Storage compounds like oil and storage proteins accumulate both in the endosperm and in the embryo of Arabidopsis seeds. The MYB118 transcription factor is specifically expressed in the endosperm and represses storage compound synthesis in this tissue, thus contributing to establishment of the embryo as the main deposition site for reserves in the seed.
Abstract
In the exalbuminous species Arabidopsis thaliana, seed maturation is accompanied by the deposition of oil and storage proteins and the reduction of the endosperm to one cell layer. Here, we consider reserve partitioning between embryo and endosperm compartments. The pattern of deposition, final amount, and composition of these reserves differ between the two compartments, with the embryo representing the principal storage tissue in mature seeds. Complex regulatory mechanisms are known to prevent activation of maturation-related programs during embryo morphogenesis and, later, during vegetative growth. Here, we describe a regulator that represses the expression of maturation-related genes during maturation within the endosperm. MYB118 is transcriptionally induced in the maturing endosperm, and seeds of myb118 mutants exhibit an endosperm-specific derepression of maturation-related genes associated with a partial relocation of storage compounds from the embryo to the endosperm. Moreover, MYB118 activates endosperm-induced genes through the recognition of TAACGG elements. These results demonstrate that the differential partitioning of reserves between the embryo and endosperm in exalbuminous Arabidopsis seeds does not only result from developmental programs that establish the embryo as the preponderant tissue within seeds. This differential partitioning is also regulated by MYB118, which regulates the biosynthesis of reserves at the spatial level during maturation.
INTRODUCTION
In angiosperms, seed development comprises two phases. Once embryo morphogenesis is achieved, the seed enters a maturation phase characterized both by the accumulation of storage compounds in zygote tissues and by the acquisition of dormancy and desiccation tolerance by the embryo, which becomes metabolically quiescent (Vicente-Carbajosa and Carbonero, 2005). The relative proportion of storage compounds stored in the embryo and in the endosperm varies greatly depending on the species considered. In exalbuminous species, these compounds are mostly deposited in a large embryo structure acquired at the expense of the endosperm, which is partially degraded as the embryo develops and persists only as a thin peripheral layer in mature seeds. Arabidopsis thaliana seeds thus accumulate seed storage proteins (SSPs; 2S albumins and 12S globulins) and triacylglycerols (TAGs; Baud et al., 2002), and dissection of mature dry seeds has shown that the embryo contains roughly 90% of these TAGs, while the remaining 10% are located in the residual endosperm (Penfield et al., 2004; Li et al., 2006).
Accumulation of storage compounds, as part of the seed maturation program, is a tightly controlled process. Genetic studies, mostly performed in Arabidopsis, have delineated a complex network of transcription factors (TFs) that are essential for completion of seed maturation (Santos-Mendoza et al., 2008) with a set of positive regulators triggering SSP and TAG biosynthesis early in seed maturation. Master regulators of the maturation program include ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEAFY COTYLEDON2 (LEC2), which belong to the B3 domain superfamily of DNA binding proteins. Members of the AFL (ABI3/FUS3/LEC2) network bind RY motifs (CATGCATG; http://arabidopsis.med.ohio-state.edu/AtcisDB/bindingsites.html) in the promoters of their target genes. AFL proteins act in concert with LEC1, a protein homologous to the HAP3 subunit of the CAAT box binding proteins (Lotan et al., 1998). Next to these master regulators, basic leucine zipper (bZIP) TFs like bZIP53 or bZIP67 bind G-boxes (CACGTG), cis-regulatory elements often associated with RY motifs, conferring correct expression patterns to maturation-associated genes (Alonso et al., 2009; Mendes et al., 2013). More recently, newly identified activators of the maturation process like the MADS domain TF AGAMOUS-LIKE15, the WD repeat motif-containing TANMEI/EMB2757, and the homeobox GLABRA2 have been characterized (Baud and Lepiniec, 2010). Two related members of the MYB family, namely, MYB118 and MYB115, have also been proposed to participate in this regulatory machinery (Wang et al., 2009; Zhang et al., 2009).
In contrast to the AFL TFs that redundantly activate the maturation program, a set of three closely related VIVIPAROUS1/ABSCISIC ACID INSENSITIVE3-Like (VAL) B3 domain factors from a sister clade shut down this program prior to and during seed germination (Tsukagoshi et al., 2007; Suzuki and McCarty, 2008; Suzuki et al., 2007). The functional symmetry of the AFL and VAL B3 factors may be related to alternating chromatin states of the corresponding B3 genes (Berger et al., 2011 and cited references). Repression of the maturation program by the VAL clade is mediated by transcriptional silencing of the embryonic AFL network and/or by direct repression of maturation-associated genes through competitive binding to RY elements. The trihelix transcriptional repressor ARABIDOPSIS 6B-INTERACTING PROTEIN 1-LIKE1 (ASIL1) binds GT elements (GTGATT) overlapping G-boxes in close proximity to RY motifs in the promoters of maturation-associated genes and also competes with the binding of AFL activators (Gao et al., 2009). Aside from these TFs, other described repressors of maturation and/or embryonic traits during germination downregulate transcription by acting at the chromatin level. These include histone deacetylases like HDA6 and HDA19, the latter interacting with VAL2 (also known as HSL1) through the zinc-finger CW domain of the TF (Zhou et al., 2013). Polycomb group proteins, SWI/SNF2 chromatin remodelers (e.g., BRAHMA), and CHD3-chromatin remodeling factors like PICKLE (Ogas et al., 1999; Zhang and Ogas, 2009; Barr et al., 2012) also participate in the repression of the maturation program. Recent studies have shown that some of the above-mentioned repressors like HDA6, ASIL1, and its closest homolog ASIL2 not only repress the seed maturation program during germination but also prevent its premature onset during early embryogenesis (Willmann et al., 2011). Similarly, microRNAs are known to be critical negative regulators of seed maturation during early embryogenesis, although the exact molecular mechanism remains to be fully elucidated (Barr et al., 2012).
The fine characterization of these two sets of antagonist regulators and the study of their complex interactions constituted a first breakthrough in our understanding of the transitions between vegetative development and seed maturation. The vast majority of these studies were focused on the activation and repression of the embryo maturation program. By contrast, the regulation of endosperm maturation has scarcely been investigated in Arabidopsis.
Here, we examine storage compound metabolism in the two zygotic tissues comprising the seed, namely, the embryo and the endosperm. We show that Arabidopsis stores different amounts of lipid and protein in these two compartments and that the composition of these reserves and their pattern of deposition differ depending on the tissue concerned. Within the framework of a survey aimed at identifying maturation-related TFs, we then describe the identification of MYB118, which is specifically induced in the endosperm at the onset of the maturation phase. We provide evidence that MYB118 functions as a negative regulator of maturation-related genes limiting the amount of oil and protein stored in the endosperm. Aside from this negative regulation, MYB118 also directly activates endosperm-induced genes through the cis-element TAACGG. Together, these data position MYB118 as a newly identified component of the regulatory machinery controlling seed maturation. Through its specific expression pattern, MYB118 contributes to the control of differential reserve allocation between the two zygotes composing the exalbuminous seed of Arabidopsis.
RESULTS
MYB118 Is Induced in the Endosperm at the Onset of the Maturation Phase
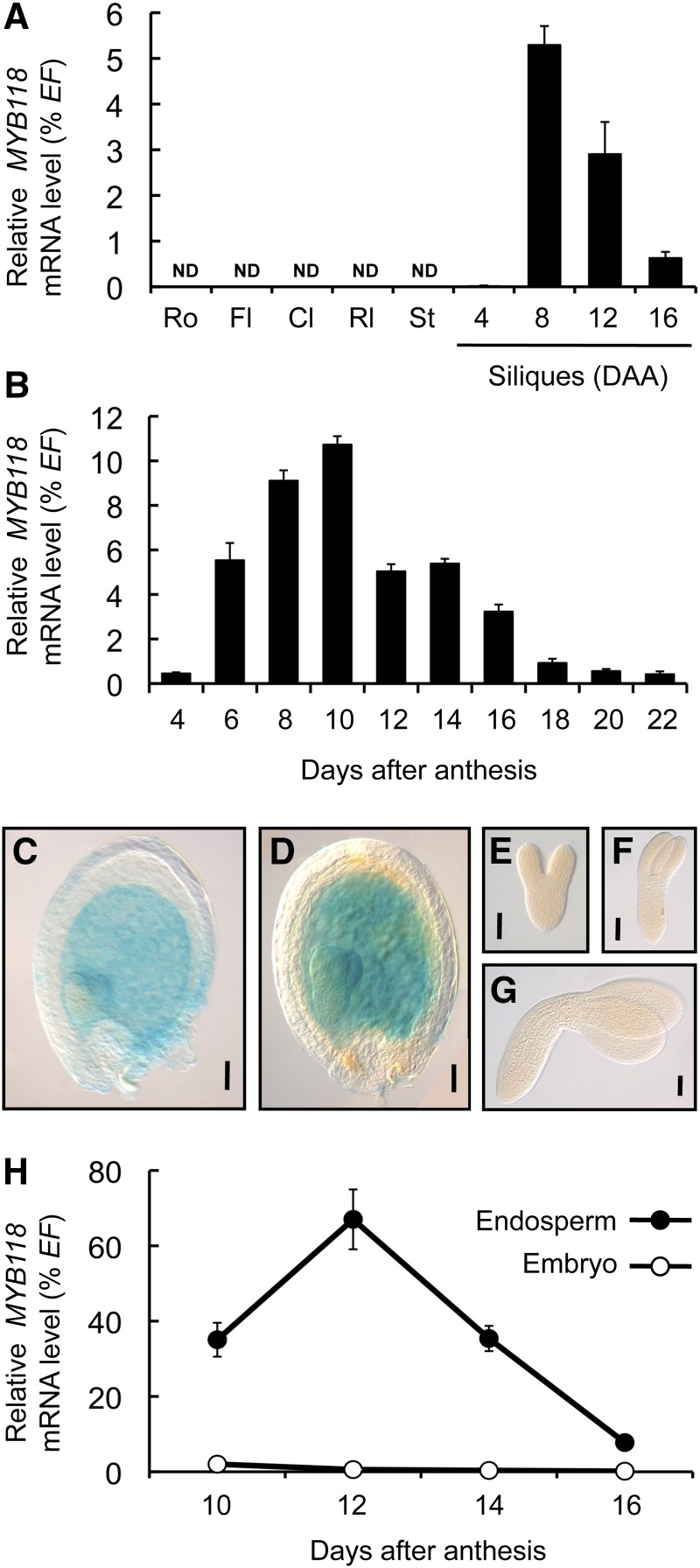
A transcriptome survey for TFs expressed during seed maturation was performed using the Expression Angler tool (http://bar.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi). Genes exhibiting a pattern similar to that of the maturation-related WRINKLED1 (WRI1) were looked for in the AtGenExpress Plus Extended Tissue Compendium data set. Candidates encoding putative TFs and exhibiting a seed-specific expression pattern (intensity <100 in non-seed tissues according to the AtGenExpress Visualization Tool; http://jsp.weigelworld.org/expviz/expviz.jsp) were retained for systematic analysis. MYB118 (At3g27785), a member of the R2R3-MYB family of TF, was the best hit obtained. The expression pattern of this gene was investigated in detail by quantitative RT-PCR (qRT-PCR) on a set of cDNA prepared from various tissues of the wild-type accession Columbia-0 (Col-0). As a first approach, a range of plant organs was considered and MYB118 appeared to be specifically expressed in siliques (Figure 1A). To further characterize the expression pattern of MYB118, a time-course analysis of MYB118 mRNA abundance was performed in developing seeds excised from the siliques, which revealed a peak of transcript accumulation at the onset of seed maturation (Figure 1B). To gain complementary information about the expression pattern of MYB118, the spatiotemporal activity of the MYB118 promoter was investigated. A 2.5-kb promoter fragment was translationally fused to the uidA reporter gene. The corresponding construct was assayed for the resulting uidA expression pattern in transgenic Arabidopsis lines (Figures 1C to 1G). β-Glucuronidase (GUS) staining was observed only in developing seeds. A closer examination of this material showed that the endosperm was stained, whereas the seed coat and the embryo were not. Similar results were obtained with a 970-bp promoter fragment (Supplemental Figure 1).
Figure 1.
Expression Pattern of MYB118.
(A) and (B) Analysis of relative mRNA accumulation of MYB118 was performed in different plant organs (A) and in developing seeds (B). The results obtained are standardized to the constitutive EF1αA4 (EF) gene expression level. Values are the means and se of three to six replicates performed on cDNA dilutions obtained from three independent mRNA extractions. Cl, cauline leaves; Fl, flowers; Rl, rosette leaves; Ro, roots; St, stems; ND, not detected.
(C) to (G) Pattern of activity of the ProMYB118(-2.5 kb):uidA cassette in developing seeds harvested 6 (C) or 8 (D) DAA and in early maturing embryos harvested 8 (E), 10 (F), or 12 (G) DAA. For histochemical detection of GUS activity, tissues were incubated overnight in a buffer containing 0.2 mM each of potassium ferrocyanide and potassium ferricyanide. Bars = 50 µm.
(H) Analysis of relative mRNA accumulation of MYB118 was performed in a developmental series of endosperm and embryo fractions. The results obtained are standardized to the constitutive EF1αA4 (EF) gene expression level. Values are the means and se of three to six replicates performed on cDNA dilutions obtained from three independent mRNA extractions.
To confirm the tissue specificity of MYB118 expression, maturing seeds were dissected and the two fractions obtained, namely, embryo and endosperm/seed coat, were independently analyzed. To evaluate possible contamination of the two fractions considered, cDNA was prepared from the embryo and endosperm fractions and specific marker genes were quantified using qRT-PCR. Transcripts for ZHOUPI (ZOU) are detected only in the endosperm by in situ hybridization experiments (Yang et al., 2008). In accordance, ZOU expression was detected only in cDNA prepared from the endosperm fraction (Supplemental Figure 2), confirming that there was no significant contamination of the embryo sample with endosperm tissues. Conversely, transcripts of an embryo-specific gene (At2g23230; Le et al., 2010) were detected only in cDNA from the embryo, confirming that no significant contamination occurred between fractions. Finally, MYB118 mRNA abundance was high in the endosperm fraction during early maturation and hardly detected in the embryo (Figure 1H). These observations were fully consistent with the GUS histochemical patterns.
Transcriptional Activation of MYB118 by LEC2
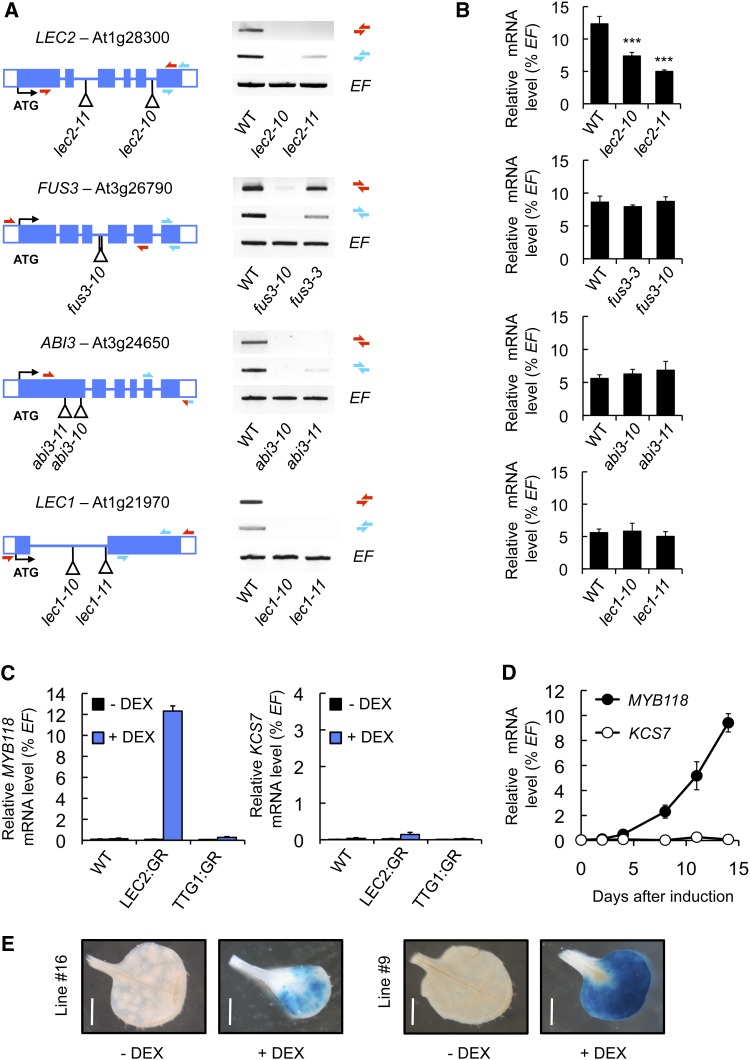
Master regulators coordinately activate the transcription of maturation-related genes at the onset of seed maturation. This regulation can be direct or indirect, and secondary regulators like WRI1 specify the action of some master regulators toward certain metabolic pathways (Baud et al., 2007b). To test whether any known master regulator of seed maturation influences the transcriptional activation of MYB118, a systematic analysis of MYB118 mRNA abundance was performed in mutant backgrounds for LEC1 and members of the AFL network. A collection of lec1, abi3, fus3, and lec2 T-DNA insertion alleles (all in Col-0 background; Supplemental Figure 3) was obtained and characterized at the molecular level (Figure 2A). MYB118 transcripts were analyzed by qRT-PCR on cDNA prepared from seeds 10 d after anthesis (DAA) for each homozygous mutant (Figure 2B). MYB118 transcript steady state levels were significantly reduced in lec2 alleles, suggesting the downregulation of MYB118 expression in the absence of LEC2. To further evaluate the effect of LEC2 on MYB118 promoter activity, a ProMYB118(-2.5 kb):uidA construct was introduced into lec2 mutant alleles and the resulting uidA expression pattern assayed (Supplemental Figure 4). Both the proportion of stained seeds and the intensity of GUS staining were drastically reduced in lec2 seeds, showing the importance of LEC2 for correct MYB118 promoter activity in maturing seeds.
Figure 2.
Transcriptional Regulation of MYB118.
(A) Molecular characterization of mutations affecting master regulators of the seed maturation program. Structure of the LEC2, FUS3, ABI3, and LEC1 genes showing the position of T-DNA insertions in lec2-10, lec2-11, fus3-10, abi3-10, abi3-11, lec1-10, and lec1-11 are presented. For each T-DNA insertion considered, confirmed flanking sequence tag(s) are anchored in the gene structure and represented by vertical bar(s). Closed boxes represent exons and open boxes untranslated regions (UTR). Accumulation of LEC2, FUS3, ABI3, and LEC1 mRNA in wild-type and corresponding mutant backgrounds was studied by RT-PCR on developing seeds harvested 10 DAA. EF1αA4 (EF) gene expression was used as a constitutive control. Primers used for this study are indicated as arrows (Supplemental Table 4).
(B) Accumulation of MYB118 mRNA in mutant seeds was quantified 10 DAA by qRT-PCR and presented as the percentage of the constitutive EF1αA4 (EF) gene expression. Values are the means and se of three to six replicates performed on three independent cDNA preparations obtained from batches of seeds dissected from four to five siliques. The three silique sets were harvested on distinct individuals. ***Significant difference from the wild type according to t test, P < 0.001.
(C) Accumulation of MYB118 and KCS7 (used as a negative control) mRNA in leaves of transgenic Pro35S:LEC2:GR, Pro35S:TTG1:GR, or wild-type 10-d-old plants cultured in vitro on a medium with (+ DEX) or without 10−5 M DEX (- DEX) for two additional weeks, was quantified by qRT-PCR and presented as percentage of the constitutive EF1αA4 (EF) gene expression. Values are the means and se of three to four replicates performed on three independent cDNA preparations.
(D) Time-course analysis of MYB118 and KCS7 (used as a negative control) mRNA accumulation in leaves of 10-d-old Pro35S:LEC2:GR plants transferred to a growth medium containing 10−5 M DEX (induction) and grown 2 weeks in vitro. Accumulation of mRNA was determined by qRT-PCR and presented as the percentage of the constitutive EF1αA4 (EF) gene expression. Values are the means and se of three to six replicates performed on three independent cDNA preparations.
(E) Transgenic ProMYB118(-2.5 kb):uidA (lines #9 and #16) × Pro35S:LEC2:GR seedlings were transferred 10 d after germination on a DEX-containing medium (10−5 M; + DEX). Rosette leaves were analyzed 2 weeks after induction. For histochemical detection of GUS activity, tissues were incubated overnight in a buffer containing 0.2 mM each of potassium ferrocyanide and potassium ferricyanide. Bars = 0.2 cm.
MYB118 mRNA is accumulated to high levels in developing wild-type seeds. As a consequence, testing the transcriptional activation of MYB118 by LEC2 overexpression in maturing seeds was not appropriate. As an alternative, we used a dexamethasone (DEX; a synthetic glucocorticoid that activates the rat glucocorticoid receptor [GR]) inducible system (Santos Mendoza et al., 2005) to quantify the relative expression level of MYB118 in vegetative organs of Pro35S:LEC2:GR plants. In rosette leaves of transgenic plants treated for 2 weeks with DEX, a specific and significant accumulation of MYB118 mRNA was observed (Figure 2C). KCS7, used as a negative control, was hardly induced upon DEX treatment. A time-course analysis of MYB118 mRNA accumulation in rosette leaves treated with DEX revealed a marked increase of MYB118 mRNA levels from 4 d after induction onwards, whereas no induction could be detected for the KCS7 control gene (Figure 2D). Finally, the ProMYB118(-2.5 kb):uidA construct was introduced into transgenic Pro35S:LEC2:GR lines. The seedlings obtained were grown for 14 d on a DEX-containing medium and were then assayed for the resulting uidA expression pattern (Figure 2E). GUS staining was detected in rosette leaves of these seedlings, confirming the ability of the LEC2:GR fusion protein to trigger MYB118 transcription.
Description of the Maturation Process in the Endosperm and in the Embryo of Arabidopsis Seeds
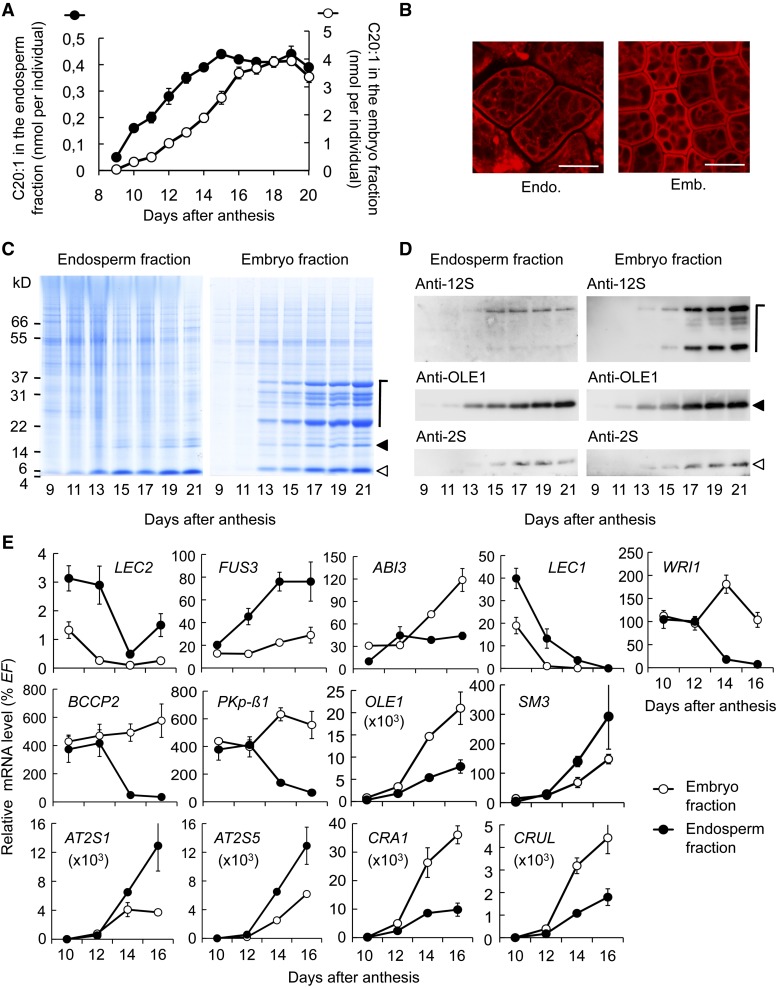
Before addressing the role of MYB118 in the maturing endosperm, we first characterized the maturation process in the two zygotic tissues comprising the seed. Wild-type seeds of the Col-0 accession were dissected during the course of maturation and the two fractions obtained, namely, embryo and endosperm/seed coat, were subjected to various biochemical, cytological, and transcriptional analyses. The endosperm tissue was thus analyzed with the seed coat attached (Penfield at al., 2004). However, this procedure did not bias our evaluation of the endosperm maturation process since the integuments of the seed do not accumulate storage compounds and in fact undergo programmed cell death early during maturation (Beeckman et al., 2000; Li et al., 2006). To analyze oil accumulation within each of the fractions, total fatty acids were quantified by gas chromatography, and eicosenoic acid (20:1), which is specific to TAG, was used as a marker for fatty acids stored as oil (Lemieux et al., 1990). Eicosenoic acid was stored in the embryo between 9 and 19 DAA (Figure 3A). Accumulation followed a sigmoidal pattern equivalent to that of TAG deposition in whole seeds (Baud et al., 2002). In the endosperm fraction, the oil deposition window was narrower, with accumulation occurring between 9 and 15 DAA. The pattern of oil accumulation was linear during this period. Using confocal microcopy, cells were observed in maturing endosperm and embryo. TAG stained with Nile Red was observed in both cell types as noncoalescent cytosolic droplets (Figure 3B).
Figure 3.
Seed Maturation in the Arabidopsis Accession Col-0.
(A) Time-course analysis of eicosenoic acid (C20:1) accumulation in embryo and endosperm fractions of developing seeds. Values are the means and se of five replicates performed on batches of 20 seeds from five distinct individuals.
(B) Sections of maturing embryo and endosperm tissues observed by confocal microscopy. Triacylglycerols accumulated in oil bodies were stained with Nile Red. Bars = 10 µm.
(C) Time-course analysis of storage protein accumulation in embryo and endosperm fractions in developing seeds. A representative SDS-PAGE gel stained with Coomassie Blue G 250 is presented. Each lane contains an amount of proteins equivalent to 16 (endosperm fraction) or four individuals (embryo fraction).
(D) Pattern of seed storage protein and oleosin deposition in the two zygotic tissues. Immunoblots of a developmental series of embryo and endosperm fractions using primary antibodies raised against 12S cruciferins (anti-12S serum), OLE1 (anti-rS3N serum), or 2S napins (anti-2S serum) and anti-rabbit secondary antibodies conjugated with horseradish peroxidase are presented. The amount of proteins loaded in each lane of the gels corresponding to embryo fractions was equivalent to 0.05 (anti-2S and anti-12S sera) or 0.01 (anti-RS3N serum) embryo. The amount of protein loaded in each lane of the gels corresponding to endosperm fractions was equivalent to 0.2 (anti-2S and anti-12S sera) or 0.04 (anti-RS3N serum) endosperm fraction. Symbols on the left of immunoblots refer to the position of corresponding proteins on SDS-PAGE gels presented in (C).
(E) Time-course analysis of mRNA abundance for genes involved in storage compound accumulation. Values are the means and se of three to six replicates performed on cDNA dilutions obtained from three independent mRNA extractions. EF1αA4 (EF) gene expression was used as a constitutive control.
Emb., embryo fraction; Endo., endosperm fraction.
We then examined the profiles of protein deposition in maturing endosperm and embryo fractions (Figure 3C). Accumulation of the precursors of 12S globulins (cruciferins), 2S albumins (napins), and oleosins occurred between 13 and 21 DAA. The protein profile in the endosperm fraction was strikingly different from that of the embryo, with a decreased abundance of 12S globulins, which were hardly detectable in comparison to other major classes of proteins. Immunoblot analyses confirmed both the identity of the proteins considered and the patterns observed by total protein staining after SDS-PAGE (Figure 3D).
To further compare embryo and endosperm maturation, the accumulation of representative mRNA was studied by qRT-PCR in a developmental series of each fraction (Figure 3E). The relative transcript abundance of genes associated with fatty acid metabolism (PKp-β1, BCCP2, and WRI1) was equivalent in the two fractions during early maturation (10 to 12 DAA). These transcripts were still detected at high levels in late maturing embryos, whereas their abundance strongly diminished in the endosperm fraction. This was consistent with the longer oil deposition period described in the embryo (Figure 3A). Genes encoding oleosins and SSP were induced from 12 DAA onwards in the two fractions. Transcript abundance for the precursors of 12S globulins (CRA1 and CRUL) was higher in the embryo fraction. Reciprocally, transcripts for the precursors of 2S albumins (AT2S1 and AT2S5) were more abundant in the endosperm fraction. These observations were consistent with the profiles of protein deposition previously described (Figures 3C). Transcript accumulation profiles for the master regulators of the seed maturation program (LEC2, FUS3, ABI3, and LEC1) were also analyzed. Corresponding transcripts were detected in both fractions, but they exhibited different accumulation profiles.
Role of MYB118 in the Regulation of Storage Compound Partitioning between the Endosperm and the Embryo
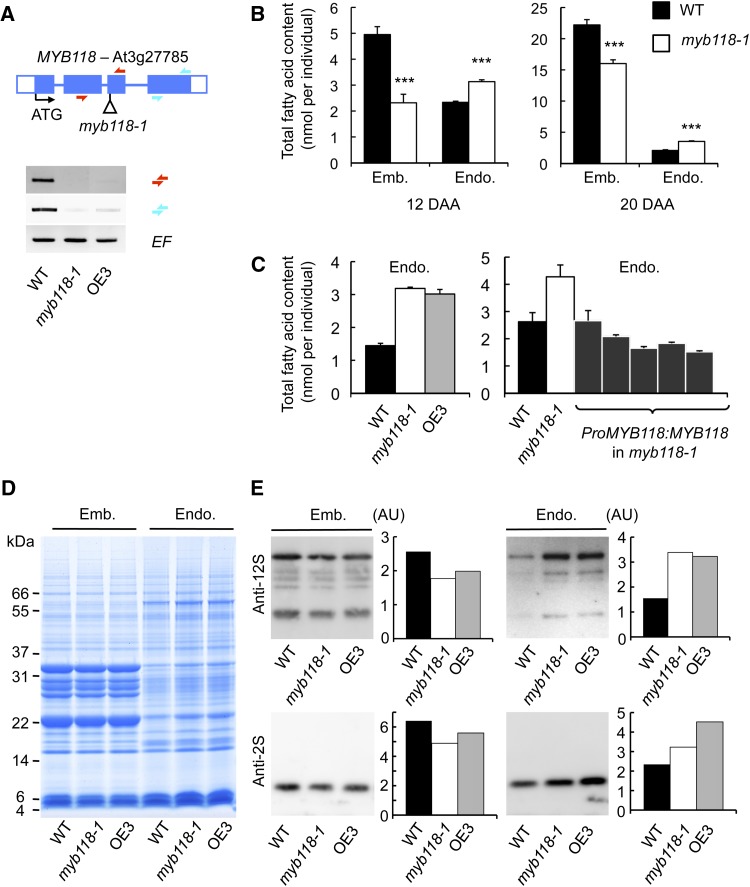
To investigate the function of MYB118 during endosperm maturation, homozygous plants were obtained for a T-DNA insertion line. Vegetative development of myb118-1 was unaffected and cDNA could be prepared from early maturing seeds (10 DAA). RT-PCR analyses showed that MYB118 full-length transcripts were absent in this material (Figure 4A). Pro35Sdual:MYB118 transgenic plants were also generated. However, these lines exhibited altered vegetative development and were sterile, thus preventing us from analyzing the effect of MYB118 overexpression in seeds (Supplemental Figure 5). Among the transformants, one line, named OE3, exhibited normal development: RT-PCR analyses performed in early maturing seeds revealed that MYB118 expression was strongly repressed in this line, which was thus considered as a second myb118 allele (Figure 4A). To elucidate the role of MYB118 in seeds, developing seeds were observed during embryo morphogenesis and early maturation. The structure and development of the three tissues composing the seed were unaffected in myb118 (Supplemental Figure 6). Likewise, microscopic observation of 14-DAA-old peeled endosperms suggested that the organization of the monolayer of endosperm cells was unmodified in myb118 (Supplemental Figure 6).
Figure 4.
Role of MYB118.
(A) Molecular characterization of myb118-1 and OE3 lines. Structure of the MYB118 gene showing the position of the T-DNA insertion in myb118-1 is presented. Confirmed flanking sequence tag is anchored in the gene structure and represented by a vertical bar. Closed boxes represent exons and open boxes untranslated regions (UTR). Accumulation of MYB118 mRNA in wild-type and transgenic backgrounds was studied by RT-PCR on developing seeds harvested 10 DAA. EF1αA4 (EF) gene expression was used as a constitutive control. Primers used for this study are indicated as arrows (Supplemental Table 4).
(B) Total fatty acid content of endosperm and embryo fractions dissected from wild-type or myb118-1 seeds during early (12 DAA) or late maturation (20 DAA). Values are the means and se of five replicates performed on batches of 20 individuals from five distinct plants. ***Significant difference according to t test, P < 0.001.
(C) Total fatty acid content of endosperm fractions dissected from wild-type, mutant, or complemented mature dry seeds. Values are the means and se of five replicates performed on batches of 20 individuals from five distinct plants.
(D) Storage protein content in embryo and endosperm fractions of wild-type and mutant dry seeds. A representative gel stained with Coomassie Blue G 250 is presented. Each lane contains an amount of proteins equivalent to 16 (endosperm fraction) or to four individuals (embryo fraction).
(E) Immunoblots of embryo and endosperm fractions using primary antibodies raised against 12S cruciferins (anti-12S serum) or 2S napins (anti-2S serum) and anti-rabbit secondary antibodies conjugated with horseradish peroxidase are presented. Relative protein contents (in arbitrary units [AU]) among genotypes and for a given fraction were then analyzed with the MultiGauge software. The amount of protein loaded in each lane of the gels corresponding to embryo fractions was equivalent to 0.05 embryo. The amount of protein loaded in each lane of the gels corresponding to endosperm fractions was 0.2 endosperm equivalents.
Emb., embryo fraction; Endo., endosperm fraction; WT, the wild type (Col-0).
[See online article for color version of this figure.]
To evaluate the effect of the myb118 mutation on seed filling, entire mature dry seeds were first subjected to detailed biochemical analyses. Seed dry weight, total fatty acid, and SSP contents were not modified in myb118 (Supplemental Figure 7). As the expression of MYB118 was endosperm-specific, dissected endosperm and embryo fractions were also analyzed separately. These experiments revealed an increased fatty acid content in the endosperm fraction of myb118-1 in both early maturing (12 DAA) and dry seeds (20 DAA; Figure 4B). Conversely, the embryo fraction was depleted of fatty acids, so that the total oil content was not significantly modified in whole dry seeds. This overaccumulation of fatty acids in the endosperm fraction was confirmed in the OE3 line (Figure 4C). Reversion of the myb118-1 phenotype could be obtained by introgression of a wild-type copy of the MYB118 gene into the mutant background (Figure 4C). To test whether the myb118 mutation resulted in a similar imbalance in SSP partitioning between the endosperm and the embryo, SDS-PAGE analysis of seed proteins was performed (Figure 4D). Although the protein profile observed in the endosperm fraction of myb118-1 and OE3 was not altered, the intensity of the staining observed for SSP and the main oleosins was higher in the mutant background. Conversely, a slight decrease in the protein content was observed in the mutant embryo fraction. These observations were confirmed by immunoblot analyses using specific anti-2S and anti-12S antibodies (Figure 4E).
MYB118 Activates Endosperm-Induced Genes
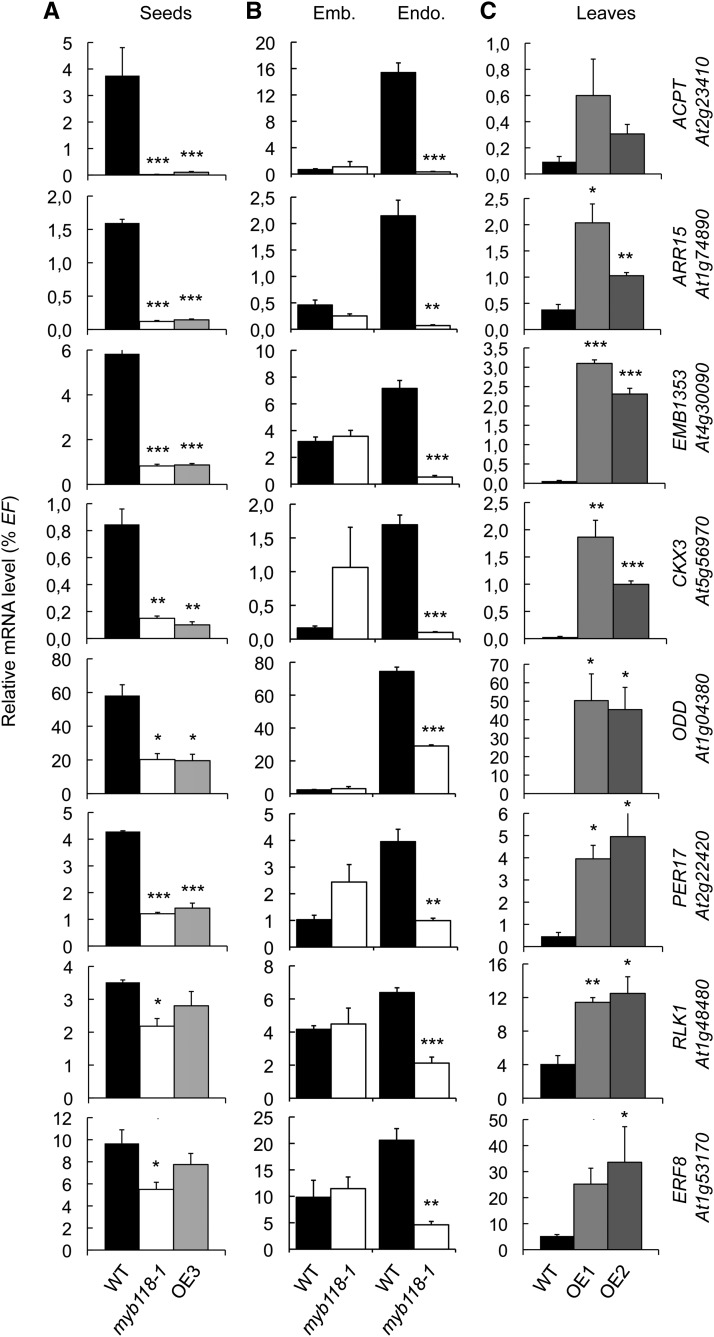
To obtain a comprehensive view of the regulatory network that is affected by MYB118 activity, we performed microarray analysis of myb118-1, OE3, and wild-type seeds. Total RNA were isolated from 10-DAA seeds. Labeled cDNA was then hybridized to the CATMA whole-genome array that represents 33,282 annotated genes (according to TAIR). Comparison of gene expression between the wild type and the mutants resulted in the identification of 177 genes that were downregulated (P ≤ 0.05) in both myb118-1 and OE3 lines (Supplemental Data Set 1 and Supplemental Table 1). This group comprised genes involved in various functional categories, including biotic and abiotic stress responses, hormone biosynthesis, and signaling. To validate the response of these genes to MYB118 deregulation, we analyzed the expression patterns of eight representative genes of this group by qRT-PCR in three independent experiments. First, their downregulation was confirmed in whole mutant seeds (10 DAA; Figure 5A). Second, analyses performed on dissected seed fractions showed that their downregulation was specific to the endosperm fraction (Figure 5B). Third, measurements performed on rosette leaves of Pro35Sdual:MYB118 lines (OE1 and OE2) demonstrated the ability of MYB118 to ectopically activate these putative target genes (Figure 5C).
Figure 5.
Genes Positively Regulated by MYB118.
qRT-PCR analysis of transcript abundance in cDNA prepared from myb118 mutant seeds (myb118-1 and OE3) harvested 10 DAA (A), from embryo and endosperm fractions isolated from myb118-1 mutant seeds harvested 10 DAA (B), or from rosette leaves of MYB118 overexpressing lines (OE1 and OE2) (C). Values are the means and se of three replicates performed on cDNA dilutions obtained from three independent mRNA extractions. Emb., embryo fraction; Endo., endosperm fraction; WT, wild type (Col-0). ***, **, and * indicate significant difference from the wild type according to t test at P < 0.001, 0.01, and 0.05, respectively.
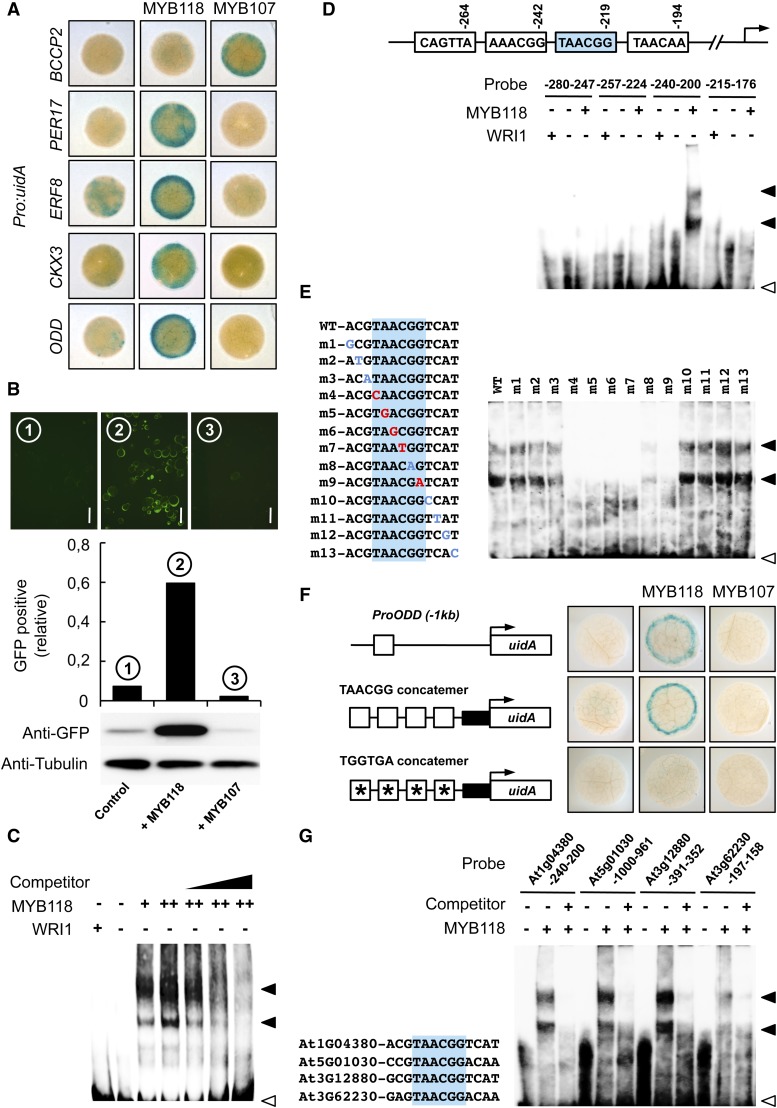
To test the ability of MYB118 to directly activate expression, we used reporter constructs containing 1 kb of upstream sequences from four putative target genes fused to GUS in transactivation assays in Nicotiana benthamiana leaves. Promoter sequences of 2-OXOGLUTARATE-DEPENDENT DIOXYGENASE (ODD), CYTOKININE OXYDASE3 (CKX3), ETHYLENE RESPONSIVE FACTOR8 (ERF8), and PEROXYDASE17 (PER17) were thus cloned upstream of the uidA reporter gene. The promoter sequence of BCCP2 was used as a negative control. Corresponding reporter constructs were infiltrated alone or in combination with a vector allowing the expression of MYB118 or MYB107 (negative control) in young leaves of N. benthamiana (Figure 6A). MYB118 was able to specifically activate ProODD:uidA, ProCKX3:uidA, ProERF8:uidA, and ProPER17:uidA reporter constructs, showing a strong increase in GUS activity compared with the reporters alone or the reporters cotransfected with MYB107.
Figure 6.
Transcriptional Activation of MYB118 Targets.
(A) Transactivation assay in leaves of N. benthamiana. Pro:uidA reporter constructs alone or in combination with a vector allowing the expression of MYB118 or MYB107 (negative control) were coinfiltrated in young leaves of N. benthamiana with a vector allowing the expression of the p19 protein of tomato bushy stunt virus (TBSV) that prevents the onset of posttranscriptional gene silencing (Voinnet et al., 2003). Leaf discs were assayed for GUS activity 3 d after infiltration. Tissues were incubated 5 h in a buffer containing 0.2 mM each of potassium ferrocyanide and potassium ferricyanide. Representative discs (diameter = 0.8 cm) are presented.
(B) Transactivation assay in protoplasts of Arabidopsis. Protoplasts were transformed with the ProODD:GFP reporter construct alone or in combination with a vector allowing the expression of MYB118 or MYB107 (negative control). Whole mounts of protoplasts were observed 12 h after transformation and the relative proportion of GFP-expressing protoplasts was calculated (between 800 and 1000 protoplasts were scored). Immunoblots of a batch of 0.5 mL transformed protoplasts for each combination of constructs assayed and using primary antibodies raised against GFP (anti-GFP serum) or α-tubulin (anti-Tubulin serum) are presented. Bars = 50 µm.
(C) Binding of MYB118 to the proximal upstream region of ODD. EMSA of a probe covering a region from −280 to −191 bp upstream from the ATG codon of ODD with increasing amounts of MYB118 (+ = 0.4 µg and ++ = 0.6 µg). WRI1 was used as a negative control. Competition of MYB118 binding was performed in the presence of 1-, 5-, and 10-fold amounts of the unlabeled ProODD(-280 to -191 bp) fragment. Positions of free probe (open arrowhead) and the shifted bands (closed arrowheads) are indicated.
(D) Identification of the binding site of MYB118 in the proximal upstream region of ODD. A schematic representation of the 280-bp ODD promoter showing the position of putative MYB-core cis-regulatory elements is presented. Indicated positions correspond to the distance of the end of these elements relative to the ATG start codon in base pairs. The EMSA of four partially overlapping sequences containing one of these elements each is shown below the representation.
(E) Binding site sequence specificity of MYB118. The EMSA of MYB118 binding to ProODD(-240 to -201 bp) probes containing single mutations near the MYB-core element (shaded in blue) is presented. Mutations that affected MYB118 binding are indicated in red, while mutations that did not affect MYB118 binding are indicated in blue.
(F) Transactivation by MYB118 of a promoter sequence made of a concatemer of TAACGG elements separated by 10 nucleotides. Schematic representations of the reporter constructs used are presented. Open boxes indicate TAACGG elements, and asterisks indicate that the MYB-core element was modified as indicated. Closed boxes represent the 35S cauliflower mosaic virus minimal promoter. Reporter constructs alone or in combination with a vector allowing the expression of MYB118 or MYB107 (negative control) were coinfiltrated in young leaves of N. benthamiana with a vector allowing the expression of the p19 protein of tomato bushy stunt virus (TBSV). Leaf discs were assayed for GUS activity 3 d after infiltration. Tissues were incubated 2 h in a buffer containing 2 mM each of potassium ferrocyanide and potassium ferricyanide. Representative discs (diameter = 0.5 cm) are presented.
(G) Binding of MYB118 to the proximal upstream regions of genes downregulated in myb118-1 seeds.
Before realizing in-depth MYB118-DNA binding studies with one of those promoter sequences, namely, ODD, transactivation of the ODD promoter by MYB118 was also validated in protoplasts derived from Arabidopsis cell suspension cultures. A 1-kb region of the ODD promoter was cloned upstream to the green fluorescent protein (GFP) reporter gene, and the construct was transfected into protoplasts, alone or in combination with MYB118 or MYB107 constructs driven by the 35S promoter (Figure 6B). Transfection with ProODD:GFP alone resulted in limited GFP expression. Cotransfection with MYB107 did not enhance expression, while cotransfection with MYB118 led to a significant increase in GFP expression. This activation was specific since MYB118 was not able to trigger expression of the ProBCCP2:GFP reporter construct (Supplemental Figure 8).
MYB118 Can Bind TAACGG Elements
The binding of MYB118 to the ODD promoter sequence was examined in vitro by electrophoretic mobility shift assay (EMSA). Purified recombinant MYB118 was incubated with a 90-bp promoter fragment containing four putative MYB-core cis-elements (according to PLACE; Higo et al., 1999), and binding was determined using biotin-labeled DNA probes. Addition of MYB118 to the DNA fragment resulted in the formation of shifted bands (Figure 6C). The signal intensity increased with the concentration of MYB118 in the assay, indicating that the protein binds to the DNA fragment. The binding was specific since addition of recombinant WRI1 protein did not result in the apparition of shifted bands. Furthermore, in competition experiments, addition of increasing amounts of unlabeled oligonucleotides suppressed the binding of MYB118 to the labeled probe.
The 90-bp ODD promoter sequence was then divided into four partially overlapping fragments containing one putative MYB-core cis-element each. MYB118 was able to bind only the fragment harboring the TAACGG element (Figure 6D). To examine the specificity of the MYB118 binding sequence, we prepared various mutant forms of the site surrounding this MYB-core element with single-base substitutions (Figure 6E). Mutations in five out of the six nucleotides composing the MYB-core element resulted in a loss of MYB118 binding, whereas mutations outside of the motif showed no effect on MYB118 binding.
The ability of MYB118 to bind to the TAACGG box in vitro prompted us to determine whether MYB118 could activate a promoter sequence made of a concatemer of this motif in planta. Transient activation assays were performed in leaves of N. benthamiana infiltrated with a reporter construct made of four repeats of the element identified fused to the 35S cauliflower mosaic virus minimal promoter upstream of the uidA reporter gene (Figure 6F). MYB118 was able to specifically activate the reporter construct, showing a strong increase in GUS activity compared with the reporter alone or to the reporter and MYB107. Nucleotide substitutions within the TAACGG element impaired this activation.
Using EMSA, we finally confirmed the binding of MYB118 to the promoters of three additional genes upregulated by MYB118 and harboring the TAACGG motif in 1-kb sequences upstream from the ATG codon (Figure 6G). These results strongly suggest that these genes are transcriptionally regulated by MYB118 in planta.
MYB118 Negatively Regulates Maturation Genes in the Endosperm
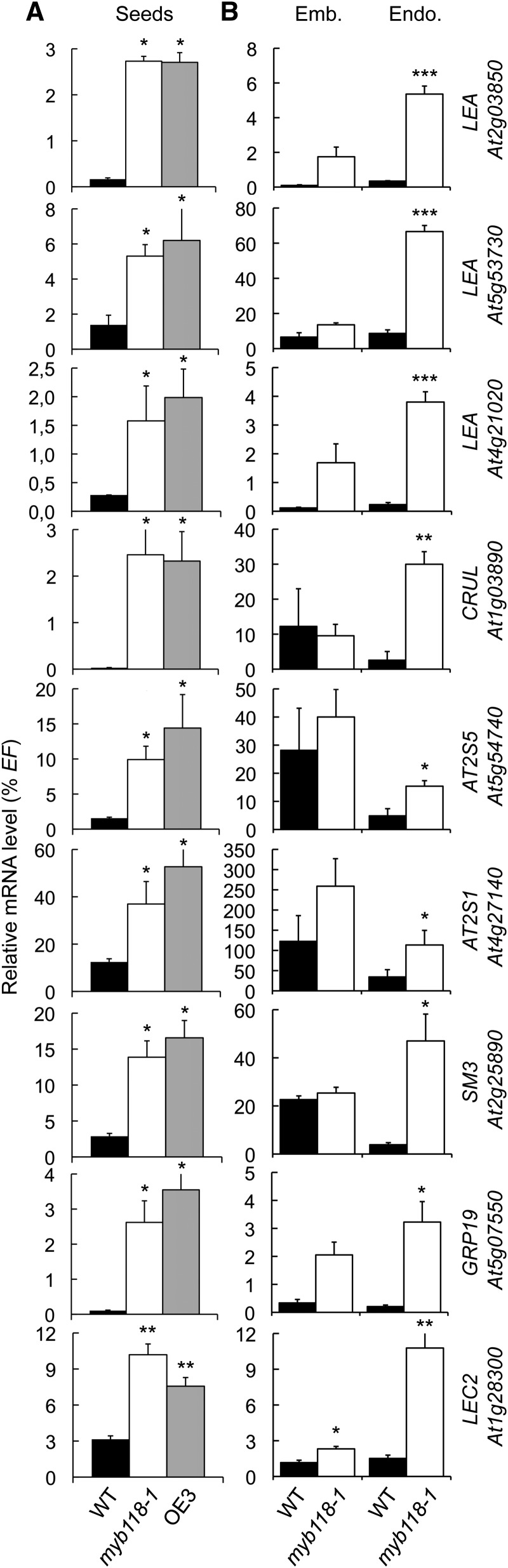
Analysis of the microarray data also allowed the identification of a second set of 230 genes upregulated (P ≤ 0.05) both in myb118-1 and in OE3 lines (Supplemental Data Set 1 and Supplemental Table 2). This group included several putative target genes implicated in seed maturation, like genes encoding 2S albumins and 12S globulins, late abundant embryogenesis (LEA) proteins, oleosins, and the master regulator LEC2. To validate the response of these genes to MYB118 deregulation and to assess the robustness of this response, we analyzed the expression patterns of nine maturation-related genes belonging to this group by qRT-PCR in two independent experiments. First, their upregulation was confirmed in whole mutant seeds (10 DAA; Figure 7A). Second, analyses performed on dissected seed fractions showed that upregulations always occurred in the endosperm fraction (Figure 7B). To further test the involvement of MYB118 in repressing the maturation program in the endosperm, similar experiments were performed with other well-known maturation-related genes that were not highlighted by our transcriptome approach on whole seeds (Supplemental Figure 9). They appeared to be specifically upregulated in the endosperm fraction of myb118.
Figure 7.
Genes Induced in myb118.
qRT-PCR analysis of gene transcript abundance in cDNA prepared from seeds harvested 10 d after flowering (A) or from embryo and endosperm fractions isolated from these seeds (B). Values are the means and se of three replicates performed on cDNA dilutions obtained from three independent mRNA extractions. Emb., embryo fraction; Endo., endosperm fraction; WT, wild type (Col-0). ***, **, and * indicate significant difference from the wild type according to t test at P < 0.001, 0.01, and 0.05, respectively.
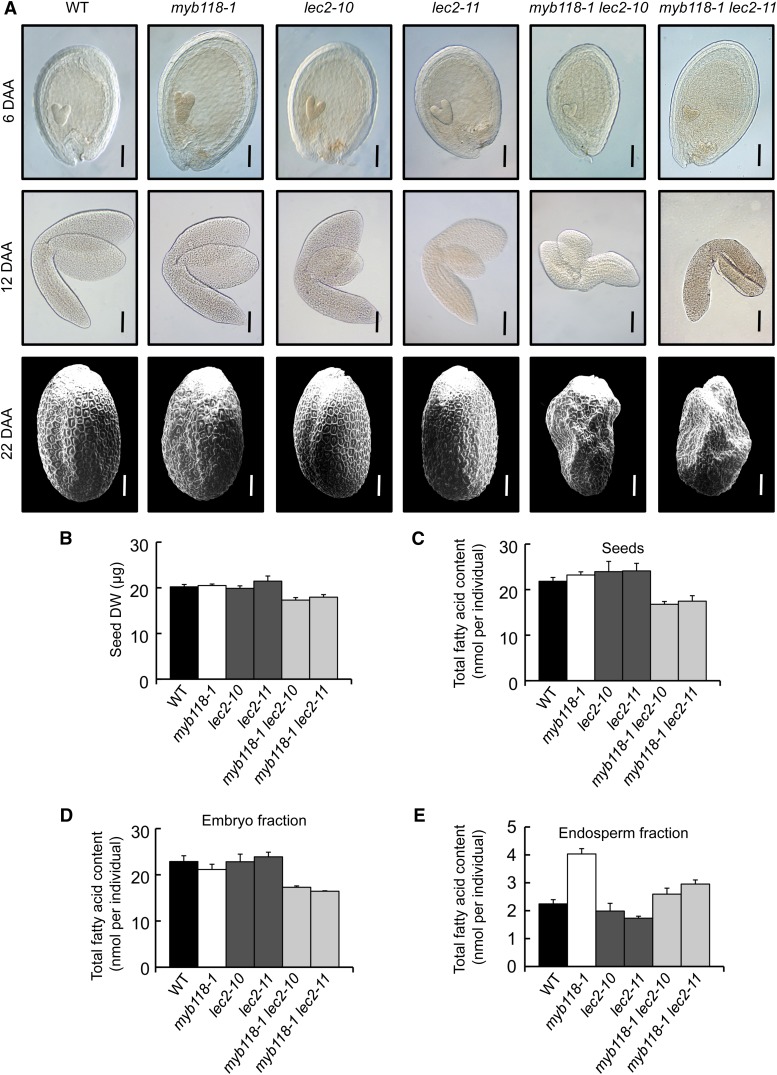
Collectively, these results demonstrate that MYB118 is a negative regulator of the maturation process in the endosperm. Since LEC2, a well-known activator of the maturation process, is repressed by MYB118, the question of a LEC2-independent repression of the maturation process by MYB118 arose. We consequently decided to generate myb118 lec2 double mutants so as to test whether or not the lec2 mutation was epistatic to the myb118 mutation. Double mutants were grown under controlled conditions, together with wild-type control and myb118 and lec2 single mutants, and both seed development and maturation were monitored. Unexpectedly, the combination of the two mutations affected embryo development. At the onset of the maturation phase, 6 DAA, young torpedo-shaped embryos in myb118 lec2 seeds appeared smaller than control embryos (Figure 8A). This phenotype was aggravated during the elongation phase of the embryo, yielding shorter and stunted embryos at 12 DAA that often failed to bend and adopt an upturned-U shape. This defect in embryo elongation was not compensated for during late maturation, so that mature myb118 lec2 seeds exhibited a severely wrinkled phenotype (Figure 8A). This phenotype was associated with a strong decrease in seed dry weight (Figure 8B). Determination of total fatty acid content of dry seeds confirmed that the double mutation severely affected seed oil content (Figure 8C). Embryos were most affected by this decrease in oil content (Figure 8D). As for the total fatty acid content in endosperm fractions of the myb118 lec2 seeds, it was intermediate between myb118 and lec2 contents, showing that the two regulators exert antagonistic and partially independent regulatory actions over oil accumulation in this fraction (Figure 8E).
Figure 8.
Characterization of myb118-1 lec2 Double Mutants.
(A) Observation of seed development. Whole mounts of early developing seeds (6 DAA) and of maturing embryos (12 DAA) were observed with Nomarski optics. Dry seeds were observed by scanning electron microscopy. Representative seeds and embryos are presented. Bars = 100 µm.
(B) Seed dry weight (DW). Values are the means and se of five replicates performed on batches of 20 individuals from five distinct plants.
(C) to (E) Total fatty acid content of mature seeds (C), embryo (D) and endosperm fractions (E) dissected from mature seeds. Values are the means and se of five replicates performed on batches of 20 individuals from five distinct plants.
[See online article for color version of this figure.]
DISCUSSION
Accumulation of storage compounds in seeds, as part of the maturation program, is subject to fine control mediated by various developmental regulators (Baud and Lepiniec, 2010; Barr et al., 2012). These regulators ensure that maturation-related programs are not induced before (during embryo morphogenesis) and after maturation (during vegetative growth) and that they are coordinately deployed during the transition phase between embryogenesis and seed maturation. Maturing seeds of Arabidopsis consist of two compartments accumulating storage compounds: the embryo and endosperm. We separately considered reserve accumulation in the two compartments and observed that the pattern of deposition, final amount, and composition of these reserves differ in the embryo and endosperm. We also demonstrated that the MYB118 TF represses reserve deposition in the endosperm. From these results, it appears that the differential partitioning of reserves between the embryo and endosperm in Arabidopsis seeds is not only the consequence of developmental programs that establish the embryo as the preponderant tissue within these exalbuminous seeds. Differential partitioning is also regulated by MYB118, which controls reserve biosynthesis at the spatial level during the maturation phase, adding another level of complexity to the regulatory network controlling seed maturation.
The Maturation Process Is Dissimilar in the Two Zygotes Composing Arabidopsis Seeds
Like in many dicots, seeds of Arabidopsis are exalbuminous: the endosperm is largely consumed during embryo development. Yet, a single cell layer of endosperm remains in dry seeds and the dissection of mature seeds has revealed that this endosperm layer accumulates approximately one-tenth of the oil present in whole seeds (Penfield et al., 2004) (Figure 3A). Using similar techniques to dissect the seeds into two parts, we observed that the endosperm fraction also accumulated significant amounts of SSP (Figure 3C). The results of these metabolic analyses are in agreement with the developmental profiles of mRNA populations showing extensive overlaps in the expression programs of maturation-related genes in the embryo and the endosperm subregions (Belmonte et al., 2013) (Figure 3E). This concerns both the actors of maturation metabolism (enzymes and SSP) and known regulators controlling genes expressed during the maturation phase (TF). Together, these data suggest substantial similarities between the maturation processes occurring in embryo and endosperm regions.
However, a closer examination of the reserves stored in the two zygotic tissues also highlighted several differences. First, a strong imbalance between the amounts of reserves stored in the two compartments is observed in dry seeds. This imbalance is consistent with the contrasted volumes occupied by the respective tissues within seeds (Baud et al., 2002). In addition, the quality of these reserves differs between the two compartments considered. Significant differences have been reported between the fatty acid profiles in these two tissues, with the endosperm exhibiting higher proportions of n7 long-chain fatty acids like vaccenic acid (18:1n7) and paullinic acid (20:1n7) (Penfield et al., 2004). Likewise, the detailed characterization of SSP accumulation in the two zygotic tissues revealed marked differences, with the endosperm synthesizing proportionally less 12S globulins and consequently exhibiting a striking enrichment in the proportion of 2S albumins (Figure 3C). Relative quantification of SSP mRNA in the two fractions composing the seed is consistent with these metabolic analyses (Figure 3E). Beyond differences affecting the quantity and quality of reserves present in dry seeds, the patterns of reserve deposition also differed between the two zygotic tissues. A detailed time-course analysis of oil deposition showed that the window for oil deposition was narrower in the endosperm fraction. This can be correlated with an earlier decrease in the mRNA levels of several fatty acid biosynthetic genes and their direct regulator WRI1 in the endosperm (Figure 3E). These observations suggest that beyond the coordinated activation of the maturation process in the two zygotes, tissue-specific regulatory processes exist that fine-tune reserve biosynthesis in these two compartments.
Transcriptional Activation of MYB118 in the Endosperm
To identify new regulators of seed maturation, we exploited publicly available transcriptome resources to search for seed-specific TFs induced during the maturation phase and thereby isolated MYB118. Detailed characterization of the MYB118 expression pattern based on complementary approaches like qRT-PCR and promoter:GUS analyses confirmed the spatio-temporal regulation of its transcription (Figure 1). The accumulation of MYB118 mRNA dramatically increased in the endosperm at the onset of the maturation phase, before decreasing during the course of seed maturation. These data are fully consistent with those obtained in different transcriptome analyses of developmental series of siliques, seeds, and seed tissues (Schmid et al., 2005; Le et al., 2010; Day et al., 2008; Belmonte et al., 2013). It should be noted that MYB118 promoter activity in maturing embryos, described by Wang et al. (2009), was not detected with our reporter construct. To clarify these discrepancies, we reproduced the GUS assay with the transgenic lines of Wang et al. and confirmed with this material the predominant activity of MYB118 promoter in the endosperm (Supplemental Figure 1). We propose that previous detection in the embryo could result from the diffusion of the GUS product from the endosperm as whole seeds were used in the assay, whereas seeds were dissected before incubating embryo and endosperm fractions separately in this study. If traces of MYB118 transcripts can be detected in the embryo, as indicated by our qRT-PCR results and by in situ hybridization experiments (Zhang et al., 2009), our observations clearly demonstrate that the embryo is not the primary site of action of this TF (see below).
The pattern of MYB118 promoter activity matches MYB118 mRNA accumulation both at the spatial and temporal levels. These data strongly suggest that MYB118 tissue-specific expression is largely controlled at the transcriptional level. The data presented in this article establish that MYB118 is a target of LEC2. Interestingly, MYB118 transcription is not completely abolished in a lec2 mutant background, suggesting that other TFs act redundantly with LEC2 to activate MYB118 (Supplemental Figure 10). Although the expression profile of LEC2 during the course of seed maturation matches well that of the accumulation of MYB118 mRNA at the temporal level, the spatial localization of the two TF is more surprising: LEC2 is expressed in both zygotic seed tissues. The pattern of MYB118 may result from the activity of endosperm-specific activators and/or embryo-specific repressors that cooperate with LEC2. The involvement of molecular controls related to imprinting mechanisms may also explain the endosperm-specific activation of MYB118 (Berger and Chaudhury, 2009).
MYB118 Represses Maturation of the Endosperm
Several repressors of seed maturation gene expression were previously identified. They were shown to be active during embryo morphogenesis (Willmann et al., 2011) or during germination and seedling establishment (Suzuki et al., 2007; Tsukagoshi et al., 2007; Gao et al., 2009). In contrast to previously identified “temporal” repressors of seed maturation that influence the timeframe of storage compound deposition, MYB118 appears rather to be a “spatial” repressor expressed during the maturation process to regulate allocation of storage compounds between the different zygotic tissues of the seed. Activation of MYB118 expression within the endosperm represses maturation-related genes in this tissue and therefore limits oil and SSP deposition in this secondary zygote. Additionally to mechanisms controlling cell proliferation and degeneration within the endosperm, and therefore the relative importance of this tissue within the seed (Olsen, 2004; Sreenivasulu and Wobus, 2013), MYB118 thus constitutes another level of control leading to the formation of exalbuminous seeds in Arabidopsis by limiting reserve accumulation in the endosperm.
Considering the relatively mild phenotypes of myb118 and the functional redundancy frequently occurring between related TFs in Arabidopsis, the question of functional redundancy between MYB118 and close homologs arose. MYB115, the closest homolog of MYB118, was previously shown to have similar overexpression phenotypes including stunted growth and sterility (Wang et al., 2009). However, analysis of seeds from the leaky myb115-1 allele and from the myb115-1 myb118-1 double mutant did not allow establishing a significant redundancy between the two TF in seeds (Supplemental Figure 11).
Repression of endosperm maturation seems important to promote embryo filling, as suggested by the depletion in storage compounds observed in myb118-1 embryos. This lower accumulation of reserves is not correlated with decreases in the mRNA levels of maturation related genes in this tissue. On the contrary, a weak trend upward in the mRNA levels of several maturation related genes is observed in embryos. This could be the consequence of the faint expression of MYB118 in this tissue (see above). Two hypotheses, not mutually exclusive, can be put forward to explain these apparent discrepancies. (1) An increased accumulation of storage compounds in the endosperm may negatively affect embryo filling by rerouting part of the nutrients usually allocated to the embryo, the sink strength of which is affected by this competitive sink within the seed. Consistent with this hypothesis is the embryo phenotype of myb118 lec2 double mutants: the lack of LEC2 in these embryos may further decrease their sink strength (the effect of the lec2 mutation in the endosperm is not so strong since it is compensated for by the derepression of maturation-related genes due to the myb118 mutation). (2) As suggested by recent reports (Xing et al., 2013), altered endosperm development may indirectly affect the embryo through signaling pathways coordinating their respective development. Complementary studies are now required to further elucidate these complex interactions between seed tissues.
MYB118 Antagonistically Regulates Distinct Gene Subcircuits
Gene expression profiling of myb118 mutant seeds using microarray and qRT-PCR approaches revealed that MYB118 is required both for the repression of maturation-related genes in the endosperm and for the activation of endosperm-induced genes. Several genes repressed by MYB118 are directly involved in storage reserve accumulation. These gene products encode SSP, enzymes associated with oil metabolism, oleosins, or LEA proteins. Other MYB118-repressed transcripts encode LEC2, a well-known regulator of reserve accumulation. Among the set of genes induced by MYB118, factors participating in various hormone biosynthetic pathways or stress responses, as well as some regulators of growth and development have been identified. It is noteworthy that many of these genes exhibit a high level of expression in the endosperm compared with other seed tissues (Supplemental Figure 12). The antagonistic regulation of these two sets of genes by MYB118 raises the question of the molecular mechanisms involved.
Together, transactivation of upregulated gene promoters in various plant systems and in vitro binding evidence of MYB118 to the corresponding nucleotidic sequences strongly suggest that MYB118 directly binds to some of these target genes in planta. MYB TFs are characterized by a highly conserved DNA binding domain (MYB domain) consisting of up to four repeats (R) of ∼50 residues (Dubos et al., 2010). MYB118 encodes an R2R3-type MYB TF harboring two adjacent R repeats, which is a common feature of plant MYBs. Although many R2R3-MYB TFs recognize AC elements (DNA motifs enriched in adenosine and cytosine residues), the range of R2R3-MYB binding sites remains poorly defined in planta (Prouse and Campbell, 2012; Wang et al., 2013). Our observations suggest that the TAACGG element is part of the in vivo MYB118 binding site. This element is also found in the cis-element TATAACGGTTTTTT bound by the soybean (Glycine max) GmMYBs (Liao et al., 2008) and is very similar to the in vivo DNA binding sequences of mammalian c-Myb proteins (YAACNG, where Y = C/T, and N = any nucleotide) (Oda et al., 1998) and to the MYB recognition site found in the promoter of the dehydration-responsive gene rd22 in Arabidopsis (YAACKG, where Y = C/T and K = G/T) (Abe et al., 2003). Furthermore, the reverse complement of the element identified, namely, CCGTTA, also fits with the MYB-core CNGTTR (where N = any nucleotide, and R = G/A), defined as the binding site for animal MYBs, and subsequently for several plant MYB proteins (Solano et al., 1995). Site-directed mutagenesis experiments also indicated that five conserved nucleotides within this element are critical for MYB118 binding (Figure 6E). Nevertheless, the bordering nucleotides of the cis-element should not be neglected since they may modulate DNA binding affinities between closely related MYB TFs. A more detailed structure-function study using techniques like SELEX or Biacore will be required to fully characterize the MYB118 DNA binding matrix.
The molecular mechanism involved in the repression of negatively regulated targets of MYB118 remains unknown. Since the lec2 mutation is not epistatic to the myb118 mutation, LEC2-independent mechanisms may exist that mediate the regulatory action of MYB118 over maturation metabolism. So far unidentified factors (e.g., transcriptional repressors activated by MYB118) may repress maturation-related genes in the endosperm and further studies will be required to identify these factors.
In conclusion, these results clearly show that the maturation process and associated metabolism for storage reserve accumulation differ in the endosperm and in the embryo of Arabidopsis seeds. The identification and comprehensive characterization of the MYB118 TF provide evidence of the existence of factors regulating maturation at the spatial level within maturing seeds of Arabidopsis (Supplemental Figure 10). By repressing maturation-related genes within the endosperm, MYB118 limits storage compound accumulation in this tissue and indirectly contributes to the establishment of the embryo as the main sink within the maturing seed. Aside from this function, MYB118 also directly activates a set of endosperm-induced genes through the TAACGG element. Further work will be required to determine the molecular mechanisms that allow this TF to antagonistically regulate distinct gene sets.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana seeds of the Col-0 accession were obtained from the Arabidopsis thaliana resource center for genomics at the Institut Jean-Pierre Bourgin (http://www-ijpb.versailles.inra.fr/) and T-DNA mutant lines (abi3-10, N659724; abi3-11, N677134; fus3-10, GABI_612E06; lec1-10, N631219; lec1-11, N500450; lec2-10, N515228; lec2-11, N873714; myb115-1, N544168; myb118-1, N611812) were ordered from the Salk Institute (http://signal.salk.edu/). ProMYB118:uidA lines (Wang et al., 2009) were provided by J. Zuo. Plants were cultured as described previously (Baud et al., 2007a). DEX induction experiments using the Pro35S:LEC2:GR construct were performed as described by Santos Mendoza et al. (2005). To sample embryo and endosperm fractions, seeds excised from siliques were dissected using a scalpel and dissecting tweezers under an optical glass binocular magnifier. Material used for RNA extraction was frozen in liquid nitrogen immediately after harvest and then stored at −80°C.
Molecular Characterization of T-DNA Mutants
Plant genomic DNA flanking the T-DNA borders of the Salk mutants were amplified by PCR (Supplemental Table 3) and sequenced to confirm the flanking sequence tags identified. Homozygous lines were then isolated (Supplemental Table 4) for further characterization (Figures 2 and 4A).
Constructs and Plant Transformation
Construction of the ProMYB118:uidA Transgenes
Regions −970 to −1 bp or −2535 to −1 bp relative to the MYB118 translational start codon were amplified with the proofreading Pfu Ultra DNA polymerase (Stratagene) from Col-0 genomic DNA using either 5′-attB1-AAATCACATATATTATTTAGATAAAGGG-3′ and 5′-attB2-ATGATTATGATGGCAAAAA-3′ (−970-bp fragment) or 5′-attB1-TCCCCATTTTAAATGATTCTC-3′ and 5′-attB2-ATGATTATGATGGCAAAAA-3′ (−2.5-kb fragment), attB1 and attB2 referring to the corresponding Gateway recombination sequences. The PCR products were introduced by BP recombination into the pDONR207 entry vector (Invitrogen) and transferred into the destination vector pBI101-R1R2-GUS (Baud et al., 2007b) by LR recombination. The resulting binary vectors were electroporated into Agrobacterium tumefaciens C58C1 strain and used for agroinfiltration of flower buds of Arabidopsis (Bechtold et al., 1993). Primary transformants were selected on Murashige and Skoog (MS) medium containing kanamycin (50 mg⋅L−1) and transferred to soil for further characterization. Between 15 and 27 independent transgenic lines were analyzed, depending on the construct considered.
Construction of the ProODD:uidA, ProCKX3:uidA, ProERF:uidA, and ProPER17:uidA Transgenes
Regions −1000 to −1 bp relative to the translational start codon were amplified using 5′-attB1-AACTTGATGAAAATGTTTGAAC-3′ and 5′-attB2-TTTTGTGTTTTGTTTATCTCTTTCTTT-3′ (ODD), 5′-attB1-TCCAAGGGTCAAGATTTATTG-3′ and 5′-attB2-TTTTTTTGAGAAATAAGTCTTTTTATTTTATG-3′ (CKX3), 5′-attB1-ACTAAAACATATAGTTTAAAAGGGTG-3′ and 5′-attB2-GGTGTAATTAGGAAGAAGAATGG-3′ (ERF8), and 5′-attB1-TTGGTTCTAGCTAGATCATTCG-3′ and 5′-attB2-ACTTTTTTCTTTTTTGGTGTTG-3′ (PER17). The PCR products were introduced by BP recombination into the pDONR207 entry vector (Invitrogen) and transferred into the destination vector pGWB3 (Nakagawa et al., 2007) by LR recombination, as previously described. Construction of the ProBCCP2:uidA transgene was described by Baud et al. (2009). Construction of the reporter constructs comprising concatemers of MYB elements is described in Supplemental Table 5.
Construction of the Pro35Sdual:MYB118 Transgene
MYB118 cDNA was amplified with 5′-attB1-ATGGAGTTCGAGTCAGTG-3′ and 5′-attB2-CTAAAGACGACCATGAGC-3′, cloned into the pDONR207, and finally transferred to the binary vector pMDC32 (Curtis and Grossniklaus, 2003) as previously described (see above). Primary transformants were selected on MS medium containing hygromycin (50 mg⋅L−1).
Construction of the Pro35Sdual:MYB107 Transgene
MYB107 cDNA was amplified with 5′-attB1-ATGGGGAGATCACCGTGT-3′ and 5′-attB2-CTATTCACGAAATGGCCA-3′ and cloned in the binary vector pMDC32 as previously described (see above).
Construction of the ProMYB118:MYB118 Transgene
MYB118 genomic sequence including a promoter region of 2535 bp relative to the translational start codon was amplified with 5′-attB1-TCCCCATTTTAAATGATTCTC-3′ and 5′-attB2-CTAAAGACGACCATGAGC-3′, cloned into the pDONR207, and finally transferred to the binary vector pBIB-Hyg-GTW. Primary transformants were selected on MS medium containing hygromycin (50 mg⋅L−1).
Construction of the ProODD:GFP and ProBCCP2:GFP Transgenes
Promoters previously cloned into the pDONR207 entry vector (see above) were transferred into the destination vector pBS TPp-B (Thévenin et al., 2012) by LR recombination.
Transactivation Assays in Arabidopsis Protoplasts
Protoplast preparation and transformation were performed as described by Fülöp et al. (2005). For each transformation, 30 µg each plasmid was mixed with 150 μL concentrated protoplast suspension and 450 μL polyethylene glycol solution [PEG 6000 25% (w/v), 0.45 M mannitol, and 0.1 M Ca(NO3)2, pH 9].
RNA Analyses
RNA extraction, reverse transcription, RT-PCR, and real-time qRT-PCR were performed as previously described (Baud et al., 2004). The sequences of primers used for RT-PCR and real-time qRT-PCR are indicated in Supplemental Table 6.
Lipid Analyses
Total fatty acid analyses were performed as previously described (Li et al., 2006) on pools of 20 seeds or seed fractions.
Protein and Immunoblot Analyses
Seed proteins were extracted from batches of 20 seeds, 20 embryos, or 200 endosperm fractions. Protoplast proteins were extracted from 0.5 mL of transformed protoplasts. Electrophoresis and immunoblot experiments were performed as described by d’Andréa et al. (2007a). Briefly, proteins were extracted in reducing Laemmli buffer, resolved on a 12% NuPAGE (seed proteins) or SDS-PAGE (protoplast proteins) gel, transferred to polyvinylidene fluoride membrane, and probed with antibody serum. Preparation of anti-2S and anti-12S sera was previously described by Jolivet et al. (2011). Anti-rS3N was produced as described by D’Andréa et al. (2007b). Rabbit sera dilutions were as follows: 1:5000 (anti-12S, anti-rS3N), 1:2000 (anti-2S), 1:2000 (anti-alpha-Tubulin; Sigma-Aldrich), and 1:1000 (anti-GFP; Roche). Detection was performed using the Enhanced ChemiLuminescence kit (GE Healthcare) according to the manufacturer’s instructions on an ImageQuant LAS 4000 system (GE Healthcare). Relative quantification of the proteins was performed with the MultiGauge software (Fujifilm).
EMSAs
The expression plasmid was constructed by transferring MYB118 cDNA from the pDONR207 to the expression vector pETG10A (http://www.embl-hamburg.de/). The resulting vector was electroporated into Escherichia coli RosettaBlue(DE3)pLysS strain (Novagen) for expression. After induction by 0.5 mM isopropyl-β-d-thiogalactopyranoside in Luria-Bertani buffer, cells were grown 1 h at 17°C. Cell lysis and protein purification were performed as previously described (Baud et al., 2009). To prepare DNA probes, complementary biotin-labeled (at the 5′ end) oligonucleotides (Eurofins MWG Operon) were annealed. For DNA binding assays, MYB118 recombinant protein was incubated with 20 fmol probe in binding buffer [20 mM Tris-HCl, pH 8, 250 mM NaCl, 2 mM MgCl2, 1% glycerol (v/v), 1 mg⋅mL−1 BSA, 1 mM DTT, 20 ng⋅µL−1 poly(dI-dC)]. For competition assays, the unbiotinylated competitor was incubated briefly with the recombinant protein before the biotinylated probe was added. After addition of the biotinylated probe, reactions were incubated 30 min at room temperature, then fractionated at 4°C by 6% PAGE. Electrophoretic transfer to nylon membrane and detection of the biotin-labeled DNA were performed according to the manufacturer’s instructions (Chemiluminescent Nucleic Acid Detection Module; Pierce) using an ImageQuant LAS 4000 system (GE Healthcare).
Microscopy
To obtain scanning electron micrographs, dry seeds were mounted onto a Peltier cooling stage using adhesive discs (Deben) and observed with a SH-1500 tabletop scanning electron microscope (Hirox). Histochemical detection of GUS activity and bright-field microscope observations were performed as described by Baud et al. (2007a). Observations of oil bodies in embryo and endosperm cells were performed as described by Miquel et al. (2014).
CATMA Array Design and Hybridization
Design and hybridization of CATMAv6.2 arrays are described in Supplemental Data Set 1. Briefly, the CATMAv6.2 array contains 73,228 primers in triplicate corresponding to 38,383 associated annotations coming from FLAGdb++ (http://urgv.evry.inra.fr/FLAGdb), an integrative database around plant genomes (Dèrozier et al., 2011), and 34,845 primers that are the reverse of previous ones. Details regarding experiment design and differential analyses are accessible in the CATdb database (http://urgv.evry.inra.fr/CATdb/; Project:RA12-06_myb118) (Gagnot et al., 2008). Microarray data were deposited in the international repository at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession number GSE48180).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ABI3, At3g24650; ACPT/CPT3, At2g23410; ARR15, At1g74890; AT2S1, At4g27140; AT2S5, At5g54740; BCCP2, At5g15530; CKX3, At5g56970; CRA1, At5g44120; CRUL, At1g03890; EF1αA4, At5g60390; EMB1353, At4g30090; ERF8, At1g53170; FUS3, At1g26790; GRP19, At5g07550; KCS7, At1g71160; LEC1, At1g21970; LEC2, At1g28300; L1L, At5g47670; MYB107, At3g02940; MYB115, At5g40360; MYB118, At3g27785; ODD, At1g04380; OLE1/S3, At4g25140; PER17, At2g22420; PKp-β1, At5g52920; RLK1, At1g48480; SM3, At2g25890; WRI1, At3g54320; and ZOU/RGE1, At1g49770.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complementary Results for the Characterization of the Expression Pattern of MYB118.
Supplemental Figure 2. Purity of the Different Seed Fractions Dissected.
Supplemental Figure 3. Phenotypes of Mutant Seeds and Embryos Affected in Master Regulators of the Maturation Program.
Supplemental Figure 4. Complementary Results for the Characterization of the Regulation of MYB118 by LEC2.
Supplemental Figure 5. Characterization of MYB118 Overexpressing Lines.
Supplemental Figure 6. Microscopic Observation of myb118-1 Seed Development.
Supplemental Figure 7. Characterization of myb118-1 and OE3 Seeds.
Supplemental Figure 8. Negative Control for Transcriptional Activation Studies in Protoplasts of Arabidopsis.
Supplemental Figure 9. Complementary Results for the Study of the Expression Pattern of Maturation-Related Genes in myb118-1 Mutant Background.
Supplemental Figure 10. A Model for the Transcriptional Regulation of Seed Maturation in Arabidopsis.
Supplemental Figure 11. Characterization of myb115-1 and myb115-1 myb118-1 Mutants.
Supplemental Figure 12. Characterization of the Expression Pattern of ODD.
Supplemental Table 1. Selected Genes Activated by MYB118 as Identified by Microarray Analyses.
Supplemental Table 2. Selected Genes Repressed by MYB118 as Identified by Microarray Analyses.
Supplemental Table 3. Primers Used for Molecular Characterization of T-DNA Insertions.
Supplemental Table 4. Primers Used for Mutant Characterization.
Supplemental Table 5. Details of Reporter Constructs Comprising Concatemers of MYB Elements and Used for Transactivation Assays in Nicotiana benthamiana.
Supplemental Table 6. Primers Used for Quantitative RT-PCR.
Supplemental Data Set 1. Results of Microarray Experiments Performed on myb118-1 and OE3 Mutant Seeds 12 Days after Anthesis.
Supplementary Material
Acknowledgments
We thank H. North for critical reading of the article, J. Zuo for sharing plant material, C. Larré and Sabine d’Andréa for the gift of antibodies, and the EMBL for providing the pETG-10a vector. We also thank S. Balzergue, M. Miquel, K. Deruyffelaere, D. De Vos, and C. Boulard for their technical assistance. This work was supported by the French National Research Agency (SOLAR, Grant ANR-10-GENM-009, and CERES, Grant ANR-BLAN-1238).
AUTHOR CONTRIBUTIONS
A.T., C.M., V.B., L.S.-T., N.B., and B.D. performed the research and analyzed the data. L.L. designed the research and analyzed the data. G.B. and S.B. designed and performed the research, analyzed the data, and wrote the article.
Glossary
- SSP
seed storage protein
- TAG
triacylglycerol
- TF
transcription factor
- qRT-PCR
quantitative RT-PCR
- Col-0
Columbia-0
- DAA
days after anthesis
- DEX
dexamethasone
- EMSA
electrophoretic mobility shift assay
- MS
Murashige and Skoog
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, R., Oñate-Sánchez, L., Weltmeier, F., Ehlert, A., Diaz, I., Dietrich, K., Vicente-Carbajosa, J., Dröge-Laser, W. (2009). A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21: 1747–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, M.S., Willmann, M.R., Jenik, P.D. (2012). Is there a role for trihelix transcription factors in embryo maturation? Plant Signal. Behav. 7: 205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud, S., Lepiniec, L. (2010). Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 49: 235–249 [DOI] [PubMed] [Google Scholar]
- Baud, S., Boutin, J.P., Miquel, M., Lepiniec, L., Rochat, C. (2002). An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 40: 151–160 [Google Scholar]
- Baud, S., Vaultier, M.-N., Rochat, C. (2004). Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J. Exp. Bot. 55: 397–409 [DOI] [PubMed] [Google Scholar]
- Baud, S., Wuillème, S., Dubreucq, B., de Almeida, A., Vuagnat, C., Lepiniec, L., Miquel, M., Rochat, C. (2007a). Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J. 52: 405–419 [DOI] [PubMed] [Google Scholar]
- Baud, S., Mendoza, M.S., To, A., Harscoët, E., Lepiniec, L., Dubreucq, B. (2007b). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Baud, S., Wuillème, S., To, A., Rochat, C., Lepiniec, L. (2009). Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J. 60: 933–947 [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., Pelletier, G. (1993). In planta infiltration of adult Arabidopsis plants. C. R. Acad. Sci. Paris Life Sci. 316: 1194–1199 [Google Scholar]
- Beeckman, T., De Rycke, R., Viane, R.,Inze, D. (2000). Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. 113: 139–148 [Google Scholar]
- Belmonte, M.F., et al. . (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, F., Chaudhury, A. (2009). Parental memories shape seeds. Trends Plant Sci. 14: 550–556 [DOI] [PubMed] [Google Scholar]
- Berger, N., Dubreucq, B., Roudier, F., Dubos, C., Lepiniec, L. (2011). Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 23: 4065–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D., Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Andréa, S., Canonge, M., Beopoulos, A., Jolivet, P., Hartmann, M.A., Miquel, M., Lepiniec, L., Chardot, T. (2007a). At5g50600 encodes a member of the short-chain dehydrogenase reductase superfamily with 11β- and 17β-hydroxysteroid dehydrogenase activities associated with Arabidopsis thaliana seed oil bodies. Biochimie 89: 222–229 [DOI] [PubMed] [Google Scholar]
- D’Andréa, S., Jolivet, P., Boulard, C., Larré, C., Froissard, M., Chardot, T. (2007b). Selective one-step extraction of Arabidopsis thaliana seed oleosins using organic solvents. J. Agric. Food Chem. 55: 10008–10015 [DOI] [PubMed] [Google Scholar]
- Day, R.C., Herridge, R.P., Ambrose, B.A., Macknight, R.C. (2008). Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 148: 1964–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dèrozier, S., Samson, F., Tamby, J.-P., Guichard, C., Brunaud, V., Grevet, P., Gagnot, S., Label, P., Leplé, J.C., Lecharny, A., Aubourg, S. (2011). Exploration of plant genomes in the FLAGdb++ environment. Plant Methods 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Fülöp, K., Pettkó-Szandtner, A., Magyar, Z., Miskolczi, P., Kondorosi, E., Dudits, D., Bakó, L. (2005). The Medicago CDKC;1-CYCLINT;1 kinase complex phosphorylates the carboxy-terminal domain of RNA polymerase II and promotes transcription. Plant J. 42: 810–820 [DOI] [PubMed] [Google Scholar]
- Gagnot, S., Tamby, J.-P., Martin-Magniette, M.L., Bitton, F., Taconnat, L., Balzergue, S., Aubourg, S., Renou, J.-P., Lecharny, A., Brunaud, V. (2008). CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 36: D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M.-J., Lydiate, D.J., Li, X., Lui, H., Gjetvaj, B., Hegedus, D.D., Rozwadowski, K. (2009). Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21: 54–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo, K., Ugawa, Y., Iwamoto, M., Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet, P., Boulard, C., Bellamy, A., Valot, B., d’Andréa, S., Zivy, M., Nesi, N., Chardot, T. (2011). Oil body proteins sequentially accumulate throughout seed development in Brassica napus. J. Plant Physiol. 168: 2015–2020 [DOI] [PubMed] [Google Scholar]
- Le, B.H., et al. . (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux, B., Miquel, M., Somerville, C., Browse, J. (1990). Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor. Appl. Genet. 80: 234–240 [DOI] [PubMed] [Google Scholar]
- Li, Y., Beisson, F., Pollard, M., Ohlrogge, J. (2006). Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Liao, Y., Zou, H.-F., Wang, H.-W., Zhang, W.-K., Ma, B., Zhang, J.-S., Chen, S.-Y. (2008). Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 18: 1047–1060 [DOI] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A., Lo, R., Kwong, R.W., Yamagishi, K., Fischer, R.L., Goldberg, R.B., Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Mendes, A., Kelly, A.A., van Erp, H., Shaw, E., Powers, S.J., Kurup, S., Eastmond, P.J. (2013). bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell 25: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel, M., Trigui, G., d’Andréa, S., Kelemen, Z., Baud, S., Berger, A., Deruyffelaere, C., Trubuil, A., Lepiniec, L., Dubreucq, B. (2014). Specialization of oleosins in oil body dynamics during seed development in Arabidopsis seeds. Plant Physiol. 164: 1866–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., Toyooka, K., Matsuoka, K., Jinbo, T., Kimura, T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Oda, M., Furukawa, K., Ogata, K., Sarai, A., Nakamura, H. (1998). Thermodynamics of specific and non-specific DNA binding by the c-Myb DNA-binding domain. J. Mol. Biol. 276: 571–590 [DOI] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, O.-A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16 (suppl.): S214–S227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield, S., Rylott, E.L., Gilday, A.D., Graham, S., Larson, T.R., Graham, I.A. (2004). Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16: 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouse, M.B., Campbell, M.M. (2012). The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta 1819: 67–77 [DOI] [PubMed] [Google Scholar]
- Santos Mendoza, M., Dubreucq, B., Miquel, M., Caboche, M., Lepiniec, L. (2005). LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 579: 4666–4670 [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza, M., Dubreucq, B., Baud, S., Parcy, F., Caboche, M., Lepiniec, L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Schölkopf, B., Weigel, D., Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Solano, R., Nieto, C., Avila, J., Cañas, L., Diaz, I., Paz-Ares, J. (1995). Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 14: 1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu, N., Wobus, U. (2013). Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu. Rev. Plant Biol. 64: 189–217 [DOI] [PubMed] [Google Scholar]
- Suzuki, M., McCarty, D.R. (2008). Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 11: 548–553 [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Wang, H.H.Y., McCarty, D.R. (2007). Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenin, J., Dubos, C., Xu, W., Le Gourrierec, J., Kelemen, Z., Charlot, F., Nogué, F., Lepiniec, L., Dubreucq, B. (2012). A new system for fast and quantitative analysis of heterologous gene expression in plants. New Phytol. 193: 504–512 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi, H., Morikami, A., Nakamura, K. (2007). Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 104: 2543–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa, J., Carbonero, P. (2005). Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 49: 645–651 [DOI] [PubMed] [Google Scholar]
- Wang, H., Guan, S., Zhu, Z., Wang, Y., Lu, Y. (2013). A valid strategy for precise identifications of transcription factor binding sites in combinatorial regulation using bioinformatic and experimental approaches. Plant Methods 9: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Niu, Q.-W., Teng, C., Li, C., Mu, J., Chua, N.-H., Zuo, J. (2009). Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 19: 224–235 [DOI] [PubMed] [Google Scholar]
- Willmann, M.R., Mehalick, A.J., Packer, R.L., Jenik, P.D. (2011). MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 155: 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Q., Creff, A., Waters, A., Tanaka, H., Goodrich, J., Ingram, G.C. (2013). ZHOUPI controls embryonic cuticle formation via a signalling pathway involving the subtilisin protease ABNORMAL LEAF-SHAPE1 and the receptor kinases GASSHO1 and GASSHO2. Development 140: 770–779 [DOI] [PubMed] [Google Scholar]
- Yang, S., Johnston, N., Talideh, E., Mitchell, S., Jeffree, C., Goodrich, J., Ingram, G. (2008). The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 135: 3501–3509 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Cao, G., Qu, L.-J., Gu, H. (2009). Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis. Plant Cell Rep. 28: 337–346 [DOI] [PubMed] [Google Scholar]
- Zhang, H., Ogas, J. (2009). An epigenetic perspective on developmental regulation of seed genes. Mol. Plant 2: 610–627 [DOI] [PubMed] [Google Scholar]
- Zhou, Y., et al. . (2013). HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25: 134–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.