Leaf shape is dynamic and influenced by numerous factors. Here, the authors quantify shape differences in leaves across the leaf series and during their development, determining their genetic basis and the effects of environment, in a population resulting from a cross between tomato and a wild desert relative.
Abstract
Leaf shape is mutable, changing in ways modulated by both development and environment within genotypes. A complete model of leaf phenotype would incorporate the changes in leaf shape during juvenile-to-adult phase transitions and the ontogeny of each leaf. Here, we provide a morphometric description of >33,000 leaflets from a set of tomato (Solanum spp) introgression lines grown under controlled environment conditions. We first compare the shape of these leaves, arising during vegetative development, with >11,000 previously published leaflets from a field setting and >11,000 leaflets from wild tomato relatives. We then quantify the changes in shape, across ontogeny, for successive leaves in the heteroblastic series. Using principal component analysis, we then separate genetic effects modulating (1) the overall shape of all leaves versus (2) the shape of specific leaves in the series, finding the former more heritable than the latter and comparing quantitative trait loci regulating each. Our results demonstrate that phenotype is highly contextual and that unbiased assessments of phenotype, for quantitative genetic or other purposes, would ideally sample the many developmental and environmental factors that modulate it.
INTRODUCTION
Leaf shape, across disparate species, is staggeringly diverse. Yet, such a statement grossly underestimates the spectrum of leaf morphology and fails to capture the true diversity of shapes exhibited by leaves. If we were to compare the leaves of different species, to reduce leaf shape to an archetypal form representing each, the ideal leaf would be truly multivariate, modulated by phylogeny as well as by developmental and environmental context (Jones, 1992; Nicotra et al., 2011). Similarly, any model of leaf shape we estimate for quantitative genetic purposes must be equally sophisticated to reflect the underlying reality.
Goethe was one of the first (and the most poetic) to describe the serial homology between lateral organs of the shoot, declaring the leaf “the true Proteus who can hide or reveal himself in all vegetal forms” (Goethe, 1817; trans. Goethe, 1952). Although referring to the more dramatic “metamorphosis” of lateral organs from leaves to floral organs within a single individual (Friedman and Diggle, 2011), “protean” aptly describes the mutable succession of not only shape but epidermal features and size of leaves iteratively produced by the shoot apical meristem during the transitions from juvenile to adult to reproductive development (Kerstetter and Poethig, 1998). The succession of leaf types produced during plant development is termed “heteroblasty,” reflecting internal changes in the state of the shoot apical meristem manifest in lateral organ development (Goebel, 1900; Ashby, 1948; Poethig, 1990, 2010). Beyond changes in the developmental program of successive leaves, the shape of any single leaf is in flux, reflecting allometric expansion during its ontogeny (i.e., age), a phenomenon quantified as early as the Vegetable Staticks (Hales, 1727).
Additionally, leaf morphology is phenotypically plastic and responsive to changes in environment. Light intensity and quality both modulate leaf shape, size, and thickness (Givnish, 1988; Smith and Whitelam, 1997; Yano and Terashima, 2001). Often, the manner in which leaves respond to different environmental conditions converges upon heteroblastic changes in leaf shape (Allsopp, 1954; Jones, 1995; Diggle, 2002).
Previously, we analyzed the genetic basis of morphometric differences in leaf shape between domesticated tomato (Solanum lycopersicum) and a wild relative originating from the coastal deserts of Peru (Solanum pennellii) using a set of near-isogenic introgression lines (which we refer to as the ILs; Eshed and Zamir, 1995; Chitwood et al., 2013a). Mature leaves were measured in a field and arose after the transition to reproductive development, representing a homogenous population with respect to development. We found that leaf shape is polygenic and highly heritable, consistent with previous studies in snapdragon (Antirrhinum majus) (Langlade et al., 2005), maize (Zea mays; Tian et al., 2011), and grape (Vitis vinifera; Chitwood et al., 2014).
However, the above studies account for only a small subset of leaf shapes present during the development of an individual plant. In a separate study, we quantified the effects of the heteroblastic series and ontogeny on leaf shape in eight accessions each of Solanum arcanum, Solanum habrochaites, and Solanum pimpinellifolium (Chitwood et al., 2012a). In addition, the effects of the proximal-distal axis on leaflet shape were measured and were consistent with the known basipetal development of tomato leaves.
While the aforementioned studies measured a genetic basis for leaf shape in field-grown ILs and described changes in leaf shape throughout development in wild tomato species, a genetic basis for heteroblastic changes in leaf shape remains undescribed. Here, we present a genetic analysis of leaf shape throughout vegetative development in the S. pennellii ILs. We first contextualize these leaf morphs in a combined analysis including previously published tomato leaves. We then analyze leaf shape with respect to the heteroblastic series and ontogeny (age) in the ILs, revealing that leaf shape is highly dynamic during development and abruptly modulated by the transition to reproductive growth. Using a principal component analysis (PCA) to decompose patterns in leaf shape across the heteroblastic series, we find that uniform changes in shape across all leaflets are highly heritable, but heteroblastic changes in shape specific to particular leaflets less so. Finally, we describe changes in leaf shape between chamber and field-grown conditions, indicating that environment significantly alters leaf morphology and its genetic basis. Together, our analyses quantitatively analyze the shape of >55,000 leaflets, revealing how genetics, development, and environment interactively modulate a complex trait.
RESULTS
Comparison of Leaf Shape between Studies
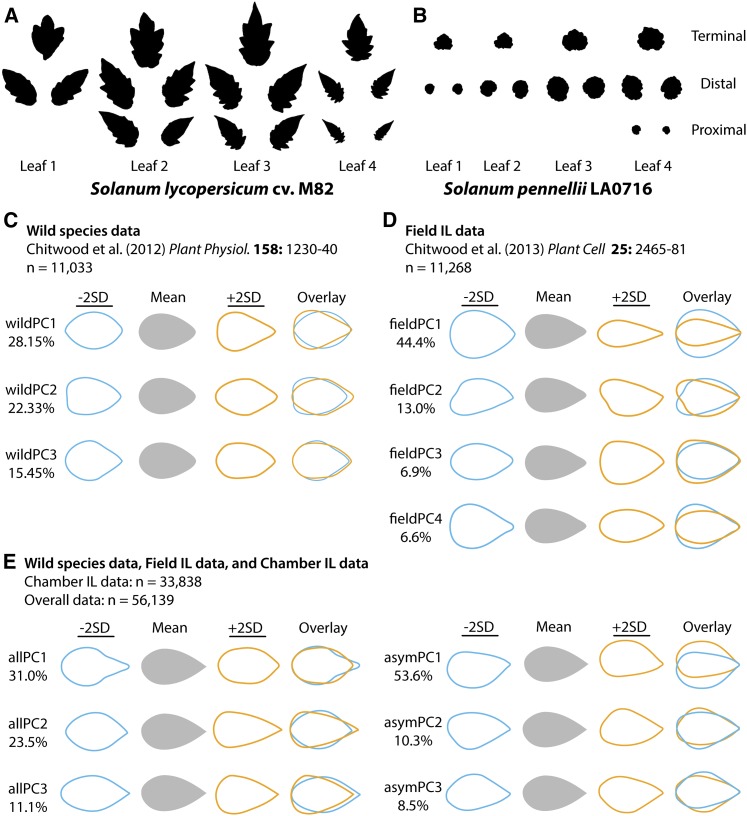
The leaves of domesticated tomato, S. lycopersicum, and one of the earliest diverging members of the tomato complex that originates from the coastal deserts of Peru, S. pennellii, are strikingly different. Domesticated tomato leaves are larger, more lanceolate, lobed, serrate, and complex (Figure 1A) compared with the smaller, rounder, more entire, sinuate, and less complex leaves of S. pennellii (Figure 1B). As one progresses through the first four leaves of domesticated tomato, the leaflets transition from a rounder to more lance-like shape, increasing in serration and slowly losing the trifoliate shape of the lobes variably present in the first two leaves. In contrast, S. pennellii leaflets appear more constant in shape through the leaf series.
Figure 1.
PCs Describing Shape Variance in Leaves of Tomato and Wild Relatives.
(A) and (B) Example of leaflets collected from (A) S. lycopersicum (domesticated tomato) and (B) S. pennellii. Leaflets from chamber-grown S. pennellii ILs were similarly collected, sampling the first four leaves and across the proximal-distal axis of each leaf.
(C) to (E) PCs resulting from EFD analysis from this and previous studies. allPC1 is associated with the trifoliate nature of tomato leaflets created by three prominent lobes, allPC2 in part describes the distribution of laminar outgrowth along the proximal-distal axis, and allPC3 aligns with shape variance producing bulging at the base of the leaflet. Blue and orange outlines represent eigenleaves (theoretical leaf outlines representing shape variance for each PC) −2 and +2 sd, respectively, along each PC axis. See Supplemental Table 1 for descriptions and prefixes for different PCAs used in this study.
(C) PCs from Chitwood et al. (2012a) (“wildPCs”) sampling leaflets from the first four leaves of eight accessions each of S. arcanum, S. habrochaites, and S. pennellii.
(D) PCs from Chitwood et al. (2013a) (“fieldPCs”) sampling field-grown leaflets of the S. pennellii ILs, and
(E) PCs resulting from a combined analysis of >33,000 chamber-grown leaflets from the S. pennellii ILs new to this study and leaflets from Chitwood et al. (2012a) and (2013a) (“allPCs,” measuring symmetric shape variance and “asymPCs” measuring asymmetric shape variance).
Seventy-six near-isogenic ILs, each harboring a relatively small, defined introgressed region from S. pennellii in an otherwise domesticated tomato background, are a powerful way to dissect the genetic architecture governing morphological differences between these two species (Eshed and Zamir, 1995). To understand the genetic basis of not only leaf shape but the heteroblastic series, we morphometrically analyzed >33,000 leaflets, sampling the first four leaves and proximal-distal axis of the leaf from chamber-grown ILs and parents.
It is important that the new results we are presenting are compared with those we previously published, to provide a complete perspective on leaflet shape variation through developmental time, across varied environments, phylogenetic distances, and across the proximal-distal axis of leaves (Chitwood et al., 2012a, 2013a). The main morphometric technique we use to quantify tomato leaflet shape is elliptical Fourier descriptor (EFD) analysis followed by PCA. Briefly, EFD analysis expands a complex outline (a leaflet in this case) into a shape spectrum by obtaining the Fourier coefficients from chain code, a lossless compression method that describes outlines as a series of integers (Kuhl and Giardina, 1982). The harmonics of the resulting series approximate the outline of the original shape. A PCA is then used to quantify orthogonal (i.e., uncorrelated), distinct shape components together comprising the harmonics of the originally described shape (Iwata et al., 1998; Iwata and Ukai, 2002). We previously published EFD-PCAs of leaflets from wild species (Chitwood et al., 2012a; wildPCs, Figure 1C) and field-grown IL leaflets (Chitwood et al., 2013a; fieldPCs, Figure 1D), which we have included for convenience and clarity.
To compare the shape of new leaflets measuring the heteroblastic series in the ILs with previous studies, an EFD-PCA encompassing all leaflets needs to be performed. “allPCs” describe symmetric shape variance and “asymPCs” asymmetric shape variance in >55,000 leaflets, representing leaflets from our two previously published works on leaf shape and the >33,000 newly measured leaflets (Figure 1E). We have not always measured both symmetric and asymmetric variance in previously published works, depending on the study’s focus, and we use “all” as a prefix here instead of the more logical “sym” simply because we used “sym” in other articles and wish to avoid confusion with our previous works. PCA is a powerful data transformation technique to elegantly explain multivariate data, and we employ it often in this article. To avoid further confusion between different PCAs, we denote different PCAs with prefixes and have described each in Supplemental Table 1.
In addition to EFD-PCA, classic morphometric parameters have been estimated for leaflets. Length-to-width ratio is estimated using aspect ratio (the ratio of the major to minor axis of the best-fitted ellipse to a leaflet, 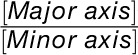 ) and roundness (
) and roundness (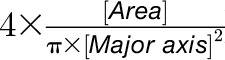 ). Circularity (a ratio of area to perimeter;
). Circularity (a ratio of area to perimeter;  ) and solidity (a ratio of the object area to the total area of the convex hull,
) and solidity (a ratio of the object area to the total area of the convex hull, 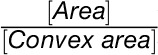 ) are used to estimate serration and lobing but are also sensitive to how round versus oblong the object is.
) are used to estimate serration and lobing but are also sensitive to how round versus oblong the object is.
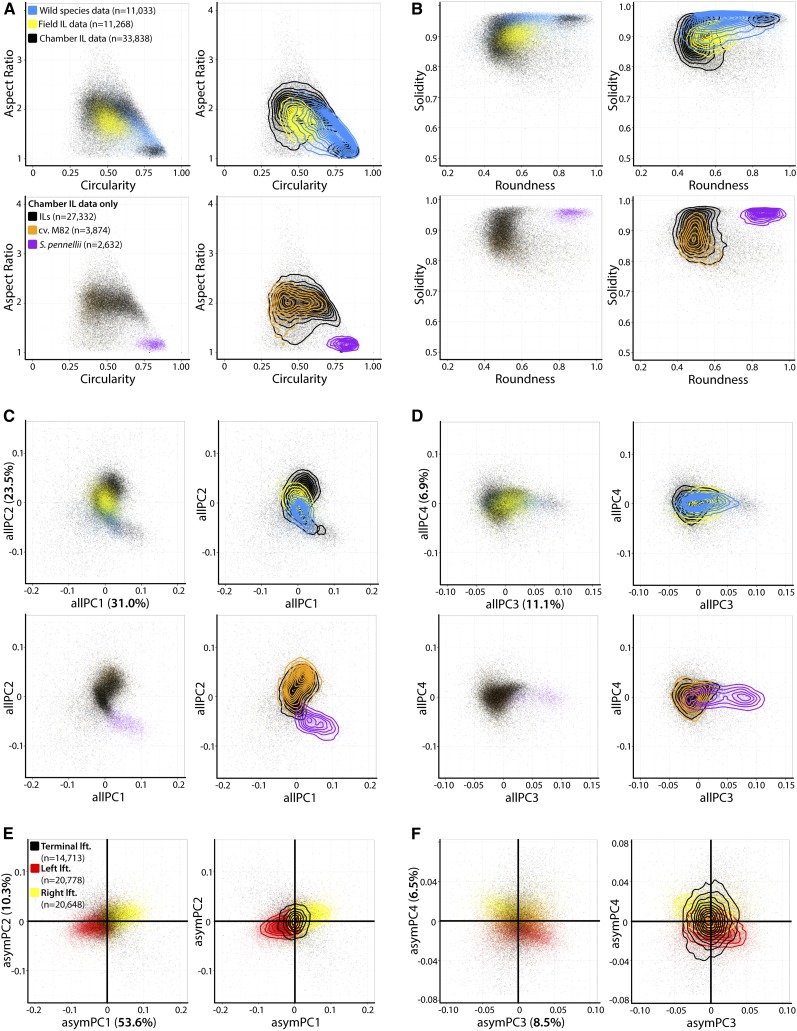
Plotting wild tomato species, field IL, and chamber IL leaflet data by aspect ratio, circularity, solidity, roundness, and allPCs 1-4, it becomes clear that domesticated tomato (S. lycopersicum cv M82) and S. pennellii leaflets occupy extreme positions in the tomato complex morphospace (Figures 2A to 2D; Supplemental Data Set 1). The wild species data, representing accessions from S. arcanum, S. habrochaites, and S. pimpinellifolium, largely span intermediate values between S. lycopersicum and S. pennellii. This demonstrates that the S. pennellii ILs are ideally situated for the genetic analysis of leaf shape, representing extreme morphological positions within the tomato complex.
Figure 2.
The Morphospace of Tomato Leaves Measured across Evolutionary Distance, Developmental Context, and Different Environments.
(A) to (D) Distributions of morphometric trait values for leaflets from multiple studies. (A) Aspect ratio and circularity, (B) solidity and roundness, (C) allPC1 and allPC2, and (D) allPC3 and allPC4. In the top panels, wild species data (blue; n = 11,033 leaflets; Chitwood et al., 2012a), IL field-grown leaflets (yellow; n = 11,268 leaflets; Chitwood et al., 2013a), and chamber-grown IL leaflets new to this study (black; n = 33,838) are plotted. Bottom panels represent only new data for chamber-grown IL and parent leaflets in this study (IL leaflets, black; S. lycopersicum cv M82 leaflets, orange; S. pennellii leaflets, purple). Left panels are scatterplots and right panels are contour plots in addition to the scatterplot to help visualize distributions. For all morphometric traits, S. pennellii occupies an extreme position in morphospace and often the shapes of wild species leaves are intermediate between S. lycopersicum and S. pennellii.
(E) and (F) Asymmetric variance is explained by whether a leaflet originates from the left (red) or right (yellow) side of a leaf. Terminal leaflets (black) on average are symmetrical. (E) Scatterplot of asymPC1 and asymPC2 and (F) asymPC3 and asymPC4.
Comparing field IL data to chamber IL data, subtle differences in shape can be discerned. Field IL leaflets have lower aspect ratio and higher roundness values compared with chamber IL leaflets, indicating that they are less elongated (Figures 2A and 2B). Field IL leaflets also have lower allPC2 and higher allPC3 values compared with chamber IL leaflets (Figures 2C and 2D), shape attributes associated with a rounder, less oblong leaflet (Figure 1E).
With respect to asymmetric shape variance, leaflets from all data sets show expected left-right asymmetries with respect to the side of the leaf from which the leaflet is derived, and terminal leaflets, on average, are symmetrical (Figures 2E and 2F). A portion of asymmetric variance in leaflets (especially in terminal leaflets) is attributable to phyllotactic handedness as we have previously shown (Chitwood et al., 2012b, 2012c), but was averaged out in this study, as leaves were not separated by the direction of their spiral.
Heteroblastic and Ontogenetic Effects on Leaf Shape
Leaf shape is heavily modulated by developmental context. Plants produce different types of leaves, by virtue of their place in the heteroblastic series (Goebel, 1900; Ashby, 1948; Poethig, 1990, 2010). Each leaf in the series continuously changes its shape, through allometric expansion, as it develops (Hales, 1727). Therefore, when describing the shape of a leaf, at least two variables must be accounted for (1) placement in the heteroblastic series and (2) age, which we refer to as “ontogeny.”
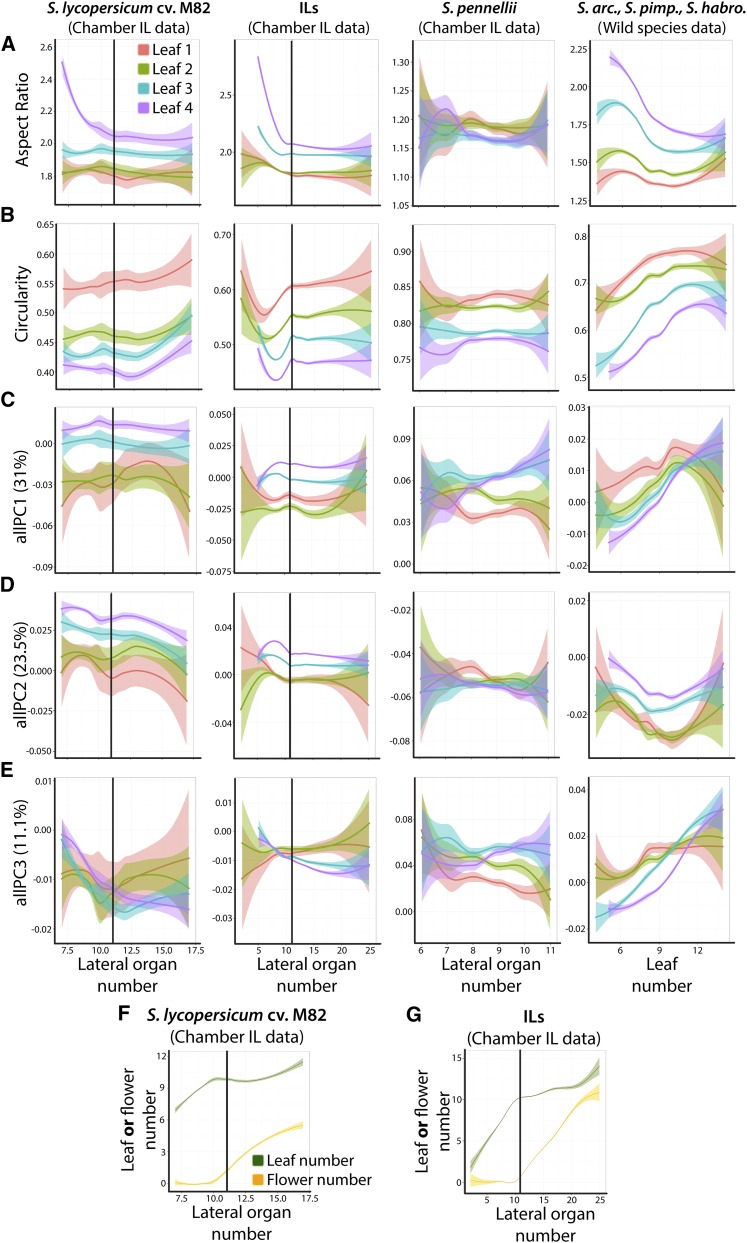
In order to accurately record the outlines of tens of thousands of leaflets, individual plants are terminally harvested. In doing so, the effects of heteroblasty and ontogeny are partially confounded. In the chamber IL data set, for example, we sample the first four leaves of the ILs and their parents. Leaf 1 is not only the first leaf in the heteroblastic series, but it is always the most mature leaf. Likewise, leaf 4 is both the farthest leaf in the series and the youngest. In our previous analysis of heteroblasty and ontogeny in wild tomato species, we found naturally segregating differences in growth rate, in addition to within genotype variability, as a useful way to begin separating the effects of heteroblasty and ontogeny (Chitwood et al., 2012a). Growth rate is measured as the total lateral organ number (including both leaves and flowers) on the plant from which leaves were harvested (including organ primordia a few millimeters in length; Figure 3).
Figure 3.
Heteroblasty and Ontogeny Modulate Leaf Shape.
(A) to (D) Loess models for (A) aspect ratio, (B) circularity, (C) allPC1, (D) allPC2, and (E) allPC3 values for leaflets originating from each of the first four leaves against developmental age, as measured by lateral organ number. Leaves 1-4 are indicated by red, green, blue, and purple, respectively, with 95% confidence bands. Left to right, shape attributes for leaflets from the chamber data set for (1) domesticated tomato, (2) the ILs, and (3) S. pennelli are plotted. Far right, pooled data from the wild species data set.
(F) and (G) An analysis of the number of leaves and leaf primordia (green) and flowers and floral primordia (yellow) comprising lateral organ number values for (F) domesticated tomato and (G) the ILs from the chamber data. Vertical lines in plots indicate a lateral organ number of 11 for comparison. Because S. pennellii initiates lateral organs slowly, and wild species data were collected differently, the vertical line is not included in these plots. Note: y axis scales are not consistent between genotypes. Please refer to Figure 2 to observe the extreme difference in S. pennellii traits relative to the cv M82 parent and ILs.
As the heteroblastic series progresses from leaf 1 to 4, leaflets become more elongated (as measured by aspect ratio and roundness; Figures 3A; Supplemental Figure 1A). Changes in the shape of leaflets derived from leaves 1 and 2 are generally stable across both young and old individuals, as measured by lateral organ number. This is to be expected, as these leaves are relatively mature and have reached a plateau in their ontogeny with respect to shape. Shape in leaflets from leaves 3 and 4, by contrast, is much more dynamic, reflecting that these leaflets are extremely early in their ontogeny in young plants and are approaching maturity in older plants. From leaves 3 and 4, we can infer that leaflet shape is becoming rounder as leaves age.
With respect to circularity and solidity (Figure 3B; Supplemental Figure 1B), which reflect lobing and serration, leaflets arising from leaves farther in the series are more serrated than their predecessors.
allPCs pick up on more specific shape attributes. allPC1 explains a “trifoliate” shape factor, which is produced by three prominent lobes (Figure 1E). Qualitatively, the trifoliate lobing is conspicuous in early terminal leaflets and subsides in leaflets found further in the heteroblastic series (Figure 1A), a phenomenon quantitatively captured by allPC1 (Figure 3C). Interestingly, the trend of increasing allPC1 values is reversed in young plants in the wild species data set but resembles the trend in domesticated tomato and the ILs in older plants. The persistence of this tendency in different species can be explored further in the raw data (Supplemental Data Set 1) but demonstrates that developmental changes in leaflet shape are varied in domesticated tomato and its wild relatives. allPC2, which explains the distribution of blade outgrowth along the proximal-distal axis of leaflets (Figure 1E), increases along the heteroblastic series (Figure 3D), again reflecting that leaflets are becoming more elongated in successive leaves.
Compared with all other genotypes, S. pennellii often exhibits (1) extreme shape values and (2) lower variance between leaves and across developmental ages, a trend that is qualitatively apparent to those who have worked with this species (Figure 1B).
Generally, changes in leaf shape with respect to lateral organ number are gradual and smooth. For some traits, however, there is an abrupt change in shape values. This is most conspicuous for circularity data in the ILs, in which starting at ∼11 lateral organs (indicated by the vertical line, Figure 3B) circularity values in all leaves abruptly become stagnant. For the IL data, this is true for most morphometric parameters, and the abrupt stagnation in shape is particularly apparent for leaf 4. It is also generally true for S. lycopersicum cv M82 data but less discontinuous than in the IL data. If the contribution of leaves and flowers to the lateral organ number is analyzed in more detail (Figures 3F and 3G), lateral organ number = 11 corresponds to the value that flowers first start making an appreciable contribution to the lateral organ count. The data suggest that the shape attributes in fully expanded leaves, across the heteroblastic series and at different stages in their ontogeny, are synced such that leaves sharply stabilize their shape at a time roughly corresponding to the production of floral primordia. Although a mechanistic cause for this phenomenon remains to be determined, one hypothesis is that diverting resources to reproductive growth may precipitously slow the growth of all leaves.
Resolving Distinct Genetic Attributes of Leaf Shape
We previously measured the genetic basis of leaf shape in the S. pennellii ILs in a field setting (Chitwood et al., 2013a). Such leaves arise after the transition to reproductive development and, unlike leaves within the heteroblastic series during vegetative development (Figure 3), are developmentally homogenous. Patterns of shape change during development are potentially revealing with respect to the genetic basis of shape. Genes may confer a uniform shape difference in all leaves, regardless of developmental context. Alternatively, genes may modulate the heteroblastic series itself, changing shape in a leaflet-specific manner. Such genetic regulation, reminiscent of heterochronic effects, would be detectable as differences in shape among leaflets in the heteroblastic series, rather than uniform changes in all leaflets.
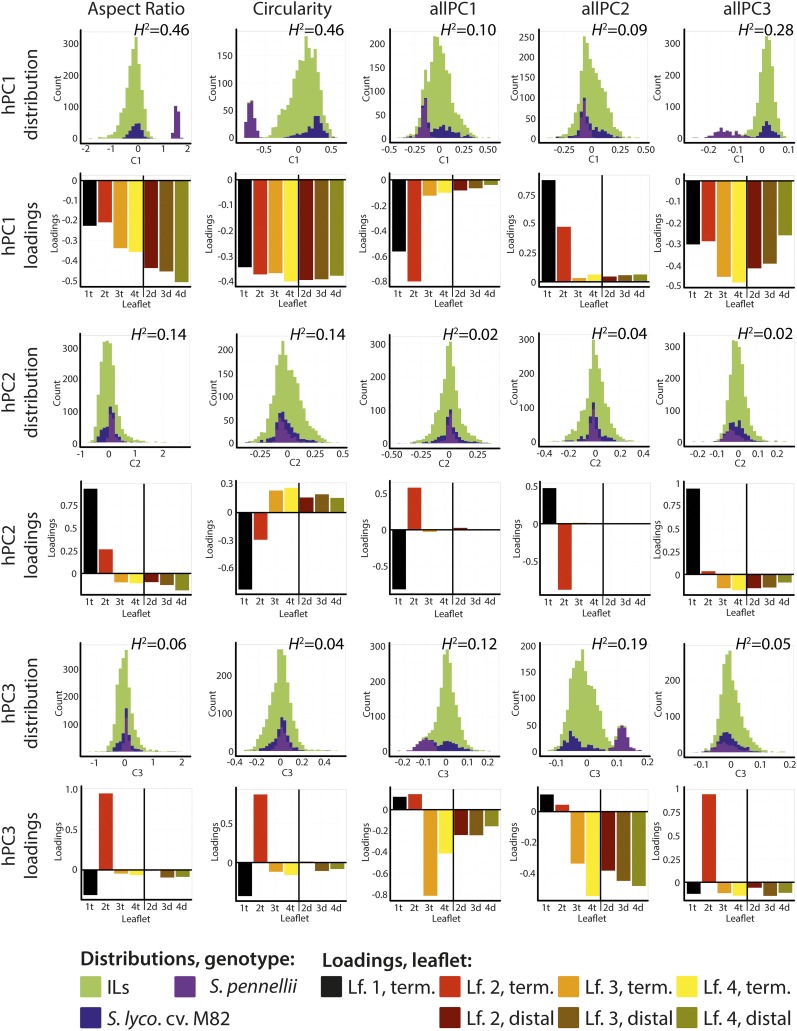
To quantify the genetic basis of patterns of leaflet shapes across vegetative development, we performed a PCA on terminal leaflet shape traits in leaves 1-4 and distal lateral leaflets in leaves 2-4 (Supplemental Data Set 2). Many more leaflets than these were collected (Figures 1A and 1B) but almost all samples had these leaflets, preventing biases from genotype-specific dropout. Principal components (PCs) resulting from the patterns of shape traits across the leaf series are named “hPCs” for “heteroblasty PCs” (Supplemental Table 1). When distinguishing “allPCs” from “hPCs” it is helpful to remember what each analysis measures. allPCs describe strictly shape attributes. allPC1 is a shape trait, as much as aspect ratio or circularity. hPCs strictly describe the patterns of shape traits across the heteroblastic series. It is in this way that allPC1, a specific shape attribute, is decomposed into multiple hPCs, each explaining a particular manner in which allPC1 varies across leaflets. Aspect ratio has its own hPCs as well. The pattern of shape variance that each hPC represents across leaflets for its corresponding shape trait can be found in the loadings for each hPC. Loadings are the weights associated with each variable contributing to a PC. The magnitude of the loading is proportional to its contribution to a PC, and its sign indicates its directional relationship to the PC.
Similar to our previous studies, hPCs are modeled using mixed-effect linear models to estimate genetic effects of introgressions and other random environmental effects (Chitwood et al., 2013a). As plant age clearly affects the pattern of leaflet shape across the series (Figure 3), lateral organ number is included as a random effect in these models to account for variance in hPCs attributable to differences in growth rate, a technique we have successfully implemented in the past (Chitwood et al., 2012d). By including a variable for growth rate in the models, hPCs more accurately reflect heteroblastic changes rather than ontogenetic effects.
hPC1 values for aspect ratio, circularity, roundness, and solidity (note: hPC1 values for each of these shape attributes are distinct from each other) are all highly heritable traits (H2 ≥ 0.43; Figure 4; Supplemental Figure 2). Note that loadings for all these hPC1 traits are consistently of the same sign and approximately the same magnitude across all leaflets measured. This indicates that these hPCs represent uniform changes, in the aspect ratio, circularity, roundness, and solidity, of all leaflets across the heteroblastic series. It is perhaps not surprising that the heritability of these hPCs is so high, as aspect ratio, circularity, roundness, and solidity all had H2 values > 0.6 under field conditions (Chitwood et al., 2013a). Under those conditions, there was no developmental context to measure, and here the primary contribution to the pattern of these traits across the leaf series is essentially uniform and represents magnitude. This suggests that in the ILs, during both vegetative and reproductive development, shape changes invariant with respect to development occur and are highly heritable. Based on the percent variance explained by the hPC1s for these traits (Supplemental Data Set 3), uniform changes in leaflet shape in all leaflets account for a majority (>50%) of the shape variance across the leaf series measured.
Figure 4.
Genetic Effects Modulating the Overall Shape of All Leaves versus the Shape of Specific Leaves in the Heteroblastic Series.
PCA on the distribution of shape attributes across the leaf series (terminal leaflets from leaves 1-4, distal lateral leaflets for leaves 2-4) results in “heteroblasty PCs” (hPCs). Left to right, analysis of hPCs for aspect ratio, circularity, allPC1, allPC2, and allPC3. Top to bottom, analysis of hPCs 1-3 for each trait are shown. Note: hPCs between shape attributes are not comparable and hPCs for each shape attribute are derived from separate PCAs. For each hPC for each shape attribute, the distribution of hPC values for S. lycopersicum (blue), S. pennellii (purple), and the ILs (green) is shown as a histogram and broad sense heritability (H2) values provided. Loadings for each hPC are also provided, showing the weighted contribution of shape attribute values for different leaflets to the hPC score. Black, leaf 1; red, leaf 2; orange, leaf 3; yellow, leaf 4; lighter shades of red, orange, and yellow represent terminal leaflets; darker shades of red, orange, and yellow represent distal lateral leaflets.
The hPC2 traits for aspect ratio, circularity, roundness, and solidity, explaining no more than 22% of the overall shape variance in the series, reflect differences in their respective shape attributes across the series rather than uniform changes in shape in all leaflets (Figure 4). The hPC2 loadings for these four traits behave in manner predictable from studies of leaf development. They show trends, increasing or decreasing, across successive terminal leaflets and with distal lateral leaflets exhibiting a distinct influence on shape. The heritabilities for these developmentally modulated hPC2s are low (between 13 and 15%). However, the heritabilities are not nonexistent, as they are for hPC3 traits for these shape attributes, the loadings of which do not behave predictably with respect to development (for example, high loading values for terminal leaflet 2 only). Note also that unlike hPC1 for these four shape attributes, the parental distributions for the less heritable hPCs largely overlap, demonstrating that there are softer differences for these hPCs between the parents, perhaps explaining the lower heritabilities for these traits.
The hPCs explaining variance in allPC values across the leaf series behave similarly to those for aspect ratio, circularity, roundness, and solidity, except that allPCs do not possess highly heritable hPCs explaining uniform shape differences in the leaf series for all leaflets (except allPC3 hPC1). They do possess hPCs explaining developmental patterns in shape across the leaf series with low heritabilities ranging from between ∼10 to 20% (e.g., allPC1 hPC1, allPC2 hPC1, and allPC2 hPC3).
The above data suggest that (1) most shape variance in the leaf series is modulated equally across all leaflets with high heritability (>40%) and (2) that there are separate, orthogonal patterns of shapes across the leaf series, modulated in a predictable developmental fashion, with lower heritability (∼10 to 15%).
Comparing Quantitative Trait Loci Regulating Distinct Shape Attributes
The developmental context of leaves provides a unique opportunity to view the genetic basis of a complex trait from multiple perspectives. The genetic regulation of leaf shape can be calculated in multiple ways. If the population of leaves is developmentally homogeneous and only varies in shape by genetics and environment, as in the IL field data (Chitwood et al., 2013a), the calculation of shape quantitative trait loci (QTL) is straightforward. If there are multiple leaves of different shapes because of developmental context (as with leaves collected from a heteroblastic series), a naïve approach is to calculate QTL for each leaflet type. Alternatively, if heteroblastic effects can be resolved from shape effects uniformly affecting all leaves, QTLs regulating overall leaf shape can be calculated separately from those regulating the temporal patterning of shape.
Shape QTL calculated for individual leaflets in the heteroblastic series (terminal leaflets 1-4 and distal lateral leaflets 2-4, seven different leaflets total) largely colocalize (Figure 5; Supplemental Figure 3). Less significant QTLs are detected for those leaflets with low calculated heritabilities, but prominent QTLs can be observed for most shape attributes across different leaflets. One exception is allPC1, in which the direction of QTLs between terminal and lateral leaflets reverses, which is expected considering that this allPC explains the “trifoliate” phenotype (Figure 1E) that varies more between different leaflet types than genotypes.
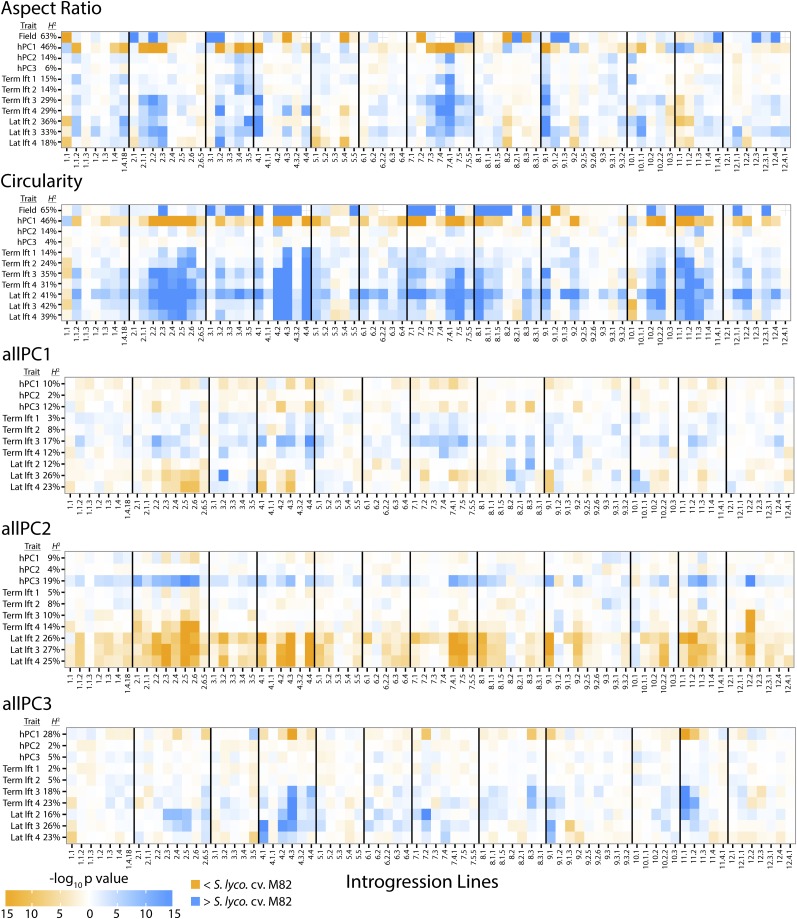
Figure 5.
Overlapping and Distinct QTLs Regulating Leaf Shape.
P values of QTLs regulating leaf shape in the S. pennellii ILs are shown by color. For each shape attribute, QTLs for leaf shape calculated from hPCs 1-3 (see Figure 4) and terminal leaflets from leaves 1-4 (“term. lft.”) and distal lateral leaflets 2-4 (“lat. lft.”) are provided. Because of the genetic structure of the IL population, QTLs are shown on a per IL basis (i.e., QTL in this context refers to the direction and significance of the IL trait value relative to the cv M82 parent). Broad-sense heritability (H2) for each trait is shown. For aspect ratio and circularity, shape QTLs calculated in Chitwood et al. (2013a) under field conditions are shown for comparison. Orange, P values for QTLs decreasing trait values relative to the S. lycopersicum cv M82 parent; blue, P values for QTLs increasing trait values relative to the domesticated parent.
Those shape attributes for which hPC1 explains uniform shape changes across leaflet types (Figure 4) have overlapping QTLs for hPC1 and QTLs calculated for each leaflet (Figure 5). For example, the QTLs for aspect ratio and circularity calculated for hPC1 and for individual leaflets largely overlap (even if the directionality of different traits, indicated with orange and blue in Figure 4, is not the same). As the uniform changes in shape across leaflet types explain most the variance in leaflet shape for these shape attributes, it makes sense that the calculation of QTLs for each individual leaflet would correspond to those for hPC1. hPCs explaining smaller amounts of shape variance attributable to the heteroblastic series have less significant QTLs overall, reflective of their decreased heritability. However, significant QTLs for these hPCs are mostly distinct from those for hPCs regulating uniform shape changes in leaflets. This is not only expected from the orthogonality of different PCs, but from the distinct genetic pathways regulating leaf morphology and developmental timing.
QTLs calculated from field conditions (Chitwood et al., 2013a) are largely distinct from other QTLs regulating leaf shape (Figure 5; Supplemental Figure 3). This is an important distinction: Both field QTLs and chamber hPC1 QTLs regulating overall leaf shape have (1) high heritability values and (2) describe shape variance invariant with respect to developmental context. It might be assumed that the same loci regulate overall leaf shape under field and chamber conditions, but this is not the case, and distinct QTLs, which are highly heritable, regulate leaf shape under different environmental conditions and developmental contexts.
Leaf Shape Difference between Chamber- and Field-Grown ILs
A homozygous genetic resource, such as the ILs, is a powerful way to measure and compare the genetic architecture of traits across different stages of development and environments. By measuring the same traits in the same genotypes, we can now compare the shape of vegetative leaves grown under chamber conditions to leaves arising after the reproductive transition grown in the field (Chitwood et al., 2013a).
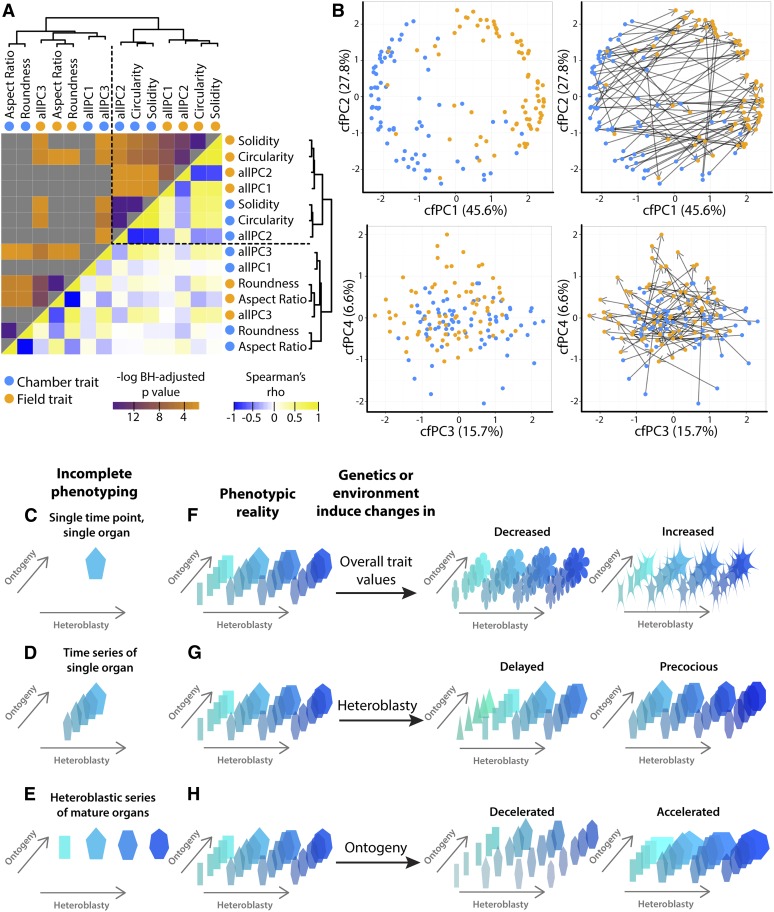
If field and chamber traits are hierarchically clustered, both trait and condition influence the resulting groups (Figure 6A). One cluster includes traits sensitive to serration and lobing (solidity and circularity) and allPC2, and the other cluster length-to-width ratio traits (aspect ratio and roundness) and allPC3. allPC1 straddles both of these groups. Within each group, traits then partition by condition.
Figure 6.
Comparison of Shape Attribute Profiles for ILs Grown in Chamber and Field Settings and a Model for the Developmental Context of Phenotype.
(A) Hierarchical clustering and correlation matrix for shape attribute profiles across the ILs, where each shape attribute is represented by a chamber (blue) and field (orange) profile. Hierarchical clustering is based on absolute Spearman’s rho values. Upper half: -log(BH-adjusted) P values for Spearman’s correlation between trait profiles (low to high significance, orange to purple; not significant, gray). Lower half: Spearman’s rho for correlation between traits (negative, blue; positive, yellow; neutral, white).
(B) Comparative PCA for field and chamber traits (cfPCs). The shape attribute profile for each IL is represented by two points in the cfPCA space: one point representing chamber shape (blue) and the other point field shape (orange). cfPCs 1-4 are shown. Left, scatterplot. Right, scatterplot with vectors for each IL, where vector base represents chamber trait shape profile and the vector tip field shape profile.
(C) to (H) Model for a heteroblastic and ontogenetic context for vegetative phenotypes. In this model, phenotype is represented as a matrix of leaf shapes. Incomplete phenotyping may measure (C) a single time point for a single organ, (D) a time series of a single organ, or (E) the heteroblastic series of only mature organs. All these scenarios are only part of a greater phenotypic reality to be measured, of the ontogeny (y axis) of the heteroblastic series (x axis) for a trait. Genetic changes induced by evolution or environment alter the ontogenetic/heteroblastic phenotype of a trait through (F) changes in shape for all organs over all time points, (G) shifts in the heteroblastic series, in which the precious or delayed appearance of shapes in organs is observed, or (H) ontogenetic changes in the speed by which an organ obtains its mature phenotype.
Another way of visualizing the relationship between chamber and field traits is using PCA. We previously used a PCA method to compare gene expression data measured in the same organs in two different species (Chitwood et al., 2013b). The approach can be used here to compare the overall trait profiles of each IL between two conditions. We call this particular PCA the “chamber-field” PCA, with “cfPCs” (Supplemental Table 1). In the cfPCA space, two points represent each IL: one representing the trait profile of the IL under chamber conditions and the other the trait profile of the IL under field conditions. Together, the two points form a vector, representing the overall change in the trait profile for the IL between the conditions.
Trait profiles of ILs under chamber and field conditions form distinct clusters in the cfPCA space (Figure 6B). Chamber → field IL vectors traverse mostly in the cfPC1 direction, which explains 45.6% of the variation. The direction of the vectors indicates that cfPC1 mostly explains differences between chamber and field IL leaves and that chamber/field differences in shape constitute most of the shape variance, whereas cfPC2 mostly explains genotypic differences between ILs that are persistent between the two conditions. Aspect ratio is the most prominent trait modulating cfPC1, suggesting that IL leaves are consistently rounder under field conditions after the reproductive transition (Supplemental Data Set 4). This contrasts with the increasing progression toward narrower leaves in the heteroblastic series (Figure 3) and suggests that the differences in leaf shape between chamber and field conditions are mostly environmental rather than developmental. By contrast, cfPC2 is more influenced by circularity, suggesting that this trait explains differences in ILs that are more persistent between conditions. These relationships are consistent with the comparison between distributions of aspect ratio and circularity values in chamber- and field-grown ILs (Figure 2A).
Our data demonstrate that chamber- and field-grown leaves differ in shape with respect to specific shape attributes that are most likely environmentally modulated. Highly heritable QTLs regulating leaf shape differ between chamber and field conditions, calling into question the relevancy of studies under controlled conditions as a proxy for those in a field setting.
DISCUSSION
Natural variation in leaf shape has been described as being polygenic, mostly additive, and highly heritable (Langlade et al., 2005; Tian et al., 2011; Chitwood et al., 2013a, 2014). In all these studies, leaves were measured in a manner so as to reduce or eliminate variance attributable to development and/or environmental context. It is convenient to reduce the problem of studying leaves to a homogeneous population, but to do so ignores the true diversity of leaf forms. Depending on genetic background, environment, and developmental stage, leaves from domesticated tomato and its wild relative occupy diverse regions of morphospace (Figure 2). Leaves within individuals vary, not only as a result of the heteroblastic progression, but because of allometric growth during their ontogeny (Figure 3). Moreover, when analyzed in the context of the heteroblastic series, distinct attributes of leaf shape, either regulating the shape of all leaves uniformly or specific leaves, can be separated genetically (Figure 4). Finally, the shapes of chamber-grown leaves vary from those of field-grown leaves and have a distinct genetic basis (Figures 5 and 6).
As previously indicated, Goethe’s prescient insight into the serial homology of lateral organs in plants was neither evolutionary nor developmental (Friedman and Diggle, 2011). Rather, in metaphorically declaring the leaf “Proteus,” Goethe was demonstrating that the underlying, archetypal leaf is mutable, rather than singular and unchanging (Goebel, 1900; Ashby, 1948). Even earlier than Goethe, Hales (1727) studied the dynamic, ever-changing morphology of a developing leaf. And yet, when describing the quantitative genetic basis of morphological traits, rarely is developmental context provided (Figure 6C), and often purposefully eliminated. Time series data begin to resolve ontogenetic information of single organs (Figure 6D), but the iterative production of organs, which dominates plant development compared with animal, often remains unaccounted for, especially in a quantitative fashion (Figure 6E).
An accurate quantitative description of vegetative phenotype would at the very least account for (1) an ontogenetic time course of morphological development and (2) the continuum of successive organ types produced during the lifetime of a plant (Figures 6F to 6H). All vegetative organs in plants have an ontogenetic history and heteroblastic context. In this framework, rather than a single value, a trait is a matrix. Environmental or genetic effects further modulate the heteroblastic-ontogenetic matrix of morphology. Indeed, one of the principle ways environmental plasticity manifests in plants is through modulating the heteroblastic progression and the ontogeny of lateral organs (Allsopp, 1954; Jones, 1995; Diggle, 2002). It remains to be seen the degree to which similar heterochronic-like mechanisms are used during evolution to achieve morphological diversity in plants. Such a multivariate model of phenotypic reality extends beyond morphology and should be applied to the quantitative genetic study of gene expression, protein levels, and metabolite accumulation as well (Chitwood and Sinha, 2013). As phenotyping continues to expand the types of traits measured, an equally expansive realm to explore is the developmental and environmental context of each.
METHODS
Plant Materials, Growth Conditions, and Experimental Design
Data from previous studies on leaf shape in wild tomato species (Chitwood et al., 2012a) and field-grown Solanum pennellii introgression lines (Chitwood et al., 2013a) are included in this study. Growth conditions and details about data collected from those experiments can be found in the respective publications. Briefly, a synopsis of the overall aims and design of those experiments is included here. A total of 11,033 leaflets were measured by Chitwood et al. (2012a). In this study, eight accessions each of Solanum arcanum, Solanum habrochaites, and Solanum pimpinellifolium were grown in chambers under simulated sun and foliar shade (i.e., supplemental far-red lighting). By genotype, each accession was sampled up to 20 times. From each individual, leaflets from the first four leaves were dissected and photographed. A total of 11,268 leaflets were measured by Chitwood et al. (2013a). In this study, the 76 S. pennellii ILs were grown in the field in Davis, CA in 2010. Each IL was represented 10 times in a randomized block design. Five leaves were sampled from each plant, from each of which the terminal and left and right distal lateral leaflets were sampled, resulting in a pseudoreplication of 15. Images of leaflets photographed in Chitwood et al. (2012a) and (2013a) are available in the database section of chitwoodlab.org.
For this study, 33,838 new leaflets were measured. The 76 S. pennellii ILs were originally obtained from the Tomato Genetics Resource Center (Davis, CA) and as a kind gift from Dani Zamir (The Hebrew University, Rehovot, Israel). Seed was washed in 50% household bleach for ∼2 min, rinsed, and placed onto water-soaked paper towels in Phytatrays (Sigma-Aldrich). Seeds were placed into darkness for 3 d at room temperature before moving to a walk-in Conviron chamber at 22°C, 16:8 light-dark cycle under simulated sun condition (explained below) for 4 d. Eight days after planting, seedlings were then transplanted into eight pots in 11 × 22-inch trays, one plant per pot. Plants then resumed their growth in one of two different Conviron chambers under either simulated sun or foliar shade conditions. Lighting consisted of alternating fluorescent (F48T12CWHO) and far-red (F48T12FRHO, peak emission 750 nm; Interlectric) bulbs. High red-to-far-red ratios of light were created under the simulated sun condition by blocking far-red bulbs with sleeves. To balance overall PAR, shade cloth was perpendicularly placed over bulbs in the simulated foliar shade treatment to match irradiance under the simulated sun treatment. Plants were grown in 12 staggered replicates, in each of which all ILs and parents were represented twice, once each for simulated sun and foliar shade conditions. Thus, each IL is potentially sampled up to 24 times. Each replicate of a lighting condition occupied two growth chamber shelves. Halfway through the experiment, simulated sun and simulated foliar shade shelves were swapped. Detailed information about replicate, chamber, light treatment, shelf, tray, and tray position is provided for each plant (Supplemental Data Set 2).
At 36 d after planting, plants were harvested for phenotyping. Plants were first visually inspected to count the number of lateral organs. Leaf and flower counts included small primordia visible to the naked eye (down to 1 to 3 mm in length). Leaflets from the first four leaves were then dissected and flattened under nonreflective glass, arranged around the rachis and small intercalary leaflets that were photographed as well. Photos were taken using a copy stand (Adorama 36-inch Deluxe Copy Stand) and an Olympus SP-500 UZ camera operated remotely from a laptop computer using Cam2Com software (Sabsik). Rulers were included in photos for scale. More than 2700 photos of raw data from which leaflet outlines are derived are available for download at chitwoodlab.org under the database section.
Image Processing and Morphometric Analysis
Photos were thresholded and converted into binary images in ImageJ (Abramoff et al., 2004), after which, using custom macros, individual leaflets were selected and named appropriately as individual files. Leaflet outlines were batch processed in ImageJ to obtain aspect ratio, circularity, roundness, and solidity values. As in previous studies, the program SHAPE was used to analyze outlines using EFDs (Iwata and Ukai, 2002). Following the method from Kuhl and Giardina (1982), SHAPE extracts contours as chain-code, which is used to calculate EFDs on leaflets oriented along the proximal-distal axis. PCA was performed on EFDs resulting from the first 20 harmonics. Symmetrical shape variance principal components were obtained by analyzing a and d coefficients, whereas asymmetric variance was analyzed using b and c coefficients (Iwata et al., 1998).
Data Visualization and Analysis
The ggplot2 package (Wickham, 2009) in R (R Core Team, 2014) was used for visualization. Functions used include: geom_point for scatterplots, stat_density2d for contour plots, stat_smooth (method=loess) for loess model generation, geom_histogram for histograms, geom_bar for bar graphs, geom_tile for heat maps, and geom_segment to create vectors connecting paired points.
The genetic basis of leaf shape within a heteroblastic context was first analyzed by performing a PCA on shape attribute values across the first four terminal leaflets and averages of the left and right distal lateral leaflets of leaves 2-4 without scaling using the prcomp function in R. The resulting PCs (“hPCs”) were then modeled using mixed effect linear models with the lmer function in the lme4 package (Bates et al., 2014). Genotype was modeled as a fixed effect, whereas spatial, replicate, and measurer factors were modeled as random effects. As we have done previously, lateral organ number was modeled as a random effect to control for its effects on the progression of shape attributes in the heteroblastic series (Chitwood et al., 2012d). Models were selected using a backward selection method, determining the significance of each term to explain variance in the model by comparing to a model without the term. After comparing all terms, the least significant term (if any, with a P value threshold of 0.05) was removed and the cycle repeated.
Hierarchical clustering was performed on shape attribute profiles (under chamber and field settings) across ILs using the absolute value of Spearman’s correlation with the Ward method using the hclust function in R. Correlation matrices with rho and P values from Spearman’s correlation were calculated using the rcorr function from the Hmisc package. In the cfPCA, each IL is represented by two points: one representing its shape attribute profile under chamber settings and the other point the shape attribute profile under field settings. Before performing the PCA, each shape attribute was scaled across ILs followed by scaling of shape attributes within ILs.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Heteroblasty and Ontogeny Modulate Leaf Shape.
Supplemental Figure 2. Genetic Effects Modulating the Overall Shape of All Leaves versus the Shape of Specific Leaves in the Heteroblastic Series.
Supplemental Figure 3. Overlapping and Distinct Quantitative Trait Loci Regulating Leaf Shape.
Supplemental Table 1. Principal Component Analyses Used in This Study.
The following materials have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.4r267.
Supplemental Data Set 1. Morphometric Traits for >55,000 Leaflets Analyzed in This Study.
Supplemental Data Set 2. Heteroblasty and Experimental Design Information for S. lycopersicum, S. pennellii, and IL Leaflets Measured in the Chamber Experiment.
Supplemental Data Set 3. hPC Variance Values.
Supplemental Data Set 4. Loading Values for cfPCA.
Supplementary Material
Acknowledgments
This work was supported through a Gordon and Betty Moore Foundation Life Sciences Research Fellowship to D.H.C., a postdoctoral fellowship from the Spanish Ministry of Science to J.A.A.-M. and a National Science Foundation grant (IOS-0820854) to N.R.S., J.N.M., and J.P.
AUTHOR CONTRIBUTIONS
D.H.C., J.P., J.N.M., and N.R.S. conceived of the experiment. D.H.C., L.R.H., and S.B. designed the research. D.H.C., A.R., R.K., Y.I., K.Z., L.R.H., E.O.-G., J.A.A.-M., S.B., L.C., D.F., and C.C.M. performed research. D.H.C. analyzed data and wrote the article.
Glossary
- IL
introgression line
- PCA
principal component analysis
- EFD
elliptical Fourier descriptor
- QTL
quantitative trait loci
- PC
principal component
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abramoff, M.D., Magalhaes, P.J., Ram, S.J. (2004). Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Allsopp, A. (1954). Juvenile stages of plants and the nutritional status of the shoot apex. Nature 173: 1032–1035 [Google Scholar]
- Ashby, E. (1948). Studies in the morphogenesis of leaves. I. An essay on leaf shape. New Phytol. 47: 152–176 [Google Scholar]
- Bates, D., Maechler, M., Bolker, B., and Walker, S. (2014). lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-5. http://CRAN.R-project.org/package=lme4
- Chitwood, D.H., Headland, L.R., Kumar, R., Peng, J., Maloof, J.N., Sinha, N.R. (2012a). The developmental trajectory of leaflet morphology in wild tomato species. Plant Physiol. 158: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, D.H., Naylor, D.T., Thammapichai, P., Weeger, A.C., Headland, L.R., Sinha, N.R. (2012b). Conflict between intrinsic leaf asymmetry and phyllotaxis in the resupinate leaves of Alstroemeria psittacina. Front. Plant Sci. 3: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, D.H., Headland, L.R., Ranjan, A., Martinez, C.C., Braybrook, S.A., Koenig, D.P., Kuhlemeier, C., Smith, R.S., Sinha, N.R. (2012c). Leaf asymmetry as a developmental constraint imposed by auxin-dependent phyllotactic patterning. Plant Cell 24: 2318–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, D.H., Headland, L.R., Filiault, D.L., Kumar, R., Jiménez-Gómez, J.M., Schrager, A.V., Park, D.S., Peng, J., Sinha, N.R., Maloof, J.N. (2012d). Native environment modulates leaf size and response to simulated foliar shade across wild tomato species. PLoS ONE 7: e29570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, D.H., Kumar, R., Headland, L.R., Ranjan, A., Covington, M.F., Ichihashi, Y., Fulop, D., Jiménez-Gómez, J.M., Peng, J., Maloof, J.N., Sinha, N.R. (2013a). A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell 25: 2465–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, D.H., Maloof, J.N., Sinha, N.R. (2013b). Dynamic transcriptomic profiles between tomato and a wild relative reflect distinct developmental architectures. Plant Physiol. 162: 537–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, D.H., Sinha, N.R. (2013). A census of cells in time: quantitative genetics meets developmental biology. Curr. Opin. Plant Biol. 16: 92–99 [DOI] [PubMed] [Google Scholar]
- Chitwood, D.H., et al. (2014). A modern ampelography: a genetic basis for leaf shape and venation patterning in grape. Plant Physiol. 164: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle, P.K. (2002). A developmental morphologist’s perspective on plasticity. Evol. Ecol. 16: 267–283 [Google Scholar]
- Eshed, Y., Zamir, D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, W.E., Diggle, P.K. (2011). Charles Darwin and the origins of plant evolutionary developmental biology. Plant Cell 23: 1194–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish, T.J. (1988). Adaptation to sun and shade: a whole-plant perspective. Aust. J. Plant Physiol. 15: 63–92 [Google Scholar]
- Goebel, K. (1900). Organography of Plants. I. General Organography. (New York: Hafner Publishing Co.). [Google Scholar]
- Goethe, J.W. (1817). Italienishe Reise. Goethe’s Werk. (Stuttgart, Germany: Dreizehnter Band). [Google Scholar]
- Goethe, J.W. (1952). Botanical Writings. Trans. B Mueller, introduction C.J. Engard. (Honolulu, HI: University of Hawaii Press). [Google Scholar]
- Hales, S. (1727). Vegetable Staticks or, an Account of Some Statistical Experiments on the Sap in Vegetables. (London: W. and J. Innys). [Google Scholar]
- Iwata, H., Niikura, S., Matsuura, S., Takano, Y., Ukai, Y. (1998). Evaluation of variation of root shape of Japanese radish (Raphanus sativus L.) based on image analysis using Elliptic Fourier Descriptors. Euphytica 102: 143–149 [Google Scholar]
- Iwata, H., Ukai, Y. (2002). SHAPE: a computer program package for quantitative evaluation of biological shapes based on elliptic Fourier descriptors. J. Hered. 93: 384–385 [DOI] [PubMed] [Google Scholar]
- Jones, C.S. (1992). Comparative ontogeny of a wild cucurbit and its derived cultivar. Evolution 46: 1827–1847 [DOI] [PubMed] [Google Scholar]
- Jones, C.S. (1995). Does shade prolong juvenile development? A morphological analysis of leaf shape changes in Cucurbita argyrosperma Subsp. Sororia (Cucurbitaceae). Am. J. Bot. 82: 346–359 [Google Scholar]
- Kerstetter, R.A., Poethig, R.S. (1998). The specification of leaf identity during shoot development. Annu. Rev. Cell Dev. Biol. 14: 373–398 [DOI] [PubMed] [Google Scholar]
- Kuhl, F.P., Giardina, C.R. (1982). Elliptic Fourier features of a closed contour. Comput. Graph. Image Process. 18: 236–258 [Google Scholar]
- Langlade, N.B., Feng, X., Dransfield, T., Copsey, L., Hanna, A.I., Thébaud, C., Bangham, A., Hudson, A., Coen, E. (2005). Evolution through genetically controlled allometry space. Proc. Natl. Acad. Sci. USA 102: 10221–10226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra, A.B., Leigh, A., Boyce, C.K., Jones, C.S., Niklas, K.J., Royer, D.L., Tsukaya, H. (2011). The evolution and functional significance of leaf shape in the angiosperms. Funct. Plant Biol. 38: 535–552 [DOI] [PubMed] [Google Scholar]
- Poethig, R.S. (1990). Phase change and the regulation of shoot morphogenesis in plants. Science 250: 923–930 [DOI] [PubMed] [Google Scholar]
- Poethig, R.S. (2010). The past, present, and future of vegetative phase change. Plant Physiol. 154: 541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2014). R: A language and environment for statistical computing. (Vienna, Austria: R Foundation for Statistical Computing). [Google Scholar]
- Smith, H., Whitelam, G.C. (1997). The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant Cell Environ. 20: 840–844 [Google Scholar]
- Tian, F., Bradbury, P.J., Brown, P.J., Hung, H., Sun, Q., Flint-Garcia, S., Rocheford, T.R., McMullen, M.D., Holland, J.B., Buckler, E.S. (2011). Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis. (New York: Springer). [Google Scholar]
- Yano, S., Terashima, I. (2001). Separate localization of light signal perception for sun or shade type chloroplast and palisade tissue differentiation in Chenopodium album. Plant Cell Physiol. 42: 1303–1310 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.