This work describes the role of a small noncoding RNA in the regulation of photosynthetic gene expression in cyanobacteria. Advanced computational target prediction combined with extensive experimental data reveal the photosynthesis regulatory RNA 1 as a key modulator of photosynthetic functions upon high-light exposure.
Abstract
Little is known so far about RNA regulators of photosynthesis in plants, algae, or cyanobacteria. The small RNA PsrR1 (formerly SyR1) has been discovered in Synechocystis sp PCC 6803 and appears to be widely conserved within the cyanobacterial phylum. Expression of PsrR1 is induced shortly after a shift from moderate to high-light conditions. Artificial overexpression of PsrR1 led to a bleaching phenotype under moderate light growth conditions. Advanced computational target prediction suggested that several photosynthesis-related mRNAs could be controlled by PsrR1, a finding supported by the results of transcriptome profiling experiments upon pulsed overexpression of this small RNA in Synechocystis sp PCC 6803. We confirmed the interaction between PsrR1 and the ribosome binding regions of the psaL, psaJ, chlN, and cpcA mRNAs by mutational analysis in a heterologous reporter system. Focusing on psaL as a specific target, we show that the psaL mRNA is processed by RNase E only in the presence of PsrR1. Furthermore, we provide evidence for a posttranscriptional regulation of psaL by PsrR1 in the wild type at various environmental conditions and analyzed the consequences of PsrR1-based regulation on photosystem I. In summary, computational and experimental data consistently establish the small RNA PsrR1 as a regulatory factor controlling photosynthetic functions.
INTRODUCTION
Regulatory, noncoding RNA molecules are ubiquitous posttranscriptional regulators of gene expression in all phyla of life. In plants, microRNAs, one particular class of RNA regulators, exert an important control on many aspects of gene expression (Cuperus et al., 2011). However, the functions of other classes of potential RNA regulators, such as long noncoding RNAs and cis-natural antisense transcripts, are scarcely understood (De Lucia and Dean, 2011). Noncoding RNAs are not only transcribed in the plant nucleus but are also abundant in plant chloroplasts (Hotto et al., 2011; Zhelyazkova et al., 2012). With regard to the possible involvement of noncoding RNAs in plastid-related regulatory networks, it is unclear at present whether these have evolved from their cyanobacterial ancestors and to what extent such regulators play a role in cyanobacteria. As a first step, small RNA (sRNA) regulators of gene expression with an impact on photosynthesis should be identified, using simple unicellular cyanobacteria as model organisms. Furthermore, in view of their ecological relevance and current efforts to develop cyanobacteria into viable systems for biofuel production, the identification and functional characterization of cyanobacterial sRNAs is of great direct interest.
Recent transcriptome analyses have demonstrated a high number of noncoding transcripts in different groups of cyanobacteria, and many of them respond to different environmental conditions (Steglich et al., 2008; Mitschke et al., 2011a, 2011b). However, direct or indirect evidence for functional interactions with target mRNAs has been presented for only a single trans-encoded cyanobacterial sRNA thus far (Nakamura et al., 2007; Richter et al., 2010). In contrast, several cis-encoded antisense RNAs (asRNAs) impacting very specific photosynthesis-related aspects have been described. In the model organism Synechocystis sp PCC 6803 (hereafter Synechocystis 6803), IsrR regulates the stress-induced accumulation of the CP43 homolog IsiA in a threshold linear response (Dühring et al., 2006a; Legewie et al., 2008; Georg and Hess, 2011). Another negatively acting asRNA in Synechocystis 6803 is As1_flv4, which prevents the premature expression of the flv4-2 operon upon a shift in inorganic carbon supply (Eisenhut et al., 2012). The flavodiiron proteins encoded by the flv4-2 operon have a pivotal function in photoprotection of photosystem II (PSII) against oxidative stress (Zhang et al., 2012). By contrast, a positive regulatory function was assigned to two asRNAs that protect the psbA2 and psbA3 5′ leader sequences from premature degradation by ribonuclease E (RNase E) (Sakurai et al., 2012).
Given the high susceptibility of the photosynthetic process to dynamic environmental changes, the involvement of a set of versatile trans-encoded sRNA molecules in the regulation of photosynthesis and inorganic carbon acquisition seems advantageous. In the bacterium Rhodobacter sphaeroides, which is facultatively capable of anoxygenic photosynthesis, the trans-acting sRNA PcrZ has a direct regulatory function in the formation of photosynthetic complexes (Mank et al., 2012). However, no trans-encoded sRNAs regulating oxygenic photosynthesis in plant chloroplasts or cyanobacteria have been described thus far.
Biocomputational prediction (Voss et al., 2009), tiling microarrays (Georg et al., 2009), and pyrosequencing of Synechocystis 6803 transcripts (Mitschke et al., 2011a) revealed the existence of several hundred candidate sRNAs in this cyanobacterial model organism. One of the most abundant sRNAs in these screens was SyR1 (for Synechocystis RNA1), a 131-nucleotide-long transcript from the intergenic region between the fabF (slr1332) and hoxH (sll1226) genes. More detailed investigation revealed that this sRNA is upregulated under high-light treatment and CO2 depletion (Georg et al., 2009; Kopf et al., 2014). In addition, a strain overexpressing SyR1 exhibited a bleaching phenotype with considerably reduced amounts of photosynthetic pigments (Mitschke et al., 2011a). Based on the data presented in this study, the previous name of this sRNA, SyrR1, was replaced by PsrR1 (for photosynthesis regulatory RNA1).
RESULTS
The sRNA PsrR1 Is Widely Distributed within the Cyanobacterial Phylum and Affects Photosynthetic Properties
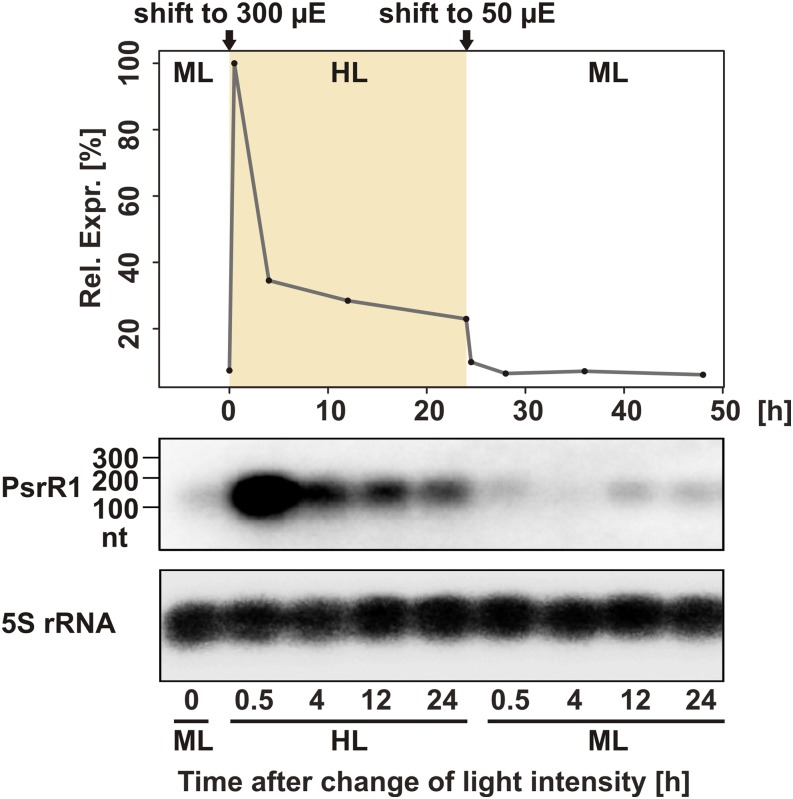
Homologs of the psrR1 gene can be found in the genomes of several cyanobacteria belonging to morphologically and phylogenetically distant groups, including unicellular cyanobacteria such as Synechocystis, Cyanothece, and Microcystis and filamentous cyanobacteria capable of cell differentiation such as Nostoc and Anabaena species. Based on morphological complexity, five subsections of cyanobacteria have been defined (Rippka et al., 1979). We found putative homologs of psrR1 in strains belonging to four of these subsections (Supplemental Figure 1). This broad occurrence suggests a widely conserved function for PsrR1. Expression of the PsrR1 homologs has been detected in transcriptomic data sets from Synechocystis 6803 (Georg et al., 2009; Mitschke et al., 2011a), Synechococcus sp PCC 7002 (Ludwig and Bryant, 2012), and Anabaena sp PCC 7120 (Mitschke et al., 2011b). Sequence alignments suggest the presence of a highly conserved central region, likely involved in target interaction, and of a second highly conserved region toward the 3′ end (Figure 1). By contrast, the 5′ ends of the PsrR1 homologs differ substantially from each other. Despite the sequence conservation of PsrR1, there is no conservation of its genomic context. In Synechocystis 6803, psrR1 expression appears strongly high-light dependent. The transcript level increased dramatically after a shift from moderate light (50 μmol photons m−2 s−1) to high-light conditions (300 μmol photons m−2 s−1) and then slowly declined to an elevated steady state level with ∼4-fold higher transcript accumulation compared with moderate light conditions. After a shift to moderate light conditions, PsrR1 accumulation dropped to the initial level within 30 min (Figure 2).
Figure 1.
Selected PsrR1 Homologs in the Cyanobacterial Phylum.
A sequence alignment of putative PsrR1 homologs from unicellular and multicellular cyanobacteria is shown. Only the conserved part is shown, including a highly conserved region involved in interaction with target mRNAs and an imperfect palindromic region able to fold into a hairpin secondary structure, followed by a T-rich sequence, hallmarks of a Rho-independent terminator of transcription. Experimentally verified transcripts are indicated by asterisks and are in boldface. In M. aeruginosa NIES-843, two copies of PsrR1 were found. The alignment includes the nucleotides 50 to 131 from the Synechocystis 6803 PsrR1 homolog.
Figure 2.
Accumulation of PsrR1 sRNA.
The expression kinetic of PsrR1 is shown after a shift from 50 μmol photons m−2 s−1 (moderate light; ML) to 300 μmol photons m−2 s−1 (high light; HL) and back to moderate light. The bottom part shows PsrR1 RNA gel blot analysis, and the top part shows densitometric analysis of the RNA gel blot. nt, nucleotides.
[See online article for color version of this figure.]
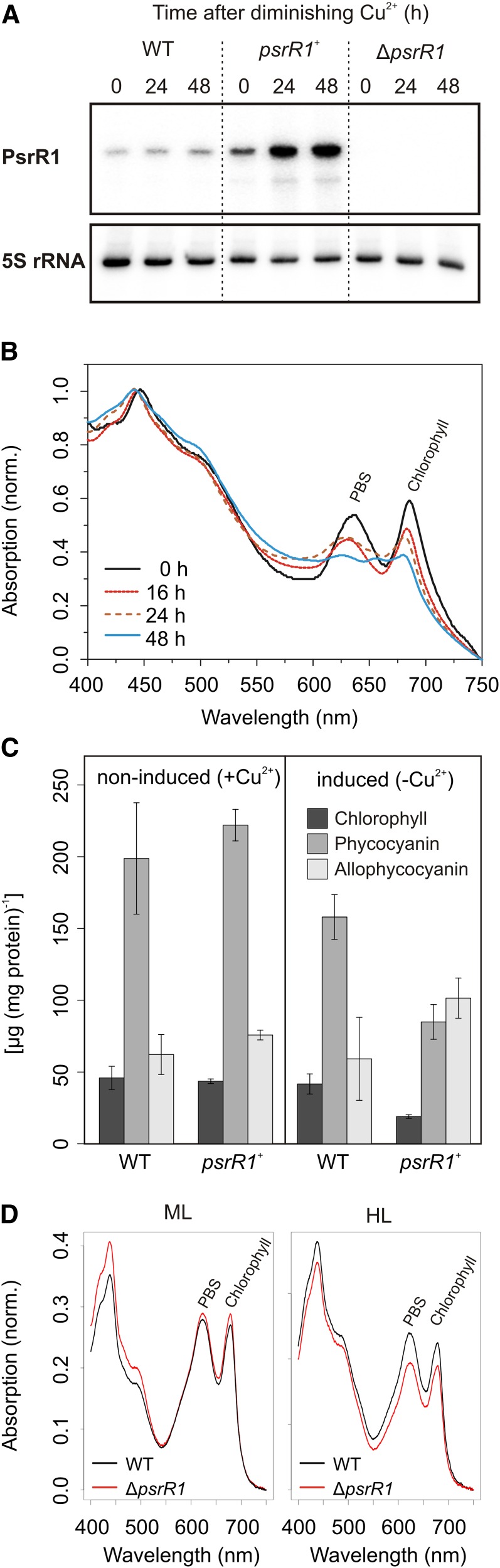
For an initial study of the phenotypic effects of altered PsrR1 abundance, we constructed a knockout strain (ΔpsrR1) by replacing the psrR1 gene with an antibiotic resistance cassette as well as an overexpressor strain (psrR1+). Ectopic expression of psrR1 was induced by copper limitation using the PpetJ promoter. Verification of overexpression as well as absence of the PsrR1 transcript in the respective strains are shown in Figure 3A. Consistent with previous reports (Mitschke et al., 2011a), a time course of PsrR1 overexpression led to stepwise cell bleaching (Figure 3B) within 48 h. In the wild type even after 5 d of growth under copper limitation, no such bleaching was observed (Supplemental Figure 2). Analysis of the pigment content in cell extracts normalized to total protein concentration of the cells revealed a major decrease in chlorophyll as well as phycocyanin upon PsrR1 overexpression in comparison with the wild type under inducing (copper-limiting) conditions (Figure 3C). Interestingly, allophycocyanin content appeared to be increased in psrR1+. As we observed a massive decrease in the content of phycocyanin, which is one of the most abundant soluble proteins in the cell, the allophycocyanin level was most probably overestimated by the normalization to total protein content. Under the same moderate light conditions, no alteration in the absorption spectrum of the ΔpsrR1 strain was evident. In order to reveal smaller differences, whole cell absorption spectra were measured using a spectrophotometer with an integrating sphere (Figure 3D). Under moderate light, a very small pigment increase was detected in ΔpsrR1 cells, whereas high-light illumination led to an ∼20% decrease of phycocyanin and chlorophyll as calculated from the spectra by the method of Myers et al. (1980).
Figure 3.
Phenotypes of the psrR1 Mutant Strains.
(A) RNA gel blot analysis of PsrR1 accumulation in the psrR1 overexpressor strain (psrR1+) upon removal of copper after 24 and 48 h in comparison with the wild type and the knockout strain (ΔpsrR1) grown under the same conditions. 5S rRNA was used as a loading control.
(B) Overexpression of psrR1 leads to cell bleaching. Whole cell absorption spectra of a psrR1+ culture are shown before as well as 16, 24, and 48 h after induction of psrR1 expression from the PpetJ promoter by copper removal. At 0 h, cells were harvested, washed twice with copper-free medium, and then incubated for a further 48 h in copper-free medium. The peak at 625 nm resembles mainly absorption from phycocyanin, whereas allophycocyanin absorbs at 655 nm. The peak at 680 nm originates from chlorophyll. Absorption spectra were normalized to OD750. PBS, phycobilisomes.
(C) Pigment analysis of the psrR1+ mutant strain under inducing (5 d under copper-limiting conditions) and noninducing conditions in comparison with the isogenic wild-type control. Data are means ± sd from three replicate analyses.
(D) Whole cell absorption spectra of wild-type and ΔpsrR1 strains grown under moderate light conditions (ML) and then shifted to high-light conditions (HL) for 48 h. Spectra were measured using a spectrophotometer equipped with an integrating sphere to reduce light scattering and then normalized to cell density.
Advanced Computational Target Prediction and Functional Enrichment Analysis Suggest Photosynthesis-Related mRNAs as Targets of PsrR1
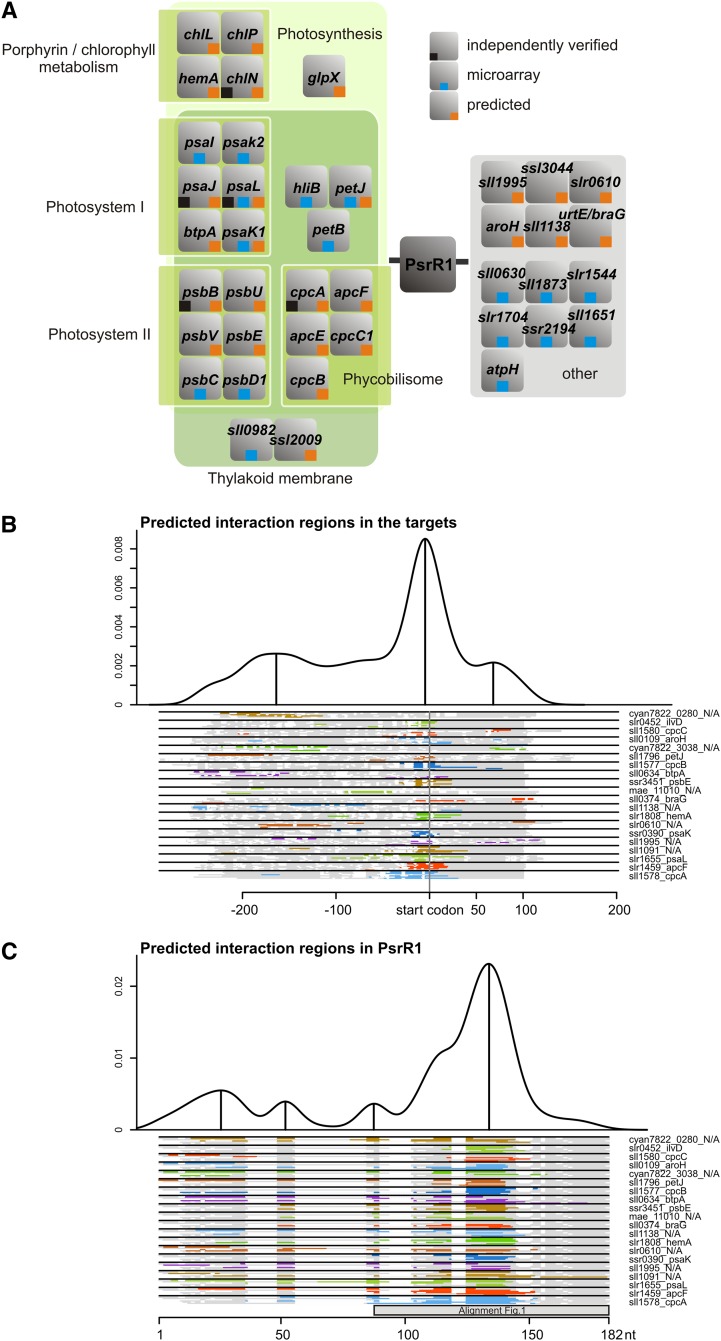
The prediction of bacterial sRNA targets can be highly improved when comparative phylogenetic information is taken into account (Wright et al., 2013). As PsrR1 has homologs throughout the cyanobacterial phylum, we applied the recently developed CopraRNA tool, which detects RNA targets that are conserved among a selected set of organisms (Wright et al., 2014). In this analysis, we included five homologs, one each from Microcystis aeruginosa NIES-843, Stanieria cyanosphaera PCC 7437, and Cyanothece sp PCC 7822, PCC 8801, and ATCC 51142, in addition to the one from Synechocystis 6803. Individual bacterial sRNAs often regulate related sets of genes (e.g., genes involved in iron homeostasis [RyhB] or amino acid metabolism [GcvB]) (Wright et al., 2013). For that reason, we arbitrarily took the top 85 predictions, including 60 targets present in Synechocystis 6803, and subjected them to a functional enrichment analysis (Huang et al., 2009) to analyze if functionally related genes are also enriched in this prediction. The complete list of predicted targets is presented in Supplemental Data Set 1. Strikingly, 20 of the 60 predicted targets are involved in photosynthesis or are thylakoid associated (Supplemental Figure 3). The final list was restricted to 26 targets, including the 15 most supported predictions and those of the top 85 targets that were functionally enriched (Figure 4A; Supplemental Table 1). The prediction of PsrR1 targets includes mRNAs for several photosystem I (PSI), PSII, and phycobilisome proteins as well as for enzymes of the chlorophyll biosynthetic pathway. The top three computationally predicted targets are all related to photosynthesis, namely the phycocyanin α-subunit CpcA, the phycobilisome core component ApcF, and the PSI reaction center protein subunit XI, PsaL. These findings suggest PsrR1 as a posttranscriptional regulator of photosynthesis, which is consistent with the observed phenotypes of psrR1 mutants (Figure 3). Although these target predictions were performed with the full-length PsrR1 sequences, the interaction domains clustered in the central conserved region of the sRNA (Figure 4C). Within the mRNAs, the vast majority of sequences predicted as PsrR1 targets are close to or overlapping with the ribosomal binding site (RBS) or the translational start codon (Figure 4B), consistent with the prevalent mechanism of sRNA function in bacteria.
Figure 4.
PsrR1 Is Predicted to Control Genes for Proteins Involved in Photosynthesis and Tetrapyrrole Metabolism.
(A) Visualization of the functional enrichment analysis for targets of PsrR1 (orange squares) predicted by the CopraRNA tool (Wright et al., 2013). All top 15 CopraRNA target predictions are shown plus selected predictions from the top 85 candidates that were functionally enriched and three additional candidates (ccmk1, ilvB, and livf). Targets subsequently verified experimentally (Figure 5) are labeled by black squares, and targets detected by microarray analysis (Supplemental Table 1) are labeled with blue squares.
(B) Visualization of the predicted interaction domains within the predicted mRNA targets of PsrR1.
(C) Visualization of the predicted interaction domains in PsrR1 for the different targets.
In (B) and (C), the density plots at the top give the relative frequency of a specific nucleotide position in the predicted PsrR1–target interactions. The plots combine all predictions with a CopraRNA P value ≤ 0.01 in all included homologs. Local maxima indicate distinct interaction domains and are marked with vertical lines. The schematic alignments of PsrR1 homologs and of targets at the bottom show the predicted interaction domains. The aligned regions are displayed in gray, gaps in white, and predicted interaction regions in color (color differences are for contrast only). The positions of start codons are annotated, and locus tags and gene names of a representative cluster member are given on the right, if available from Synechocystis 6803. The region covered by the alignment in Figure 1 is indicated by the gray box. The numbering at the x axes refers to the alignment position.
Genome-Wide Identification of PsrR1 Targets in Synechocystis 6803 Based on Microarrays
As a second, independent approach to find PsrR1 targets, we performed transcriptome analysis using microarrays. Although sRNAs mainly function at the posttranscriptional level, target mRNA levels frequently, but not invariably, decrease when protein synthesis or transcript stability is affected (Vogel and Wagner, 2007). Initial tests using quantitative RT-PCR monitoring the kinetics of psrR1 overexpression revealed a distinct overaccumulation of PsrR1 under the control of the PpetJ promoter 24 h after induction (Supplemental Figure 4). For the induction of expression from the PpetJ promoter, the copper-containing medium has to be replaced, after which it takes time before the cells have depleted their internal copper pool and start expressing the sRNA. This explains the relatively long delay in maximal PsrR1 expression after induction. Consequently, we compared the overexpressor strain psrR1+ with an empty-vector control strain (designated here as the wild type) by microarray after 24 h of psrR1 induction. Total RNA was sampled immediately before (time 0) as well as 24 h after copper step down in order to favor the detection of direct target candidates of PsrR1. This strategy was chosen to minimize the number of false positives that might result from secondary and pleiotropic effects upon long-term overexpression of the sRNA. A decrease of phycocyanin and chlorophyll absorption was observed upon psrR1 overexpression (Figure 3B), confirming that regulatory interactions were effectively taking place. Quantification of the microarray data demonstrated a 13.6-fold increase of PsrR1 abundance in comparison with the wild type at 24 h. Microarray results were hence analyzed in terms of differential transcript accumulation between the wild type and psrR1+ at 24 h after copper step down (psrR1+/wild type). Transcripts with a log2 fold change ≥ 0.8 and an adjusted P value ≤ 0.05 were taken as significantly differentially expressed. The results are summarized in Supplemental Table 1 and visualized in a whole genome expression plot (Supplemental Data Set 2).
Besides PsrR1, the levels of 25 RNA features were identified as significantly affected by PsrR1 overexpression (Supplemental Table 1). This includes the coding regions of 17 mRNAs plus eight 5′ untranslated regions (UTRs), most of which show the same tendency as their corresponding mRNAs. An inverse effect was observed for psaL, whose 5′ UTR accumulated to higher levels in the psrR1+ strain, whereas the coding region showed the opposite behavior. The changes in transcript levels were relatively small, with a maximal fold change of ∼2−1.4 (psaK1, hliB-5′ UTR). Again, the set of transcripts showing differential accumulation pointed strongly toward the regulation of photosynthesis, as it included the mRNAs encoding the PSI components PsaK1 and PsaK2, the dicistronic message for the subunits PsaL and PsaI, as well as the mRNAs encoding HliB, PetJ, PsbC, and some unknown or hypothetical proteins. The majority of these mRNAs (12 of 17) and 5′ UTRs (5 of 8) were negatively influenced by psrR1 overexpression. Only sll0982, an uncharacterized thylakoid-associated protein (Srivastava et al., 2005), the psbD1C operon, cytochrome b6 (petB), atpH, as well as the 5′ UTRs of psaL and srrA accumulated to higher levels in the mutant. Certainly, this microarray-based approach could not identify PsrR1 targets that were not expressed under our standard growth conditions. Furthermore, regulation by sRNAs at the posttranscriptional level does not necessarily lead to a significant change in the accumulation of the target mRNAs. Therefore, it is advantageous to have the results of an independent computational target prediction that is complementary to the microarray analysis, as summarized in Figure 4A.
Verification of PsrR1 Targets
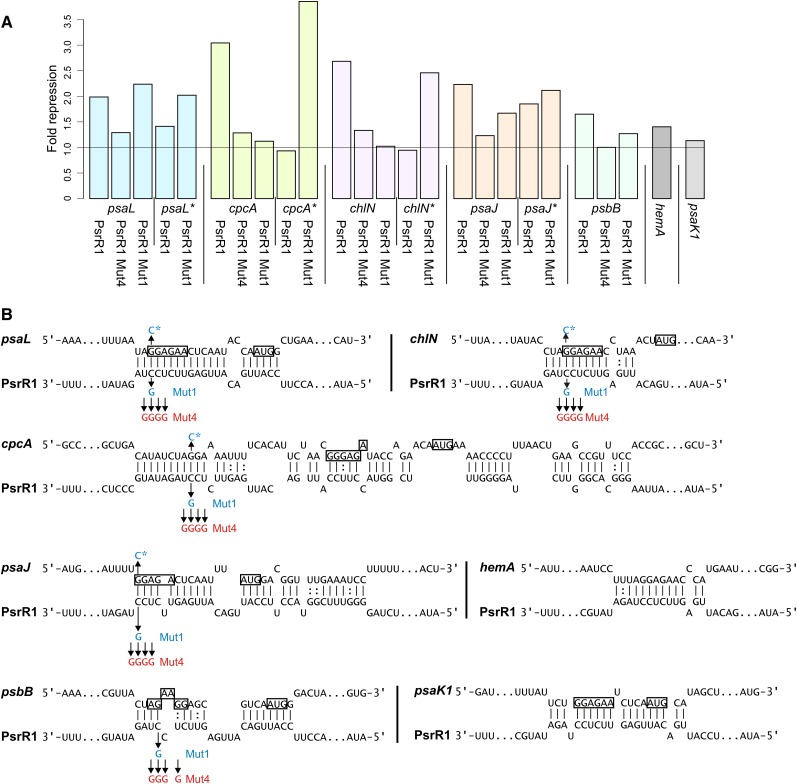
To reject or verify computational target predictions and targets suggested by the microarray experiment, we selected seven mRNAs for experimental analysis. These were cpcA, psaL, psaK1, hemA, chlN, psbB, and psaJ, which were ranked on positions 1, 3, 6, 8, 34, 38, and 41 of the CopraRNA prediction (Supplemental Table 1 and Supplemental Data Set 1). For this purpose, we employed a reporter assay tailored to investigate posttranscriptional regulation (Corcoran et al., 2012), in which the 5′ UTR of the putative target is fused to the superfolder green fluorescent protein gene (sgfp) and is coexpressed with PsrR1 or a control sRNA in Escherichia coli. The presence of PsrR1 led to significantly reduced GFP fluorescence in strains carrying reporter fusions with the 5′ UTRs of psaL (1.99-fold; Figure 5A), cpcA (3.04-fold), chlN (2.68-fold), psaJ (2.23-fold), and psbB (1.65-fold). Only a minor effect and no effect were detected for the hemA (1.40-fold) and psaK1 (1.13-fold) UTRs, respectively (Figure 5A). This latter result may be caused by the fact that cyanobacterial mRNAs are not always compatible with the translation machinery in E. coli or may indicate that hemA and psaK1 mRNAs are not genuine targets of PsrR1. The raw fluorescence values are shown in Supplemental Figure 5.
Figure 5.
Verification of PsrR1–mRNA Interactions in a Heterologous Reporter Assay.
(A) Translational repression of 5′ UTR–sgfp fusions when overexpressing PsrR1 in E. coli. The wild-type form of PsrR1 was compared against two versions with point mutations in the targeting region (Mut4 and Mut1 [B]) in its interaction with the 5′ UTR of genes psaL, psaL*, cpcA, cpcA*, chlN, chlN*, psaJ, psaJ*, and psbB. The 5′ UTR–sgfp fusions for genes psaK1 and hemA were tested with the wild-type PsrR1 only. Point mutations in the UTRs are indicated by asterisks. The fold reduction is the ratio of the GFP fluorescence of the respective translational 5′ UTR–sgfp fusion in the presence of the control plasmid pJV300 and a plasmid for the overexpression of the respective PsrR1 variant, after subtraction of the background fluorescence.
(B) Predicted interactions of the tested targets with PsrR1. Point mutations are indicated by arrows and red and blue letters. The putative ribosome binding sites and the start codons are boxed.
A change of four nucleotides (UCCU to GGGG, Mut4) (Figure 5B) within the most conserved central part of PsrR1 drastically reduced the negative impact on the expression of sgfp when fused to the 5′ UTRs of psaL (1.99- to 1.29-fold), cpcA (3.04- to 1.28-fold), chlN (2.68- to 1.33-fold), psaJ (2.23- to 1.23-fold), and psbB (1.65- to 1.00-fold) (Figure 5A). We further scrutinized the interactions by the substitution of a single nucleotide in PsrR1 (C to G, Mut1) (Figure 5B) and of the predicted complementary nucleotide in the 5′ UTRs of psaL, psaJ, cpcA, and chlN. The effects of single nucleotide substitutions in combination with changes of the respective complementary nucleotide in the 5′ UTRs are shown in Figure 5A. The single point mutations in either PsrR1 or the UTRs reduced the repression of GFP fluorescence in all cases, except the combination of the psaL wild-type 5′ UTR and PsrR1 Mut1, where this single substitution appeared insufficient to disturb the interaction. For all tested UTRs, we could reestablish the repression by PsrR1 Mut1 by the respective compensatory mutations (Figure 5A). In summary, testing seven predicted targets in a heterologous reporter system confirmed five as likely targets of PsrR1. Four targets are linked to photosynthesis and one to chlorophyll biosynthesis. The point mutations in PsrR1 as well as in the UTR produced additional evidence for a direct sRNA–target interaction.
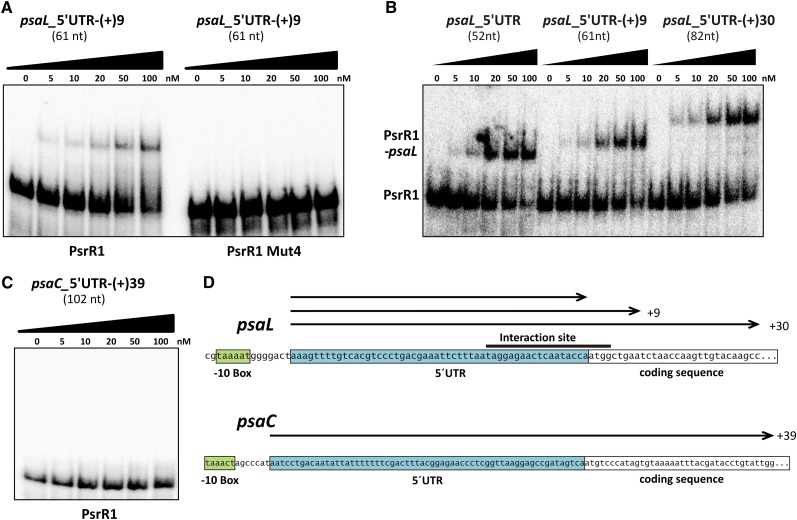
In Vitro Interaction of PsrR1 with the 5′ UTR of psaL mRNA
The psaL mRNA was suggested as a target by all three methods (bioinformatic prediction, microarray analysis, and the E. coli reporter system) used. This indicates that psaL is a promising direct target. To address the in vitro properties of the predicted interaction of PsrR1 and the psaL 5′ UTR, electrophoretic mobility shift assay (EMSA) was performed using PsrR1 as the 5′ end 32P-labeled probe at a constant final concentration (1 nM). In order to exclude possible effects from artificial secondary structures of in vitro–transcribed 5′ UTR fragments, we assayed three different versions of the psaL leader RNA, each covering the predicted interaction site around the RBS (Figure 6). The minimal version (psaL-5′UTR) contained the complete experimentally validated 5′ UTR (Mitschke et al., 2011a) from nucleotides −52 to −1, whereas the longer fragments extended to nucleotide +9 [psaL-5′UTR-(+)9] and +30 [psaL-5′UTR-(+)30] of the coding sequence. The PsrR1 probe was incubated at 30°C with increasing concentrations of unlabeled psaL fragments in the range of 0 to 100 nM. RNA–RNA duplex formation was detected as retardation of PsrR1 migration in native gel electrophoresis. In this approach, PsrR1–psaL interaction could be demonstrated for all tested psaL variants with comparable sensitivity (Figure 6A). In order to confirm the specificity of this in vitro interaction, a mutated version of PsrR1 (PsrR1 Mut4) containing four consecutive base substitutions within the predicted interacting region (Figure 5B) was analyzed, using psaL-5′UTR-(+)9 as bait RNA. Even at a psaL leader concentration of 100 nM, the electrophoretic mobility of PsrR1 Mut4 showed no retardation at all (Figure 6B), proving the significance of the highly conserved region of PsrR1 (Figures 1, 4C, and 5B) for functional interactions with its targets. The 5′ UTR of psaC has not been predicted to interact with PsrR1 (prediction rank 278; P = 0.06). Therefore, a psaC leader fragment covering all 63 nucleotides of the 5′ UTR up to nucleotide +39 within the coding sequence [psaC-5′UTR-(+)39] appeared to be an appropriate negative control for the PsrR1 binding studies. Figure 6C clearly demonstrates that psaC-5′UTR-(+)39 does not interact with PsrR1 in vitro. Together, our results provide strong evidence for specific direct interactions between PsrR1 and the region around the RBS of psaL mRNA.
Figure 6.
Interaction of PsrR1 with the 5′ UTR of psaL mRNA in Vitro.
(A) EMSA of PsrR1 with the 5′ UTR of psaL mRNA. At a constant final concentration of 1 nM, labeled PsrR1 was incubated for 30 min at 30°C with increasing concentrations (0 to 100 nM) of unlabeled psaL RNA segments.
(B) Comparative EMSA of PsrR1 and a PsrR1 mutant version (PsrR1 Mut4) with psaL-5′UTR-(+9). For the in vitro synthesis of PsrR1 Mut4, a nucleotide exchange was introduced to the anti-RBS sequence as depicted in Figure 5B.
(C) EMSA of PsrR1 with the 5′ UTR of psaC mRNA as a negative control. PsaC was not predicted as a target of PsrR1.
(D) Schematic representation of the fragments tested in (A) to (C). The 5′ UTR and the proposed −10 region of the promoter are indicated by boxes. The black bar indicates the predicted interaction site. Numbers denote nucleotide positions with respect to the first nucleotide of the start codon.
[See online article for color version of this figure.]
Analysis of Photosynthetic Complexes
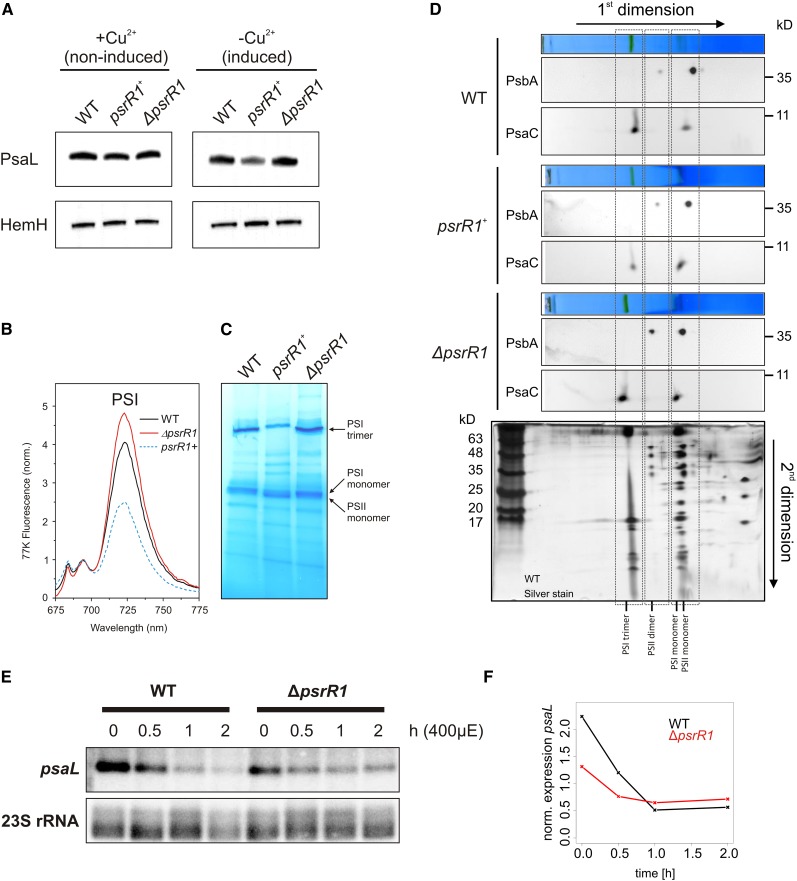
Considering the large set of putative targets of PsrR1, we expected pleiotropic and overlapping effects in mutants with altered PsrR1 abundance. These could make the comprehensive characterization of PsrR1 functions potentially challenging. Therefore, we focused in the following on the specific function of PsaL, as a verified target, in the accumulation and trimerization of PSI. First, we confirmed the microarray data on the repression of psaL mRNA in the psrR1+ by RNA gel blot analysis (Supplemental Figure 6) as well as on protein level by detecting PsaL in the psrR1+ strain by immunoblot analysis under inducing and noninducing conditions (Figure 7A). As expected, overexpression of PsrR1 resulted in reduced amounts of PsaL compared with the wild type and the ΔpsrR1 strain grown under the same conditions.
Figure 7.
Analysis of PSI Complexes from PsrR1 Mutant Strains.
(A) Immunoblot analysis of PsaL and HemH in cell extracts of psrR1 mutant strains. The Synechocystis 6803 isogenic wild type as well as psrR1 overexpressor (psrR1+) and knockout (ΔpsrR1) strains were grown in BG-11 medium to exponential growth phase and incubated in medium lacking CuSO4 (inducing psrR1 expression) or containing 5 µM CuSO4 (repressing psrR1 expression) for 48 h. Proteins of whole cell extracts were subjected to immunoblot analysis of PsaL accumulation. Probing for ferrochelatase (HemH) served as a loading control. For independent quantification, see Supplemental Figure 7.
(B) The 77K fluorescence emission spectra of psrR1+ cultures after induction of PsrR1 overaccumulation for 48 h in comparison with the wild-type and ΔpsrR1 strains. Spectra were normalized to the peak at 695 nm, reflecting fluorescence originating mainly from PSII. The peak at 725 nm originates from PSI complexes.
(C) BN-PAGE of solubilized thylakoids isolated from the wild type as well as psrR1 overexpressor (psrR1+) and ΔpsrR1 mutant strains grown in copper-free medium 48 h after copper step down. Green protein complexes that were seen on the unstained gel are labeled.
(D) After separation of the protein complexes on the blue-native gel (shown above each respective blot), the lanes were excised and subjected to Tris-Tricine PAGE (second dimension). Gels from the second dimension were electroblotted and probed sequentially with PsbA and PsaC antisera. For identification of the protein complexes, a Tris-Tricine gel that originated from the same first dimension of a wild-type sample was subjected to silver staining (bottom panel).
(E) Wild-type and ΔpsrR1 cells were grown under moderate light conditions (time 0) and then shifted for at least 2 h to high light. The isolated RNA was subjected to RNA gel blot analysis and probed with psaL and 23S rRNA as a control.
(F) Signals from the RNA gel blot were quantified with Quantity One software. Levels of psaL at 0, 0.5, 1, 1.5, and 2 h after a shift from 40 to 400 μmol photons m−2 s−1 are shown.
Furthermore, the accumulation of photosynthetic complexes was examined by recording whole cell 77K fluorescence emission spectra of wild-type, ΔpsrR1, and induced psrR1+ cultures. For comparison, all strains were grown for 48 h under copper-limiting conditions at moderate light intensity. Excitation was at 440 nm (Soret absorption band for chlorophyll a; Figure 7B). The spectra show that the peak at 725 nm representing fluorescence from PSI complexes decreased remarkably in psrR1+ cells when spectra were normalized to emission at 680 nm (mainly representing fluorescence from PSII). By contrast, ΔpsrR1 accumulated slightly more PSI complexes than the wild-type and psrR1+ strains, indicated by the stronger peak at 725 nm. These findings point to an impact of PsrR1 on the photosynthetic apparatus, in particular on pigmentation and PSI:PSII ratio. In immunoblot analysis, where the samples were normalized to total protein amount, the small difference in PSI accumulation in ΔpsrR1 as revealed by 77K fluorescence spectra (normalization to PSII fluorescence) was hardly detected (Figure 7A; see quantification in Supplemental Figure 7).
Since PsaL plays a key role in the trimerization of PSI (Chitnis and Chitnis, 1993), possible effects of PsrR1 on the PSI trimer:PSI monomer ratio were examined by blue-native PAGE (BN-PAGE) of isolated thylakoid membranes. The BN-PAGE analysis shown in Figure 7C mainly detected three chlorophyll-containing complexes, whose identities were determined on the basis of subunit composition by Tricine SDS-PAGE analysis in the second dimension followed by silver staining (Figure 7D). Indeed, the psrR1+ strain exhibited a lowered accumulation of the PSI trimer (Figure 7C). Using an antiserum against PsaC, immunoblot analysis of the second dimension further validated a lower PSI trimer:PSI monomer ratio in the psrR1+ overexpressor strain (Figure 7D). While no such pronounced differences were found in the ΔpsrR1 strain, a slight increase in the overall accumulation of PSI complexes was indicated in our BN-PAGE analysis (Figure 7C) as well as from the 77K fluorescence spectra (Figure 7B). Surprisingly, the higher amount of PSI as detected by 77K fluorescence spectroscopy and especially of the PSI trimer does not match the lower amount of the psaL mRNA we detected in the ΔpsrR1 mutant (Figures 7E and 7F). This contradictory result might be a consequence of the complete lack of PsrR1 in the ΔpsrR1 strain. However, upon illumination of the cells with high-intensity light (conditions that naturally induce PsrR1 accumulation), the amount of the psaL mRNA decreased rapidly in the wild type, while in the ΔpsrR1 strain the accumulation of psaL decreased more slowly (Figure 7F). In addition, the amplitude of the psaL high-light response is significantly smaller in the ΔpsrR1 strain (∼−1.8-fold) than in the wild type (∼−4-fold). Both facts are consistent with our view that PsrR1 is required for the dynamic adaptation of the photosynthetic apparatus during a shift to high light. However, as we showed that PsrR1 targets not only psaL but also many other mRNAs related to photosynthesis, possible pleiotropic effects of the permanent lack of PsrR1 might be considered as well.
In summary, analysis of the accumulation of photosynthetic complexes suggests that downregulation of PsaL by the overexpression of PsrR1 leads to a lower PSI content and a specific change in the PSI monomer-to-trimer ratio, whereas deletion of psrR1 leads to slightly increased amounts of PSI. Thus, we conclude also from physiological data that the psaL mRNA is one of the primary targets of PsrR1. In the following, we reveal mechanistic aspects of PsrR1 function using the example of psaL mRNA.
Posttranscriptional Destabilization of psaL mRNA upon PsrR1 Overexpression
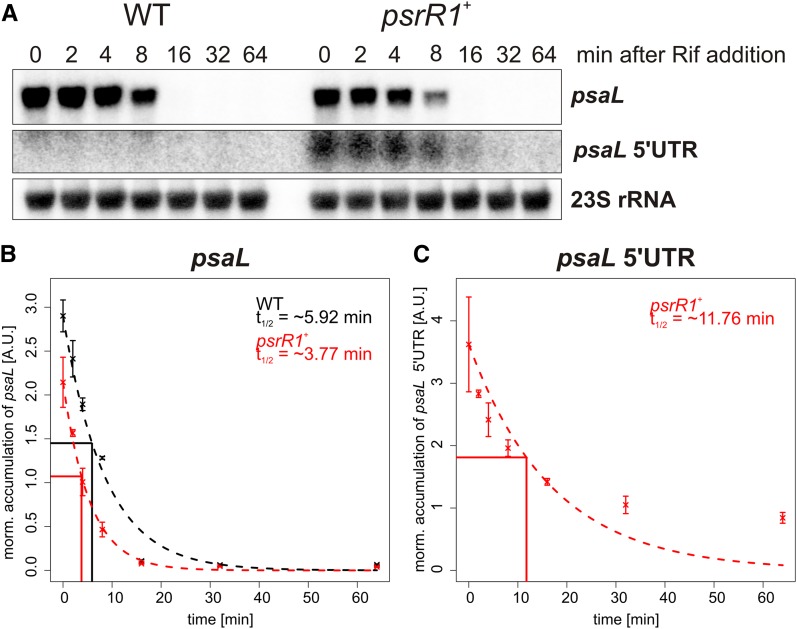
To test if the reduced transcript accumulation of psaL in psrR1+ cells is due to a disturbed transcriptional regulation or to a posttranscriptional destabilization, we estimated the half-life of psaL in wild-type and psrR1+ cells after 24 h of PsrR1 overexpression by copper depletion. In addition to the full-length transcript, we investigated also the psaL 5′ UTR fragment, which was shown to accumulate differently in the microarray analysis (Supplemental Table 1). Rifampicin, which prevents the de novo initiation of new transcripts by binding to the β-subunit of RNA polymerase (Campbell et al., 2001; Saramago et al., 2014), was added and samples were taken prior to (0 min) and 2, 4, 8, 16, 32, and 64 min after rifampicin addition. The amounts of psaL and the 5′ fragment were quantified by RNA gel blot analysis (Figure 8A), and differences in the loaded RNA amounts were normalized to a 23S rRNA control hybridization. The half-life of psaL in the mutant (∼3.77 min) is ∼1.5 times lower than in the wild type (∼5.92 min) (Figure 8B), which indicates a posttranscriptional destabilization of the mRNA and is in the correct range to explain the decrease in the psaL mRNA amount as detected in our microarray analysis. In agreement with the microarray data, the psaL 5′ fragment had an enhanced half-life of ∼11.76 min in psrR1+ cells (Figure 8C) and remained below the detection limit in wild-type cells.
Figure 8.
Half-Life Estimation of psaL and psaL 5′ UTR RNAs.
(A) RNA gel blot analysis for psaL and the psaL 5′ UTR (same probe) and the 23S rRNA control hybridization. Wild-type and psrR1+ cells were treated with rifampicin (final concentration of 300 μg/mL) after 24 h of copper depletion under moderate light conditions. RNA was extracted prior to and 2, 4, 8, 16, 32, and 64 min after rifampicin addition.
(B) The intensity of the psaL signal was quantified with Quantity One software and normalized with the respective 23S rRNA signal for the wild type and the psrR1+ mutant. The half-life time (t1/2) was estimated by fitting the formula N(t) = N0 × 0.5^(t/t1/2), where t = time after rifampicin addition, N(t) = normalized amount of transcript at time point t, and N0 = normalized amount of transcript at time point t = 0, to the experimental data with the nonlinear least squares function of R. The figure gives the experimental data points for the wild type (black) and the mutant (red) with the sd from two biological replicates. The theoretical curve for the calculated t1/2 is plotted as a broken line. The reference lines indicate the respective half-lives for RNAs in the wild type and mutant.
(C) Quantified data for the psaL 5′ UTR. The t1/2 was calculated only for the psrR1+ mutant, because the UTR does not accumulate in the wild type under standard conditions.
PsrR1-Dependent Processing of psaL by RNase E
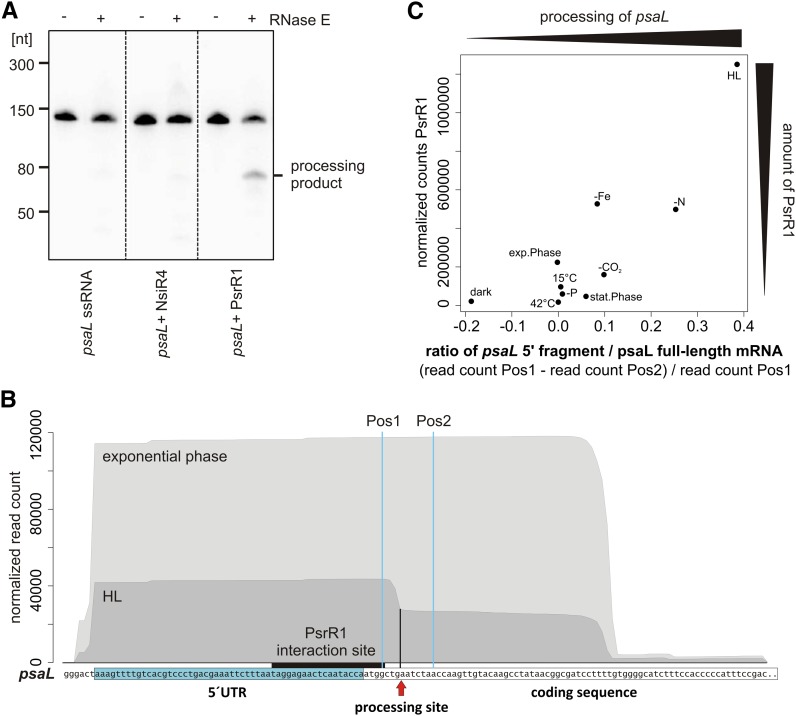
The appearance of an individual psaL 5′ fragment in psrR1+ (Figure 8A; Supplemental Table 1) hints at a specific processing of psaL mRNA in this strain. In bacteria, sRNA-guided processing of mRNAs is frequently facilitated by RNase E (Saramago et al., 2014). Using an in vitro assay, we investigated whether a 130-nucleotide psaL 5′ fragment (Figure 9B; Supplemental Figure 8B) could be cleaved by RNase E alone or in a PsrR1-dependent manner. The sRNA NsiR4 (Kopf et al., 2014) from Synechocystis 6803 was used as a negative control. Reaction products were analyzed by gel electrophoresis. The ethidium bromide–stained gel showed that there is no processing of the single-stranded psaL 5′ fragment and no processing in the presence of NsiR4 (Supplemental Figure 8). By contrast, the psaL 5′ fragment–PsrR1 duplex clearly is a substrate for RNase E cleavage. RNA gel blot analysis with a specific oligonucleotide probe against the 5′ part of the psaL in vitro transcript demonstrated a specific processing product (Figure 9A).
Figure 9.
Conditional Processing of the psaL Transcript.
(A) PsrR1 mediates the RNase E–dependent destabilization of psaL mRNA in vitro. Shown is an 8 M urea, 10% PAA gel of in vitro–transcribed single-stranded psaL RNA (left panel), psaL RNA + NsiR4 (middle panel), and duplex psaL RNA:PsrR1 (right panel) incubated without (−) and with (+) recombinant Synechocystis 6803 RNase E. psaL RNA cleavage fragments were identified by RNA gel blot hybridization using a 5′ [γ-32P]ATP–labeled oligonucleotide probe complementary to the 5′ end of the psaL mRNA. The psaL fragment used is shown in (B). An ethidium bromide–stained version of the gel is shown in Supplemental Figure 8.
(B) The processed psaL 5′ fragment is present in vivo under high-light conditions (HL). Normalized Solexa transcriptome sequencing reads are for the psaL 5′ region for the exponential growth phase and high-light conditions (470 µmol photons m−2 s−1 for 30 min; raw data from Kopf et al., 2014). The 5′ UTR of psaL and the interaction site with PsrR1 are indicated. Sequence coverage in the data from high light shows a sharp drop, which indicates the processing site, marked by the arrow and the black vertical line.
(C) Accumulation of the psaL 5′ fragment correlates with the abundance of PsrR1 in vivo under different stress conditions. As a measure for the amount of processed 5′ fragment, we compared the read coverage from a nucleotide position 5′ of the processing site with the read coverage from a nucleotide position 3′ of the processing site (Pos1 and Pos2 in [B], respectively). To get roughly the relative amount of 5′ fragments from all psaL transcripts (i.e., the amount of processed psaL), we divided the difference from the read count at Pos1 and Pos2 by the read count at Pos1. This value is calculated for all 10 conditions tested in the Kopf et al. (2014) transcriptome study and plotted against the respective read count for PsrR1. The Pearson correlation of this ratio with the PsrR1 abundance is ∼0.86 (Spearman correlation of ∼0.7).
[See online article for color version of this figure.]
Processing of psaL Occurs Also under Physiological PsrR1 Induction in the Wild Type
The prior experiments showed that psaL is specifically processed in psrR1+ cells but not in the wild type grown in the same conditions (Figure 8A). This processing depends both on PsrR1 and RNase E (Figure 9A). We wondered if the psaL 5′ fragment, as an indicator of PsrR1-dependent regulation, can also be observed in high-light conditions in vivo, when PsrR1 is naturally induced. To address this possibility, we used data from another study where the Synechocystis 6803 transcriptome from 10 different growth conditions was sequenced (Kopf et al., 2014). The authors used the dRNAseq method, which allows the identification of primary transcripts with a triphosphate 5′ end. The maximum read length of 100 nucleotides was defined by the sequencing technique used. Thus, for unprocessed transcripts with a length of >100 nucleotides, reads of ∼100 nucleotides were obtained, starting from transcript position +1. This is what we detected for the psaL transcript from exponential phase conditions (Figure 9B), where cells were grown at ∼50 µmol photons m−2 s−1. However, an additional smaller fragment of 60 nucleotides appeared in the transcriptome of cells stressed at ∼470 µmol photons m−2 s−1 for 30 min, leading to a characteristic edge in the visualized data (Figure 9B). The size of this fragment fits the length of the cleaved psaL 5′ UTR fragment inferred from the in vitro RNase E assay (Figure 9A) and the RNA gel blot analysis of RNA from the psrR1+ strain (Figure 8A). This fragment is also visible in other stress conditions (e.g., CO2 or nitrogen depletion) (Supplemental Figure 9). To get a measure for the percentage of the processed reads, we calculated the difference of the read count from a nucleotide position 5′ (Pos1 in Figure 9B) and from a position 3′ (Pos2 in Figure 8B) from the processing site and divided it by the read count at Pos1. In high-light conditions, as well as with nitrogen and CO2 depletion, ∼39, ∼25, and ∼10% of all psaL reads were in the processed form. The transcriptome data also provided information about the expression level of PsrR1 in the respective conditions. In Figure 9C, the read number of the PsrR1 transcript is plotted versus the percentage of processed psaL mRNA. There is a clear correlation (Pearson correlation = ∼0.86) between the abundance of PsrR1 and the amount of cleaved psaL 5′ fragment.
In summary, we conclude that psaL is regulated in the wild type by PsrR1 in high light but also in other conditions where PsrR1 is naturally induced (e.g., CO2, nitrogen, and iron depletion stress) (Figure 9C).
DISCUSSION
Bioinformatic and Experimental Prediction of PsrR1 Targets
In all studied bacterial phyla, sRNAs have been found as versatile posttranscriptional regulators of gene expression that frequently target multiple mRNA molecules (Papenfort and Vogel, 2009). However, the identification of true targets of trans-encoded sRNAs is challenging, even in Synechocystis 6803 with its well-established systems for genetic manipulation. Overexpression of sRNAs has proven to be a valuable tool for functional studies of RNA regulators, so far mainly in enterobacteria (Vogel and Wagner, 2007). Here, we demonstrate that this approach is also useful for the identification of targets of cyanobacterial sRNAs. The observed alterations caused by PsrR1 overexpression on physiological and transcriptome levels may reflect a mixture of direct and indirect effects. Indirect effects may result from a PsrR1-dependent alteration of the amount of a regulatory protein, feedback responses to the caused dysregulation, or general stress responses. For that reason, microarray analysis upon pulse expression of PsrR1 was combined with a new approach of target prediction for sRNAs. CopraRNA is a comparative prediction algorithm that combines individual whole genome target predictions from several different species, thereby dramatically increasing the reliability of the obtained results (Wright et al., 2013). The PsrR1 sRNA was an excellent candidate for the application of this strategy, as it is conserved in a wide range of different cyanobacteria. After postprocessing, the prediction yielded 26 putative targets. The majority could be functionally linked to photosynthesis or thylakoid membrane function (Figure 4A). Five of seven tested targets could be verified in a reporter gene test system (Figure 5A), showing that CopraRNA works also for cyanobacteria. Microarray and prediction produced rather complementary results with psaL, psaK1, and petJ as overlaps, together indicating multiple-target regulation by PsrR1.
Mechanism of PsrR1 Posttranscriptional Regulation
Trans-acting regulatory RNAs have multiple mechanisms for posttranscriptional regulation. Most of the described sRNAs from other bacteria act through direct base pairing with short sequence segments within 5′ UTRs or coding regions of their target mRNAs, whereas others bind to more distantly located sites. They interfere with translation or alter transcript stability and thus can regulate the expression of the corresponding genes positively or negatively. Thereby, sRNAs constitute important mediators of posttranscriptional gene regulation in bacteria (Papenfort and Vogel, 2009; Waters and Storz, 2009; Liu and Camilli, 2010). Certainly, control of the initiation of translation and stability-based mechanisms are not mutually exclusive and can appear simultaneously. Essentially, different mechanisms might be used for different targets of a particular sRNA, which is also indicated for PsrR1 by our data.
Our bioinformatic analysis demonstrates that most predicted interactions of PsrR1 with its putative targets resemble the classical binding proximal to the translational start site (Figures 4B and 4C). The expression of the psaL, cpcA, chlN, and psaJ 5′ UTR sgfp fusions was clearly repressed by PsrR1 in the reporter gene assay (Figure 5), whereas the microarray experiment revealed for the latter three only minor effects of PsrR1 on the accumulation of these mRNAs (Supplemental Table 1). This suggests an inhibition of the initiation of translation without a strong effect on transcript stability at least for some targets, as is known from many other examples (Vogel and Wagner, 2007). By contrast, there is a clear effect on the RNA level for psaL. A priori, this can be caused either directly by posttranscriptional regulation through PsrR1 or indirectly by transcriptional regulation. To distinguish between these possibilities, we estimated the half-life of psaL in the wild type and in the psrR1+ strain (Figure 8) and confirmed a posttranscriptional destabilization (Figure 8). One reason for the reduced half-life of the mRNA could be a higher accessibility of the RNA to RNases if not covered by ribosomes. However, for psaL, we demonstrated specific PsrR1- and RNase E–dependent processing of the transcript (Figure 9). These data explain the effect on psaL accumulation as detected in the microarray and reporter gene assays, but they do not rule out an additional function of PsrR1 in translation inhibition. Interestingly, we observe higher PSI levels and also more PsaL protein in the ΔpsrR1 strain, although at the same time the cells contain less psaL mRNA (Figure 7). This contradictory finding may be explained by an enhanced translation efficiency of psaL in the absence of PsrR1, suggesting an additional, PsrR1-mediated mechanism of translation control. A potential candidate for an indirect target mRNA is psaK1, which has a reduced transcript accumulation in psrR1+ (Supplemental Table 1) but was not repressed by PsrR1 in the reporter gene assay (Figure 5). However, the specific psaK1/PsrR1 mechanisms might not work in the heterologous E. coli system, and a final classification is not possible at the moment.
Physiological Implications of PsrR1-Mediated Regulation
Overexpression of PsrR1 leads to a gradual decrease of chlorophyll and phycocyanin contents in psrR1+ cells; moreover, 48 h after psrR1 induction, a peak at 650 nm (representing allophycocyanin) becomes apparent (Figures 3B and 3C). Hence, the phycocyanin-to-allophycocyanin ratio decreased dramatically in the mutant. This phenotype is consistent with the bioinformatic prediction of five potential mRNA targets that encode phycocyanin subunits (CpcA and CpcB) and phycobilisome linker proteins (CpcC1, ApcE, and ApcF), whereas allophycocyanin subunits (ApcA and ApcB) were not predicted as potential targets (Figure 4A). The negative influence of PsrR1 on cpcA was verified in our study using an E. coli reporter gene assay. In addition, four mRNAs encoding subunits of enzymes involved in porphyrin/chlorophyll biosynthesis were highly ranked in the bioinformatics prediction of PsrR1 targets but not identified in our microarray approach. ChlN, encoding a subunit of the light-independent protochlorophyllide reductase, was independently verified by a gene reporter assay in E. coli. Thus, bleaching of cells could be also explained by translational repression of these mRNAs upon overexpression of PsrR1.
Besides the photosynthetic pigments, PSI was particularly affected by the overexpression of PsrR1. Analysis of thylakoid complexes clearly indicated a considerable decrease of the PSI trimer-to-monomer ratio (Figure 7). This phenotype further supports the regulatory interaction of PsrR1 with the psaL mRNA. The microarray results indicated that the whole psaLI operon is affected by PsrR1. Thus, we conclude that psaI is also affected and regulated via the psaL–PsrR1 interaction. psaJ is another verified direct target (Figure 5). The whole psaFJ operon is slightly affected at the transcript level; however, the fold change of ∼2−0.5 was below our threshold of 2±0.8. Alterations in the amount of these PSI subunits will have additional effects on the stability and assembly of PSI complexes. psaK1 was predicted as a high-ranking PsrR1 target by CopraRNA. Interestingly, both psaK1 and psaK2 (together with its adjacent gene sll0630) were among the most strongly affected transcripts in psrR1+ (Supplemental Table 1). These genes encode two alternative PSI subunits that are involved in state transitions and high-light adaptation of the PSI complex (Fujimori et al., 2005; Dühring et al., 2007). PsaK is incorporated as the final subunit during PSI assembly (Dühring et al., 2007), supporting the idea that PsrR1 causes a retardation in trimer accumulation upon changes in light intensity, by regulating the final steps of complex assembly. Note, however, that PsaL is still the limiting factor, since neither PsaK1 nor PsaK2 is essential for trimerization (Naithani et al., 2000). In view of the upregulation of PsrR1 accumulation under high-light illumination (Figure 2) (Mitschke et al., 2011a), this observation is in line with previous data showing that in high light–acclimated cells the amount of trimeric PSI complexes is strongly downregulated (Kopečná et al., 2012). In addition, these authors showed that newly synthesized chlorophyll mainly serves the PSI trimer but not PSI and PSII monomers, although they are synthesized faster in high-light conditions. Indeed, three enzymes involved in chlorophyll metabolism were predicted to be targeted by PsrR1 (Figure 4A), supporting the idea that trimeric PSI complexes are affected to a greater extent upon overexpression of PsrR1 and also under high-light conditions. This is consistent with the idea that the function of the PSI trimer in cyanobacteria is to provide a larger antenna system under low-light conditions (Grotjohann and Fromme, 2005) and also for state transitions (Aspinwall et al., 2004). Contrary to that, Wang et al. (2008) discussed data implying that the PSI trimer-to-monomer ratio increases in response to high-light treatment. This could be due to different CO2 supply and light conditions in their experimental setup. Furthermore, inactivation of psrR1 leads to a slight increase of the PSI amount, as detected by 77K emission fluorescence spectra, and specifically also to a higher PSI trimer-to-monomer ratio revealed by BN-PAGE analysis. According to a proposed role of PSI trimers under low-light illumination, a higher PSI trimer content could be a disadvantage for cells after a shift to high-light conditions. Indeed, ΔpsrR1 cells bleach more strongly than the wild type when shifted to high-light conditions. But we are cautious with simple explanations regarding the ΔpsrR1 phenotype, as the PsrR1 RNA has many targets and the permanent lack of PsrR1 should have pleiotropic effects, which might be accelerated under high-light stress. As chlorophyll and phycocyanin synthesis seem to be negatively targeted by PsrR1, a higher pigment content would actually be expected in ΔpsrR1 cells, especially under high light, when PsrR1 accumulates in wild-type cells. Nevertheless, under high-light conditions, ΔpsrR1 cells accumulate less pigment, implying a general stress effect in the mutant.
Our analysis of the PsrR1-dependent processing of psaL mRNA in the wild type under different growth conditions (Figure 9C) illustrates that, in addition to high light, PsrR1 regulates psaL expression also under other environmental stress conditions like iron and nitrogen limitation and possibly also under CO2 limitation. All these stress factors are linked to light-dependent regulation, and the corresponding environmental signals are certainly sensed, transduced, and integrated by a complex regulatory network.
In this respect, we want to point out that we also observed several differences between published high-light acclimation responses in the wild type and effects induced upon artificial upregulation of PsrR1. First, both psaK1 and psaK2 are likely negatively targeted by PsrR1, whereas high-light conditions lead to upregulation of the psaK2 mRNA (Hihara et al., 2001). Second, the transcript levels of the high light–inducible protein HliB (encoded by ssr2595), together with the cotranscribed slr1544, appeared strikingly diminished in psrR1+ over the control. Nevertheless, since this effect was also observed at time 0, we cannot exclude a pleiotropic effect caused by the introduced plasmid. On the other hand, in our experience the petJ promoter is not completely repressed under copper-replete conditions when using a self-replicating plasmid, as indicated by the readily (∼2-fold) increased PsrR1 levels in the mutant before copper depletion (Supplemental Figure 4). Whether or not it is directly affected by PsrR1 itself, as HliB is known to be involved in stabilization of the trimeric PSI complex under high-light conditions (Wang et al., 2008) and probably in the assembly and repair of PSII (Promnares et al., 2006), it fits very well with the other confirmed candidate targets.
Role of PsrR1 in the High-Light Response
Major acclimation responses to growth under high-intensity light include a reduction in the number of photosystems and phycobilisome complexes (Muramatsu and Hihara, 2012). Most notably, the synthesis of PSI is suppressed upon transfer from low- to high-intensity light, consequently leading to a decrease in the PSI:PSII ratio (Fujita et al., 1994), which we also demonstrated for the psrR1+ strain.
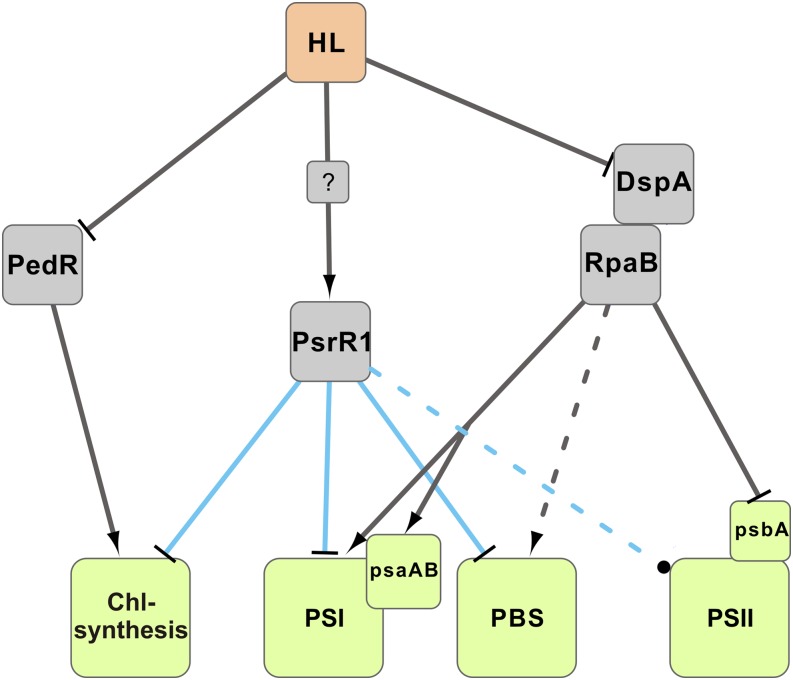
The transcriptional and translational regulation of high-light acclimation includes many different factors, and PsrR1 seems to be one of them (Figure 8). The response regulator RpaB was shown to be a transcriptional activator of PSI genes (Seino et al., 2009) and a repressor of hliB under low-light conditions (Kappell and van Waasbergen, 2007) by binding to the HLR1 sequence. Hanaoka and Tanaka (2008) suggested that the upregulation of such high light–inducible genes as hliB (another potential target of PsrR1) and the downregulation of PSI genes are induced upon the release of RpaB from the HLR1 sequence directly after high-light shift. Interestingly, the transcriptional regulation of at least one RpaB-regulated gene, psaL, was compromised in the ΔpsrR1 mutant (Figures 7E and 7F).
This indicates that a permanent lack of posttranscriptional regulation by PsrR1 may have an impact on the transcription of psaL. As psaL is part of the RpaB regulon (Seino et al., 2009), a general disturbed transcriptional regulation of further photosynthesis components may be assumed. RpaA, another response regulator, was recently shown to regulate the accumulation of monomeric PSI under high-light conditions by an as yet unknown mechanism (Majeed et al., 2012). The transcriptional regulator PedR activates the expression of chlN, chlB, and chlL mRNAs (Nakamura and Hihara, 2006), which encode subunits of the light-independent protochlorophyllide reductase under low-light conditions. We propose a model where the regulatory RNA PsrR1 acts as an additional important factor for high-light acclimation (Figure 10). The posttranscriptional regulator might ensure the rapid control of translation initiation after high-light exposure of the cells, thereby possibly also affecting RNA stability. Interestingly, the putative regulon of PsrR1 overlaps with both the RpaB and the PedR regulons and furthermore includes additional targets (Figures 4 and 8).
Figure 10.
Simplified Scheme of the High-Light Response in Synechocystis 6803.
The model for the activity of transcription factors and their targets explains most of the physiological observations during acclimation to high light. The two-component system DspA-RpaB activates PSI and likely also phycobilisome (PBS) gene expression under low-light conditions, whereas it represses psbA. PedR activates some chlorophyll (Chl) biogenesis genes under low-light conditions. Under high-light (HL) conditions, PsrR1 is induced by a so far unknown mechanism and posttranscriptionally represses chlorophyll biogenesis and phycobilisome and PSI genes. The effect of PsrR1 on PSII is so far unclear. High-light acclimation is achieved by the interplay of transcriptional regulation by transcription factors (gray lines), σ-factors (not included in the model), and posttranscriptional regulation by PsrR1 (blue lines).
Whether mRNAs encoding PSII subunits are targeted is still unclear. Two PSII genes (psbC and psbD) that are transcribed in an operon that is not responsive to high-light treatment (Muramatsu and Hihara, 2012) were found to be upregulated after induction of PsrR1. Four PSII mRNAs, but not psbC and psbD, were predicted by CopraRNA to be targets of PsrR1. The predictions include psbB and psbV, which are known to be repressed under high-light conditions (Muramatsu and Hihara, 2012). Our physiological analysis did not show a specific effect on PSII in both psrR1 mutant strains. Nevertheless, a weak psbB–PsrR1 interaction was indicated by the reporter gene assay (Figure 5), and further analysis is needed to reveal whether PsrR1 is also involved in the posttranscriptional regulation of PSII mRNAs.
However, the precise molecular mechanisms regulating the levels of different components of the photosynthetic apparatus, as well as the integration of other stress responses, remain largely unclear. Here, we add a factor to this very complex regulatory network by suggesting that, in addition to known protein factors, the accumulation of PsaL, CpcA, PsaJ, ChlN, PsbB, PsaI, PsaF, and possibly several other photosynthesis-related gene products is controlled at the posttranscriptional level by the regulatory RNA PsrR1.
METHODS
Culture Conditions
The Synechocystis sp PCC 6803 strain PCC-M (Trautmann et al., 2012) used in this study was provided by S. Shestakov (Moscow State University) and was propagated on BG-11 (Rippka et al., 1979) containing 0.75% (w/v) agar (Bacto agar; Difco) plates. Liquid cultures of wild-type and mutant strains were grown in BG-11 medium containing 10 mM TES buffer, pH 8.0, under continuous illumination with white light at 40 to 50 μmol photons m–2 s–1 at 30°C. Media for mutant strains were supplemented with 40 μg/mL kanamycin and 10 μg/mL streptomycin. Alternative growth conditions are indicated in the corresponding figure legends or in the text. For induction of the PpetJ promoter, CuSO4 was omitted from the medium and cells were first washed twice and then resuspended in copper-free medium. Cultures were incubated in copper-free medium for the indicated times. Repression of the promoter PpetJ was achieved by adding 5 μM CuSO4 to the medium.
Mutagenesis
The psrR1+ strain was constructed as described previously (Mitschke et al., 2011a). The psrR1 gene was replaced by a streptomycin resistance gene cassette generating the ΔpsrR1 mutant strain. For that purpose, upstream and downstream regions of psrR1 were amplified with primer combinations fabF-fw/BglII-psrR-rev and BglII-psrR1-fw/hoxH-rev, respectively (Supplemental Table 2). Both products were digested with BglII, ligated, and used as template for the second PCR. The PCR product was ligated into pDrive cloning vector (Qiagen), and a streptomycin resistance cassette was finally ligated into the blunted BglII site. This vector was used to transform the Synechocystis 6803 wild-type strain. Transformants were selected on 10 μg/mL streptomycin.
Computational Prediction of PsrR1 Targets
PsrR1 target prediction was done with CopraRNA (Wright et al., 2013) using the PsrR1 homologs from genomes of Microcystis aeruginosa NIES-843, Stanieria cyanosphaera PCC 7437, Cyanothece sp PCC 7822, Cyanothece sp PCC 8801, Cyanothece sp ATCC 51142, and Synechocystis 6803. The prediction was done on UTRs including 200 nucleotides upstream and 100 nucleotides downstream of the annotated start codon. Genomic coordinates of PsrR1 sequences are listed in Supplemental Data Set 4.
Functional Enrichment Analysis
We tested whether specific functional terms connected to the genes in the top 85 CopraRNA predictions are significantly enriched over the background (i.e., all Synechocystis 6803 genes involved in the target prediction). The enrichment analysis was done with the functional annotation clustering tool on the DAVID web server (Huang et al., 2009) and the following settings: classification stringency = high; initial and final group membership = 2. All genes belonging to clusters with a DAVID enrichment score of ≥1.3 were considered as functionally enriched.
RNA Preparation, Microarray Analysis, and RNA Gel Blot Hybridization
Synechocystis 6803 liquid cultures were collected by quenching on ice and immediate centrifugation at 4°C. RNA was isolated using the method described by Pinto et al. (2009) with an additional phenol:chloroform:isoamyl alcohol (25:24:1, v/v) extraction preceding the RNA precipitation. Subsequent quantification, electrophoresis, and hybridization experiments were performed essentially as described previously (Dienst et al., 2008) using radioactively labeled in vitro transcripts as probes. For microarray analysis, 20-μg aliquots of RNA were treated with Turbo DNase (Invitrogen) according to the manufacturer’s protocol and precipitated with ethanol/sodium acetate. Labeling and hybridization were performed as described (Georg et al., 2009), with 3 μg used for the labeling and 1.65 μg of RNA used for hybridization. The full data set is accessible in the Gene Expression Omnibus database with the accession number GSE41411.
Immunoblot and BN-PAGE Analyses
Crude extracts of Synechocystis 6803 were prepared as described previously (Dühring et al., 2006b). Proteins were separated by Tricine SDS-PAGE (Schägger, 2006), SDS-PAGE (Laemmli, 1970), or BN-PAGE (Dühring et al., 2006b) using precast blue-native gels (Serva) and transferred electrophoretically onto nitrocellulose membranes. Membranes were incubated with specific primary antibodies and then with a secondary antibody (goat anti-rabbit IgG peroxidase conjugate; Sigma-Aldrich). Immunolabeled bands were visualized using the Immobilon Western membrane chemiluminescence system (Millipore).
Absorption, 77K Fluorescence Spectra, and Determination of Pigment and Protein Contents
Absorption spectra of whole cells were recorded using a UV-2401 PC spectrophotometer (Shimadzu). For measurement of 77K fluorescence emission spectra, cyanobacterial cultures were adjusted to the same chlorophyll contents and glycerol was added to a final concentration of 70% (v/v). The suspensions were frozen dropwise in liquid nitrogen to form transparent beads. Spectra were measured using a Fluoromax Mark II spectrofluorimeter (SPEX Jobin-Yvon) with excitation at 440 nm (10 nm slit width) and a 550-nm cutoff filter in the emission beam to block scattered excitation light. Spectra were normalized to the peak at 695 nm. Chlorophyll contents of thylakoid membranes were measured in 80% acetone according to MacKinney (1941). Phycocyanin and allophycocyanin contents were determined in the soluble protein fraction of cell extracts (Tandeau de Marsac and Houmard, 1988). The total protein concentration from whole cell extracts was assayed by a modified Lowry method (Bensadoun and Weinstein, 1976) and used for normalization of the pigment content. Pigment contents were also quantified from absorption spectra recorded with an integrating sphere (Shimadzu) according to Myers et al. (1980).
Half-Life Estimation
Wild-type and psrR1+ cells were copper depleted at OD750 = 0.6 to 0.8 (described above) for 24 h to induce PsrR1 overexpression in the psrR1+ strain. After this 24 h, de novo RNA transcription was inhibited by the addition of rifampicin to a final concentration of 300 μg/mL, and RNA was extracted prior to (0 min) and 2, 4, 8, 16, 32, and 64 min after rifampicin addition. Each sample was taken from two identically treated biological replicates. The relative amounts of the psaL and psaL 5′ fragments were calculated by the quantification of an RNA gel blot signal with Quantity One software (Bio-Rad) and normalized with the respective relative amount of 23S rRNA. The half-life time (t1/2) was estimated by fitting the formula N(t) = N0 × 0.5^(t/t1/2), where t = time after rifampicin addition, N(t) = normalized amount of transcript at time point t, and N0 = normalized amount of transcript at time point t = 0, to the experimental data with the nonlinear least-squares function of R.
Overexpression and Purification of Synechocystis 6803 Recombinant RNase E
pQESlr1129 (courtesy of M. Asayama; Horie et al., 2007) was used for the production of a C-terminal hexahistidine fusion of Synechocystis 6803 RNase E. Expression of recombinant Synechocystis 6803 RNase E and affinity purification under native conditions were performed following the instructions published by Sakurai et al. (2012).
RNase E in Vitro Cleavage of psaL RNA
RNA was in vitro transcribed and depleted from residual DNA according to Stazic et al. (2011). Full-length in vitro RNA was purified by gel extraction. Briefly, RNA was excised from 8 M urea, 10% PAA gels and extracted by incubation overnight at 4°C in RNA elution buffer (0.3 M sodium acetate, pH 5.2, 1 mM EDTA, pH 8.0, and 0.1% SDS). Subsequently, RNA was concentrated by ethanol precipitation. The protocol for the in vitro RNase E assay is described by Stazic et al. (2011) and was modified as follows: 2 pmol of single-stranded RNA and 4 pmol of duplex RNA were treated with Synechocystis 6803 recombinant RNase E for 30 min at 30°C and separated on an 8 M urea, 10% PAA gel. Following gel transfer to a Hybond-N nylon membrane (Amersham), psaL RNA exclusive cleavage fragments were identified by hybridizing the membrane with a 5′ [γ-32P]ATP–labeled oligonucleotide probe directed against the 5′ UTR of psaL RNA. 5′ radiolabeling and purification of the oligonucleotide probe and RNA gel blot hybridizations were performed according to Stazic et al. (2011). Oligonucleotides used for template generation for the in vitro synthesis of psaL RNA, PsrR1, and SyR12 and for RNA gel blot analysis of psaL RNA are listed in Supplemental Table 2.
Gel Mobility Shift Analyses
RNA was prepared by in vitro transcription of PCR fragments carrying the T7 promoter at the 5′ terminus of the coding sequence using specific oligonucleotides as described (Supplemental Table 2). Oligonucleotides T7-PsrR1-fw-GS and PsrR1-rev-GS1 were used for PsrR1, covering its region from position +1 to +125. Primer GS-PsrR1-Mut4-rev was used to replace nucleotides 94 to 97 (TCCT) by a G4 stretch, resulting in PsrR1-Mu. For psaL-5′UTR, psaL-5′UTR-(+)9, and psaL-5′UTR-(+)30, primers GS-T7-UTR-psaL-fw and GS-UTR-psaL-rev/GS-UTR+9-psaL-rev/GS-UTR+30-psaL-rev were used. In vitro transcription reactions were performed with the AmpliScribe T7-Flash Transcription Kit (Epicentre) followed by DNase I digestion. After extraction with phenol:chloroform:isoamyl alcohol (25:24:1, v/v) and overnight precipitation at −20°C with 3 volumes of ethanol:NaOAc (30:1), the size and integrity of the synthesized RNA were checked using urea-polyacrylamide gel electrophoresis. The RNA was eluted from the excised gel region by overnight incubation in CSS (500 mM ammonium acetate, 0.1% [w/v] SDS, and 0.1 mM Na-EDTA) at 4°C using an overhead shaker (Intelli Mixer RM-2M). This was followed by another extraction and precipitation step as described above. Quantification of the purified RNA was conducted with a NanoDrop ND-1000 spectrophotometer (PeqLab Biotechnology). For 5′ end labeling, 5 pmol of the RNA was dephosphorylated with FastAP (MBI Fermentas) following the manufacturer’s instructions. A further purification step was followed by the labeling reaction using 20 µCi of [γ-32P]ATP (Hartmann Analytic) for each 5 pmol of dephosphorylated RNA in a mixture containing 10 units of polynucleotide kinase (MBI Fermentas) following the manufacturer’s recommendations. Unincorporated nucleotides were removed by gel filtration in G-50 microspin columns (GE Healthcare) followed by the extraction and precipitation procedure as before. Labeled RNA was used at a final concentration of 1 nM in 10-μL reaction volumes containing 10 mM Tris-HCl, pH 7.5, 20 mM NaCl, 200 mM KCl, and 2 mM MgCl2. Unlabeled RNA was added to ascending final concentrations ranging from 5 to 100 nM. After incubation at 30°C for 30 min, the RNA complexes were separated by electrophoresis at 4°C using 5% nondenaturing polyacrylamide gels in 0.5× TBE buffer. Signals were detected on a Personal Molecular Imager FX system with Quantity One software (Bio-Rad).
In Vivo GFP Reporter Assays
For these measurements, we used the reporter system described by Urban and Vogel (2007) and the sGFP plasmid introduced by Corcoran et al. (2012). The primers used and the resulting plasmids are given in Supplemental Tables 2 and 3. Testing was done as described previously (Wright et al., 2013).
Accession Numbers
Sequence data relevant to this work can be found in the CyanoBase and GenBank/EMBL databases under the following accession numbers: NC_005230 and CP003265 to CP003272 (PCC-M wild type).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Unrooted Neighbor-Joining Phylogenetic Tree of 28 PsrR1 Homologs.
Supplemental Figure 2. Absorption Spectrum of the psrR1+ Overexpressor Strain.
Supplemental Figure 3. Visualization of the Functional Enrichment of the PsrR1 Target Prediction.
Supplemental Figure 4. Induction Pattern of PsrR1 under Control of the petJ Promoter in the psrR1+ Strain upon Copper Depletion.
Supplemental Figure 5. Raw Data of sGFP Reporter Gene Assay.
Supplemental Figure 6. Verification of the PsrR1 Impact on psaL Expression.
Supplemental Figure 7. Protein Gel Blot Analysis of Wild-Type (WT), ΔpsrR1, and psrR1+ Cell Extracts Using PsaL and HemH Antisera.
Supplemental Figure 8. PsrR1-Dependent Processing of psaL by RNase E.
Supplemental Figure 9. Normalized Solexa Transcriptome Sequencing Reads for the psaL 5′ Region for 10 Growth and Stress Conditions.
Supplemental Table 1. Combined List of mRNAs and 5′ UTRs Significantly Affected by PsrR1 Overexpression.
Supplemental Table 2. Oligonucleotides Used in This Study.
Supplemental Table 3. Plasmids Used in This Study.
Supplemental Data Set 1. CopraRNA Prediction for PsrR1.
Supplemental Data Set 2. Whole Genome Expression Plot from the Microarray Results.
Supplemental Data Set 3. FASTA File of the PsrR1 Alignment Corresponding to Figure 1 and Supplemental Figure 1.
Supplemental Data Set 4. Genomic Coordinates and Sequences of PsrR1 Homologs.
Supplementary Material
Acknowledgments
We thank Ulrike Ruppert, Gudrun Krüger, and Werner Bigott for excellent technical assistance, Conrad Mullineaux for proofreading the article, and Thomas Börner for valuable discussions and generous support. This work was supported by the Deutsche Forschungsgemeinschaft Focus program Sensory and Regulatory RNAs in Prokaryotes (Grant SPP1258 to A.W. and W.R.H.) and by the Federal Ministry of Education and Research (Grant e:bio RNAsys 0316165 to W.R.H.).
AUTHOR CONTRIBUTIONS
A.W. and W.R.H. designed and supervised the project. J.G. executed target prediction and hybridization data analyses of the microarray experiment, reporter gene experiments, and determination of RNA half-lives. D.D. and N.S. designed and implemented mutagenesis strategies. N.S. and S.K. performed RNA extraction and RNA gel blot hybridization. E.K. conducted immunoblot and pigment analyses. D.D. performed EMSA analysis. T.W. performed BN-PAGE analysis. D.S. performed RNase E cleavage assays. H.L. measured 77K fluorescence spectra and interpreted and wrote the respective part of the article. D.K. performed physiological and quantitative RT-PCR analyses. A.W., D.D., J.G., N.S., and W.R.H. interpreted the experimental data and wrote the article.
Glossary
- sRNA
small RNA
- asRNA
antisense RNA
- PSII
photosystem II
- PSI
photosystem I
- RBS
ribosomal binding site
- UTR
untranslated region
- EMSA
electrophoretic mobility shift assay
- BN-PAGE
blue-native PAGE
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Aspinwall, C.L., Sarcina, M., Mullineaux, C.W. (2004). Phycobilisome mobility in the cyanobacterium Synechococcus sp. PCC7942 is influenced by the trimerisation of photosystem I. Photosynth. Res. 79: 179–187 [DOI] [PubMed] [Google Scholar]
- Bensadoun, A., Weinstein, D. (1976). Assay of proteins in the presence of interfering materials. Anal. Biochem. 70: 241–250 [DOI] [PubMed] [Google Scholar]
- Campbell, E.A., Korzheva, N., Mustaev, A., Murakami, K., Nair, S., Goldfarb, A., Darst, S.A. (2001). Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104: 901–912 [DOI] [PubMed] [Google Scholar]
- Chitnis, V.P., Chitnis, P.R. (1993). PsaL subunit is required for the formation of photosystem I trimers in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 336: 330–334 [DOI] [PubMed] [Google Scholar]
- Corcoran, C.P., Podkaminski, D., Papenfort, K., Urban, J.H., Hinton, J.C., Vogel, J. (2012). Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 84: 428–445 [DOI] [PubMed] [Google Scholar]
- Cuperus, J.T., Fahlgren, N., Carrington, J.C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia, F., Dean, C. (2011). Long non-coding RNAs and chromatin regulation. Curr. Opin. Plant Biol. 14: 168–173 [DOI] [PubMed] [Google Scholar]
- Dienst, D., Dühring, U., Mollenkopf, H.J., Vogel, J., Golecki, J., Hess, W.R., Wilde, A. (2008). The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology 154: 3134–3143 [DOI] [PubMed] [Google Scholar]
- Dühring, U., Axmann, I.M., Hess, W.R., Wilde, A. (2006a). An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. USA 103: 7054–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dühring, U., Irrgang, K.D., Lünser, K., Kehr, J., Wilde, A. (2006b). Analysis of photosynthetic complexes from a cyanobacterial ycf37 mutant. Biochim. Biophys. Acta 1757: 3–11 [DOI] [PubMed] [Google Scholar]
- Dühring, U., Ossenbühl, F., Wilde, A. (2007). Late assembly steps and dynamics of the cyanobacterial photosystem I. J. Biol. Chem. 282: 10915–10921 [DOI] [PubMed] [Google Scholar]
- Eisenhut, M., Georg, J., Klähn, S., Sakurai, I., Mustila, H., Zhang, P., Hess, W.R., Aro, E.M. (2012). The antisense RNA As1_flv4 in the cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J. Biol. Chem. 287: 33153–33162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori, T., Hihara, Y., Sonoike, K. (2005). PsaK2 subunit in photosystem I is involved in state transition under high light condition in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 280: 22191–22197 [DOI] [PubMed] [Google Scholar]
- Fujita, Y., Murakami, A., Aizawa, K., and Ohki, K. (1994). Short-term and long-term adaptation of the photosynthetic apparatus: Homeostatic properties of thylakoid. In The Molecular Biology of Cyanobacteria, D.A. Bryant, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 677–692. [Google Scholar]
- Georg, J., Hess, W.R. (2011). cis-Antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 75: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg, J., Voss, B., Scholz, I., Mitschke, J., Wilde, A., Hess, W.R. (2009). Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 5: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotjohann, I., Fromme, P. (2005). Structure of cyanobacterial photosystem I. Photosynth. Res. 85: 51–72 [DOI] [PubMed] [Google Scholar]
- Hanaoka, M., Tanaka, K. (2008). Dynamics of RpaB-promoter interaction during high light stress, revealed by chromatin immunoprecipitation (ChIP) analysis in Synechococcus elongatus PCC 7942. Plant J. 56: 327–335 [DOI] [PubMed] [Google Scholar]
- Hihara, Y., Kamei, A., Kanehisa, M., Kaplan, A., Ikeuchi, M. (2001). DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie, Y., Ito, Y., Ono, M., Moriwaki, N., Kato, H., Hamakubo, Y., Amano, T., Wachi, M., Shirai, M., Asayama, M. (2007). Dark-induced mRNA instability involves RNase E/G-type endoribonuclease cleavage at the AU-box and SD sequences in cyanobacteria. Mol. Genet. Genomics 278: 331–346 [DOI] [PubMed] [Google Scholar]
- Hotto, A.M., Schmitz, R.J., Fei, Z., Ecker, J.R., Stern, D.B. (2011). Unexpected diversity of chloroplast noncoding RNAs as revealed by deep sequencing of the Arabidopsis transcriptome. G3 (Bethesda) 1: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Sherman, B.T., Lempicki, R.A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Kappell, A.D., van Waasbergen, L.G. (2007). The response regulator RpaB binds the high light regulatory 1 sequence upstream of the high-light-inducible hliB gene from the cyanobacterium Synechocystis PCC 6803. Arch. Microbiol. 187: 337–342 [DOI] [PubMed] [Google Scholar]
- Kopečná, J., Komenda, J., Bucinská, L., Sobotka, R. (2012). Long-term acclimation of the cyanobacterium Synechocystis sp. PCC 6803 to high light is accompanied by an enhanced production of chlorophyll that is preferentially channeled to trimeric photosystem I. Plant Physiol. 160: 2239–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf, M., Klähn, S., Scholz, I., Matthiessen, J.K., Hess, W.R., Voß, B. (2014). Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res., doi/10.1093/dnares/dsu018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Legewie, S., Dienst, D., Wilde, A., Herzel, H., Axmann, I.M. (2008). Small RNAs establish delays and temporal thresholds in gene expression. Biophys. J. 95: 3232–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.M., Camilli, A. (2010). A broadening world of bacterial small RNAs. Curr. Opin. Microbiol. 13: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, M., Bryant, D.A. (2012). Acclimation of the global transcriptome of the cyanobacterium Synechococcus sp. strain PCC 7002 to nutrient limitations and different nitrogen sources. Front. Microbiol. 3: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinney, G. (1941). Absorption of light by chlorophyll solutions. J. Biol. Chem. 140: 315–322 [Google Scholar]
- Majeed, W., Zhang, Y., Xue, Y., Ranade, S., Blue, R.N., Wang, Q., He, Q. (2012). RpaA regulates the accumulation of monomeric photosystem I and PsbA under high light conditions in Synechocystis sp. PCC 6803. PLoS ONE 7: e45139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank, N.N., Berghoff, B.A., Hermanns, Y.N., Klug, G. (2012). Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. Proc. Natl. Acad. Sci. USA 109: 16306–16311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschke, J., Georg, J., Scholz, I., Sharma, C.M., Dienst, D., Bantscheff, J., Voss, B., Steglich, C., Wilde, A., Vogel, J., Hess, W.R. (2011a). An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA 108: 2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschke, J., Vioque, A., Haas, F., Hess, W.R., Muro-Pastor, A.M. (2011b). Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. USA 108: 20130–20135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu, M., Hihara, Y. (2012). Acclimation to high-light conditions in cyanobacteria: From gene expression to physiological responses. J. Plant Res. 125: 11–39 [DOI] [PubMed] [Google Scholar]
- Myers, J., Graham, J.R., Wang, R.T. (1980). Light harvesting in Anacystis nidulans studied in pigment mutants. Plant Physiol. 66: 1144–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naithani, S., Hou, J.M., Chitnis, P.R. (2000). Targeted inactivation of the psaK1, psaK2 and psaM genes encoding subunits of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth. Res. 63: 225–236 [DOI] [PubMed] [Google Scholar]
- Nakamura, K., Hihara, Y. (2006). Photon flux density-dependent gene expression in Synechocystis sp. PCC 6803 is regulated by a small, redox-responsive, LuxR-type regulator. J. Biol. Chem. 281: 36758–36766 [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Naito, K., Yokota, N., Sugita, C., Sugita, M. (2007). A cyanobacterial non-coding RNA, Yfr1, is required for growth under multiple stress conditions. Plant Cell Physiol. 48: 1309–1318 [DOI] [PubMed] [Google Scholar]
- Papenfort, K., Vogel, J. (2009). Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 160: 278–287 [DOI] [PubMed] [Google Scholar]
- Pinto, F.L., Thapper, A., Sontheim, W., Lindblad, P. (2009). Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol. Biol. 10: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promnares, K., Komenda, J., Bumba, L., Nebesarova, J., Vacha, F., Tichy, M. (2006). Cyanobacterial small chlorophyll-binding protein ScpD (HliB) is located on the periphery of photosystem II in the vicinity of PsbH and CP47 subunits. J. Biol. Chem. 281: 32705–32713 [DOI] [PubMed] [Google Scholar]
- Richter, A.S., Schleberger, C., Backofen, R., Steglich, C. (2010). Seed-based INTARNA prediction combined with GFP-reporter system identifies mRNA targets of the small RNA Yfr1. Bioinformatics 26: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka, R., Deruelles, J., Waterbury, J.B., Herdman, M., Stanier, R.Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111: 1–61 [Google Scholar]
- Sakurai, I., Stazic, D., Eisenhut, M., Vuorio, E., Steglich, C., Hess, W.R., Aro, E.M. (2012). Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 160: 1000–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saramago, M., Bárria, C., Dos Santos, R.F., Silva, I.J., Pobre, V., Domingues, S., Andrade, J.M., Viegas, S.C., Arraiano, C.M. (2014). The role of RNases in the regulation of small RNAs. Curr. Opin. Microbiol. 18: 105–115 [DOI] [PubMed] [Google Scholar]
- Schägger, H. (2006). Tricine-SDS-PAGE. Nat. Protoc. 1: 16–22 [DOI] [PubMed] [Google Scholar]
- Seino, Y., Takahashi, T., Hihara, Y. (2009). The response regulator RpaB binds to the upstream element of photosystem I genes to work for positive regulation under low-light conditions in Synechocystis sp. strain PCC 6803. J. Bacteriol. 191: 1581–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, R., Pisareva, T., Norling, B. (2005). Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5: 4905–4916 [DOI] [PubMed] [Google Scholar]
- Stazic, D., Lindell, D., Steglich, C. (2011). Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res. 39: 4890–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich, C., Futschik, M.E., Lindell, D., Voss, B., Chisholm, S.W., Hess, W.R. (2008). The challenge of regulation in a minimal photoautotroph: Non-coding RNAs in Prochlorococcus. PLoS Genet. 4: e1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandeau de Marsac, N., Houmard, J. (1988). Complementary chromatic adaptation: Physiological conditions and action spectra. Methods Enzymol. 167: 318–328 [Google Scholar]
- Trautmann, D., Voss, B., Wilde, A., Al-Babili, S., Hess, W.R. (2012). Microevolution in cyanobacteria: Re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res. 19: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, J.H., Vogel, J. (2007). Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35: 1018–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., Wagner, E.G. (2007). Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 10: 262–270 [DOI] [PubMed] [Google Scholar]
- Voss, B., Georg, J., Schön, V., Ude, S., Hess, W.R. (2009). Biocomputational prediction of non-coding RNAs in model cyanobacteria. BMC Genomics 10: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Jantaro, S., Lu, B., Majeed, W., Bailey, M., He, Q. (2008). The high light-inducible polypeptides stabilize trimeric photosystem I complex under high light conditions in Synechocystis PCC 6803. Plant Physiol. 147: 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, L.S., Storz, G. (2009). Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, P.R., Georg, J., Mann, M., Sorescu, D.A., Richter, A.S., Lott, S., Kleinkauf, R., Hess, W.R., Backofen, R. (2014). CopraRNA and IntaRNA: Predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 42: W119–W123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, P.R., Richter, A.S., Papenfort, K., Mann, M., Vogel, J., Hess, W.R., Backofen, R., Georg, J. (2013). Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. USA 110: E3487–E3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Eisenhut, M., Brandt, A.M., Carmel, D., Silén, H.M., Vass, I., Allahverdiyeva, Y., Salminen, T.A., Aro, E.M. (2012). Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell 24: 1952–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova, P., Sharma, C.M., Förstner, K.U., Liere, K., Vogel, J., Börner, T. (2012). The primary transcriptome of barley chloroplasts: Numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.