Abstract
Glioblastoma (GBM) is the malignant form of glioma, and the interplay of different pathways working in concert in GBM development and progression needs to be fully understood. Wnt signaling and sonic hedgehog (SHH) signaling pathways, having basic similarities, are among the major pathways aberrantly activated in GBM, and hence, need to be targeted. It becomes imperative, therefore, to explore the functioning of these pathways in context of each other in GBM. An integrative approach may help provide new biological insights, as well as solve the problem of identifying common drug targets for simultaneous targeting of these pathways. The beauty of this approach is that it can recapitulate several known facts, as well as decipher new emerging patterns, identifying those targets that could be missed when relying on one type of data at a time. This approach can be easily extended to other systems to discover key patterns in the functioning of signaling molecules. Studies were designed to assess the relationship between significant differential expression of genes of the Wnt (Wnt/β-catenin canonical and Wnt non-canonical) and SHH signaling pathways and their connectivity patterns in interaction and signaling networks. Further, the aim was to decipher underlying mechanistic patterns that may be involved in a more specific way and to generate a ranked list of genes that can be used as markers or drug targets. These studies predict that Wnt pathway plays a relatively more pro-active role than the SHH pathway in GBM. Further, CTNNB1, CSNK1A1, and Gli2 proteins may act as key drug targets common to these pathways. While CTNNB1 is a widely studied molecule in the context of GBM, the likely roles of CSNK1A1 and Gli2 are found to be relatively novel. It is surmised that Gli2 may be antagonistic to CSNK1A1, preventing the phosphorylation of CTNNB1 and SMO proteins in Wnt and SHH signaling pathway, respectively, by CSNK1A1, and thereby, aberrant activation. New insights into the possible behavior of these pathway molecules relative to each other in GBM reveal some key interesting patterns.
Keywords: glioblastoma; Wnt and SHH signaling pathways; integrative analysis; gene expression; protein–protein interaction networks; bottleneck nodes; simultaneous targeting; β-catenin (CTNNB1); casein kinase 1, alpha 1 (CSNK1A1); glioma-associated oncogene 2 (Gli2) proteins
Introduction
Glioblastoma (GBM) is the most malignant of all the brain tumors with very low median survival time of one year, as per Central Brain Tumor Registry of the United States, 2001. Several groups have initiated high-throughput molecular profiling studies toward a better understanding of GBM.1–5 GBMs, also known as grade IV gliomas, are a heterogeneous bunch of tumors arising from astroglial cells and sometimes from oligodendrocytes and are characterized by four distinct molecular subtypes on the basis of gene expression, copy number changes, and DNA sequence alterations, viz., neural, proneural, mesenchymal, and classical subtypes.5 The neural subtype is defined by the presence of neuron markers such as NEFL and SLC12A5, whereas the proneural subtype is characterized by the expression of proneural development genes such as SOX, DLL3, OLIG2, and TCF4, as well as high levels of expression of PDGFRA and p53 mutations. Mesenchymal subtype is characterized by high-level expression of genes in NF-κB pathway, as well as tumor necrosis factor (TNF) superfamily pathway, with mutations in NF1 and PTEN tumor suppressor genes. High-level EGFR amplification with high-level expression of genes of Notch pathway, sonic hedgehog (SHH) pathway, and NES gene, and absence of p53 mutations define the classical subtype of GBM.
Among the major pathways studied in GBM tumors, aberrant activation of Wnt/β-catenin signaling pathway, as well as SHH signaling pathway has been reported.6,7 The aberrant activation of these pathways is one of the many mechanisms that lead to cellular migration, proliferation, and enhanced survival of tumor cells. Further, these two pathways are also involved in the maintenance, proliferation, and clonogenicity of glioma cancer stem cells.8 These cancer stem cells have a role to play in the initiation, proliferation, and invasion in gliomas, and therefore, can be one of the several important points of therapeutic intervention.
In normal cells, these pathways are involved in vertebrate organogenesis, morphogenesis, and other developmental roles. Several similarities between these pathways during their signal transduction events can be identified9 such as activation through a G-protein-coupled receptors (GPCRs)-related membrane protein and prevention of phosphorylation-dependent proteolysis of β-catenin (CTNNB1) effector. This effector molecule helps activate target genes through conversion of a repressor protein (TCF) into an activator protein. Several other roads and milestones in these two pathways are pathway specific.9
Studies have found an overexpression of Wnt ligands of canonical pathway, Wnt1 and Wnt3a, in high-grade gliomas.10 Non-canonical Wnt signaling pathway ligand, Wnt5a, was also found to be involved in tumor progression.11 Another study observed an overexpression of Wnt5a and Wnt7b, as well as Frizzled proteins Fzd-2, -6, and -7 in glioma cells.12 In the case of SHH pathway, expression of SHH pathway genes such as PTCH, SMO, Gli1, and Gli2 was observed in CD133-positive malignant glioma cells, and this pathway was found to be playing an important role in cellular migration of these cells.13
Keeping in view the similarities as well as the differences between these pathways and their likely co-ordinated role in GBM tumor progression, there arises a need to explore their contextual functioning in more detail, particularly the genes’ behavior in relation to each other. Further, it will be useful to discern a specific molecule or set of molecules common to these pathways that can serve as potential drug target/s so that these pathways can be targeted simultaneously. These drug targets can, more often, be “bottlenecks” in a pathway,14 ie, the bottleneck genes/gene products which connect two or more pathways together and therefore are more likely, essential genes/gene products. One of the approaches may, thus, involve cohesive integration of both gene expression data and different types of networks involving these genes or their products. Using this approach, gene/s with a potential as attractive drug target candidates that are usually overlooked when relying on differential gene expression analysis or protein–protein interaction (PPI) networks alone can be enumerated as a ranked list. This approach will also be useful in providing more insights into their behavior in context.
Materials and Methods
Dataset assembly
From The Cancer Genome Atlas (TCGA) website, level 3 normalized and processed gene expression dataset for 49 genes coding for ligands, receptors, co-receptors, destruction complex, transcriptional effectors, antagonists, downstream targets, tumor suppressors, and apoptotic genes involved in SHH, as well as Wnt/β-catenin canonical and non-canonical Wnt signaling pathways (Table 1) was compiled. In all, data belonging to a total of 431 GBM and 10 normal tissue samples were downloaded. The microarray platform used was Affymetrix HT_HG-U133A platform and the GBM samples were primary GBM samples.
Table 1.
Wnt and SHH signaling pathway genes used in this study categorized as ligands, receptors, co-receptors, destruction complex, transcriptional effectors, antagonists, downstream targets, tumor suppressors, and apoptotic genes.
| WNT PATHWAY: | |||
|---|---|---|---|
| PATHWAY COMPONENTS | ENTREZ GENE ID | GENE SYMBOL | GENE NAMES |
| Ligands | 7471 | WNT1 | Wingless-Int1 |
| 7474 | WNT5 A | Wingless-Int5A | |
| 7482 | WNT2B | Wingless-Int2B | |
| Receptors | 2535 | FZD2 | Frizzled2 |
| 7855 | FZD5 | Frizzled5 | |
| 7976 | FZD3 | Frizzled3 | |
| 8321 | FZD1 | Frizzled1 | |
| 8322 | FZD4 | Frizzled4 | |
| 8323 | FZD6 | Frizzled6 | |
| 8324 | FZD7 | Frizzled7 | |
| 8325 | FZD8 | Frizzled8 | |
| 8326 | FZD9 | Frizzled9 | |
| 11211 | FZD10 | Frizzled10 | |
| Co-receptors | 4040 | LRP6 | Low Density Lipoprotein Receptor-related Proteins-6 |
| 4041 | LRP5 | Low Density Lipoprotein Receptor-related Proteins-5 | |
| Transcriptional Activators | 1499 | CTNNB1 | Beta-Catenin |
| β-catenin destruction complex | 2932 | GSK3β | Glycogen synthase kinase 3 β |
| 324 | APC | Adenomatous polyposis coli | |
| 8312 | AXIN1 | Axin | |
| 1452 | CSNK1A1 | Casein kinase 1, alpha 1 | |
| Effectors | 6932 | TCF7 | Transcription factor 7 (T cell specific, HMG box) |
| 6934 | TCF7L2 | Transcription factor 7-like 2 (T-cell specific, HMG-box) | |
| 83439 | TCF7L1 | Transcription factor 7-like 1 (T-cell specific, HMG-box) | |
| 51176 | LEF1 | Lymphoid enhancer-binding factor 1 | |
| 1855 | DVL1 | Dishevelled-1 | |
| 1856 | DVL2 | Dishevelled-2 | |
| 1857 | DVL3 | Dishevelled-3 | |
| Canonical pathway activators | 10023 | FRAT1 | Frequently rearranged in advanced T-cell lymphomas-1 |
| 23401 | FRAT2 | Frequently rearranged in advanced T-cell lymphomas-2 | |
| Wnt antagonists | 22943 | DKK1 | Dickkopf1 |
| 6422 | SFRP1 | Secreted Frizzled-related protein 1 | |
| Downstream targets | 595 | CCND1 | Cyclin D1 |
| 652 | BMP4 | Bone morphogenetic protein 4 | |
| 891 | CCNB1 | Cyclin B1 | |
| 894 | CCND2 | Cyclin D2 | |
| 999 | CDH1 | E-cadherin | |
| 4609 | c-MYC | V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | |
| SHH PATHWAY: | |||
| PATHWAY COMPONENTS | ENTREZ GENE ID | GENE SYMBOL | GENE NAMES |
| Ligand | 6469 | SHH | Sonic Hedgehog |
| Receptors | 5727 | PTCH1 | Patched-1 |
| 8643 | PTCH2 | Patched-2 | |
| Transcriptional Activators | 2735 | GLI1 | GLI Family Zinc Finger 1 |
| 2736 | GLI2 | GLI Family Zinc Finger 2 | |
| Destruction complex | 2932 | GSK3β | Glycogen synthase kinase 3 β |
| 1452 | CSNK1A1 | Casein kinase 1, alpha 1 | |
| Effectors/Downstream targets | 2735 | GLI1 | GLI Family Zinc Finger 1 |
| 2736 | GLI2 | GLI Family Zinc Finger 2 | |
| 2737 | GLI3 | GLI Family Zinc Finger 3 | |
| 6608 | SMO | Smoothened | |
| 595 | CCND1 | Cyclin D1 | |
| 652 | BMP4 | Bone morphogenetic protein 4 | |
| 891 | CCNB1 | Cyclin B1 | |
| 894 | CCND2 | Cyclin D2 | |
| 3714 | JAG2 | Jagged 2 | |
| 4609 | c-MYC | V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | |
| 6422 | SFRP1 | Secreted Frizzled-related protein 1 | |
| Apoptotic gene | 355 | FAS | Fas Cell Surface Death Receptor |
| PATHWAY COMPONENTS | ENTREZ GENE ID | GENE SYMBOL | GENE NAMES |
| Tumor suppressors | 999 | CDH1 | E-cadherin |
| 5728 | PTEN | Phosphatase And Tensin Homolog | |
| 6598 | SMARCB1 | SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin Subfamily B Member 1 | |
| 6615 | SNAI1 | Snail Family Zinc Finger 1 | |
Significant differential gene expression analysis
Significant differential gene expression was analyzed using both the significance analysis of microarrays (SAM) and T-test modules of MultiExperiment Viewer (MeV) version 4.6. Two different statistical tests were used in order to enhance confidence in predictions of significantly differentially expressed genes. A default p-value cutoff of 0.01 was used to assess significant differential expression using T-test. Differential gene expression was considered significant if false discovery rate was <0.05 and delta-value was 1.0 using 1000 permutations in SAM, and this cutoff was used in order to enlist a majority of significantly differentially expressed genes, as well as biologically meaningful relationships. Comparative marker selection analysis with default parameters from GenePattern suite of tools was used to assess upregulation or downregulation of these genes.
Network assembly
In order to gain a comprehensive understanding, several types of networks such as PPI, co-expression, co-localization networks, and pathways were constructed using GeneMania plugin installed in Cytoscape version 3.0. GeneMania uses several quality data sources to assemble validated networks such as GEO for co-expression network, BioGrid for physical interaction (PPI) networks, PathwayCommons for pathways network, among others. In brief, the interaction/association dataset for the organism Homo sapiens was installed locally from GeneMania plugin and in combination with Cytoscape 3.0, used for the assembly of a new network for further studies. In all, this installed dataset comprised 144 networks with 21,438 genes (nodes) and thousands of interactions (edges) among these genes. From this dataset, new networks were assembled for all the genes or gene products included in the dataset under study (Table 1). Some neighboring genes, which were not part of dataset in Table 1, were added automatically by GeneMania plugin, with the top 20 related genes chosen to be added to the network with automatic weighting (default) option.
Network analysis
Network analysis for node degree, color-coded expression values, and betweenness centrality, among others, was done using Network Analyzer plugin, VistaClara, and VizMapper in Cytoscape 2.8. First neighbors of selected nodes to determine directly connected nodes were analyzed using Select menu of Cytoscape 2.8.
Node degree in a network denotes number of edges/connections in an undirected interaction network, ie, how much connected a node is to all other nodes in the network. More densely connected nodes have a higher node degree and are considered as “hub” molecules. The gene expression values, when superimposed on a network, provide a way to visualize network in terms of nodes corresponding to significantly differentially expressed genes. A green–red color gradient denotes lower–higher expression values for gene products of significantly differentially expressed genes, and green-yellow-red color gradient denotes nodes with lower to higher node degree. VistaClara plugin was used to superimpose the gene expression values on the network, and VizMapper was used to generate the color gradient. Betweenness is an important topological property of a network that defines the number of shortest paths that are non-redundant going through a particular node. Since these nodes tend to be critical points, these can be thought of as bottleneck nodes without which the information flow would be virtually impossible. Higher the betweenness, more essential and critical the molecule is likely to be. Depending upon “hubness” (node degree) and “betweenness,” the bottleneck nodes are classified as (a) hub–non-bottlenecks; (b) non-hub–non-bottlenecks; (c) non-hub–bottlenecks; and (d) hub–bottlenecks. The nodes in the network have been colored using a green-red color gradient for assessing their lower–higher betweenness centrality, using Network Analyzer to calculate the betweenness centrality and VizMapper to color the nodes according to this measure.
Results and Discussion
Majority of genes encoding ligands, receptors, co-receptors, regulators, and transcriptional effectors among others involved in SHH, as well as Wnt/β-catenin canonical and Wnt non-canonical signaling pathways are upregulated and significantly differentially expressed in GBM
Wnt/β-catenin and SHH pathway genes are aberrantly activated in GBM. Upregulation of some of these pathway genes has been reported in literature as mentioned earlier. Genes in these signaling pathways functioning as ligands, receptors, co-receptors, destruction complex, transcriptional effectors, antagonists, downstream targets, tumor suppressors, and apoptotic genes (Table 1) were studied for their expression and interaction patterns. In all, a total of 49 genes were analyzed, and on the basis of comparative marker selection analysis results, 28 genes were found to be upregulated and 9 genes downregulated in GBM (Table 2). SAM and T-test analyses both pointed to a majority of genes being significantly differentially expressed. Out of a total of 37 significantly differentially expressed genes that were enlisted using SAM and T-tests, 33 genes were observed to be significantly differentially expressed by both these tests, and three genes were found to be so by either of these. The significant differential expression is analyzed in the context of both tumor and normal tissues. Their respective q-values in percent, which is the likelihood of a false positive case, at FDR value set at <0.05 or <5% and p-values set at 0.01, are given in Table 2. It is seen from this table that q-values and p-values for all of the genes listed, except one, fall within the given cutoff. Some genes with significant differential expression may be upregulated in tumors and some may be upregulated in normal tissues (downregulated in tumors), as detailed below.
Table 2.
Significantly differentially expressed genes upregulated in tumors, false discovery rate or q-value <0.05 or <5% (likelihood of a false positive case), and delta-value 1.0 were used in SAM analyses and p-value cutoff of 0.01 was used for T-test.
| S. NO. | GENES | Q-VALUE(%) | P-VALUE |
|---|---|---|---|
| 1. | WNT5A | 0.0 | 0.0 |
| 2. | CSNK1A1 | 0.0 | 0.0 |
| 3. | FZD7 | 0.0 | 7.79E-14 |
| 4. | FZD6 | 0.0 | 0 |
| 5. | CCNB1 | 0.0 | 5.48E-10 |
| 6. | LRP5 | 0.0 | 0.0 |
| 7. | FZD1 | 0.0 | 5.46E-10 |
| 8. | TCF7L1 | 0.0 | 1.71E-07 |
| 9. | c-MYC | 0.0 | 1.73E-06 |
| 10. | FZD2 | 0.0 | 1.61E-06 |
| 11. | FAS | 0.0 | 2.27E-05 |
| 12. | DVL3 | 0.0 | 1.38E-06 |
| 13. | DVL2 | 0.0 | 1.32E-05 |
| 14. | CTNNB1 | 0.0 | 9.83E-06 |
| 15. | LEF1 | 0.0 | 1.57E-05 |
| 16. | CCND1 | 0.0 | 1.46E-05 |
| 17. | TCF7L2 | 0.0 | 5.02E-06 |
| 18. | DKK1 | 0.9 | 7.18E-04 |
| 19. | FZD5 | 0.0 | 3.50E-05 |
| 20. | SMARCB1 | 0.0 | 0.001261 |
| 21. | GLI2 | 3.4 | 4.03E-05 |
| 22. | TCF7 | 3.4 | 2.18E-04 |
| 23. | LRP6 | 0.0 | 4.94E-07 |
| 24. | FZD4 | 3.4 | 5.31E-05 |
| 25. | FZD10 | 0.0 | 1.87E-05 |
| 26. | AXIN1 | 1.0 | * |
| 27. | SMO | NaN** | * |
| 28. | CDH1 | NaN | 9.22E-04 |
| 1. | WNT1 | 0.95 | * |
| 2. | FZD9 | 0.0 | 0.004177 |
| 3. | GSK3β | 0.0 | 0.005612 |
| 4. | SFRP1 | 1.0 | 0.001744 |
| 5. | PTCH2 | 0.0 | 0.001241 |
| 6. | WNT2B | 0.0 | 5.56E-05 |
| 7. | DVL1 | 0.0 | 1.06E-05 |
| 8. | JAG2 | 0.0 | 8.05E-06 |
| 9. | APC | 0.0 | 5.15E-12 |
Notes:
Not significant.
Differential expression in Figure 1. NaN: q-value not calculated.
Significantly differentially expressed genes upregulated in normal tissue samples, false discovery rate or q-value <0.05 or <5% (likelihood of a false positive case) and delta-value 1.0 were used in SAM analyses and p-value cutoff of 0.01 was used for T-test.
Significant differential expression of members of SHH signaling pathways
Genes such as CSNK1A1, PTCH2, GSK3β, and Gli2 were found to be significantly differentially expressed, whereas SHH as well as Gli1, Gli3, and PTCH1 genes were not significantly differentially expressed. Of these, CSNK1A1 and Gli2 were found to be upregulated in tumors. Low-level expression of SHH ligand in tumors is unexpected since it may be needed for the SHH signaling pathway to proceed. However, several studies have also reported a low-level expression of SHH in tumors.15,16 Braun et al.15 found in their studies that there was no correlation between Hh activity and the levels of SHH, Gli1, and PTCH1 mRNA expression in tumor cells derived from GBM and that there was very low overall expression of SHH. Bar et al.16 reported SHH activity in some, as opposed to all, primary GBM tumors and speculated that “the SHH mRNA we detected in primary glioma samples was being generated by non-neoplastic cells and that pure tumor cultures are therefore negative.” Ehtesham et al.17 also mention similar results that SHH pathway is activated in Grade II and III gliomas, but not in Grade IV de novo GBM tumors. Taken together, this may be interpreted to mean that the Hh pathway in GBM may progress via a ligand other than SHH or in a ligand-independent manner. Further, ligand-independent function may occur due to loss-of-function mutation in PTCH or gain-of-function mutation in SMO, as mentioned in several studies.
Verhaak et al.5 using TCGA dataset in their analyses mentioned that “Sonic hedgehog (SMO, GAS1, GLI2) signaling pathways were highly expressed in the Classical subtype,” similar to studies in this current paper. Interestingly, there was no mention of SHH ligand expression in the paper by Verhaak et al.
Significant differential expression of members of Wnt signaling pathways and other genes implicated in the signaling process
Majority of members of Wnt signaling pathways were significantly differentially expressed, as well as upregulated in tumors in contrast to relatively few members of SHH signaling pathway. This shows that in comparison to SHH signaling, Wnt signaling mechanisms are more pro-active in GBM tumors. In brief, significantly differentially expressed genes such as CTNNB1, CSNK1A1, Frizzled receptors, LRP5, LRP6, TCF7L1, TCF7L2, and LEF1, among others, were upregulated in tumors. Among significantly differentially expressed Wnt ligands, non-canonical signaling molecule, Wnt5a, was found to be upregulated and canonical signaling molecules such as Wnt1 and Wnt2b downregulated in tumors. In fact, significant differential expression was highest in the case of two molecules: Wnt5a and CCNB1. These results are consistent with another study,18 where non-canonical Wnt signaling molecule Wnt5a was found upregulated in GBM, whereas canonical Wnt signaling molecules such as Wnt1 were not regulated as compared to normal brain. CCNB1 is known to contribute to cellular proliferation, lending it an important role in GBM progression. The non-canonical Wnt5a signaling pathway is a CTNNB1-independent pathway, but may also activate Wnt/CTNNB1 canonical signaling in the presence of Fzd4 and LRP5.19 The fact that Fzd4 and LRP5 are significantly differentially expressed as well as upregulated in tumors along with Wnt5a in the current study lends credence to the theory that Wnt5a may be activating the canonical pathway in GBM as well.
Other significantly differentially expressed genes found to be upregulated in tumors were SMARCB1 and FAS cell surface death receptor genes. This is interesting given the fact that SMARCB1 acts as a tumor suppressor gene in malignant rhabdoid tumors, and given its function, should be down-regulated in tumors, but its role in GBM is not fully studied. However, many tumor suppressor genes such as p16INK4a have been found to be overexpressed in a wide variety of tumors20 and may provide evidence, in part, that the upregulation of SMARCB1 in GBM observed in the current study may be related to GBM development, and therefore, needs further exploration. It is surmised that the upregulation of FAS cell surface death receptor gene, which leads to apoptosis, is circumvented, in part, by the upregulation of Wnt signaling proteins, mainly by Wnt5A, which has been shown to drive apoptosis resistance in pancreatic cancer cells.21 SHH signaling may also play a role.22
SFRP1, JAG2, GSK3β, and APC genes were found significantly upregulated in normal tissues. SFRP1 is a putative tumor suppressor gene and an antagonist of Wnt non-canonical signaling and JAG2 is a Notch ligand, both proteins being HH signaling targets. Their significant differential upregulation in normal tissue samples provides further evidence that hedgehog pathway is less active than Wnt pathway in GBM. DKK1, an antagonist of Wnt canonical signaling pathway, is upregulated in tumors and may inhibit this pathway, although Wnt5a molecule may serve to overcome this activity as has been explained above. GSK3β and APC are parts of CTNNB1 destruction complex, their downregulation in tumor cells may lead to loss of activity of destruction complex and hence, stabilization of CTNNB1, which functions as transcriptional co-activator of TCF/LEF family of transcription factors.
CSNK1A1 and Gli2 are the novel targets identified through an integration of gene expression data and network connectivity patterns
Several groups have used PPI networks to understand the patterns of connectivity between genes or gene products. Information on key gene/s or gene product/s acting as “hub” molecules with a high degree of connectivity, and which are distinct from their neighboring genes in gene expression patterns, can be used to leverage their potential as attractive drug target/s. To identify key gene products common to both pathways that can be targeted simultaneously and to minimize the chances of important genes being overlooked when relying on single type of analyses, significant differential gene expression analyses and network connectivity patterns were integrated together.
PPI network
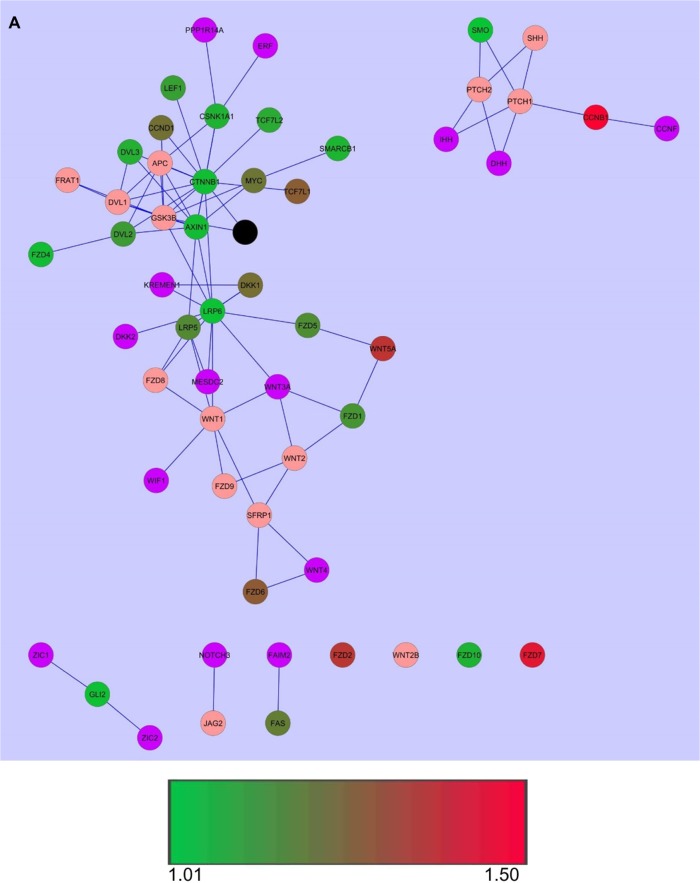
PPI networks were overlaid with gene expression data to further generate an underlying pattern (Fig. 1A and B). It was observed that CTNNB1 and CSNK1A1 were the top two genes/gene products that were both significantly differentially expressed and highly connected with the number of nodes (gene/gene product) being 40 and 26, respectively. It was further seen that for most of the other gene products, the number of connections to other nodes, ie, node degree was not related to the expression level. As an example, Wnt5a, which has highest level of significant differential expression and is upregulated in tumors, has lower node degree as compared to LRP6, which has comparatively lower level of expression but a higher node degree (Fig. 1C).
Figure 1.
PPI networks overlaid with gene expression data. (A) PPI networks were overlaid with gene expression data for each gene in tumors. (B) PPI networks were overlaid with gene expression data for each gene in normal tissues. Significantly differentially expressed nodes are colored based on expression values. (C) Nodes in PPI network sized and colored according to node degree distribution, bigger size of a node corresponds to higher node degree, while the color gradient from green to yellow to red denotes lower to higher node degrees.
Notes: The color gradient from green to red denotes lower to higher expression values, black node: CDH1, purple-colored nodes: proteins encoded by genes not present in the gene list under study, but automatically added as neighboring proteins in PPI network, pink-colored nodes: under-expressed or not expressed in a particular type of sample.
Further, the proteins encoded by genes that are not significantly differentially expressed, showed some degree of connectivity to those proteins that were encoded by significantly differentially expressed genes. As an example, Fzd8, which was not significantly differentially expressed was found connected to LRP5, LRP6, and Wnt1, all of which are significantly differentially expressed, each with its own interacting partners encoded by significantly differentially expressed genes. This suggests that Fzd8 may be playing an important role, in spite of its differential expression not being significant.
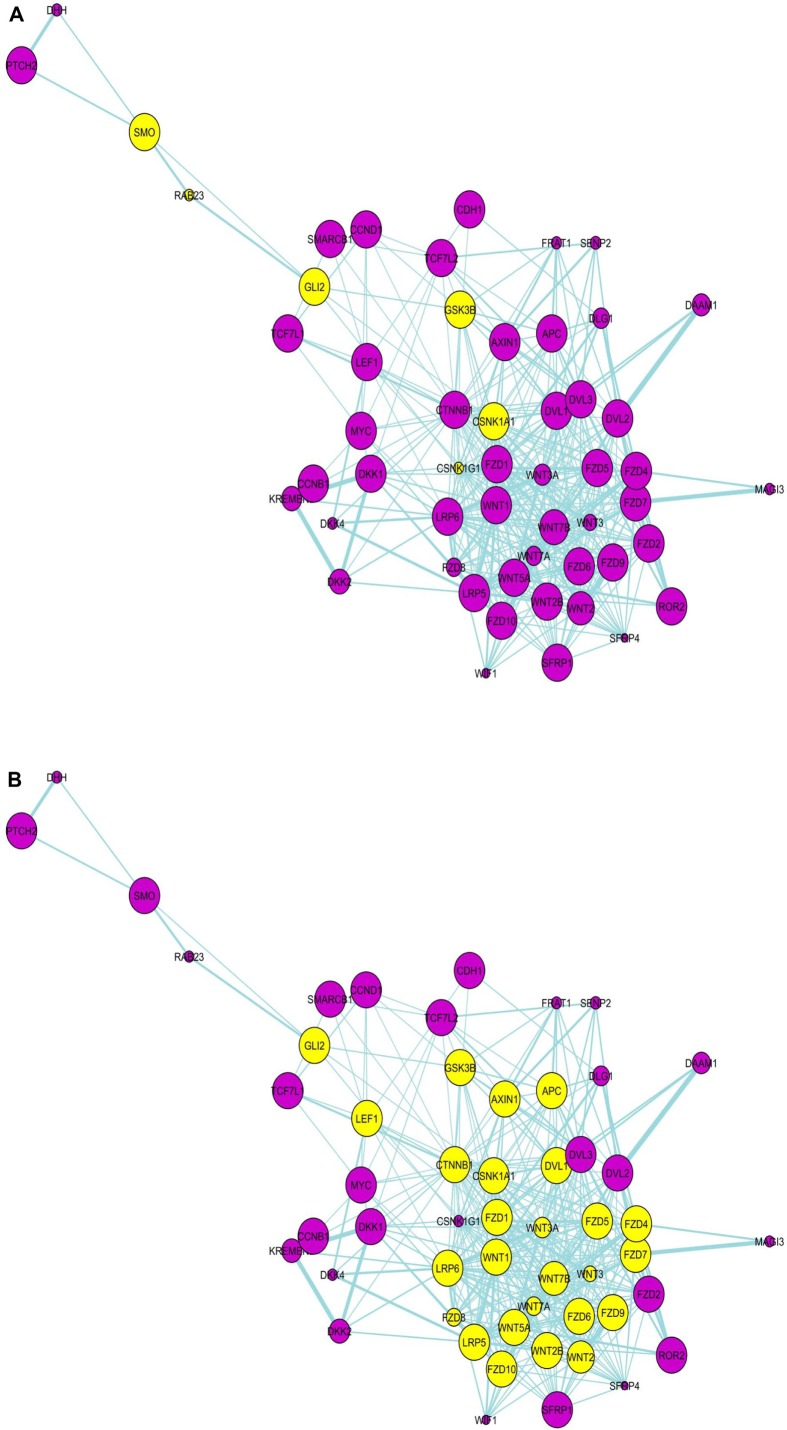
Pathway network
In the pathway network, the Wnt and SHH pathway components are seen as clear separate modules with a few molecules linking these two modules/pathways. A significant finding in the pathway network (Fig. 2A–C) is that glioma-associated oncogene 2 (Gli2) is one of the molecules connecting the two pathways together, and appears to be the central connector. It is connected to SMO, RAB23 in SHH pathway and to GSK3β, CSNK1A1 and CSNK1G1 in the Wnt pathway (Fig. 2a). While Gli2 is a primary regulator in SHH signaling and mainly functions as a transcriptional activator, it also regulates Wnt genes involved in morphogenesis. Even though it is not yet found to be involved in PPI with other molecules in the gene list in the present paper as seen in PPI network (Fig. 1), due to its significant differential expression in GBM coupled with the fact that it acts as a bridge connecting SHH and Wnt signaling in the pathways network, it is highly probable that it plays a central role. In fact, Gli2, by dint of being a connector molecule may play a much bigger role than is generally anticipated and studies should be directed toward expanding its functional role. PubMed search as on January 30, 2014 indicates that there are a total of just 613 studies covering Gli2 molecule, even as its name implies its possible crucial role in glioma development.
Figure 2.
Pathway network involving the Wnt- and SHH pathway molecules. Gli2 appears as the connector molecule of Wnt- and SHH pathway in this network, connected to CSNK1A1 and others in Wnt pathway network, and SMO and others in SHH pathway network. Yellow-colored nodes are the first neighbors (directly connected) of (a) Gli2, (b) CSNK1A1, and (c) CTNNB1.
As such, this study points out that Gli2 upregulation may be correlated with GBM progression. Since Gli2 degradation occurs via GSK3β-dependent phosphorylation and ubiquitination, increasing the activity of GSK3β may be one potential mechanism of therapy. What is more conclusive is that, GSK3β is found upregulated in normal tissues and not in tumors, hence Gli2 is not degraded in tumors, and so, may play a pro-active role in GBM tumor development.
Another molecule that appears to connect the two pathways is CSNK1A1 (Fig. 2B), and is in focus because of its significant differential expression and high node degree in PPI network overlaid with gene expression data from tumors (Fig. 1a and c). It is connected to both Gli2 and CTNNB1 in pathway network. CSNK1A1 phosphorylates CTNNB1 in Wnt pathway and SMO in SHH pathway, thereby inactivating these proteins. The mechanism by which CTNNB1 and SMO proteins are prevented from inactivation or remain activated in the presence of high levels of CSNK1A1 in GBM tumors is a matter of further experimental investigation. However, the emerging patterns in this study point to a possible antagonistic role of Gli2 in this mechanism as is explained in “Insights from key emerging patterns” section.
The gene or protein expression levels of CTNNB1, CSNK1A1, and Gli2 have been reported as prognostic and predictor factors in several types of tumors. CTNNB1 and Gli1 are found to serve as prognostic markers in GBM.23 Significant correlation was observed between high β-catenin (CTNNB1) activity and poor prognosis of the patients, and this was considered as “a strong and independent prognostic factor in breast cancer.”24 CTNNB1 has also been found to serve as a useful prognostic marker in non-small cell lung cancer and gastric cancer25,26 and in pair with CSNK1E, a prognostic marker in colorectal cancer.27
CSNK1A1 has been reported to be overexpressed at both mRNA and protein levels in melanoma cells as compared to normal cells leading to the proposition that it can serve as a useful diagnostic marker.28 High Gli2 protein expression level in hepatocellular carcinoma (HCC) was found to be associated with poor prognosis in HCC patients after hepatectomy29 and in the case of intrahepatic cholangiocellular carcinoma (ICC) was found to be associated with unfavorable overall survival prognosis.30 The gene expression (mRNA expression) level of Gli2 was found to be a negative prognostic factor in acute myeloid leukemia (AML).31
Observed among the immediate neighbors of Wnt5a in the pathway network are Fzd4 and LRP5. In the presence of these components, Wnt5a is able to activate the canonical Wnt/β-catenin pathway19 and may be functioning in the same manner in GBM.
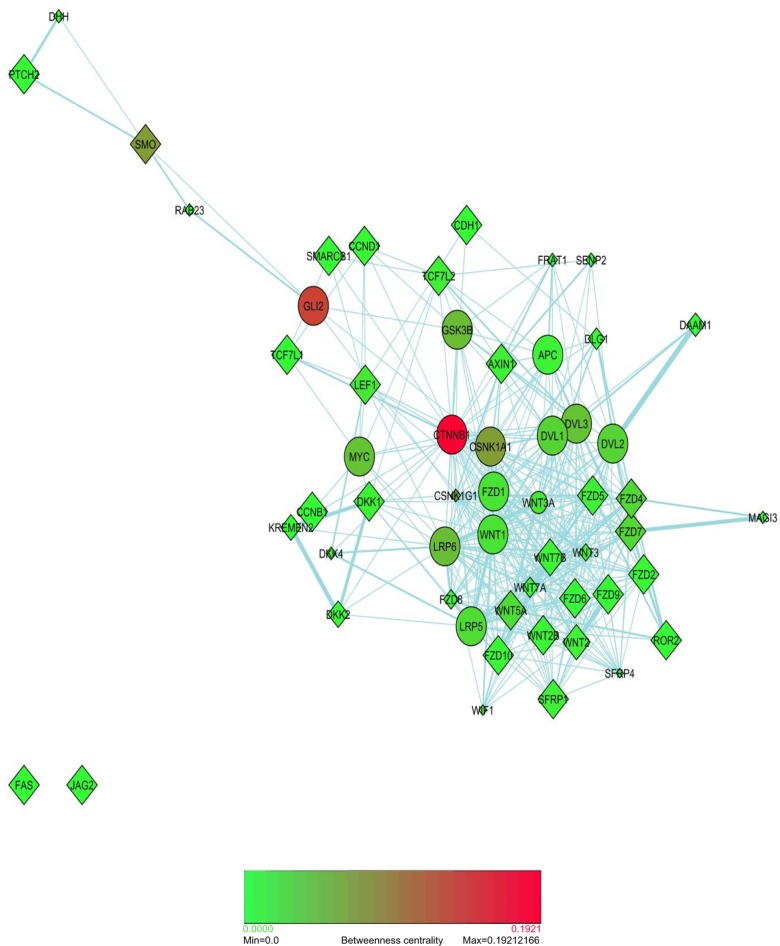
Bottleneck nodes
Betweenness centrality measure is a more significant indicator that a gene/gene product is essential to the proper functioning of a pathway network. This is measured in terms of those network nodes that have many shortest paths going through them, and the nodes with higher betweenness centrality are termed as “bottleneck” nodes.14 Bottleneck nodes are key connector nodes in a network. As an example, a transcription factor regulating several target genes may function as a bottleneck node in a regulatory network. A key protein/s that can co-ordinate two or more signal transduction pathways is another example of a bottleneck.
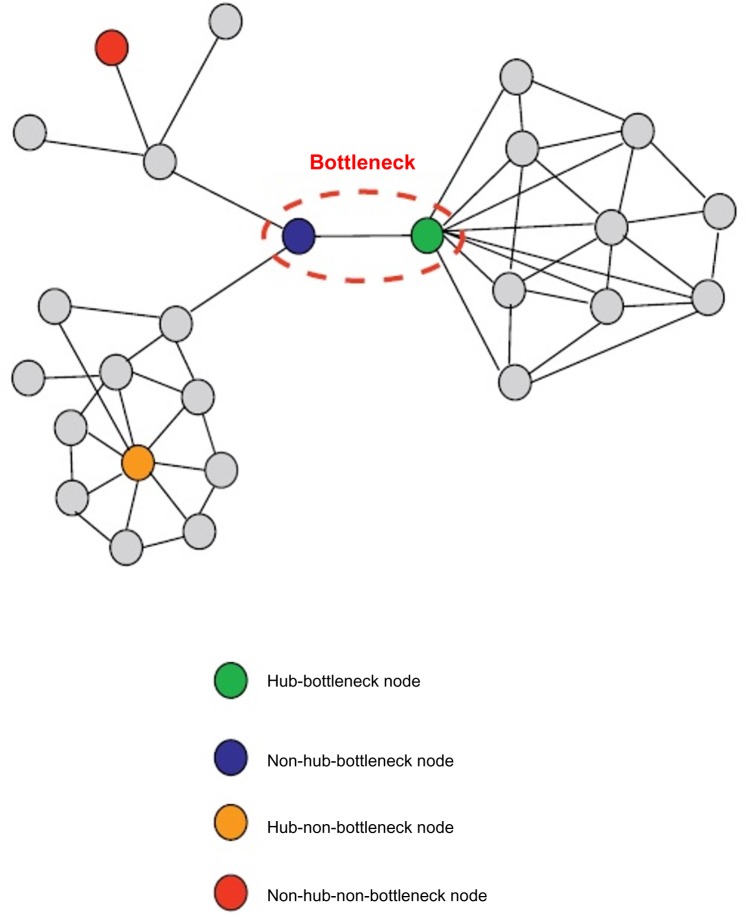
Using the convention based on Figure 3 for classification of nodes depending upon “hubness” and “betweenness,” it was observed that CTNNB1 and CSNK1A1 fit perfectly as hub–bottleneck nodes, and Gli2 as non-hub–bottleneck node connecting the two major pathways in this study (Fig. 4). The node with the highest betweenness centrality in Figure 4 is, obviously, CTNNB1. The bottleneck proteins have been found to be essential proteins in both interaction and regulatory networks with high significance.14 CTNNB1 and CSNK1A1 are well documented to be essential proteins in regulating Wnt and SHH pathways. Non-hub–bottlenecks that are involved in signal transduction pathways are also surmised to be products of essential genes. In this respect, Gli2 as a non-hub bottleneck node may be a gene essential to the overall functioning and cross-talk between these two major pathways. Connecting major pathways together, bottleneck proteins are in a state of dynamic flux for most of the time. Therefore, these are usually significantly co-expressed to a lesser degree with their neighbors and have fewer binding partners than most other nodes in the network, as is observed in the case of Gli2 in co-expression network (data not shown) and Figure 1a, respectively. The “Insights from key emerging patterns” section details the important roles these three proteins can play as potential therapeutic drug targets.
Figure 3.
Schematic depiction of bottleneck nodes. Reproduced with permission from Ref. 14.
Figure 4.
Bottleneck nodes discovered in this study. Nodes in pathway network are colored by betweenness centrality measure.
Notes: The color gradient from green to red denotes lower to higher betweenness centrality, and nodes with higher betweenness centrality are the bottleneck nodes.
Insights from key emerging patterns
Combining and integrating all of the above analyses, the picture is becoming clearer. Wnt pathway has emerged as a relatively stronger contender for involvement in the development and progression of GBM as compared to SHH pathway. SHH pathway, through the upregulation and connectivity of some of its gene/gene products to molecules in Wnt pathway, may be playing a helper role in GBM development, at those stages where Wnt pathway might face the roadblocks of inactivation or regulated activation. Even though SHH ligand is not found to be significantly differentially expressed, this pathway is able to survive in GBM. Most likely, this is not due to a ligand-independent aberrant activation, but by some other mechanism involving Wnt pathway molecules in view of SHH pathway playing a helper role, thus creating interdependencies amongst the two. Wnt5a molecule may be the major player in the aberrant activation of both Wnt canonical and non-canonical pathways. Further, in the PPI network, those genes that are not significantly differentially expressed, but are surrounded by genes that are significantly differentially expressed may also be disease associated. An example here is Fzd8, which does not appear to be significantly differentially expressed in this study, but nevertheless, may be playing an active role in GBM development solely due to its connectivity to significantly differentially expressed proteins such as LRP5, LRP6, and Wnt1. Bottleneck proteins in a network that connect different functional clusters are more likely to be product of essential genes,14 which when targeted can lead to the inactivation of all the linked clusters simultaneously. These proteins need not have a high node degree, ie, linked individually to most of the other nodes. In this respect, CSNK1A1, Gli2, and CTNNB1 are prominent in the role of a bottleneck, and therefore, may function as strong drug targets. CSNK1A1, by virtue of it being connected to both Gli2 and CTNNB1, may be a stronger target. In order to serve as a target, it would need to be overexpressed, leading to phosphorylation of CTNNB1 and SMO and subsequent inactivation of the two pathways; this activation, instead of inhibition, of a kinase molecule may present a novel approach in GBM therapy. Indeed, a FDA-approved small-molecule activator of casein kinase 1 alpha, pyrvinium, when used to treat colon cancer cells with mutation in APC or CTNNB1 gene, inhibited both Wnt signaling and proliferation.32
To the best of knowledge till date, the interplay between CSNK1A1 and Gli2 molecule is not explored and so, the effect of CSNK1A1 overexpression on Gli2 molecule is open to experimental investigation. While it is entirely possible that Gli2 molecule may also be phosphorylated, leading to its inactivation, it is more likely that Gli2 molecule may act as an antagonist of CSNK1A1. In its antagonistic role, it may diminish the effect of CSNK1A1 on CTNNB1 and SMO, and thereby aberrant activation of these pathways. This may be the reason that despite CSNK1A1 being significantly differentially expressed and upregulated in tumors, Wnt and SHH pathways still proceed as seen from the greater expression of majority of genes in tumors.
GBMs are developing resistance to temozolomide (TMZ) chemotherapy, the main treatment regimen in combination with surgery and radiotherapy. This occurs, in part, due to self-renewal capacity of glioma stem cells. Hh/Gli1 signaling axis controls the behavior of glioma stem cells,33 and inhibition of SHH pathway with cyclopamine has been shown to increase the efficacy of TMZ in CD133(+) glioma stem cells.34 Using Gli2 inhibitor Gant61, or a CTNNB1 inhibitor such as PNU74654 or BC21, or CSNK1A1 activator, pyrvinium, the same approach can be applied to increase the efficacy of TMZ in GBM therapy.
Keeping into account all of these analyses, a schematic model is proposed for the interdependent nature of the two pathways providing us with a new biological insight open to experimentation, as well as a way for simultaneous targeting in GBM (Fig. 5).
Figure 5.
A schematic model of Wnt- and SHH pathways working interdependently in GBM based upon observations in this study. As observed from PPI network and betweenness centrality measures, CSNK1A1 molecule is directly connected to both Gli2 in SHH pathway and CTNNB1 in Wnt pathway, all these three molecules having high betweenness centrality. These are considered as plausible drug targets based on this study and denoted as diamond-shaped nodes. CSNK1A1 is indirectly connected to SMO in SHH pathway. The arrows indicate that the overexpression of CSNK1A1 leads to phosphorylation of CTNNB1 and SMO (indicated by “P” in the nodes), thereby inactivating these two pathways, for which evidence is present in literature. However, the cross-talk between CSNK1A1 and Gli2 is not available to the best of knowledge, and therefore, needs to be studied further. It is surmised that since Wnt and SHH pathways appear to be aberrantly activated in GBMs in this study, despite upregulation and significant differential gene expression of CSNK1A1 in tumors, Gli2 molecule may simply be acting as an antagonist of CSNK1A1. It may diminish the effect of CSNK1A1 on CTNNB1 and SMO, or inhibit CSNK1A1 altogether, leading to aberrant activation of these pathways.
Conclusions
Using the mRNA expression patterns of Wnt and SHH pathway genes from TCGA dataset for GBM tumors integrated with interaction networks, several significantly differentially expressed and highly connected genes in the network were identified. The present studies point to the potential major role of CTNNB1, CSNK1A1, and Gli2 in both Wnt and SHH pathways aberrantly activated in GBM. Further, this integrative analysis suggests these molecules as potential therapeutic drug targets to inhibit/inactivate these pathways simultaneously. While CTNNB1 has been studied extensively as a therapeutic target, CSNK1A1 and Gli2 are found to be relatively novel and to the best of the knowledge of this author, not discovered in the context of GBM before. The interplay between CSNK1A1 and Gli2 needs to be discerned, and hence, more studies should be directed toward this end. It is speculated from the patterns derived from this study that CSNK1A1 may be antagonized by Gli2, leading to aberrant activation of Wnt and SHH signalling pathways. In their respective capacities as potential druggable targets, CTNNB1 and Gli2 need to be inhibited while CSNK1A1 requires itself to be activated. The drug-dependent activation of a kinase molecule is uncommon, and therefore, paves the avenue for novel approaches toward drug design in GBM tumors.
Acknowledgments
The author is grateful to Dr. Anjan Misra, SJHMC, Phoenix, Arizona, for providing the publicly available dataset down-loaded from TCGA, and to Prof. Subrata Sinha, National Brain Research Center, New Delhi, India for his initial guidance in another work.
Footnotes
Author Contributions
Conceived and designed the experiments: SM. Analyzed the data: SM. Wrote the first draft of the manuscript: SM. Made critical revisions: SM. The author reviewed and approved of the final manuscript.
ACADEMIC EDITOR: JT Efird, Editor in Chief
FUNDING: Authors disclose no funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties.
REFERENCES
- 1.Dunn GP, Rinne ML, Wykosky J, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–84. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Mercier M, Hastir D, Moles Lopez X, et al. A simplified approach for the molecular classification of glioblastomas. PLoS One. 2012;7(9):e45475. doi: 10.1371/journal.pone.0045475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zarkoob H, Taube JH, Singh SK, Mani SA, Kohandel M. Investigating the link between molecular subtypes of glioblastoma, epithelial-mesenchymal transition, and CD133 cell surface protein. PLoS One. 2013;8(5):e64169. doi: 10.1371/journal.pone.0064169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Mayhani MT, Grenfell R, Narita M, et al. NG2 expression in glioblastoma identifies an actively proliferating population with an aggressive molecular signature. Neuro Oncol. 2011;13(8):830–45. doi: 10.1093/neuonc/nor088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;19:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8(5):387–98. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 7.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10:319–31. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 8.Huang Z, Cheng L, Guryanova OA, Wu Q, Bao S. Cancer stem cells in glioblastoma – molecular signaling and therapeutic targeting. Protein Cell. 2010;1(7):638–55. doi: 10.1007/s13238-010-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalderon D. Similarities between the Hedgehog and Wnt signaling pathways. Trends Cell Biol. 2002;12:523–531. doi: 10.1016/s0962-8924(02)02388-7. [DOI] [PubMed] [Google Scholar]
- 10.Kaur N, Chettiar S, Rathod S, et al. Wnt3a mediated activation of Wnt/β-catenin signaling promotes tumor progression in glioblastoma. Mol Cell Neurosci. 2013;54:44–57. doi: 10.1016/j.mcn.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Augustin I, Goidts V, Bongers A, et al. The Wnt secretion protein Evi/Gpr177 promotes glioma tumourigenesis. EMBO Mol Med. 2012;4(1):38–51. doi: 10.1002/emmm.201100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamino M, Kishida M, Kibe T, et al. Wnt-5a signaling is correlated with infiltrative activity in human glioma by inducing cellular migration and MMP–2. Cancer Sci. 2011;102:540–8. doi: 10.1111/j.1349-7006.2010.01815.x. [DOI] [PubMed] [Google Scholar]
- 13.Uchida H, Arita K, Yunoue S, et al. Role of sonic hedgehog signaling in migration of cell lines established from CD133-positive malignant glioma cells. J Neurooncol. 2011;104:697–704. doi: 10.1007/s11060-011-0552-2. [DOI] [PubMed] [Google Scholar]
- 14.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3(4):e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun S, Opperman H, Mueller A, et al. Hedgehog signaling in glioblastoma multiforme. Cancer Biol Ther. 2012;13(7):487–95. doi: 10.4161/cbt.19591. [DOI] [PubMed] [Google Scholar]
- 16.Bar EE, Chaudhry A, Lin A, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25(10):2524–33. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehtesham M, Sarangi A, Valadez JG, et al. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene. 2007;26(39):5752–61. doi: 10.1038/sj.onc.1210359. [DOI] [PubMed] [Google Scholar]
- 18.Reis M, Czupalla CJ, Ziegler N, et al. Endothelial Wnt/β-catenin signaling inhibits glioma angiogenesis and normalizes tumor blood vessels by inducing PDGF-B expression. J Exp Med. 2012;209:1611–27. doi: 10.1084/jem.20111580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits β-catenin–TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Poi MJ, Tsai MD. Regulatory mechanisms of tumor suppressor P16INK4A and their relevance to cancer. Biochemistry. 2011;50:5566–82. doi: 10.1021/bi200642e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griesmann H, Ripka S, Pralle M, et al. WNT5 A-NFAT signaling mediates resistance to apoptosis in pancreatic cancer. Neoplasia. 2013;15:11–22. doi: 10.1593/neo.121312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morton JP, Lewis BC. SHH signaling and pancreatic cancer: implications for therapy? Cell Cycle. 2007;6(13):1553–7. doi: 10.4161/cc.6.13.4467. [DOI] [PubMed] [Google Scholar]
- 23.Rossi M, Magnoni L, Miracco C, et al. β-catenin and Gli1 are prognostic markers in glioblastoma. Cancer Biol Ther. 2011;11(8):753–61. doi: 10.4161/cbt.11.8.14894. [DOI] [PubMed] [Google Scholar]
- 24.Lin SY, Xia W, Wang JC, et al. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97(8):4262–6. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woenckhaus M, Merk J, Stoehr R, et al. Prognostic value of FHIT, CTNNB1, and MUC1 expression in non-small cell lung cancer. Hum Pathol. 2008;39(1):126–36. doi: 10.1016/j.humpath.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 26.Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–7. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiong KL, Chang KC, Yeh KT, et al. CSNK1E/CTNNB1 are synthetic lethal to TP53 in colorectal cancer and are markers for prognosis. Neoplasia. 2014;16:441–50. doi: 10.1016/j.neo.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun D, Zhou M, Kowolik CM, et al. Differential expression patterns of capping protein, protein phosphatase 1 and casein kinase 1 may serve as diagnostic markers for malignant melanoma. Melanoma Res. 2011;21(4):335–43. doi: 10.1097/CMR.0b013e328346b715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D, Cao L, Li Y, Lu H, Yang X, Xue P. Expression of glioma-associated oncogene 2 (Gli 2) is correlated with poor prognosis in patients with hepatocellular carcinoma undergoing hepatectomy. World J Surg Oncol. 2013;11:25. doi: 10.1186/1477-7819-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang L, Tan YX, Jiang BG, et al. The prognostic significance and therapeutic potential of hedgehog signaling in intrahepatic cholangiocellular carcinoma. Clin Cancer Res. 2013;19(8):2014–24. doi: 10.1158/1078-0432.CCR-12-0349. [DOI] [PubMed] [Google Scholar]
- 31.Wellbrock J, Koehler JM, Wagner K, et al. Expression of hedgehog pathway mediator Gli2 represents a clinically negative prognostic marker in acute myeloid leukemia and its inhibitor GANT61 exerts anti-leukemic effects in vitro. Blood. 2013;122(21) doi: 10.1158/1078-0432.CCR-14-1059. [DOI] [PubMed] [Google Scholar]
- 32.Thorne CA, Hanson AJ, Schneider J, et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α. Nat Chem Biol. 2010;6(11):829–36. doi: 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santoni M, Burattini L, Nabissi M, et al. Essential role of Gli proteins in glioblastoma multiforme. Curr Protein Pept Sci. 2013;14(2):133–40. doi: 10.2174/1389203711314020005. [DOI] [PubMed] [Google Scholar]
- 34.Ulasov IV, Nandi S, Dey M, Sonabend AM, Lesniak MS. Inhibition of Sonic hedgehog and Notch pathways enhances sensitivity of CD133(+) glioma stem cells to temozolomide therapy. Mol Med. 2011;17(1–2):103–12. doi: 10.2119/molmed.2010.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]