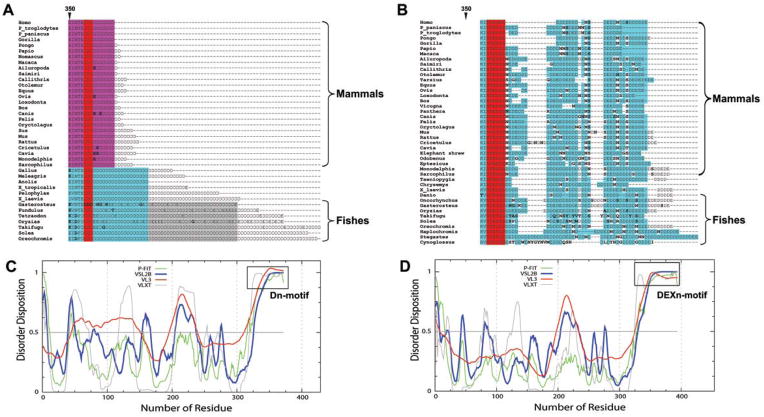
Figure 1. The C-terminal tail is distinctly different between CASQ isoforms.
Multiple sequence alignment of the C-terminal residues of various CASQ sequences from all the species. Alignment of the whole protein is presented in Supplemental Figure 1. (A) The CASQ1 tail possesses only negative residues, mostly Asp and rarely Glu. Among mammalian species (shown in pink), the conservation is very high, only very rarely Glu is found and referred to as Dn-motif. The length of the Dn-motif decreased with evolution as fishes have the longest, mammals have the shortest and amphibians and reptiles have intermediate length. (B) The CASQ2 has longer tail that has non-negative residues like Asn, Gly, and Ser repeating after every few negatively-charged residues, Asp and Glu; therefore designated as DEXn-motif. Red indicates total conservation of the residue and cyan indicates conservative substitution of the residue. Unique residues that are not conserved are shown in bold font. The probability of disordered region dispersion in rat CASQ1 (C) and CASQ2 (D) sequences. All the prediction programs unanimously show that the C-terminus (Dn and DEXn motifs) of CASQ isoforms are intrinsically disordered regions, while the domain I, II and III regions of either isoform do not show high probability of intrinsically disorderedness. CASQ sequences from other organisms also show similar disordered pattern.