Abstract
Human short-chain dehydrogenases/reductases (SDRs) protein family has been the subject of recent studies for its critical role in human metabolism. Studies also found that single nucleotide polymorphisms of the SDR protein family were responsible for a variety of genetic diseases, including type II diabetes. This study reports the effect of sequence variation on the structural and functional integrities of human SDR protein family using phylogenetics and correlated mutation analysis tools. Our results indicated that (i) tyrosine, serine, and lysine are signature protein residues that have direct contribution to the structural and functional stabilities of the SDR protein family, (ii) subgroups of SDR protein family have their own signature protein combination that represent their unique functionality, and (iii) mutations of the human SDR protein family showed high correlation in terms of evolutionary history. In combination, the results inferred that over evolutionary history, the SDR protein family was able to diverge itself in order to adapt with the changes in human nutritional demands. Our study reveals understanding of structural and functional scaffolds of specific SDR subgroups that may facilitate the design of specific inhibitor.
Keywords: human short-chain dehydrogenases/reductases (SDRs), correlated mutation, mutational variability, consensus sequence, phylogeny, multiple sequence alignment
Introduction
Short-chain dehydrogenases/reductases (SDRs) belong to one of the largest enzyme superfamily, which has 122,940 members.1 In 2009, at least 140 different enzymes had been sequenced and about 70 of them were found to belong to the human SDR family. In 2013, 47 human SDR proteins, corresponding to 75 genes, have been identified. Most SDR proteins are nicotinamide adenine dinucleotide (NAD) or nicotinamide adenine dinucleotide phosphate (NADP)-dependent oxidoreductases, which share similar, sequence motifs and functional mechanism.2,3 This SDR protein family was found to be present in both prokaryotic and eukaryotic organisms including human and plays important role in a variety of key metabolic processes such as lipid, amino acid, carbohydrate, hormone, etc. Notably, SDR protein family was found to contribute to human metabolic diseases including type II diabetes.2–5 There have been attempts to design inhibitor toward controlling hormone signaling through using SDR protein family as target because of its important function in human metabolism.6,7 Previous studies also found that members of the human SDR protein family have a highly diverged relationship with only 15–30% sequence similarity.1,2 With such high divergence, SDR protein family can be divided into two groups, classical SDR and extended SDR, based on their differences in glycine (GLY) binding motifs, coenzyme binding motifs, and chain length (250 residues in length for the classical SDR group and 350 residues in length for the extended SDR group).8 These two SDR groups share similarity in sequence motifs such as the cofactor binding site (TGxxxGxG) and the catalytic tetrad (NSYK).8,9 Of the catalytic tetrad, three residues (SYK) exhibit highest conservation within the active site10 because of their important role in the formation of structural motif with Asn through hydrogen bonding with other residues. The bonding between Asn of the tetrad results in a sharp shrinking at the helix position, which forces the protein backbone into a position where it can bind to a water molecule. This binding in return connects Asn to the active site residue Lys instead of binding Asn to the main chain as expected from the helix structure. However, the SDR’s structures that replace Asn with Ser will allow Lys to bind to the interacting water molecule in the same binding mode.9 Moreover, the three-dimensional (3D) structure of all human SDRs shares common features such as an alpha or beta folding motif characterized by a central beta sheet. This central beta sheet is a typical formation of Rossmann fold with helices on either side.8 Owing to these structural similarities, it is necessary to study the evolutionary history of the SDR superfamily to extend the understanding upon the particular 3D structural formation with such low sequence similarity. It has been hypothesized that these common binding motifs could have been conserved over evolutionary time to maintain the structural and functional properties that differentiate human SDR family from other protein families.9 While the variability of the SDR family occurs at the level of sequence, the effects of these mutations are noticeable at the structural and functional levels. A study on comparative sequence and structure alignments of different human SDRs in terms of evolutionary context may reveal information about the diversification of human SDR family.
In order to improve our understanding of the diversification of human SDR family, we performed a rigorous comparative analysis of the homologous sequence and the relationship between sequence in structure and function of this protein family using bioinformatics tools. Our goal was to identify and compare the convergent and divergent residues of the human SDR at catalytic sites. We hypothesized that evolutionarily conserved regions in the human SDR family would reside close to the location of the active and binding sites of the protein for maintaining functional integrity at these sites is utmost important for the protein–protein and protein–ligand interactions and ultimately proper metabolic activity. We expected to find variable regions in the human SDR family that are near the nucleotide binding sites because of the specific variation in substrate enzyme interactions of human SDR proteins. These variable regions may have evolved to allow human SDR proteins obtain their specific enzymatic functionality. Such particular structural and functional features are important in the design of novel human SDR protein inhibitor with higher specificity and efficacy.11,12
Materials and Methods
Multiple sequence alignment of human SDR proteins and alignment verification
In all, 75 sequences of human SDR enzymes were collected from UniProtKB database (http://www.uniprot.org). Sequences were initially aligned with ClustalW,13 T-Coffee,14 Muscle,15 and Kalign16 using the template sequence Q14376 (UDP-glucose-4-epimerase). In order to create the most robust alignment, initial alignments using each method were compared against one another and the most differing sequences were removed from subsequent analyses. Finally, the multiple sequence alignment was checked and verified with the aid of the genetic semihomology algorithm from Geisha 3.0 software17–20 in order to obtain the reliable Multiple Sequences Alignment (MSA). The potential evolutionary relationship between the corresponding non-identical positions from the four different multiple alignments was verified separately using the genetic semihomology algorithm implemented in Geisha 3.0.17 Geisha 3.0 is freely accessible from the web site http://atama.wnb.uz.zgora.pl/~jleluk/linki.html. Verifying multiple sequence alignments using Geisha helps to identify and reduce potential mismatches that may occur during the initial alignment process. ClustalW, T-Coffee, Muscle, and Kalign are based on the hidden Markov model. Geisha 3.0 improves alignment accuracy by completing the alignment while considering point mutations. Geisha 3.0 assumes that the probability of the replacement of one amino acid with another depends significantly on the amino acid that occupied the same position previously.
Human SDR consensus sequence construction and Basic Local Alignment Search Tool (BLAST) search
As a way of summarizing the verified human SDR multiple sequence alignments, a single consensus sequence for the entire human SDR superfamily was established. The consensus sequence was obtained using the consensus sequence constructor with default parameter values. This is an original application designed by our Polish collaborators and is freely available for non-commercial academic purposes from the website http://atama.wnb.uz.zgora.pl/~jleluk/linki.html. The most robust consensus sequence was then used to identify two types of specificity for all members of the human SDR superfamily: (1) the general specificity, which indicates common features of the entire enzyme superfamily and (2) the individual specificity, which distinguishes the unique structural properties of each grouping within the human SDR superfamily separately. Consequently, the general specificity is concerned with the more conservative regions of human SDR proteins, while the individual specificity highlights the more variable regions. By investigating both types of specificity, our results may be of better use for future work on developing inhibitors and can be directed to only one or a few enzymes without affecting the activity of the others. Lastly, the optimal consensus sequence was also used in a BLAST search for potential new members of the human SDR family. The new sequences supplemented the original 75 SDR family members (about 100 sequences) and were aligned in the same way as described above.
Phylogenetic tree construction and comparison of phylogenetic consensus sequences
The results of our multiple sequence alignments were used as input data for constructing phylogenetic trees that would outline the interrelationships of the various members of the human SDR superfamily. In this study, two independent software were used to construct the phylogenetic trees – PHYLIP (Felsenstein, J. 1989. PHYLIP manual, version 3.2. University of California Herbarium, Berkeley, California.) and SSSSg (database: UniProt, matrix: Blosum45, number of matches: 10, and E upper value: 5.0). PHYLIP is a free package of programs for inferring phylogenies accessible at http://www.phylip.com. SSSSg is our original software, and is freely accessible at http://atama.wnb.uz.zgora.pl/~jleluk/software/wlasne/ssssg/ssssg.zip. PHYLIP uses Fitch’s maximum parsimony algorithm, and constructs the phylogenetic tree that requires the least amount of evolutionary change to fit the input data. To supplement our parsimony analyses, we also applied the maximum likelihood algorithm to our data using the program SSSSg. Maximum likelihood is an optimality criterion, like maximum parsimony, for the reconstruction of phylogenies. Maximum likelihood methods differ from the non-parametric parsimony approach because it uses an explicit model of character evolution for tree construction. Both maximum parsimony and maximum likelihood methods recovered the same five, high-level branching events within the human SDR family, and lower level topological differences were negligible. As such, we arbitrarily chose to use the maximum likelihood tree for all subsequent analyses.
Using consensus sequence constructor, we identified a single consensus protein sequence for each of the five human SDR subgroups. Comparative analysis was carried out on the five resultant consensus sequences in order to identify the conservative and variable sequence regions in human SDR enzymes.
To further elucidate patterns of conservation and variation in human SDR enzymes, comparative analysis of the 3D protein structure of each of the five consensus sequences was also conducted. We identified a representative structure for each of the five groups recovered in the phylogenetic reconstructions using the Protein Data Bank. Basically, all 3D protein structures within each group were compared with each other and the selected structure was the one that shares the highest level of similarity compared to the others. The selection criteria focused on the maximum similarity of the sequence alignment from all members in each group and the highest degree of similarity at the tertiary structural level.
Mutational variability of human SDRs
We used the five identified representative structures (see below) together with all protein sequences available in each group identified in our phylogenetic analyses to study the mutational variability within the five subgroups of the human SDR family. ConSurf (available at consurf.tau.ac.il) and Talana (available at http://www.bioware.republika.pl) were used to identify conservative and variable residues of functional regions in the aligned homologous sequences. ConSurf and Talana are designed to estimate the evolutionary conservation of amino acids based on the phylogenetic relations between homologous sequences.
Both programs analyze the evolutionary conservation of amino acids based on the sequences and produce conservation scores that correspond to the rate of evolution at each site. The two programs systematically plot these conservation scores onto the protein structures. The scores are divided into nine grades for the visualization of differing rates of evolution in ConSurf: grade 1 is the most variable position and is colored turquoise, grade 5 is the intermediately conserved position and is colored white, and grade 9 is the most conserved position and is colored maroon. Alternatively, in Talana, the conservation scores are divided into 12 grades: grade 1 is the most conserved position (darkest blue), grade 6 is the intermediately conserved position (white), and grade 12 is the most variable position (darkest red).
The variable and conservative human SDR regions that were recovered from both ConSurf and Talana were plotted on the five template SDR structures (see below) and visualized using Rastop2.2 (http://www.geneinfinity.org/rastop). The results of these two approaches were compared mutually for verification of their compatibility.
| GROUPS | PDB CODE AND NAME OF REPRESENTATIVE STRUCTURES |
|---|---|
| 1 | 3edm chain A, Short-chain dehydrogenase from Agrobacterium tumefaciens |
| 2 | 1hdc chain A, 3-ALPHA-(20-BETA)-HYDROXYSTEROID DEHYDROGENASE |
| 3 | 1yb1 chain A, 17-beta hydroxysteroid dehydrogenase 11 |
| 4 | 3rd5 chain A, a putative uncharacterized oxidoreductase protein from Mycobacterium Paratuberculosis |
| 5 | 1q7b chain A, beta-ketoacyl-[ACP] reductase from E. coli |
The active site residues are not directly exposed to the protein surface but rather hidden into the substrate binding pocket since the substrates are considerably small molecules. Therefore, the buried water molecules must play a significant role in the catalytic activity of the SDR enzymes. However, the variability and correlation of the residues interacting with the buried water molecules is a separate problem to be studied later in future research.
Analysis of correlated mutations
Lastly, investigation on the tendency of different amino acids along human SDR proteins to mutate together was conducted. There is a high probability that many residues within the same protein have evolved to form specific molecular complexes, and the specificity of this interaction is essential for their function. This network of interactive residues may contain divergence of the protein sequence. To maintain functionality, it is reasonable to assume that mutation event during the evolution of one of the interactive residues must be accompanied by mutation compensation in the other residues.21,22
Correlated mutations in representative protein structures and corresponding consensus sequences in each group of human SDR family were identified, localized, and analyzed with the aid of Talana and Corm (freely available for non-commercial academic purposes at http://atama.wnb.uz.zgora.pl/~jleluk/software/wlasne/corm.jar). The program FEEDBACK was implemented in Corm, which is designed to analyze the aligned protein sequences for the occurrence of correlated mutations. It returns all possible residues occurring at all sequence positions of aligned proteins for each residue occurring at each position. Talana not only produces a similar set of results but also highlights correlated sequence mutations in the corresponding protein structures. The candidate correlated sequence and structure mutations that were recovered using both software packages were compared and then visualized on the SDR template structure of the five groups within the human SDR family using DSVisualizer1.7 of Accelrys (http://accelrys.com/products/discoverystudio/visualization-download.php) and/or Rastop2.2 (http://www.geneinfinity.org/rastop). The visualization of the protein sequence mutation correlation results from Talana, and Corm provides an addition method of investigating potential correlated mutations in protein structure.
Availability of original software generated by authors
The original applications of Geisha 3.0, Consensus Constructor, SSSSg, Talana, and Corm are freely available at the addresses listed above. They are also available directly upon any request sent to the authors. Additionally, the authors are willing to assist in the appropriate, effective running of all these applications.
Results
Multiple sequence alignment, consensus sequence generation, and analysis of human SDR specificity
The collection of homologous SDR sequences has started from BLAST search using the selected SDR (Q14376) sequence as the query sequence. After collecting the first set of sequences revealing the significant similarity, they were aligned with the aid of ClustalX, and separately with T-Coffee, Muscle, and Kalign. The alignments were compared with each other to construct the sequence alignment as the result of all those approaches. Then the multiple sequence alignment was thoroughly verified with the algorithm of genetic semihomology for correction of some doubtful fragments and for proper gap location. On the basis of the obtained multiple sequence alignment, there was constructed the consensus sequence (with the aid of Consensus Constructor), and the BLAST search was run again with the consensus sequence as the query sequence. The obtained sequence set was once again aligned, verified with genetic semihomology algorithm, and another consensus sequence was constructed. The constructed consensus sequence was used again for the final BLAST search, which gave in result the final set of homologous SDR sequences.
After multiple sequence alignment and verification, we identified four sequences (P49327, P14060, P56159, and P56937) that shared an unusual low sequence similarity with the rest of the members of the human SDR family, and they were removed from subsequent analyses. We constructed the consensus sequence from the remaining 71 sequences, and used it to identify features of general and individual specificities.
Our comparative analyses reveal a discrete amount of general specificity but a high level of individual specificity among human SDR sequences (Fig. 1). Figure 1 presents the consensus sequence that describes the overall SDR superfamily. It does not specify the regions such as substrate binding pocket or active site. The consensus sequence defines the conserved and variable regions within the family, and shows the essential features that define the homologous superfamily at the level of primary structure. Among 306 positions in the consensus sequence, only 5 positions (1.6%) are occupied by the same residue in more than 70% of sequences, whereas 105 positions (34.3%) are occupied by the same residue in at least 30% of sequences. In all, 196 positions (64.1%) are occupied by any particular residue in more than 30% of sequences.
Figure 1.
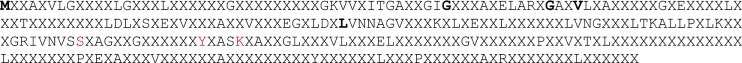
Complete consensus sequence of 71 human SDR sequences. Consensus sequences for the human SDR family, constructed using consensus sequence constructor. The highly conserved positions (>70% identity) are marked with bolded black letters as M, G, G, V, and L. Intermediate conservation (>30% identity) is indicated with black characters corresponding to the most commonly occurring residue. The positions marked as X are the variable positions that are occupied by any particular residue in more than 30% of sequences. As a whole, this figure displays the highly variable characteristics of the human SDR family.
Sequence specificity and interrelationships of the human SDR family
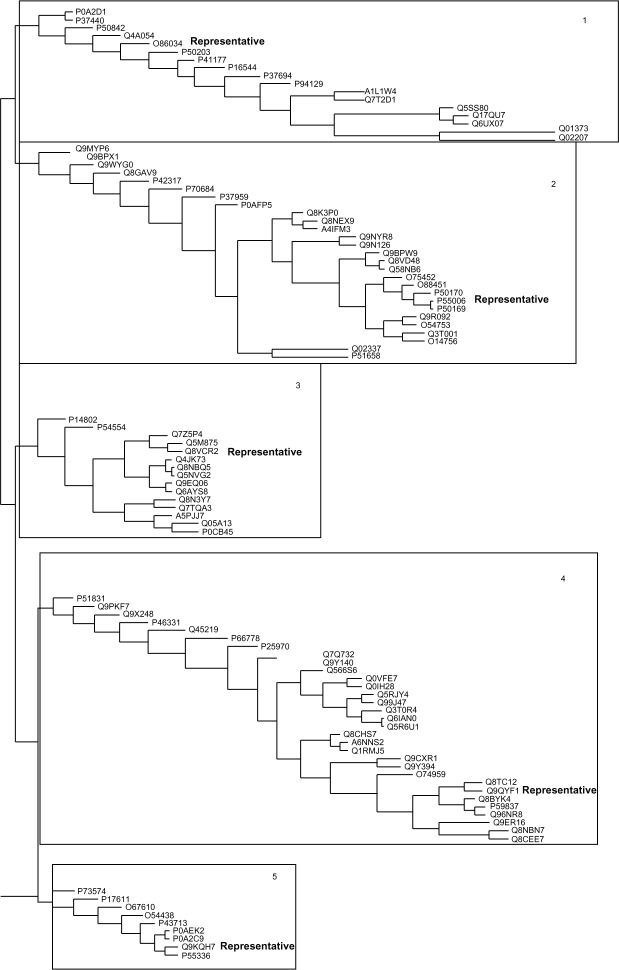
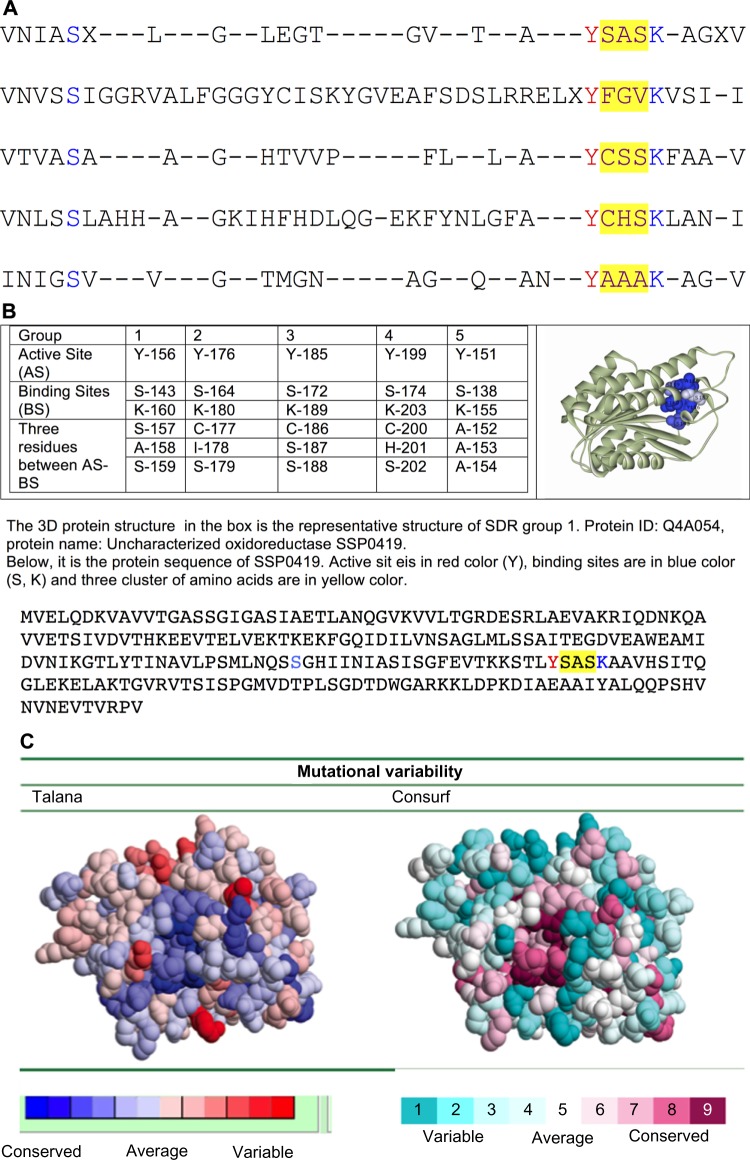
We recovered five distinct subgroups within the human SDR family (Fig. 2). The consensus sequence for each of these five groups is shown in Figure 3A. The positions that form the binding site between substrates and the enzymes (K and S), and the active site (Y) are marked with red letters (Fig. 3A). Based on a comparison of the consensus sequences for each of the five groups of human SDRs family, the binding and active sites typically exhibit highly conserved residues, and occupy the same type of residues in all five groups. In contrast to the highly conserved nature of the active and binding sites, three clusters of amino acid located directly adjacent to the active site and next to one of the binding sites (marked with red letters in Fig. 3A) show substantial variability.
Figure 2.
Phylogenetic tree – a phylogram constructed in SSSSg. The SDRs family can be phylogenetically grouped into five distinct clades. The protein members in each group are considered to share the similar physiochemical properties to respond to the changing environment. Thus, during evolution, these members adopt their own features that are unique compared to other groups.
Figure 3.
Comparison of the five consensus human SDR sequences. (A) The binding sites (K, S) and the active site (Y) of the enzymes are among the characters marked with blue color. These locations were found to be conserved residues and outline the common sequence features within the human SDR family. In contrast, the three clusters of amino acids marked in red (such as SAS, FGV, CSS, CHS, and AAA) indicate the presence of variable residues directly adjacent to the conserved residues. These locations determine the narrow specificity within each group. (B) The location of the conserved and variable residues in the template structure of group 1 of human SDR was identified by Talana. For example, conservative residues included active site and binding sites (Y-156, S-143 and K-160) both of which are located in a conserved region (grade 1 in color scheme of Talana). In contrast, the three clusters of residues (S-157, A-158 and S-159) are clearly located in a more variable region. (C) The identification of functional regions within group 1 using ConSurf and Talana. Group 1 expressed the full grade of coloring scheme in ConSurf: the continuous conservation scores are partitioned into a discrete scale of nine bins for visualization, such that bin 9 contains the most conserved position and bin 1 contains the most variable position. The color grades (1–9) are assigned as follows: the most conserved regions are on the darkest maroon color and the least variable regions are on the lightest turquoise color on the visualization. Similarly, using Talana, group 1 also expressed the full grade of coloring. Grade 1 to grade 12 show the most conserved regions in the darkest blue color to the least variable regions in the lightest rose color on visualization. Therefore, both tools displayed similar results for the identification of functional regions in protein structure.
Mutational variability of human SDRs
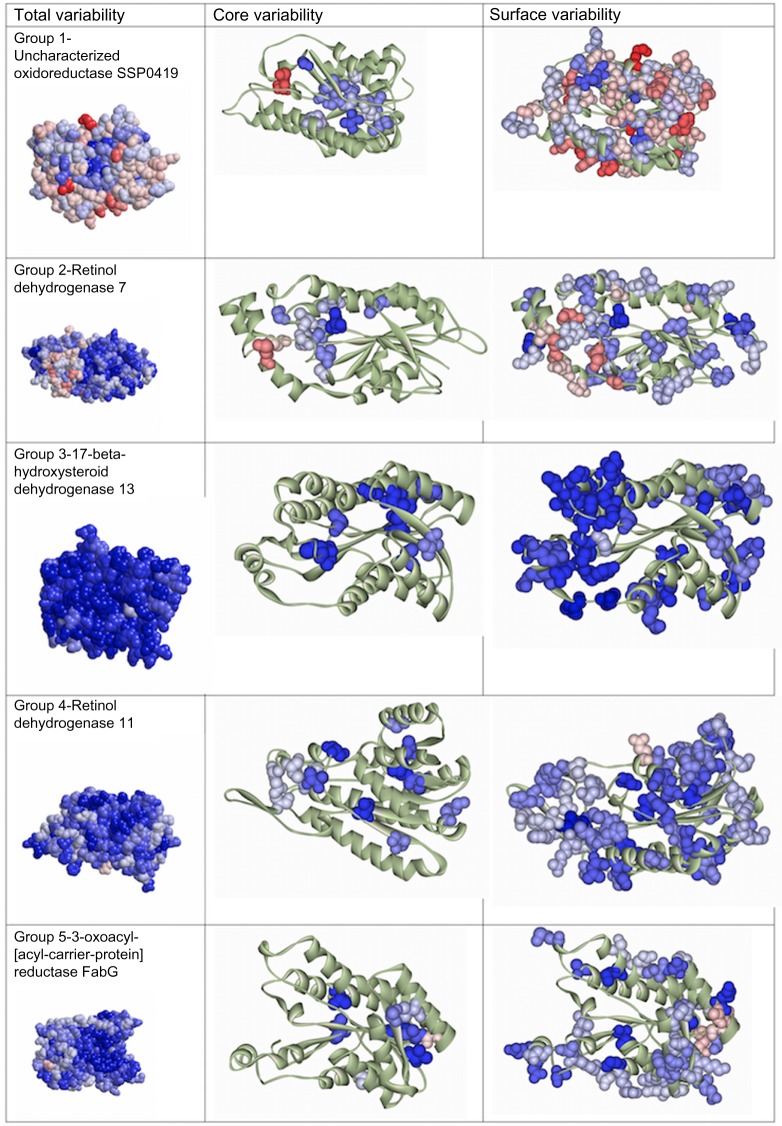
The two programs (Talana and ConSurf) used to analyze the mutation variability of both sequence and structure of the protein templates in each of the five human SDR groups yielded similar results (Fig. 3C). The identification of conservative and variable sequences and structure regions within the human SDR family is presented in Figure 4. The conservative and variable sequences and structures differ not only among the five human SDR groups but also within each group.
Figure 4.
Variability profiles for each of the five groups of human SDRs. Total, core, and surface variability profiles are displayed for each group based on the distribution of residues on the protein structure. Group 3 displayed the most conservative level (grade 1 of the color scheme is dominant in the entire structure) compared to groups 4 and 5. Group 1 showed the most variable level (full grade of color scheme, from grade 1 to grade 12 in the structure), and group 2 showed an intermediate level of variability.
Across the different groups of human SDRs, the protein structure of group 1 contains a mixture of conserved and variable regions with the variable level (full grades in color scheme of Talana) being dominant in the whole structure. In contrast, group 3 displayed the most conservative level (grade 1 in color scheme of Talana is dominant in the protein structure) compared to the others. Group 4 displayed an intermediately conservative level, whereas group 2 displayed an intermediate level of variability (Fig. 4).
In addition, conservative and variable structures were detected within each group. With few exceptions, the conserved residues occurred within active and substrate binding sites, whereas the variable residues (a cluster of three amino acids that are located directly adjacent to the active site and next to one of the binding sites) were found at random locations in the protein structure (Fig. 3B). For example, in group 1, the active site (Y-156) and the binding sites (S-143 and K-160) are found at a conserved region in the protein structure, whereas a cluster of three amino acids (S-157, A-158, S-159) are located at a variable region next to the conserved region in the protein structure (Fig. 3B). Similar patterns exist in each of the other four groups of human SDR, but involve clusters of different amino acids.
Correlated mutations within the human SDR family
Our analyses on correlated mutation within the human SDR protein family using both Corm and Talana revealed similar outcomes. Based on the distribution of mutations mapped onto protein structure, the correlated mutations can be classified into two groups: the core group and surface group. The core group includes all mutations that show core molecular contact (Table 1A) with most mutations located in conserved regions of the protein structures (the core variability in Fig. 4). The surface group includes all mutations that appear on the surface of the protein structure (Table 1B) with most mutations located at variable regions within the protein structure (the surface variability in Fig. 4). Table 2 summarizes the number of observed sets of correlated mutations for group 5 of human SDRs.
Table 1A.
Core and surface residues in five groups of human SDRs identified by Corm.
| Group 1 | V11, I85, I87, M95, T119, H137, I141, V163, V181, T182, S183, I184, G187, A214 |
| Group 2 | I115, V117, G120, V133, V141, M166, G208, V213, L219, S245, M247 |
| Group 3 | V73, V89, V104, V117, A123, G147, I151, V173, C174, I211 |
| Group 4 | I42, V43, L67, G75, M129, K160, A195, T224, V227, T231, S234, Q248, V252 |
| Group 5 | V7, A57, V69, G79, V80, V158, V174, T178, A220 |
Table 1B.
Surface residues in five human SDR groups are identified by Corm.
| Group 1 | E3, Q5, V8, A20, S21, I22, T25, Q29, D39, S41, R42, E45, V46, K48, I50, Q51, N53, Q55, V57, E59, S61, I62, D64, H67, E69, T72, E73, E80, Q84, I87, M95, S98, A99, I100, E102, E109, A110, M111, D113, I116, K117, G118, T119, Y121, S129, N132, H137, I144, E148, V149, T150, L155, S157, A161, V163, I166, Q168, E171, R180, V181, T182, S183, G187, M188, S194, G195, T197, W199, K204, L205, K208, I210, E212, A213, A214, I215, Y216, Q219, Q220, H223, V224, N225, E228, T230, V231, R232, P233 |
| Group 2 | K64, R70, S71, D75, E78, I81, V91, E99, R100, N103, I115, V117, M119, N122, R126, F130, A131, S132, L134, D135, L139, N147, R153, M166, T195, Y196, G208, V213, T214, M216, S220, D221, L223, A230, V234, I237, K241, F242, D244, S245, M247, A249, E251, N255, C257, G259, D266, C275, H276, S282, W285 |
| Group 3 | T35, Q59, R62, V86, V89, N102, D105, Q106, R109, E115, A123, P126, L130, S131, K133, E135, E136, T138, I145, L155, S158, R161, R162, G177, I179, Y181, I183, P184, A201, D204, K208, V219, T226, R232, P235, L237, R244, S245, I247, N248, N253, Y262, N264, I268, K271 |
| Group 4 | Q35, L36, V43, E53, K56, L67, V72, D73, G75, L77, R80, Q83, A84, V85, G87, Q90, F92, K95, A99, D100, T101, K109, D110, H117, M129, S133, A136, H142, H155, K160, E163, L175, H178, L179, R181, I182, H183, H185, E190, F192, A195, L197, H201, K211, K218, S220, T224, Y225, V227, S234, S241, I242, M243, W245, W247, Q248, F251, V252, Q258, Y266, C267, L269 |
| Group 5 | E4, L24, R28, K31, E39, Q43, S46, D47, Y48, G50, A57, T61, N62, P63, K71, A72, T74, G79, M96, S104, I106, E108, M126, K128, Q130, A149, V174, V179, K190, A191, N193, D194, E195, A202, A206, D211, P212, R213, E226, I244 |
Table 2.
Selected correlated mutations in human SDRs identified by Corm. Correlated mutation in group 5 was analyzed by the Talana program, indicating that if a mutation happened at one specific location, it led to mutation in other positions. For example, if mutation occurred at position 6 (I), the other mutations occurred at the same time at positions 61 (D), 73 (EKNR), 78 (ADEP), and 129 (ACFGH).
| REFERENCE POSITION | AA at | SEQUENCE COUNTS | CORRELATED MUTATIONS AND AMINO ACIDS | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| REFERENCE POSITION | ||||||||||||||||||||||
| 6 | I | 5 | 61: D | 73: EKNR | 78: ADEP | 129: ACFGH | ||||||||||||||||
| V | 4 | 61: NS | 73: LQTY | 78: GQR | QSW | |||||||||||||||||
| 70 | E | 5 | 23: EKT | 56: EMV | 71: EHKNQ | 73: KQRY | 129: CFHW | 192: PST | ||||||||||||||
| K | 4 | 23: AL | 56: A | 71: -AT | 73: ELNT | 129: AQQS | 192: NR | |||||||||||||||
| 79 | I | 4 | 30: FTV | 46: AE | 62: EKV | 68: ADLT | 71: EQT | 73: KLNY | 107: DKNQ | 129: AFSW | 201: EQS | |||||||||||
| V | 4 | 30: IK | 46: DG | 62: FP | 68: V | 71: -AKN | 73: ERT | 107: AE | 129: CGHQ | 201: AD | ||||||||||||
| 105 | I | 4 | 3: -E | 27: KR | 38: E | 45: S | 47: Y | 49: G | 60: T | 73: KNRT | 95: M | 125: M | 127: K | 177: TV | 193: D | 197: AT | 201: AS | 205: A | 212: KQR | 219: AV | 225: DE | |
| V | 4 | 3: QT | 27: ADLT | 38: AFGS | 45: ALV | 47: EQT | 49: -EIT | 60: LQS | 73: ELQY | ALV | 125: AILV | 127: -AQ | 177: A | 193: AE | 197: -EQ | 201: DEGQ | 205: LM | 212: DE | 219: GLRS | 225: GILP | ||
| I | 4 | 129: ACFHQ | 177: TV | 193: D | 197: AT | 201: AS | 205: A | 210: DEG | 212: KQR | 219: AV | 225: DE | |||||||||||
| V | 4 | 129: GSW | 177: A | 193: AE | 197: -EQ | 201: DEGQ | 205: LM | 210: QRS | 212: DE | 219: GLRS | 225: GILP | |||||||||||
| 148 | A | 4 | 42: DKQ | 73: LRT | 78: EGR | 107: DE | 129: CHQS | 201: AQ | ||||||||||||||
| T | 4 | 42: AE | 73: EKNQ | 78: ADPQ | 107: ANQ | 129: AFGW | 201: DGS | |||||||||||||||
| 157 | L | 4 | 71: EHKN | 73: QRY | 129: CHW | 189: AR | 192: PS | |||||||||||||||
| V | 5 | 71: -AQT | 73: EKLNT | 129: AFGQS | 189: EKM | 192: NRT | ||||||||||||||||
| 173 | I | 4 | 23: ET | 56: MV | 71: HKNQ | 73: KQR | 78: DEQ | 211: A | ||||||||||||||
| V | 5 | 23: AKL | 56: AE | 71: -AET | 73: ELNTY | 78: AGPR | 211: P | |||||||||||||||
| 194 | D | 4 | 73: ENR | 78: AEP | 103: DN | 129: ACGH | 243: QV | |||||||||||||||
| E | 4 | 73: KLTY | 78: DGR | 103: EFQS | 129: FQSW | 243: FIS | ||||||||||||||||
| 211 | A | 4 | 23: ET | 56: MV | 71: HKNQ | 73: KQR | 78: DEQ | 173: I | ||||||||||||||
| P | 5 | 23: AKL | 56: AE | 71: -AET | 73: ELNTY | 78: AGPR | 173: V | |||||||||||||||
Discussion
Structural and functional variabilities within the human SDR family
In this study, we sought to elucidate the evolution of sequence and structure within the human SDR protein family. Our results indicated that the human SDR protein family possesses a discrete degree of overall sequence conservation (Fig. 1). This proves that evolutionary differentiation has led to the formation of narrow specificity in individual members of the family, rather than preservation in sequence similarity. This conclusion is further supported by the results of our phylogenetic analysis (Fig. 2). Low overall sequence similarity has led to the grouping of the human SDR family into five distinct clusters, with each group potentially further classified into two subgroups (conserved and variable) based on the results of mutational variability and correlated mutation analyses.
Conserved residues are found near the active and binding sites, which are located on the protein structure next to the binding pocket, for instance, Y-156 (active site) and S-143 and K-160 (binding site) in group 1 (Fig. 3B). Furthermore, the results from our mutational variability confirm that the conserved residues are located in the conserved region in the protein structure (such as Y, S, and K of group 1 in Figure 3B). These results complement prior studies on the identification of conserved residues Y, S, and K. According to several previous studies, Y, S, and K residues are considered, together, as a catalytic triad present in the active sites of the majority of human SDR proteins.10 For example, tyrosine (Y) functions as the catalytic base, whereas serine (S) stabilizes the substrate and lysine (K).10 We interpret the presence and location of these conserved residues (Y, S, K) as an evolutionary constraint at the level of sequence and structure that leads to the retention of similar physiochemical characteristics, thus maintaining a given function in the human SDR family. These conserved residues displayed the general specificity that defines common characteristics of the entire human SDRs family.
Variable groups, essentially, occurring only as three particular clusters of amino acids were found to be located directly adjacent to the binding pocket (between the active site and one of the binding sites in Figure 3A). As with the conserved residues, we found that these variable residues occur near a conserved region of the protein structure as well. Additionally, the three clusters of amino acids form a narrow cluster on the binding pocket such as S-157, A-158, and S-159 in group 1 (Fig. 3B). Particularly, we were able to observe several events of serine and alanine transitions via single point mutations, knowing that serine can be encoded with six codons (UCU, UCC, UCA, UCG, AGU, and AGC) while alanine can be encoded with four codons (GCU, GCC, GCA, and GCG). Hence, mutation that leads to the conversion of serine into alanine can be accomplished by replacement of uracil with guanine at the first position of the codon; for instance, serine (UCU) changed to alanine (GCU). Thus, single point mutations could potentially be the mechanism underlying the marked variability of group 1, the least conservative group overall. In contrast, the three clusters of amino acids in group 3 are cysteine–serine–serine, but unlike serine and alanine, cysteine can be encoded with only two codons (UGU and UGC). Although single point mutation could also be the main mechanism for mutation in group 3, cysteine and two serine residues can form a salt bridge, which may increase overall protein stability.22 Hence, group 3 of human SDRs shows the most sequence conservation compared to the others. The analysis of all five different groups of the SDRs family revealed both different and similar features of their sequences and structures. The different features may be related to the adaptive mechanism used to respond to environmental changes during the evolution of human SDRs. Additionally, during particular divergent evolution, adaptive specificity has permitted each family to adapt its own specific targets. In contrast, the similarities are related to the functional conservation used by each group to maintain the metabolic functions (hormone, mediator, and xenobiotic metabolism) of the human SDRs family.23 These similarities and differences in structure and function of the human SDR family are important for the future design of specific inhibitors to target only a particular group within the human SDR protein family.
Correlated mutation analysis
Our analysis on the correlated mutation of each position along the SDR protein sequences shows that particular fragments are highly variable. The surface variability of the SDR protein family indicated that these positions are random in direct contact with each other but maintain contact with conserved positions. For example, according to the results of correlated mutations in group 5 (Table 2), a mutation at position 23 is accompanied by mutation at 70 and other positions. There is no obvious relationship between the positions of correlated mutations and their contact with each other (surface variability in group 5, Fig. 4) because such correlated mutations are generally in positions that are very distant from each other. According to the currently assumed model, positive mutations (ones that improve fitness) do not occur independently. Instead, the occurrence of one mutation depends on other locally occurring mutations. In this way, the nature of correlated mutations reflects the protein–protein interaction and the necessity of preserving the biological activity and structural properties of the molecules.24 Therefore, the correlated mutations revealed in our study provide useful information for further study of complex protein–protein interactions. In previous study, it was hypothesized that protein–protein interactions only happen to proteins in close proximity.25 However, our findings show that such interactions may also occur when proteins are distant from one another. Thus, our findings suggest that the correlated and distant mutations were selectively conserved to maintain protein–protein complex interactions. These interactions may act as a potential adaptive mechanism within the human SDRs family, which allows them to gradually and adaptively change during evolution in response to fluctuating external conditions and functional demands.
Furthermore, we found that, in each human SDRs group, there are core residues that form a narrow correlated cluster on protein structures, and most of them are in a conserved region (core variability, Fig. 4, Table 1A). There is evidence to suggest that these core residues tend to mutate together to maintain proper functioning.26 Our results support the claim that these centralized residues tend to mutate together to preserve the biological function of the SDR proteins. Moreover, the differences in core variability may explain the reason the human SDR family shares a low level of similarity in sequences (15–30%) but not in protein structures. In contrast to core residues, surface residues were found to randomly scatter over the protein structure and were not directly in contact with each other (surface variability, Fig. 4, Table 1B). Interestingly, our results indicated that the surface residues of human SDR proteins do seem to be interacting with one another, despite the distance between them (Table 1B and C).
Table 1C.
Correlated mutation sets include the core and surface residues in group 5.
| GROUP 5 POSITIONS | CORE AND SURFACE RESIDUES CORE | SURFACES | ||
|---|---|---|---|---|
| 6 | Val-7 | Gin-130 Thr-74 |
Gly-179 Asn-62 |
|
| 70 | Asn-193 Thr-74 Lys-71 Leu-24 |
Gin-130 Ala-72 Ala-57 |
||
| 79 | Val-80 | Val-69 | Ala-202 Glu-108 Ala-72 Asp-47 |
Gin-130 Thr-74 Pro-63 Lys-31 |
| 105 | Thr-178 | Ala-220 | Glu-226 Asp-211 Asp-194 Lys-128 He-106 Thr-74 Gly-50 Ser-46 Arg-28 |
Arg-213 Ala-206 Gin-130 Met-126 Met-96 Thr-61 Tyr-48 Glu-39 Glu-4 |
| 148 | Gly-79 | Ala-202 GLn-130 Thr-74 |
Ala-149 Glu-108 Gln-43 |
|
| 157 | Val-158 | Asn-193 Gin-130 Ala-72 |
Lys-190 Thr-174 |
|
| 173 | Val-174 | Ala-57 | Pro-212 Thr-74 Leu-24 |
Gly-79 Ala-72 |
| 194 | Thr-74 Ser-104 Glu-195 |
Gly-79 Gin-130 Ile-244 |
||
Notes: (A) It displays the core residues identified by the Talana program. The residues in each group are located at the core of the protein structure. The occurrences of valine and isoleucine are more frequent compared to other amino acids, showing that these hydrophobic amino acids potentially play a more vital role in stabilizing the chemical structure of the proteins. (B) It displays the surface residues identified using Talana. These residues are located on the surface of protein structures and are distant from each other. (C) It shows the identification of correlated mutation sets and their core and surface characteristics for group 5.
The mechanism of the mutations that cannot be explained by single transition/transversion has not been fully investigated and understood. Here, we suggest a few potential explanations of such phenomenon. One possibility is that we do not know all members of the homologous protein family and those potential unknown members could contain the residues that make the “bridge” enabling the variability occurrence by single transition/transversion. For example, if the corresponding positions in two homologous sequences contain E (GAA) and P (CCA) amino acids, respectively, there could be a third unknown sequence that contain A (GCA) or Q (CAA) amino acid at that position. A second possibility is that the mechanism of variability at these sites is different from a single point mutation. Although single point mutations are major contributors to the acquisition of beneficial mutations through evolution, the correlation of surface mutations does not seem to be adequately explained by the occurrence of single point mutations alone. Using the data presented here as a springboard, further investigation of correlated mutations in distantly located positions may help researchers gain further insight into the causes, prevention, and treatment of diseases caused by genetic or protein structure mutations. The advantage of the correlated mutations analysis with the aid of the Corm program is that it depends neither upon the charge of the correlated residues nor upon their direct interaction. Therefore, it is possible to identify and locate the correlated mutations that are very distant from each other. For this reason, the involvement in salt bridge formations, the charge of the side chain, and the direct contact between resides were not taken into consideration while searching for correlated mutations. In such a way, we avoided the loss of significant and complete data about all the existing correlated mutations. The characteristics of the side chains and the distance between residues were the subject of further detail analysis after the correlations were identified. However, the involvement of the correlated positions in salt bridge formation and stabilization was not the subject of our study.
The correlated mutations that occur in homologous proteins are potential rich sources of information about the biological activity of the protein, about the structure–function relationship, as well as about interaction mechanisms with other proteins. For example, identifying mutational correlated clusters enables the discovery of the regions that are responsible for the unknown activity. They also help explain the mechanism of the action of drugs directed at specific proteins (eg, viral proteins).27
The availability of original software generated by authors and coworkers
The original applications such as Consensus Constructor, Talana, Corm, and SSSSg are freely available at the addresses described above. Also, they are available directly upon request sent to the authors. Additionally, the authors are willing to assist in appropriate, effective running of all these applications in case of any problems.
Supplementary Files
Supplementary Figure 1. Multiple sequence alignment of 75 human SDR sequences by Geisha 3.0 (support for Fig. 1).
Supplementary Figure 2. Enzyme names correspond to enzyme ID (support to Fig. 2).
Acknowledgments
The authors recognize the contribution and support provided in terms of software design for sequence analysis by Dr Jacek Leluk.
Footnotes
Author Contributions
NNT designed the study, implemented the methods, performed the experiments, analyzed the results, and drafted the manuscript. LL conceived the study, participated in its design and coordination, and helped to draft the manuscript. Both authors read and approved the final manuscript.
ACADEMIC EDITOR: Jike Cui, Associate Editor
FUNDING: This work was supported by the Institute of Computational Science and Technology of Ho Chi Minh City, Vietnam, under grant number 260/TB-SKHCN. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties.
REFERENCES
- 1.Persson B, Kallberg Y. Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs) Chem Biol Interact. 2013;202(1–3):111–5. doi: 10.1016/j.cbi.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Kallberg Y, Oppermann U, Persson B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models. FEBS J. 2010;277(10):2375–86. doi: 10.1111/j.1742-4658.2010.07656.x. [DOI] [PubMed] [Google Scholar]
- 3.Kavanagh KL, Jörnvall H, Persson B, Oppermann U. Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65(24):3895–906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moeller G, Adamski J, Moeller G, Adamski J. Multifunctionality of 17beta-hydroxysteroid dehydrogenases: an update. Mol Cell Endocrin. 2008;65(24):3895–906. [Google Scholar]
- 5.Keller B, Volkmann A, Wilckens T, Moeller G, Adamski J. Bioinformatic identification and characterization of new members of short-chain dehydrogenase/reductase superfamily. Mol Cell Endocrinol. 2006;248(1–2):56–60. doi: 10.1016/j.mce.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 6.Jörnvall H, Höög JO, Persson B. SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 1999;445(2–3):261–4. doi: 10.1016/s0014-5793(99)00130-1. [DOI] [PubMed] [Google Scholar]
- 7.Kallberg Y, Oppermann U, Jörnvall H, Persson B. Short-chain dehydrogenases/reductases (SDRs) Eur J Biochem. 2002;269(18):4409–17. doi: 10.1046/j.1432-1033.2002.03130.x. [DOI] [PubMed] [Google Scholar]
- 8.Jörnvall H, Persson B, Krook M, et al. Short-chain dehydrogenases/reductases (SDRs) Biochemistry. 1995;34:6003–13. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 9.Oppermann U, Filling C, Hult M, et al. Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem Biol Interact. 2003;143–4:247–53. doi: 10.1016/s0009-2797(02)00164-3. [DOI] [PubMed] [Google Scholar]
- 10.Filling C, Berndt KD, Benach J, et al. Critical residues for structures and catalysis in SDRs. J Biol Chem. 2002;277:25677–84. doi: 10.1074/jbc.M202160200. [DOI] [PubMed] [Google Scholar]
- 11.Persson B, Kallberg Y, Oppermann U, Jörnvall H. Coenzyme-based functional assignments of short-chain dehydrogenases/reductases (SDRs) Chem Biol Interact. 2003;143–4:271–8. doi: 10.1016/s0009-2797(02)00223-5. [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Lukacik P, Kavanagh KL, Oppermann U. SDR-type human hydroxysteroid dehydrogenases involved in steroid hormone activation. Mol Cell Endocrinol. 2007;265–6:71–6. doi: 10.1016/j.mce.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Thompson J, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notredame C, Higgins DG, Heringa J. T-COFFEE: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–13. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 15.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughout. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lassmann T, Sonnhammer ELL. KALIGN: an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leluk J, Hanus-Lorenz B, Sikorski AF. Application of genetic semihomology algorithm to theoretical studies on various protein families. Acta Biochim Pol. 2001;48(1):21–33. [PubMed] [Google Scholar]
- 18.Leluk J. A non-statistical approach to protein mutational variability. Biosystems. 2000;56:83–93. doi: 10.1016/s0303-2647(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 19.Leluk J. Regularities on mutational variability in selected protein families and the Markovian model of amino acids replacements. Comput Chem. 2000;24:659–72. doi: 10.1016/s0097-8485(00)00070-x. [DOI] [PubMed] [Google Scholar]
- 20.Leluk J. A new algorithm for analysis of the homology in protein primary structure. Comput Chem. 1998;22:123–31. doi: 10.1016/s0097-8485(97)00035-1. [DOI] [PubMed] [Google Scholar]
- 21.Göbel U, Sander C, Schneider R, Valencia A. Correlated mutations and residue contacts in proteins. Proteins. 2004;18(4):309–17. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- 22.Hall BG. Spontaneous point mutations that occur more often when advantageous than when neutral. Genetics. 1990;126:5–16. doi: 10.1093/genetics/126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishikawas S, Adivinata J, Morioka H, et al. A thermoresistant mutant of Ribonuclease T1 having three disulfide bonds. Protein Eng. 1990;3:443–8. doi: 10.1093/protein/3.5.443. [DOI] [PubMed] [Google Scholar]
- 24.Oppermann UC, Filling C, Jörnvall H. Forms and functions of human SDR enzymes. Chem Biol Interact. 2001;(1–3):130–132. 699–705. doi: 10.1016/s0009-2797(00)00301-x. [DOI] [PubMed] [Google Scholar]
- 25.Waugh DF. Protein-protein interaction. Adv Protein Chem. 1954;9:325–437. doi: 10.1016/s0065-3233(08)60210-7. [DOI] [PubMed] [Google Scholar]
- 26.Pazos F, Helmer-Citterich M, Ausiello G, Valencia A. Correlated mutations contain information about protein–protein interaction. J Mol Biol. 1997;271(4):511–23. doi: 10.1006/jmbi.1997.1198. [DOI] [PubMed] [Google Scholar]
- 27.Le L, Leluk J. Study on phylogenetic relationships, variability, and correlated mutations in M2 proteins of influenza virus A. PLoS One. 2011;6(8):e22970. doi: 10.1371/journal.pone.0022970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Multiple sequence alignment of 75 human SDR sequences by Geisha 3.0 (support for Fig. 1).
Supplementary Figure 2. Enzyme names correspond to enzyme ID (support to Fig. 2).