Abstract
Given current evidence supporting a genetic predisposition for pelvic organ prolapse (POP), we conducted a systematic review of published literature on the genetic epidemiology of POP. Inclusion criteria were linkage studies, candidate gene association and genome-wide association studies (GWAS) in adult women published in English and indexed in PubMed through December 2012, with no limit on date of publication. Methodology adhered to the PRISMA guidelines. Data were systematically extracted by two reviewers and graded by the Venice criteria for studies of genetic associations. A meta-analysis was performed on all single nucleotide polymorphisms (SNPs) evaluated by two or more studies with similar methodology. The meta-analysis suggests that collagen type 3 alpha 1 (COL3A1) rs1800255 genotype AA is associated with POP, OR 4.79 (95% CI 1.91 to 11.98, p= 0.001) compared to the reference genotype GG in populations of Asian and Dutch women. There was little evidence of heterogeneity for rs1800255 (p-value for heterogeneity= 0.94; proportion of variance due to heterogeneity, I2= 0.00%). There was insufficient evidence to determine whether other SNPs evaluated by two or more papers were associated with POP. An association with POP was seen in individual studies for estrogen receptor alpha (ER-α) rs2228480 GA, COL3A1 exon 31, chromosome 9q21 (HLOD score 3.41) as well as six SNPs identified by a GWAS. Overall, individual studies were of small sample size and often of poor quality. Future studies would benefit from more rigorous study design as outlined in the Venice recommendations.
Keywords: genetic epidemiology, genome wide association study, pelvic organ prolapse, single nucleotide polymorphism
Introduction
Pelvic organ prolapse affects 40% of postmenopausal women and directly impacts bladder and bowel function, as well as quality of life(1,2). Surgical correction of pelvic organ prolapse (POP) is anticipated to increase 48% from 2010 to 2050 given the aging population in the United States(3). The pathophysiology of this prevalent disorder is believed to be multifactorial, involving vaginal parity and other obstetric risk factors(4,5), as well as advanced age, increased body-mass index, smoking, constipation and vaginal hysterectomy(6–8). Yet, even with multiple risk factors, there is a large component of risk that is not understood. This is exemplified by the fact that nulliparous women can develop prolapse, and conversely, most parous women do not develop prolapse(1). It is plausible that genetics contribute significantly to the development of prolapse. Studies show a five-fold increased risk of prolapse among siblings of women with severe prolapse as compared to the general population(9) and a high concordance of prolapse in twins(10) as well as, in nulliparous and parous sister pairs(11).
The interrelationship of epidemiologic, environmental and genetic risk factors for POP constitutes the genetic epidemiology of prolapse. With improved understanding of these relationships, there may be a role for individual risk assessment in future. Perhaps, women at high risk for prolapse may choose to prophylactically perform pelvic muscle strengthening exercises; or, following the development of prolapse, potentially opt for a primary sacrocolpopexy with mesh instead of pelvic reconstruction with native tissue. Currently, both our understanding of the genetic epidemiology of POP as well as our knowledge about the efficacy and longevity of treatment options is too limited to make definitive recommendations; but, as our knowledge advances, this information may be incorporated into patient counseling and treatment decisions. This type of personalized medicine is becoming a reality in other fields, such as cardiology and oncology, in which genetic risk stratification is more advanced(12–14). Given the preliminary data supporting a genetic component to the etiology of prolapse, this study aimed to systematically review and highlight current research in this area. We focused on genome-wide association studies (GWAS), linkage and candidate gene association studies in adult women with pelvic organ prolapse.
Methods for Review
We initially conducted a broad search on the genetics and genetic epidemiology of POP and urinary incontinence. For the analysis presented here, only those papers pertaining to the genetic epidemiology of pelvic organ prolapse were included. Methodology adhered to the PRISMA Statement guidelines(15). The search was limited to publications in English with an adult female population that were indexed in PubMed through December 2012. There were no limitations on date of publication. Controlled vocabulary terms served as the foundation of our search with one clinical term (pelvic organ prolapse, cystocele, rectocele, urinary incontinence, urge incontinence, stress incontinence, mixed incontinence, pelvic floor, uterus/uterine/vaginal/vault, urogenital/bladder/pelvic organ/genitourinary and prolapse, vaginal and defect, or enterocele) and one genetic term (genetic phenomena, genetics, genetic models, genetic techniques, polymorphism, genome, phenotype, genotype, gene, genes, variant, exome, exon, gene expression, microarray, sequencing, protein biosynthesis, protein, protein, proteomic, hereditary, familial or inherited). We excluded all newspaper articles, letters, comments, case reports, reviews, practice guidelines, news, historical articles, meta-analyses, legal cases, published erratum and congresses. References from key articles were hand-searched to identify additional studies.
We defined POP as anatomic prolapse of the vaginal walls and/or uterus and defined genetic epidemiology to include linkage studies, candidate gene association studies and GWAS. For GWAS and candidate gene studies, studies needed to include a comparator of women without prolapse. Outcomes of interest were single nucleotide polymorphisms (SNPs) associated with pelvic organ prolapse.
We utilized four reviewers: two MD clinicians and two PhD genetic epidemiologists. All reviewers evaluated the first 50 abstracts in order to ensure consistency. The remaining abstracts underwent dual review to determine inclusion or exclusion, followed by dual full-text review of all articles selected for inclusion. Discordance was resolved by third-party adjudication. Data from included articles was extracted using a standardized form and a second team member ensured the extraction was accurate, complete and consistent. Given the translational nature of the project, dual review and data extraction involved a clinician and a genetic epidemiologist at each step in the process. Individual reviewers recused themselves from the evaluation and data extraction of any study they were involved in or had co-authorship. This study did not involve human subjects and was exempt from Institutional Board Review.
We graded the quality of the GWAS and candidate gene studies using the Venice guidelines(16). These guidelines grade the cumulative evidence in support of a genetic association based on three criteria: (1) the amount of evidence, (2) whether replication was performed and (3) protection from bias. Each category can receive a grade of “A”, “B” or “C”. Studies graded AAA have the strongest evidence, “A” and “B” studies indicate moderate evidence and any study with a category “C” represents only weak evidence.
A meta-analysis was performed of all SNPs evaluated by two or more studies with similar methodology. We used odds ratios (ORs) as the effect measure of choice to report the weighted associations between SNPs of interest and pelvic organ prolapse. If any study in a meta-analysis set reported only crude numbers and a chi-square test, then crude odds ratios were calculated and reported for all studies in the set, when possible, to ensure comparability. Similarly, when studies within a meta-analysis set presented different types of ORs (dominant model vs. additive model), ORs were recalculated to accommodate the measure that was common to all studies or calculable in a set. If a study reported only adjusted ORs without reporting crude numbers to recalculate ORs, adjusted ORs were used. The ORs for the meta-analysis were estimated using inverse variance weighted fixed effect models. All analyses were performed with STATA/SE 12.0, StataCorp LP, College Station, Texas.
Results
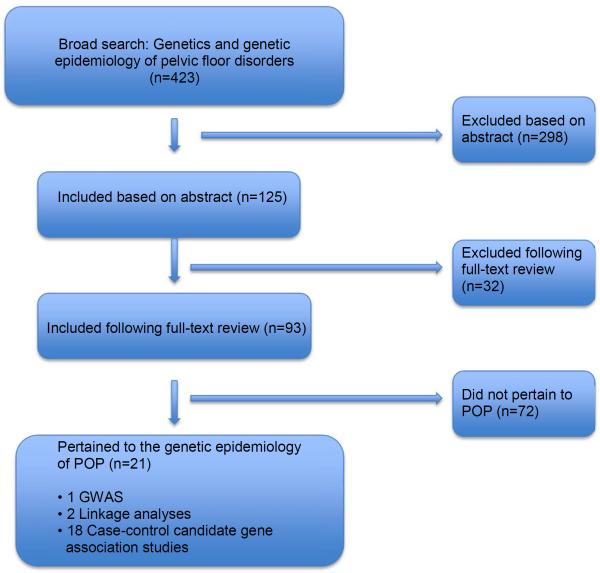
For the overarching topic of genetics and the genetic epidemiology of pelvic floor disorders, our literature search identified 423 non-duplicate articles, of which 125 met inclusion criteria based on the abstract, and 93 met inclusion after full-text review. Of these articles, 21 pertained to the genetic epidemiology of POP (Figure 1). This included one GWAS(17), two linkage analyses(18,19) and 18 case-control candidate gene association studies(20–37) involving 10 candidate genes (collagen type 1 alpha 1 (COL1A1) (n=5)(26–28,33,35), collagen type 3 alpha 1 (COL3A1) (n=4)(20,30–32), laminin gamma-1 (LAMC1) (n=3) (19,23,36), matrix metalloproteinase 9 (MMP9) (n=3)(24,28,37), matrix metalloproteinases 1 and 3 (MMP1 & 3) (n=2)(28,34), lysyl oxidase-like 1 (LOXL1) (n=1)(29), estrogen receptor alpha (ERα)(n=1)(21), estrogen receptor beta (ERβ)(n=1)(22), progesterone receptor (PGR) (n=1)(25) (Table 1). All studies were published in 2007 or later. Most had government(17,18,20,21,23,25,29,32–37) and/or university grant funding(20–22,24,25), 71% (15/21).
Figure 1.
Flowchart of reviewed and included studies
Flowchart of studies that were reviewed and included in this manuscript.
Table 1.
Studies included in systematic review
| Author, Year |
Race and Ethnicity (Study country of origin) |
Cases (with POP) |
Controls (no POP) |
Phenotype – Cases |
Phenotype - Controls |
Gene | SNPs Evaluated* |
Quality (Venice Guidelin es**) |
|---|---|---|---|---|---|---|---|---|
| GWAS | ||||||||
| Allen-Brady, 2011(17) | Cases: White (USA) Controls: white | 115 | Illumina iControl DB 2,976 | Treated for POP with a family history of prolapse or other pelvic floor disorders | Pelvic floor information not known. Excluded duplicate and closely related samples. | rs1455311, 4q21.21; rs1036819, 8q24.22; rs430794, 9q22.2; rs8027714, 15q11.2; rs1810636, 20p13; rs2236479, 21q22.3 | CBC (CCC for SNPs rs43079 4, rs18106 36 due to lack of replicati on) | |
| Linkage Analysis | ||||||||
| Allen-Brady, 2009(18) | European descent (USA) | 70 cases (Familial study: 32 families) | N/A | Treated for moderate-severe POP, usually POP-Q stage III-IV (41/66) | N/A | LOD score 3.41; Chr9: 80.35Mb–88.81Mb with HLOD>=1.86 | N/A | |
| Nikolov a, 2006(19) | NR (USA) | 6 | Prolapse evaluated by POP-Q | LAMC1 sequence variant 1q31 | rs10911193 | N/A | ||
| Case-Control, Candidate Gene Analysis | ||||||||
| Chen C, 2010(23) | Caucasia n and African America n; results reported by race (USA) | 165 (102 Caucasian, 63 AA) | 246 (163 Caucasian, 83 AA) | POP-Q stage III, IV | POP-Q stage 0-I; Cases and controls were matched on age, race, menopausal status, smoking history, BMI & parity. | LAMC1 | rs10911193; rs20563 | CCC |
| Chen H, 2008(22) | NR (Taiwan) | 69 | 141 | POP-Q stage II-IV | POP-Q stage 0-I | ER6 | rs2987983 (−13950 T/C) Promoter; rs1271572 (−12214 G/T) Promoter; rs9444599 (−1213 T/C) Promoter; rs1256049 (25652 A/G) Exon 6; rs1255998 (110943 G/C) 3'-UTR | CCB |
| Chen H, 2008(21) | NR (Taiwan) | 88 | 153 | POP-Q stage II-IV | POP-Q stage 0-I | ERα | rs17847075 (exon 1 C/T); rs2207647 (exon 1 G/A); rs2234693 (intron 1 T/C); rs3798577 (exon 8 C/T); rs2228480 (exon 8 G/A) | CCB |
| Chen H, 2008(20) | NR (Taiwan) | 84 | 147 | POP-Q stage II-IV | POP-Q stage 0-I | COL3A1 | rs1800255 (Exon 30 G>A); rs1801184 (exon 32 T>C); | CCB |
| Chen H, 2009(25) | NR (Taiwan) | 87 | 150 | POP-Q stage II-IV | POP-Q stage 0-I | PGR | rs500760 (exon 8 A/G); rs484389 (3' untranslated region C/T) | CCB |
| Chen H, 2010(24) | NR (Taiwan) | 92 | 152 | POP-Q stage II-IV | POP-Q stage 0-I | MMP-9 | rs3918242; rs17576; rs2250889 | CCB |
| Cho, 2009(26) | Korean (Korea) | 15 | 15 | POP-Q stage III-IV (women undergoing hysterectomy) | POP-Q stage 0 (hysterectomy for uterine myoma) | COLIA1 Sp1 binding site | No polymorphism seen at Sp-1 bindingsite in COLIA1 (all G/G, cases and controls) | CCC |
| Feiner, 2009(27) | Caucasian or Ashkenazi-Jewish (Israel) | 36 | 36 | POP-Q stage III-IV POP | POP-Q stage 0-I | COLIA1 Sp1 binding site | SP-1 binding site (no rs#) | CCC |
| Ferrari, 2012(28) | NR (Italy) | 137 | 96 | POP-Q stage II-IV | POP-Q stage 0-I | COL1A1, MMP1, 3,9 | SP1 site of COL1A1 point mutation (G-T) in 1st intron; neg 1562/T of MMP9; neg 1171 5A/6A of MMP3; neg 1607 1G/2G of MMP1 | CCB |
| Ferrell, 2009(29) | African American and Caucasian (USA) | 137 | 141 | POP-Q stage II-IV | POP-Q stage 0-I, matched to cases on age, race, menopausal status, smoking history, BMI and parity | LOXL1 | No rs # (labeled-659 in promoter) | CCC |
| Jeon, 2009(30) | Korean (South Korea) | 36 | 36 | POP-Q stage II-IV, Postmenopa usal and parous | POP-Q stage 0-I, no stress urinary incontinence, postmenopa usal and parous | COL3A1 | 5'-AAGTATACAA ATTTCTAGATT G-3' (forward)/5'-ATAAATGATCA GAAGGAAATC A-3' (reverse) | CCC |
| Kluivers, 2009(31) | European, Dutch (Netherlands) | 202 | 102 | POP present (not defined) | Vaginally parous, descent < 1cm above hymenal remnants, no prior POP surgery | COL3A1 | rs1800255 | CCC |
| Martins, 2011(32) | White and Non white Brazilians (Brazil) | 107 | 209 | POP-Q stage III-IV, postmenopa usal, no HRT | POP-Q stage 0-I, no documented vaginal surgery or stress incontinence, postmenopa usal,no HRT | COL3A1 | No rs# (Labeled Exon 31 G Allele) | CCC |
| Rodrigues, 2008(35) | White and Nonwhite (Brazil) | 107 | 209 | POP-Q stage III-IV | POP-Q stage 0-I | COL1A1 Sp1 binding site | COL1A1 Sp-1 binding site polymorphism (no rs#) | CCC |
| Skorupski, 2007(33) | NR (Poland) | 37 | 40 | POP-Q stage III-IV | POP-Q stage 0-I | COL1A1 | position 1240 in 1st intron; G -> T substitution; transcription factor Sp1 binding site of COL1A1 | CCB |
| Skorupski, 2012(34) | NR (Poland) | 133 | 132 | POP-Q “grade” II-IV, undergoing surgery | POP-Q “grade” 0-I, dysfunctional uterine bleeding or undergoing TAH/SCH | MMP1,3 | MMP1 polymorphism (position −1607/−1608); MMP3 polymorphism (position −1612/−1617) | CCB |
| Wu, 2012(36) | White, non-hispanic (USA) | 239 | 197 | POP-Q stage III-IV, not pregnant; no age cutoff but preferentially recruited younger women | POP-Q stage 0-I, no history of POP surgery, preferentially recruited older women | LAMC1 | rs10911193; rs1413390; rs20558; rs20563; rs10911206; rs2296291; rs12041030; rs12739316; rs3768617; rs2483675; rs10911211; rs41475048; rs1058177; rs12073936 | CCA |
| Wu, 2012(37) | White, non-hispanic (USA) | 239 | 197 | POP-Q stage III-IV, no age cutoff but preferentially recruited younger women | POP-Q stage 0-I, no history of POP surgery, preferentially recruited older women | MMP9 | rs3918253; rs3918256; rs3918278; rs17576; rs2274755; rs17577; rs2236416; rs3787268 | CCA |
NR = Not reported
N/A = Not applicable
Only SNPs with significant findings were reported for GWAS and linkage analyses. All evaluated SNPs are reported for candidate gene studies.
Venice guidelines grade (1) the amount of evidence, (2) whether replication was performed and (3) protection from bias.
The prolapse phenotype was most commonly defined by POP-Q stages II-IV(20–25,28,29,33,37), although some studies were more stringent(17,18,26–28,30,32–36), and two studies did not define the prolapse phenotype(19,31). All of the case-control studies defined the control as POP-Q stage 0 or 0-I. Many excluded women with connective tissue diseases(17,23,27,29,31,34,36,37). The GWAS(17) and both linkage analyses(18,19) were performed in families with a high rate of POP. All other studies were population based(20–25,28,30,31,36,37). Studies looked at Asian (33.3%, 7/21) (20–22,24–26,30), European (23.8%, 5/21) (27,28,31,33,34) and U.S. Caucasian populations (23.8%, 5/21(17–19,36,37); two studies included sub-analyses of African Americans(23,29) and Brazilian white and non-white (9.5%, 2/21)(32,35) populations (Table 1). When reported, the mean or median age of prolapse cases ranged from 48 to 66 years(17,18,21–24,27–37) and mean or median age of controls ranged from 49 to 69 years (17,21–24,27–37). Age was similar between cases and controls for nine studies(17,26–31,33), had a discrepancy of ≥ 5 years for six studies (23,32,34–37), a discrepancy of ≥ 10 years for three studies(21,22,24) and markedly disparate proportions of younger and older women in two studies (study did not report mean or median ages)(20,25). Two studies preferentially recruited controls from an older population(36,37); all other studies with an age discrepancy had controls that were younger than the prolapse cases.
Sample sizes were small across all of the studies (Venice category C, Table I). The GWAS included 115 cases of pelvic organ prolapse and 2,976 white controls from Illumina 550K(17). The linkage analysis showing a predisposition for pelvic floor disorders on chromosome 9q21 involved 70 affected women from 32 families and mostly evaluated sister pairs(18). The linkage analysis showing an association with LAMC1 involved genotyping of 9 individuals from one family, 6 of whom had prolapse. Prolapse cases spanned three generations(19). Of the 18 candidate gene studies, sample sizes ranged from 15(26) to 239(36,37), with 9 studies having fewer than 100 cases(20–22,24–27,30,33). Control populations for these studies ranged from 15(26) to 246(23), with 5 studies having fewer than 100 controls(26–28,30,33). Only the GWAS had replication of its findings(17) (Venice category B, Table I). Methodology was strong for two papers (36,37) and moderate for eight studies(28,33,34) (20–22,24,25) (Venice categories A and B, Table 1)1.
A meta-analysis was performed on all SNPs evaluated by two or more studies and included type III collagen (a key component of connective tissue), matrix metalloproteinases (enzymes which degrade extracellular matrix proteins and likely play a role in tissue remodeling) and laminins (a component of the basement membrane involved in the structural scaffolding of tissue). By convention, each SNP is reported by its gene and a reference SNP identification number (rs#). If an identifier has not yet been assigned, then the location of the allele is listed. The reference and effect alleles have different nucleotide sequences which can be specifically stated (adenine (A), cytosine (C), guanine(G), thymine (T)).
Two studies evaluated each of the following genetic variants
COL3A1 rs1800255, MMP9 rs17576, MMP1 position −1607/8, and LAMC1 rs20563. Three studies assessed LAMC1 rs10911193. The meta-analysis suggests that COL3A1 rs1800255 genotype AA is associated with POP, OR 4.79 (95% CI 1.91 to 11.98, p= 0.001) compared to the reference genotype GG in populations of Asian and Dutch women. There was little evidence of heterogeneity for rs1800255 (p-value for heterogeneity= 0.94; proportion of variance due to heterogeneity, I2= 0.00%). There was insufficient evidence to determine whether the following SNPs were associated with POP: MMP1 −1607/−1608 2G/2G (OR 1.41, 95% CI 0.85 to 2.33, p=0.18) and MMP9 rs17576 GG/AG (OR 0.87, 95% CI 0.57 to 1.31, p=0.50), LAMC1 rs10911193 TT/TG (OR 1.16, 95% CI 0.80 to 1.68, p=0.43) and LAMC1 rs20563 AA/AG (OR 1.22, 95% CI 0.88 to 1.68, p=0.43) (Table 2).
Table 2.
Meta-analysis of odds ratios for COL3A1 (rs1800255), MMP1 (1607/1608) and MMP9 (rs17576)
| COL3A1 (rs1800255) | ||||||||
|
| ||||||||
| Study | Effect | Ref | OR | 95% CI | Weight | p-value† | Het p-value‡ | |
| Chen (2008) | AG | GG | 0.74 | (0.41, 1.26) | 40.38% | |||
| Kluivers(2009) | AG | GG | 0.96 | (0.59, 1.58) | 59.62% | |||
| Meta-analysis | AG | GG | 0.87 | (0.59, 1.27) | 100% | 0.46 | 0.52 | 0.00% |
|
| ||||||||
| Chen (2008) | AA | GG | 4.59 | (1.17, 18.05) | 44.98% | |||
| Kluivers (2009) | AA | GG | 4.95 | (1.44, 17.06) | 55.02% | |||
| Meta-analysis | AA | GG | 4.79 | (1.91, 11.98) | 100% | 0.001 | 0.94 | 0.00% |
|
| ||||||||
| MMP1 (1607/1608) | ||||||||
|
| ||||||||
| Study | Effect | Ref | OR | 95% CI | Weight | p-value† | Het p-value‡ | |
| Ferrari (2012) | 1G/2G | 1G/1G | 2.24 | (1.16, 4.30) | 42.05% | |||
| Skorupski(2012) | 1G/2G | 1G/1G | 0.96 | (0.55, 1.67) | 57.95% | |||
| Meta-analysis | 1G/2G | 1G/1G | 1.39 | (0.90, 2.09) | 100% | 0.15 | 0.05 | 73.40% |
|
| ||||||||
| Ferrari (2012) | 2G/2G | 1G/1G | 2.81 | (1.25, 6.33) | 44.98% | |||
| Skorupski(2012) | 2G/2G | 1G/1G | 0.93 | (0.49, 1.75) | 55.02% | |||
| Meta-analysis | 2G/2G | 1G/1G | 1.41 | (0.86, 2.33) | 100% | 0.18 | 0.04 | 77.50% |
|
| ||||||||
| MMP9 (rs17576)** | ||||||||
| Study | Effect | Ref | OR | 95% CI | Weight | p-value† | Het p-value‡ | |
| Chen (2010) | GG/AG | AA | 5.67 | (1.28, 25.12) | 7.78% | |||
| Wu (2012) | GG/AG | AA | 0.74 | (0.48, 1.14) | 92.22% | |||
| Meta-analysis | GG/AG | AA | 0.87 | (0.57, 1.31) | 100% | 0.5 | 0.01 | 84.90% |
|
| ||||||||
| LAMC1 (rs10911193)** | ||||||||
| Study | Effect | Ref | OR | 95% CI | Weight | p-value† | Het p-value† | |
| Chen (2010) African Americans | TT/TG | GG | 1.83 | (0.59, 5.65) | 10.85% | |||
| Chen (2010) Caucasians | TT/TG | GG | 0.88 | (0.48, 1.62) | 37.68% | |||
| Wu (2012) | TT/TG | GG | 1.29 | (0.77, 1.68) | 51.47% | |||
| Meta-analysis | TT/TG | GG | 1.16 | (0.80, 1.68) | 100% | 0.43 | 0.46 | 0.00% |
|
| ||||||||
| LAMC1 (rs20563)* | ||||||||
| Study | Effect | Ref | OR | 95% CI | Weight | p-value† | Het p-value† | |
| Chen (2010) African Americans | AA/AG | GG | 1.43 | (0.56, 3.65) | 11.78% | |||
| Chen (2010) Caucasians | AA/AG | GG | 0.8 | (0.45, 1.46) | 29.19% | |||
| Wu (2012) | AA/AG | GG | 1.44 | (0.95, 2.19) | 59.03% | |||
| Meta-analysis | AA/AG | GG | 1.22 | (0.88, 1.68) | 100% | 0.23 | 0.28 | 22.30% |
Abbreviations: OR = Odds ratio; SNP = Single NuCleotide Polymorphism; pos = position; Het = Heterogeneity;
CI = Confidence Interval
p-value that tests the null hypothesis that the overall odds ratio = 1.
Het p-value tests if the odds ratios for the individual studies are heterogeneous.
I2 Explains the percentage of variation in the odds ratios attributable to heterogeneity.
Odds ratios for SNP rs17576 are based on a dominant model.
All ORs reported in table are re-calculated crude odds ratios, with the exception of Wu et al (2012), who provided adjusted odds ratios only.
An association with POP was seen in individual studies for ERα rs2228480 GA, PGR rs484389 CT and COL3A1 exon 31, six SNPs from the GWAS and chromosome 9q21 (HLOD score 3.41), (Table 3).
Table 3.
Individual studies with significant findings (not included in meta-analysis):
| Study | Gene or chromosome | SNP | Allele frequencies | Odds (HLOD or OR) | p-value | |||
|---|---|---|---|---|---|---|---|---|
| Allen-Brady, 2009(18) | Chromosome 9q21 | HLOD score 3.41; Chr9: 80.35Mb–88.81Mb with HLOD>=1.86 |
||||||
| Allen-Brady, 2011(17) | 4q21.21 | rs1455311 | OR 2.58 EMMAX: 7.65 × 10exp-12; Genie <1 × 10exp-9 |
|||||
| 8q24.22 | rs1036819 | OR 4.03 EMMAX: 3.57 × 10exp-21; Genie <1 × 10exp-9 |
||||||
| 9q22.2 | rs430794 | OR 0.35 EMMAX: 6.74 × 10exp-5; Genie 8 × 10exp-8(Only significant by Genie) |
||||||
| 15q11.2 | rs8027714 | OR 9.04 EMMAX: 5.65 × 10exp-43; Genie <1 × 10exp-9 |
||||||
| 20p13 | rs1810636 | OR 2.3 EMMAX: 6.06 × 10exp-8; Genie 3.3 × 10exp-7 |
||||||
| 21q22.3 | rs2236479 | OR 2.2 EMMAX: 6.61 × 10exp-9; Genie 2.8 × 10exp-7 |
||||||
| Chen H, 2008 (21) | ERα | rs2228480 (exon 8 G/A) | GG | GA | AA | GA:OR2.05(95%CI: 1.05 to 4.02), p = 0.036 AA:OR0.52(95%CI: 0.05 to 5.03), p = 0.571 |
0.015 | |
| Cases | 45 (51.1%) | 41 (46.6%) | 2 (2.3%) | |||||
| Controls | 102 (66.6%) | 44 (28.8%) | 7 (4.6%) | |||||
| Chen H, 2009(25) | PGR | rs484389 (3' untranslated region C/T) | TT | CT | CC | CT: OR 4.77 (95% CI: 1.93–11.79), p=0.0007 CC: OR 1.06 (95% CI: 0.28–5.07), p=0.9276 | 0.013 | |
| Cases | 63 (72.4%) | 19 (21.8%) | 5 (5.7%) | |||||
| Controls | 130 (86.7%) | 13(8.7%) | 7 (4.7%) | |||||
| Jeon, 2009(30) | COL3A1 | Exol 31 2092 5'-AAGTATA CAAATTTC TAGATTG-3' (forward)/5'-ATAAATG ATCAGAA GGACAAT CA-3' (reverse) | AA | GA | GG | OR 4.3 (95% CI: 1.4 to 13.3) | 0.005 | |
| Cases | 0 | 13 (36%) | 23 (64%) | |||||
| Controls | 5 (14%) | 20 (55%) | 11 (31%) | |||||
Comment
Our systematic review and meta-analysis suggest that there is a 4.79 increased odds of developing POP when the COL3A1 rs1800255 genotype AA is present. Other potential genes or SNPs of interest include MMP1, MMP9, LAMC1, ER-α rs2228480 GA and COL3A1 exon 31, but current data are either insufficient or have not been independently replicated to confirm an association with POP. There is one study showing genome-wide evidence for linkage with chromosome 9q21; no candidate gene studies have been performed from this genetic region.
It is hypothesized that alterations in the connective tissue and extracellular matrix of the pelvic organs may play a role in the development of POP. Thus genetic mutations in the components of the extracellular matrix are of particular interest, and several candidate-gene studies have focused on this area. Collagen types I and III are the two main components of pelvic connective tissue. Laminins are glycoproteins involved in the structural scaffolding of tissues and matrix metalloproteinases are involved in degradation of the extracellular matrix and likely have a role in tissue remodeling. Our review found very preliminary evidence that polymorphisms in the genes coding for these proteins may have an association with POP. Interest in the role of hormone receptors is also biologically plausible. This review found one study looking at ER-α, which suggested a role in POP.
Collagen is the main component in pelvic connective tissue. Type I fibers are well-organized and are present in the uterosacral ligaments which provide DeLancey level I support of the cervix and vaginal apex(38). Type III fibers are more prominent in the loose areolar tissue surrounding the vagina and pelvic organs. Evaluation of the expression of collagen types I and III in women with and without POP has yielded varying results, with some studies showing increased expression and others showing decreased expression(39,40). Many variables may contribute to this: different tissues being studied (such as the uterosacral ligaments versus the vaginal wall), harvesting and extraction methods, patient characteristics and the molecular makeup of the collagen. For example, uterosacral ligament resilience is four-fold less in women with prolapse compared to controls, but resilience also decreases with age, highlighting the need for age-equivalent controls (41). If there are genetic variants in the collagen, it may be the molecular structure and not the amount of collagen that is related to the development of prolapse.
Two studies found a strong association between COL3A1 rs1800255 genotype AA and POP. Polymorphisms with a strong disease association often are seen across different genetic populations; the similar finding in both Asian and Dutch populations increases the likelihood that this is a true association. Both studies were limited by potential misclassification bias. In the Chen paper, the control population was significantly younger than the cases, 30.6% of non-prolapse controls were ≥54 years of age compared to 72.6% of the prolapse cases(20). Given the widespread prevalence of POP, with most cases presenting after menopause, the relatively young age of the controls increases the risk for misclassification of women who have not yet manifested prolapse. This was successfully minimized in the Kluivers paper by having a similar median age and age distribution among cases and controls. The main potential for misclassification in the Kluivers paper was the lack of a defined prolapse phenotype(31). Additionally, women with stage II prolapse were categorized as controls (17%) and women with stage I prolapse were categorized as cases (5%). Stage I is rarely symptomatic and may represent physiologic changes associated with aging. Controls with stage II prolapse, even if not symptomatic, are certainly at risk for progression of the prolapse in the future. Using the Venice guidelines, both papers had significant limitations in methodologic quality with small sample sizes (category C), no replication performed in the original study (category C) and moderate (category B, Chen study) or weak (category C, Kluivers study) protection from bias (Table 1). These guidelines are designed to evaluate cumulative evidence and set a very high bar for “A” grade quality. Our meta-analysis validates these results and increases the quality of data regarding replication for this genetic association to a category B.
The COL3A1 rs1800255 genotype AA mutation results in an alanine to threonine amino acid substitution at position 698 in all 3 chains of the type III collagen helix. The threonine side chain is more hydrophilic and may alter the structure and function of the collagen helix. Biologically, it is plausible that this type of mutation could impact the strength of the pelvic floor.
Among the published literature, there was relative consistency in defining the POP phenotype, except for the Kluivers study, as noted above. The remaining candidate gene case-control studies restricted cases to ≥Stage II prolapse and controls to Stage 0-I prolapse. Some studies were more restrictive with POP-Q staging(26,27,35,36) and others excluded Ehlers Danlos, Marfan syndrome(17,23), Steinert disease(17), multiple sclerosis or prior stroke(17). Excluding women with rare conditions which may exacerbate prolapse, such as connective tissue disorders, optimizes the ability of a genetic study to detect alleles associated with the most common and prevalent forms of POP. Defining the phenotype of controls is just as critical as defining cases. A recent opinion piece highlights the importance of this when considering study design(42). The most common bias seen was the inclusion of controls that were younger than cases and may not yet manifest the prolapse phenotype(20–25,32,34,35). Only two studies preferentially recruited controls from an older population, thus minimizing the risk of misclassification bias related to future, as-of -yet not manifested prolapse, among controls(36,37). One study used controls from a database which did not comment on the presence of pelvic floor disorders(17).
In this review, none of the SNPs evaluated in candidate gene studies were among the significant associations observed on the GWAS. This may be a reflection of the limited number of candidate gene studies, as well as findings in other fields that GWAS may not solely predict associated genes.
As additional studies are performed, the increased volume of data will allow more sophisticated analyses evaluating the likelihood and validity of specific genetic associations with POP. In order to detect genetic associations, large numbers of subjects are needed. None of the current work in this field meets Venice level A for the amount of evidence; this requires sample sizes of over 1000 (cases and controls together, 1:1 ratio)(43). The use of DNA databanks can facilitate this process, but only if information about pelvic floor disorders is obtained when establishing the databank. Given the prevalence and impact of pelvic floor disorders on women's health, and preliminary evidence suggesting a genetic contribution, we believe this should be a priority. Additional work is needed to identify candidate genes, in addition to studies looking at the impact of non-coding regions of the genome. Studies also need to replicate their findings. Of the current literature, only the GWAS published results from replication of their findings(17); this meta-analysis provides validation for COL3A1 rs1800255.
Acknowledgments
Dr. Velez Edwards is supported by K12HD4383, Building Interdisciplinary Research Careers in Women's Health career development program
Dr. Wu is supported by K23HD068404, Eunice Kennedy Shriver National Institute of Child Health & Human Development
Poster presentation, 34th American Urogynecologic Society Scientific Meeting in Las Vegas, NV, October 16–19, 2013
There was no financial support for this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors report no conflict of interest.
References
- 1.Hendrix S. Pelvic organ prolapse in the women's health initiative: Gravity and gravidity. Am. J. Obstet. Gynecol. 2002 Jun;186(6):1160–6. doi: 10.1067/mob.2002.123819. [DOI] [PubMed] [Google Scholar]
- 2.Jelovsek JE, Maher C, Barber MD. Pelvic organ prolapse. Lancet. 2007 Mar 24;369(9566):1027–38. doi: 10.1016/S0140-6736(07)60462-0. [DOI] [PubMed] [Google Scholar]
- 3.Wu JM, Kawasaki A, Hundley AF, Dieter AA, Myers ER, Sung VW. Predicting the number of women who will undergo incontinence and prolapse surgery, 2010 to 2050. Am. J. Obstet. Gynecol. 2011 Sep;205(3):230.e1–230.e5. doi: 10.1016/j.ajog.2011.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mant J, Painter R, Vessey M. Epidemiology of genital prolapse: observations from the Oxford Family Planning Association Study. Br J Obstet Gynaecol. 1997 May;104(5):579–85. doi: 10.1111/j.1471-0528.1997.tb11536.x. [DOI] [PubMed] [Google Scholar]
- 5.Weber AM, Buchsbaum GM, Chen B, Clark AL, Damaser MS, Daneshgari F, et al. Basic science and translational research in female pelvic floor disorders: proceedings of an NIH-sponsored meeting. Neurourol. Urodyn. 2004:288–301. doi: 10.1002/nau.20048. [DOI] [PubMed] [Google Scholar]
- 6.Nygaard I, Bradley C, Brandt D, Women's Health Initiative Pelvic organ prolapse in older women: prevalence and risk factors. Obstet Gynecol. 2004 Sep;104(3):489–97. doi: 10.1097/01.AOG.0000136100.10818.d8. [DOI] [PubMed] [Google Scholar]
- 7.Swift SE, Pound T, Dias JK. Case-control study of etiologic factors in the development of severe pelvic organ prolapse. Int Urogynecol J Pelvic Floor Dysfunct. 2001;12(3):187–92. doi: 10.1007/s001920170062. [DOI] [PubMed] [Google Scholar]
- 8.Samuelsson EC, Victor FT, Tibblin G, Svärdsudd KF. Signs of genital prolapse in a Swedish population of women 20 to 59 years of age and possible related factors. YMOB. 1999 Feb;180(2 Pt 1):299–305. doi: 10.1016/s0002-9378(99)70203-6. [DOI] [PubMed] [Google Scholar]
- 9.Jack GS, Nikolova G, Vilain E, Raz S, Rodríguez LV. Familial transmission of genitovaginal prolapse. Int Urogynecol J Pelvic Floor Dysfunct. 2006 Sep;17(5):498–501. doi: 10.1007/s00192-005-0054-x. [DOI] [PubMed] [Google Scholar]
- 10.Altman D, Forsman M, Falconer C, Lichtenstein P. Genetic influence on stress urinary incontinence and pelvic organ prolapse. Eur. Urol. 2008 Oct;54(4):918–22. doi: 10.1016/j.eururo.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Buchsbaum GM, Duecy EE, Kerr LA, Huang L-S, Perevich M, Guzick DS. Pelvic organ prolapse in nulliparous women and their parous sisters. Obstet Gynecol. 2006 Dec;108(6):1388–93. doi: 10.1097/01.AOG.0000245784.31082.ed. [DOI] [PubMed] [Google Scholar]
- 12.GIANT [Internet] www.broadinstitute.org. www.broadinstitute.org [cited 2013 Jul 16]. Available from: http://www.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium.
- 13.Myocardial Infarction Genetics Consortium. Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009 Mar;41(3):334–41. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomlinson IPM, Dunlop M, Campbell H, Zanke B, Gallinger S, Hudson T, et al. COGENT (COlorectal cancer GENeTics): an international consortium to study the role of polymorphic variation on the risk of colorectal cancer. Br. J. Cancer. 2010 Jan 19;102(2):447–54. doi: 10.1038/sj.bjc.6605338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JPA, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009:e1–34. doi: 10.1016/j.jclinepi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Ioannidis JP, Boffetta P, Little J, O'Brien TR, Uitterlinden AG, Vineis P, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. International Journal of Epidemiology. 2007 Dec 3;37(1):120–32. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 17.Allen-Brady K, Cannon-Albright L, Farnham JM, Teerlink C, Vierhout ME, van Kempen LCL, et al. Identification of Six Loci Associated With Pelvic Organ Prolapse Using Genome-Wide Association Analysis. Obstet Gynecol. 2011 Dec;118(6):1345–53. doi: 10.1097/AOG.0b013e318236f4b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen-Brady K, Norton PA, Farnham JM, Teerlink C, Cannon-Albright LA. The American Journal of Human Genetics. 5. Vol. 84. The American Society of Human Genetics; May 15, 2009. Significant Linkage Evidence for a Predisposition Gene for Pelvic Floor Disorders on Chromosome 9q21; pp. 678–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikolova G, Lee H, Berkovitz S, Nelson S, Sinsheimer J, Vilain E, et al. Sequence variant in the laminin γ1 (LAMC1) gene associated with familial pelvic organ prolapse. Hum Genet. 2006 Oct 5;120(6):847–56. doi: 10.1007/s00439-006-0267-1. [DOI] [PubMed] [Google Scholar]
- 20.Chen H-Y, Chung Y-W, Lin W-Y, Wang J-C, Tsai F-J, Tsai C-H. Collagen type 3 alpha 1 polymorphism and risk of pelvic organ prolapse. International Journal of Gynecology & Obstetrics. 2008 Oct;103(1):55–8. doi: 10.1016/j.ijgo.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 21.Chen H-Y, Chung Y-W, Lin W-Y, Chen W-C, Tsai F-J, Tsai C-H. Estrogen receptor alpha polymorphism is associated with pelvic organ prolapse risk. Int Urogynecol J. 2008 Apr 3;19(8):1159–63. doi: 10.1007/s00192-008-0603-1. [DOI] [PubMed] [Google Scholar]
- 22.Chen H-Y, Wan L, Chung Y-W, Chen W-C, Tsai F-J, Tsai C-H. Estrogen receptor beta gene haplotype is associated with pelvic organ prolapse. European Journal of Obstetrics & Gynecology and Reproductive Biology. 2008 May;138(1):105–9. doi: 10.1016/j.ejogrb.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Hill LD, Schubert CM, Strauss JF, Matthews CA. YMOB. 5. Vol. 202. Elsevier Inc; May 1, 2010. Is laminin gamma-1 a candidate gene for advanced pelvic organ prolapse? pp. 505.e1–505.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H-Y, Lin W-Y, Chen Y-H, Chen W-C, Tsai F-J, Tsai C-H. European Journal of Obstetrics and Gynecology. 2. Vol. 149. Elsevier Ireland Ltd; Apr 1, 2010. Matrix metalloproteinase-9 polymorphism and risk of pelvic organ prolapse in Taiwanese women; pp. 222–4. [DOI] [PubMed] [Google Scholar]
- 25.Chen H-Y, Chung Y-W, Lin W-Y, Chen W-C, Tsai F-J, Tsai C-H. Progesterone receptor polymorphism is associated with pelvic organ prolapse risk. Acta Obstet Gynecol Scand. 2009 Jan;88(7):835–8. doi: 10.1080/00016340902822073. [DOI] [PubMed] [Google Scholar]
- 26.Cho HJ, Jung HJ, Kim SK, Choi JR, Cho NH, Bai SW. Polymorphism of a COLIA1 Gene Sp1 Binding Site in Korean Women with Pelvic Organ Prolapse. Yonsei Med J. 2009;50(4):564. doi: 10.3349/ymj.2009.50.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feiner B, Fares F, Azam N, Auslender R, David M, Abramov Y. Does COLIA1 SP1-binding site polymorphism predispose women to pelvic organ prolapse? Int Urogynecol J. 2009 May 7;20(9):1061–5. doi: 10.1007/s00192-009-0895-9. [DOI] [PubMed] [Google Scholar]
- 28.Ferrari MM, Rossi G, Biondi ML, Viganò P, Dell'Utri C, Meschia M. Type I collagen and matrix metalloproteinase 1, 3 and 9 gene polymorphisms in the predisposition to pelvic organ prolapse. Arch Gynecol Obstet. 2011 Dec 31;285(6):1581–6. doi: 10.1007/s00404-011-2199-9. [DOI] [PubMed] [Google Scholar]
- 29.Ferrell G, Minyan Lu, Stoddard P, Sammel MD, Romero R, Strauss JF, et al. A Single Nucleotide Polymorphism in the Promoter of the LOXL1 Gene and Its Relationship to Pelvic Organ Prolapse and Preterm Premature Rupture of Membranes. Reproductive Sciences. 2009 Feb 5;16(5):438–46. doi: 10.1177/1933719108330567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeon MJ, Chung SM, Choi JR, Jung HJ, Kim SK, Bai SW. JURO. 3. Vol. 181. American Urological Association; Mar 1, 2009. The Relationship Between COL3A1 Exon 31 Polymorphism and Pelvic Organ Prolapse; pp. 1213–6. [DOI] [PubMed] [Google Scholar]
- 31.Kluivers KB, Dijkstra JR, Hendriks JCM, Lince SL, Vierhout ME, Kempen LCL. COL3A1 2209G>A is a predictor of pelvic organ prolapse. Int Urogynecol J. 2009 May 15;20(9):1113–8. doi: 10.1007/s00192-009-0913-y. [DOI] [PubMed] [Google Scholar]
- 32.Martins K de F, Bella ZIK de J-D, da Fonseca AMRM, Castro RA, Guerreiro da Silva IDC, Castello Girão MJB, et al. Evaluation of demographic, clinical characteristics, and genetic polymorphism as risk factors for pelvic organ prolapse in brazilian women. Neurourol. Urodyn. 2011 May 23; doi: 10.1002/nau.21066. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 33.Skorupski P, Miotła P, Jankiewicz K, Rechberger T. Polymorphism of the gene encoding alpha-1 chain of collagen type I and a risk of pelvic organ prolapse--a preliminary study. Ginekologia polska. 2007;78(11):852. [PubMed] [Google Scholar]
- 34.Skorupski P, Jankiewicz K, Miotła P, Marczak M, Kulik-Rechberger B, Rechberger T. The polymorphisms of the MMP-1 and the MMP-3 genes and the risk of pelvic organ prolapse. Int Urogynecol J. 2012 Oct 30; doi: 10.1007/s00192-012-1970-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodrigues AM, Girão MJBC, Silva IDCG, Sartori MGF, Martins K de F, Castro R de A. COL1A1 Sp1-binding site polymorphism as a risk factor for genital prolapse. Int Urogynecol J. 2008 Jun 13;19(11):1471–5. doi: 10.1007/s00192-008-0662-3. [DOI] [PubMed] [Google Scholar]
- 36.Wu JM, Visco AG, Grass EA, Craig DM, Fulton RG, Haynes C, et al. Comprehensive analysis of LAMC1 genetic variants in advanced pelvic organ prolapse. Am. J. Obstet. Gynecol. 2012 May;206(5):447.e1–447.e6. doi: 10.1016/j.ajog.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu JM, Visco AG, Grass EA, Craig DM, Fulton RG, Haynes C, et al. Matrix Metalloproteinase-9 Genetic Polymorphisms and the Risk for Advanced Pelvic Organ Prolapse. Obstet Gynecol. 2012 Sep;120(3):587–93. doi: 10.1097/AOG.0b013e318262234b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLancey JO. Anatomic aspects of vaginal eversion after hysterectomy. YMOB. 1992 Jun;166(6 Pt 1) doi: 10.1016/0002-9378(92)91562-o. 1717–24-discussion1724–8. [DOI] [PubMed] [Google Scholar]
- 39.Mosier E, Lin VK, Zimmern P. Extracellular matrix expression of human prolapsed vaginal wall. Neurourol. Urodyn. 2009 Sep 21;29(4):582–6. doi: 10.1002/nau.20806. [DOI] [PubMed] [Google Scholar]
- 40.Connell KA, Guess MK, Chen H, Andikyan V, Bercik R, Taylor HS. HOXA11 is critical for development and maintenance of uterosacral ligaments and deficient in pelvic prolapse. J. Clin. Invest. 2008 Feb 1; doi: 10.1172/JCI34193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reay Jones NHJ, Healy JC, King LJ, Saini S, Shousha S, Allen-Mersh TG. Pelvic connective tissue resilience decreases with vaginal delivery, menopause and uterine prolapse. Br J Surg. 2003;90(4):466–72. doi: 10.1002/bjs.4065. [DOI] [PubMed] [Google Scholar]
- 42.Wu JM, Ward RM, Allen-Brady KL, Edwards TL, Norton PA, Hartmann KE, et al. Phenotyping clinical disorders: lessons learned from pelvic organ prolapse. Am. J. Obstet. Gynecol. 2012 Nov 27; doi: 10.1016/j.ajog.2012.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ioannidis JP, Boffetta P, Little J, O'Brien TR, Uitterlinden AG, Vineis P, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. International Journal of Epidemiology. 2007 Dec 3;37(1):120–32. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]