with sterilized water five times. Sterilized seeds were germinated in water for 2 d, and well-germinated plants were transferred onto 1% agar and grown for an additional 3 d. Then, 1 μL of ethanol containing BL (0 or 500 ng; Sigma-Aldrich) with/without DEX (100 ng; Sigma-Aldrich) was applied on the tip of the second leaf blade. Three days later, lamina joint angles of the second leaves were measured.
Scanning Electron Microscopy
Leaf tissues were fixed in FAA fixative solution (formalin:acetic acid:50% ethanol = 5:5:90) for 12 h at 4 °C and dehydrated through a graded series of ethanol from 70 to 100%. Samples were dried using a critical point dryer, sputter coated with gold palladium for 45 s, and observed on a Hitachi S-4700 scanning electron microscope at an accelerating voltage of 5 kV.
Histological Analysis
For conventional histological analysis, paraffin sections (10 μm thick) were prepared as described previously (Tsuda et al., 2011). For CYP734A2 RNAi plants, plastic sections were made using Technovit 7100 (Heraeus Kulzer) according to the manufacturer’s protocol. Two-micrometer-thick sections were stained with toluidine blue.
Plasmid Construction
For the construction of 35S:OSH1-GR, the OSH1 coding region was amplified using primers KT694 and KT695 from the OSH1 full-length cDNA. Rat glucocorticoid receptor domain (Aoyama and Chua, 1997) was PCR amplified using primers KT696 and KT697. These two fragments were fused by a second PCR using primers KT694 and KT697 and cloned into the XbaI-KpnI site of binary vector pBCH1 (Ito et al., 2001) and sequenced. For the CYP734A6 reporter, the promoter region 1.6 kb upstream from its start codon was amplified using primers KT604 and KT607. The GFP coding region was amplified using KT608 and KT609. The CYP734A6 3′ region (∼1 kb) was amplified using KT610 and KT611. These fragments were then PCR fused as described above. The resulting fragment was cloned into the XbaI-KpnI site of pBCH1 and sequenced. These constructs were introduced into T65 calli as described previously (Tsuda et al., 2011). For the CYP734A2 RNAi construct, a part of its coding region was PCR amplified using primers KT469 and KT470 and cloned into pENTR/D-TOPO (Invitrogen) and sequenced. The insert was transferred into the pANDA vector (Miki and Shimamoto, 2004) by LR recombination. The primers used are listed in Supplemental Table 1.
ChIP-seq
About 200 to 300 young panicles at 5-mm stage of Nipponbare were used for each biological replicate. Tissue fixation, nuclei extraction, and chromatin immunoprecipitation using α-OSH1 antibody were performed as described previously (Tsuda et al., 2011). For a negative control, normal rabbit IgG (sc2027; Santa Cruz Biotechnology) was used. Two biological replicates were prepared for each antibody. Immunoprecipitated DNA from 20 panicles was pooled into one tube for each sample. End-repair, A-tailing, and adapter ligation were performed according to Kaufmann et al. (2010) except for library amplification. The ChIP-seq DNA library was amplified by 18 cycles of PCR using KOD neo (TOYOBO), Primer PE 1.0, and Primer PE 2.0. Then, 200- to 500-bp fragments were obtained by gel-size fractionation. The ChIP-seq DNA libraries were sequenced using the Illumina GAII platform. ChIP-seq reads were aligned to the rice genome (MSU7.0) allowing two mismatches (-n 2) with the option of seed length 60 (-l 60) and maximum insert size 500 (-X 500) using Bowtie-0.12.8 (Langmead et al., 2009). MACS software (version 1.4.2) (Zhang et al., 2008) was used for peak calling with the genome size of –g 3.73e+08 and with default settings of cut off P value (<1E-05). Significant (false discovery rate <5% and fold enrichment > 50) and reproducible peaks between two biological replicates were assigned to the nearest gene using closestBed function of BEDTools (version 2.17.0) (Quinlan and Hall, 2010). The .wig files of MACS output were visualized using Integrated Genomics Viewer (v.2.3.8) (Robinson et al., 2011). For OSH1 peak distribution analysis, peaks were binned in each 100-bp window from 10 kb upstream to 10 kb downstream of the gene model normalized to the average gene length in rice. Motif enrichment analysis among OSH1 binding peaks was performed using the MEME program (Bailey et al., 2006). Functional classification of gene category was performed as described previously (Bolduc et al., 2012) using rice gene Functional Category (Rice_japonica_mapping_merged_08 download) available from the Mapman website (http://mapman.gabipd.org/).
RNA-seq
The RNA-seq experiment was performed with two biological replicates. Wild-type and 35S:OSH1-GR plants were grown on MS media in sterilized containers. Two weeks later, these containers were opened and water was added and replaced every day. Another week later, the water was replaced with 10 μM DEX water solution. Before and after 3, 6, and 24 h of DEX treatment, 1-mm-square sections of young leaves 1 mm above the shoot apices from 20 plants were dissected and immediately frozen in liquid nitrogen. Total RNA was extracted using Trizol (Invitrogen). Poly(A) RNA was purified from 2 μg of total RNA using a Dynabeads Oligo(dT) kit (Invitrogen), and this purification was repeated again to exclude rRNA. After elution of poly(A) RNA with 10 μL of water, RNA-seq libraries were prepared using the ScriptSeq v2 RNA-Seq library preparation kit (Epicentre) based on the manufacturer’s protocol. Eight libraries (including the wild type and 35S:OSH1-GR for all time points) were indexed using ScriptSeq Index PCR primers (epicenter), pooled per lane, and sequenced using Illumina Hisequation 2000 with SR50 (single ended). Reads were aligned to the rice genome (MSU7.0) using Tophat (v.2.0.7) (Trapnell et al., 2009) with the option of –library-type fr-secondstrand. A GFF3 file containing the coordinates of mRNA, exon, and coding sequence of nuclear-encoded genes for the rice genome MSU7.0 was supplied as a guide for transcript annotation. Identification of differentially expressed genes was performed by pairwise comparison between samples before and after DEX treatment using the cuffdiff function of Cufflinks (v.2.0.2) (Trapnell et al., 2010). The GEF3 file used above was provided to cuffdiff and differentially expressed genes were determined with a cutoff P value of 0.05.
qPCR
For the DEX induction studies, samples were harvested as described above. For expression analysis between the wild type and osh1 mutants, shoot apices containing the SAM, first three leaf primordia, and the stem from 1 d after imbibition were harvested and frozen in liquid nitrogen. Total RNA was extracted using Trizol and cDNA was synthesized as described previously (Tsuda et al., 2011). cDNA from a 20-μL reaction was diluted by adding 80 μL of water, and 1 μL of cDNA was used for a 20-μL reaction of qPCR as described previously (Bolduc and Hake, 2009). Three biological replicates were performed for each sample. For ChIP-seq validation, 10 young panicles at the 5-mm stage were used and ChIP DNA was prepared as described (Tsuda et al., 2011). Using 1 μL of ChIP DNA, qPCR reactions were performed as described above. The primers used are listed in Supplemental Table 1.
Confocal Microscopy
Hand sections of transgenic plants were mounted in sterilized water. GFP fluorescence was observed under LSM710 confocal microscopy (Leica) with 470-nm excitation and 535-nm emission filters. GFP images were merged with transmitted light images.
Accession Numbers
ChIP-seq raw data have been deposited in DDBJ under accession numbers DRA000206 and DRA000313. RNA-seq data have been deposited in DDBJ under accession number DRA002287.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. OSH1 ChIP-seq Data on Class I KNOX Loci.
Supplemental Figure 2. Verification of OSH1-Bound Loci Found in ChIP-seq.
Supplemental Figure 3. Motif Enrichment Analysis among OSH1-Bound Regions.
Supplemental Figure 4. Seven OSH1-Bound Genes Upregulated in All Time Points after DEX Induction in 35S:OSH1-GR.
Supplemental Figure 5. OSH1 Binding Motifs Found in ChIP-seq Peaks at Three CYP734A Loci.
Supplemental Figure 6. ChIP Assay Using α-OSH1 Antibody Followed by qPCR on Three CYP734A Loci in 2-Week-Old Shoot Apices.
Supplemental Figure 7. Expression of BR Pathway Genes Bound by OSH1.
Supplemental Figure 8. KN1 Targets CYP734A Genes in Maize.
Supplemental Table 1. Oligonucleotide Sequences Used in This Study.
Supplemental Table 2. Summary Statistics for ChIP-seq Library Alignment
Supplemental Table 3. Summary Statistics for RNA-seq Library Alignment
Supplemental Data Set 1. Putative OSH1 Direct Targets Identified by ChIP-seq.
Supplemental Data Set 2. Genes Modulated in 35S:OSH1-GR after DEX Treatment.
Supplementary Material
Acknowledgments
We thank Hake laboratory members for technical advice, discussion, and critical reading of the article. We thank Masahiro Fujita for RNA-seq data registration. The work was funded by NIFA Grant 2012-67014-19429 and JSPS Postdoctoral Fellowships for Research Abroad.
AUTHOR CONTRIBUTIONS
K.T., N.K., and S.H. designed the research. K.T. performed the research and analyzed the data. H.O. contributed to the analysis of ChIP-seq data. K.T. and S.H. wrote the article.
Glossary
- SAM
shoot apical meristem
- GA
gibberellin
- CK
cytokinin
- ChIP-seq
chromatin immunoprecipitation followed by next-generation sequencing
- BR
brassinosteroid
- RNAi
RNA interference
- DEX
dexamethasone
- BL
brassinolide
- qPCR
quantitative PCR
- MS
Murashige and Skoog
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aoyama, T., Chua, N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bailey, T.L.W., Williams, N., Misleh, C., Li, W.W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34: W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, E.M., Lin, W.C., Husbands, A.Y., Yu, L., Jaganatha, V., Jablonska, B., Mangeon, A., Neff, M.M., Girke, T., Springer, P.S. (2012). Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 109: 21146–21151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc, N., Hake, S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc, N., Yilmaz, A., Mejia-Guerra, M.K., Morohashi, K., O’Connor, D., Grotewold, E., Hake, S. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Chen, S. (1993). Brassinosteroid-induced rice lamina joint inclination and its relation to indole-3-acetic acid and ethylene. Plant Growth Regul. 16: 189–196 [Google Scholar]
- Claeys, H., De Bodt, S., Inzé, D. (2014). Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 19: 231–239 [DOI] [PubMed] [Google Scholar]
- Clouse, S.D., Sasse, J.M. (1998). BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Freeling, M., Hake, S. (1985). Developmental genetics of mutants that specify knotted leaves in maize. Genetics 111: 617–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron, J.M., Liu, J.S., Fan, M., Bai, M.Y., Wenkel, S., Springer, P.S., Barton, M.K., Wang, Z.Y. (2012). Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 21152–21157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z., Fujioka, S., Blancaflor, E.B., Miao, S., Gou, X., Li, J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig, T., Corvalan, C., Best, N.B., Budka, J.S., Zhu, J.Y., Choe, S., Schulz, B. (2012). Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS ONE 7: e36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z., et al. . (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32: 495–508 [DOI] [PubMed] [Google Scholar]
- Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., Yoshida, S., Ashikari, M., Kitano, H., Matsuoka, M. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, T., Numaguchi, K., Miura, K., Yoshida, K., Thanh, P.T., Htun, T.M., Yamasaki, M., Komeda, N., Matsumoto, T., Terauchi, R., Ishikawa, R., Ashikari, M. (2013). OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 45: 462–465, e1–e2 [DOI] [PubMed] [Google Scholar]
- Ito, Y., Eiguchi, M., Kurata, N. (2001). KNOX homeobox genes are sufficient in maintaining cultured cells in an undifferentiated state in rice. Genesis 30: 231–238 [DOI] [PubMed] [Google Scholar]
- Jackson, D. (2002). Double labeling of KNOTTED1 mRNA and protein reveals multiple potential sites of protein trafficking in the shoot apex. Plant Physiol. 129: 1423–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D., Veit, B., Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., Phillips, A., Hedden, P., Tsiantis, M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Kaufmann, K., Muiño, J.M., Østerås, M., Farinelli, L., Krajewski, P., Angenent, G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Konishi, S., Izawa, T., Lin, S.Y., Ebana, K., Fukuta, Y., Sasaki, T., Yano, M. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396 [DOI] [PubMed] [Google Scholar]
- Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., Sakakibara, H., Kyozuka, J. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Langmead, B., Trapnell, C., Pop, M., Salzberg, S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele, G., Ori, N., Sato, Y., Hake, S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, D., Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Nguyen, S.M., Malancharuvil, E.J., Fujioka, S., Noguchi, T., Seto, H., Tsubuki, M., Honda, T., Takatsuto, S., Yoshida, S., Chory, J. (1999). BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, D.L., Runions, A., Sluis, A., Bragg, J., Vogel, J., Prusinkiewicz, P., Hake, S. (2014). A division in PIN-mediated patterning during lateral organ initiation in grasses. PLoS Comput. Biol. 10: e1003447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, A.R., Hall, I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Robinson, J.T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E.S., Getz, G., Mesirov, J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M., Iwahori, S., Matsuoka, M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., Kawabe, A., Tokida-Segawa, A., Shimizu, B., Takatsuto, S., Shimada, Y., Fujioka, S., Mizutani, M. (2011). Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. Plant J. 67: 1–12 [DOI] [PubMed] [Google Scholar]
- Sentoku, N., Sato, Y., Matsuoka, M. (2000). Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev. Biol. 220: 358–364 [DOI] [PubMed] [Google Scholar]
- Sentoku, N., Sato, Y., Kurata, N., Ito, Y., Kitano, H., Matsuoka, M. (1999). Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11: 1651–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M.S., Boschke, I., Hake, S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. USA 99: 9579–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). Patterns in Plant Development. (Cambridge, UK: Cambridge University Press). [Google Scholar]
- Sun, B., Xu, Y., Ng, K.H., Ito, T. (2009). A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 23: 1791–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, B., Looi, L.S., Guo, S., He, Z., Gan, E.S., Huang, J., Xu, Y., Wee, W.Y., Ito, T. (2014). Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343: 1248559. [DOI] [PubMed] [Google Scholar]
- Sun, Y., et al. . (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester, A.W., Cande, W.Z., Freeling, M. (1990). Division and differentiation during normal and liguleless-1 maize leaf development. Development 110: 985–1000 [DOI] [PubMed] [Google Scholar]
- Trapnell, C., Pachter, L., Salzberg, S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Williams, B.A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M.J., Salzberg, S.L., Wold, B.J., Pachter, L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K., Ito, Y., Sato, Y., Kurata, N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23: 4368–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht, E., Reiser, L., Hake, S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Wada, K., Marumo, S., Ikekawa, N., Morisaki, M., Mori, K. (1981). Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol. 22: 323–325 [Google Scholar]
- Wang, Z.Y., Bai, M.Y., Oh, E., Zhu, J.Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46: 701–724 [DOI] [PubMed] [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H., Matsuoka, M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai, O., Shani, E., Dolezal, K., Tarkowski, P., Sablowski, R., Sandberg, G., Samach, A., Ori, N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Liu, T., Meyer, C.A., Eeckhoute, J., Johnson, D.S., Bernstein, B.E., Nusbaum, C., Myers, R.M., Brown, M., Li, W., Liu, X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.