Abstract
Picoinjection is a promising technique to add reagents into pre-formed emulsion droplets on chip; however, it is sensitive to pressure fluctuation, making stable operation of the picoinjector challenging. We present a chip architecture using a simple pressure stabilizer for consistent and highly reproducible picoinjection in multi-step biochemical assays with droplets. Incorporation of the stabilizer immediately upstream of a picoinjector or a combination of injectors greatly reduces pressure fluctuations enabling reproducible and effective picoinjection in systems where the pressure varies actively during operation. We demonstrate the effectiveness of the pressure stabilizer for an integrated platform for on-demand encapsulation of bacterial cells followed by picoinjection of reagents for lysing the encapsulated cells. The pressure stabilizer was also used for picoinjection of multiple displacement amplification (MDA) reagents to achieve genomic DNA amplification of lysed bacterial cells.
Introduction
In droplet microfluidics, water-in-oil droplets provide subnanoliter volume reaction vessels that enable secure containment of aqueous reagents in an inert continuous oil phase [1-3]. Droplet stability is essential to ensure both the isolated reaction environment and the fluidic operation of droplets, and a high degree of stability is achieved by the use of proper surfactants [4-5]. However, many biochemical assays require multiple steps where different reagents are added into droplets at defined times to modulate reactions within droplets. The addition of reagents to stabilized droplets is extremely challenging without an external perturbation such as a focused laser [6-7] or electric field [8-9]. Most droplet coalescence techniques are incapable of merging more than two droplets at a time and tuning the desired concentrations of multiple reagents simultaneously [8-11]. Picoinjection, a technique developed recently, allows direct injection of a reagent into a droplet from an injection channel under pressure using an electric field to locally disrupt the surfactant layer in the vicinity of the injection channel [12-14]. Direct dispensing into surfactant-stabilized droplets offers great possibilities since multi-step reactions can be done by sequential picoinjections. Despite its simplicity, picoinjection is extremely sensitive to internal pressure fluctuations and usually requires long stabilization time to balance the flow in each injection channel with the main flow channel, and this process is especially challenging for multiple injection systems. A small fluctuation in pressure within the channels may disturb the pressure balance between the picoinjection channel and the main channel, making picoinjection ineffective. While the picoinjection strategy can work well for the case of generation of a steady stream of droplets in a passive manner where there is little pressure fluctuation, it does not work well in many less steady instances. One example is on-demand droplet systems that rely on periodic pressure-driven droplet generation, which inherently produces pressure fluctuations [15]. Another example is situations where droplets in the channel generate significant additional flow resistance as they flow with the continuous phase [16]. This droplet resistance effect becomes nontrivial when the size of droplets is much larger than the channel dimension or when a channel is locally overpopulated with droplets. A third example is when a droplet system has active components to control flow of droplets where channel pressures can change considerably during operation [17].
Here, we present a pressure stabilizer module for reproducible picoinjection to facilitate multi-step bioassay applications with droplets. We show that by effectively dissipating internal pressure in the channel, the pressure stabilizer greatly improves the functionality and controllability of picoinjectors. This unique feature of system stabilization is particularly appropriate for on-demand droplet systems, since the pressure stabilizer mitigates pressure fluctuations introduced by actuation and enables the flowing droplets to make a good contact with the injection fluid for reproducible picoinjection. We demonstrate the power of this pressure stabilization technique by using a picoinjector to perform droplet-based DNA amplification of lysed cells using multiple displacement amplification (MDA). A multiple picoinjection system with a single pressure stabilizer is also demonstrated, which will simplify bioassays that require two or more discrete reagent addition steps at different locations within the device.
Materials and Methods
Device Fabrication
The microfluidic droplet processors were built with poly(dimethylsiloxane) (PDMS) by conventional soft lithography methods [18]. Rectangular flow channels were designed to have a width of 70 μm and a depth of 30 μm. The master mold with SU-8 25 (MicroChem) patterns was generated by UV exposure (Karl Suss MA/BA-6 aligner) followed by development in SU-8 developer (MicroChem). For improved mold recovery, the master molds were silanized with (tridecafluoro-1,1,2,2-tetrahydrooctyl)-1-trichlorosilane (UCT) for 15 minutes. PDMS replicas were molded using a 10:1 (w/w) mixture of RTV615A (Momentive) and curing agent, and cured at 75°C for 2 hours. Inlet and outlet holes were punched using a 0.75 mm-diameter Uni-Core biopsy punch. The PDMS replicas were irreversible bonded with a glass slide using oxygen plasma. Electrodes were constructed in empty channels by capillary wicking of low-temperature indium solder (AIM Specialty Materials) heated at 150°C. The flow channel surfaces were treated with a commercial fluorinated surface coating agent (PicoGlide®, Dolomite). Plastic collection chips were made using a laser cutter VLS2.30 (Versa Laser). Channels and chambers on collection chips were patterned in 75 μm thick Mylar films with pressure sensitive adhesive (PSA) (3M) film sandwiched by two poly(methyl methacrylate) (PMMA) layers (1.5 mm thick) (McMaster-Carr) that contain inlets and outlets.
Device Operation
Fluorinated oil (HFE-7500, 3M) containing 2% (w/w) surfactant (PicoSurf®, Dolomite) was used for the continuous phase. Its interfacial tension with water was estimated to be ~5 mN/m for calculations [11]. All aqueous and oil flows were driven by pressurizing off-chip reservoirs using 0-15 psi scalable pressure modulators (Pneutronics); fluid in reservoirs was delivered to chip inlets via polytetrafluoroethylene [PTFE] tubing (1/32” OD). The typical range of operation was 1-10 psi. Picoinjection inputs were independently controlled to adjust optimal pressures. The stability and reproducibility of picoinjection was investigated with dye injection experiments at a pre-tuned injection pressure (~ 1.07 psi). All pressures were controlled using software written in LabView. Commercial food dyes in different colors were used to differentiate aqueous phases during injection. Microscopic observations were made using an Olympus IX71 inverted microscope with Andor Clara interline CCD camera. Droplets were collected via tubing into a separate collection chip. After collection, all inlets and outlets of the collection chip were completely sealed with a UV glue (NOA 81, Norland Product) to prevent evaporation during incubation.
Cell Lysis and Multiple Displacement Amplification
GFP-expressing E. coli cells were diluted in nuclease-free water and the cells captured in droplets were lysed with the alkaline solution containing 400 mM KOH and 10mM EDTA. For thermal lysis and denaturation, the cells were heated at 95 C for 10 minutes and immediately placed on ice. Multiple displacement amplification was carried out with the amplification reaction mix containing 1X reaction buffer (40 mM Tris-Cl (pH7.5), 50 mM KCl, 10 mM MgCl2, 5 mM (NH4)2SO4), 0.5 mM dNTPs, 50 μM random hexamers, 4% PEG-400, 2% DMSO, 20 μM SYBR Green and 0.15 μM of a double-affinity purified ϕ29 DNA polymerase, which was expressed and purified in-house from a plasmid kindly provided by Dr. Paul Blainey [19]. The 200 pL droplets were transferred to a collection chip and incubated at 30 °C for 18 hours using PTC-225 DNA Engine Thermocycler (MJ Research).
Results and Discussion
The picoinjection technique is vulnerable to abrupt internal pressure changes resulting in unstable operation. Successful picoinjection requires a precise pressure balance at the intersection of the droplet (main) channel and the injection channel. The operation of typical picoinjection is illustrated in Figure 1. The picoinjection channel has a thin slit to increase the interfacial pressure development, and is pressurized at a fixed pressure. The interfacial tension at this interface results in a pressure difference between the injector and the main channel, which is approximated by the Laplace pressure,
where γ, the water/oil interfacial tension, and r the radius of curvature of the interface at the slit. When the interface is fully hemispherical, r approaches half of the slit width (w) and the maximum Laplace pressure is expected,
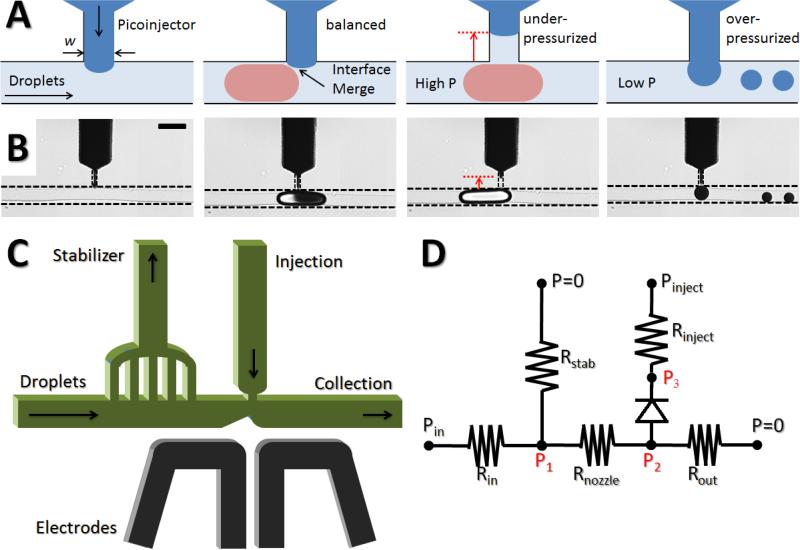
Figure 1.
(A) Illustrated pressure-sensitivity of picoinjection (not to scale); (first) Dark blue indicates aqueous injection fluid and light blue represents continuous oil phase. Injection channel is pressurized at a fixed pressure. A hemispherical interface is developed where there is little pressure fluctuation. (second) When the injection pressure is balanced with the main channel pressure and the interfacial pressure, the interface is maintained at the T-junction enabling a transient contact that leads to coalescence. (third) The injection channel becomes under-pressurized when the main channel pressure suddenly increases due to a fluctuation, and the interface retracts. (fourth) The injection channel becomes over-pressurized when the main channel pressure decreases from a fluctuation, and it starts to generate small droplets of injection fluid. (B) Micrographs of pressure-sensitive picoinjection: (first) a stable interface, (second) proper operation of picoinjection, (third) missed picoinjection due to the raised interface, (fourth) unwanted droplets generated due to over-pressurized picoinjection. The scale bar represents 100 μm. (C) A schematic of the pressure stabilizer for reproducible picoinjection (not to scale). The stabilizer dissipates internal pressure by venting continuous oil phase and is followed by the injection nozzle. Arrows show flow directions. (D) A simplified electronic circuit analogy model. The diode in the injection wire represents interfacial pressure development at the junction due to the two-phase flow.
The position and shape of the picoinjection fluid interface is determined by applied pressure to the injection channel in addition to the Laplace pressure at the T-junction. When the injection fluid pressure is in balance with the main channel pressure and the interfacial pressure, the interface remains at the T-junction enabling good contact with passing droplets for electro-coalescence. However, when a systemic fluctuation causes increased pressure in the main channel, the picoinjection channel is suddenly under-pressurized, causing the interface to retract. In contrast, when the fluctuation results in a pressure decrease in the main channel, the picoinjector overflows through the slit and begins to generate discrete droplets of the picoinjection fluid. The use of our novel pressure stabilizer (near the injection junction) to overcome this limitation is illustrated in Figure 1C. It consists of a fork-like structure with multiple thin slits connecting the main flow channel to a venting channel. The pressure stabilizer structure prevents droplets larger than the width of each slit from entering the venting route, yet it minimizes the flow resistance imposed on the structure by having multiple slits. Droplets in the continuous oil phase travel down the main channel for reagent injection. Since these droplets are stabilized with surfactants, an electric field from embedded electrodes is used to induce a transient destabilization of the droplet interface for coalescence with the injection fluid. Figure 1D shows a simplified electronic circuit analogy model corresponding to the picoinjection system with a pressure stabilizer. Droplets are generated and enter the injection system at a controlled pressure (Pin) and the injection fluid is sustained at another pressure (Pinject). These two controllable pressures then determine internal pressures (P1, P2 and P3) based on flow resistances in the channel circuit. The diode represents interfacial pressure development at the T-junction due to the two-phase flow. It normally allows the current to flow from the main channel to the injection channel with negligible resistance but prevents the current in the opposite direction except for a small leakage current. Once the voltage is over the breakdown level of the diode, it breaks and no longer prevents the reverse current, which is analogous to the breakage of interface due to a high injection pressure. The pressure stabilizer is located right before the picoinjection T-channel to modulate the pressure at the injection junction (P2). It simply dissipates internal pressure by venting continuous oil phase to an open outlet (P=0) while holding droplets in the main channel. Flow resistances are determined by channel geometry and possible tubing between the device and external pressurized reservoirs.
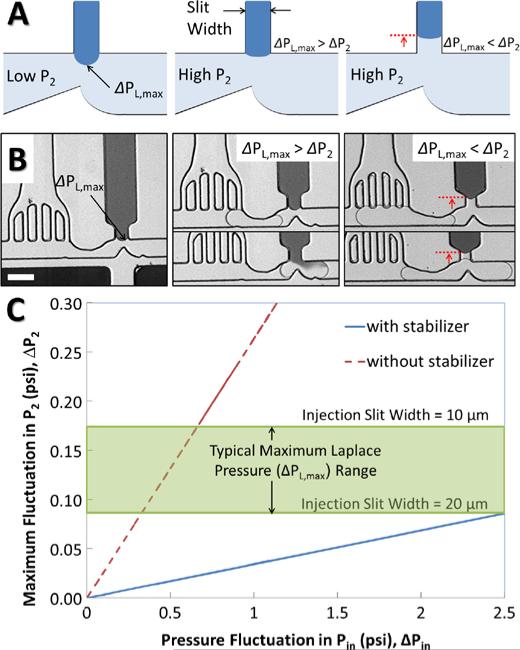
Figure 2 shows the function of the pressure stabilizer to improve resistance to pressure fluctuation. At steady state, the injection fluid pressure above the T-junction (P3) is balanced by the pressure of the flowing fluid (P2) in the main channel and the Laplace pressure (Figure 2A, left). In the event of a mild pressure fluctuation of P2 (ΔP2 < Δ PL,max), the pressure balance is maintained by an increase in the radius of curvature at the interface (Figure 2A, center). In the case of a larger fluctuation in the main channel pressure (ΔP2 > ΔPL,max), the pressure balance at the interface is lost and the fluid interface retracts into the picoinjector channel (Figure 2A, right). Considering the maximal Laplace pressure is defined by channel geometry, it is thus necessary to reduce the pressure fluctuation (Δ2) to withhold the interface at the T-junction. When the pressure stabilizer exists before the injection junction, droplets remain in the main channel while a large portion of carrier oil takes the by-pass route to an alternate outlet, resulting in significant reduction of internal pressure in the main channel after the stabilizer. Figure 2B shows the maximum Laplace pressure developed (left), a properly adjusted pressure allowing picoinjection (center), and retraction of the injection interface due to excessive pressure fluctuation preventing picoinjection (right), respectively. Disturbances upstream of the stabilizer result in transient increases or decreases if flow rate of carrier oil through the bypass, reducing the pressure fluctuation experienced at the picoinjector and improving controllability. Figure 2C presents an “operating diagram” for a picoinjector with and without the stabilizing structure: addition of the stabilizing structure reduces the magnitude of pressure fluctuations in the channel (Δ2), to less than the critical Laplace pressure (ΔPL,max) meaning that controllability of the picoinjector can be maintained over a wider range of upstream fluctuations (ΔPin).
Figure 2.
Use of the pressure stabilizer to improve resistance to pressure fluctuation. (A) Illustration of improved picoinjection with pressure stabilization. (left) For low internal main channel pressures at the nozzle (P2), maximum Laplace pressure (ΔPL,max) is developed. (center) When P2 increases, the Laplace pressure (ΔPL) decreases due to the increase of its radius of curvature. The interface is maintained at the junction if ΔP2 is lower than ΔPL,max. (right) The injection interface is retracted ΔP2 is greater than ΔPL,max. Injection does not occur due to the retracted interface. (B) Micrographs of pressure-sensitive picoinjection: (left) the maximum Laplace pressure (ΔPL,max), (center) pressure fluctuation (ΔP2) within the range of ΔPL,max, resulting in normal picoinjection, (right) ΔP2 greater than ΔPL,max and picoinjection fails due to the raised interface. The scale bar represents 100 μm. (C) Pressure fluctuations at the injection nozzle (ΔP2) estimated from the circuit model. The pressure stabilizer significantly reduces ΔP2 below the typical range of ΔPL,max (0.09-0.17 psi) whereas a small fluctuation may cause a high ΔP2(>ΔPL,max) without the pressure stabilizer.
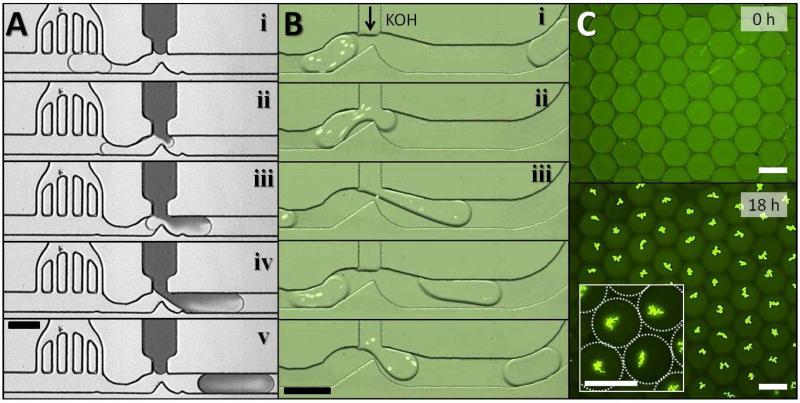
In many passive-type droplet systems, internal pressures can be modulated to minimize pressure fluctuation, although we have often found that maintaining long-term, stable operation of a picoinjector is challenging even in a purely passive system. The challenge is compounded in pressure-driven on-demand droplet generation system designed to allow selective encapsulation of individual cells [15]. This system is uniquely enabled by a rapidly actuated on-demand pressure pump which, when actuated, triggers an abrupt increase of internal pressure by up to 3 psi for ΔPin. This rapid pressure fluctuation overwhelms the delicate balance of pressure required for operation of a picoinjector downstream of droplet formation. Without a pressure stabilizer, even a mild fluctuation of Pin (~1 psi) makes δP2 far greater than the typical range of maximal replace pressures (0.09-0.17 psi) tolerated by the picoinjector with the slit widths of 10-20 μm, resulting in failure of picoinjection. Addition of the pressure stabilizer prior to the picoinjector, leads to approximately 10-fold reduction of internal pressure at the picoinjector. Since the reduced fluctuation is within the range of Laplace pressure, the injection interface remains steady and still except for a slight shape change to compensate for the pressure change, enabling good contact between the flowing droplets and the injection fluid. Figure 3A shows a sequence of micrographs for the picoinjection process with aid of a pressure stabilizer for an on-demand droplet generation system [15]. A flowing droplet slows down due to the reduced pressure after the pressure stabilizer (i). When a droplet flows into the narrowed nozzle right before the injection T-channel, the pressure drop across the nozzle momentarily increases from squeezing and the interfacial energy increases at the same time (ii). Because of this change in interfacial energy, the droplet readily ruptures due to an electrically induced instability [20]. At the same time, the momentary pressure drop at the junction helps the injection fluid move down closer to the droplet and the electrodes, facilitating electrostatic coalescence. The injection fluid in the above pressurized channel then slowly enters the partially ruptured droplet (iii). Once the droplet passes through the nozzle away from the electrodes, the linkage between the injection fluid and the droplet breaks due to its recovered stability (iv). After the injected droplet is pinched off, it flows down the channel to further processing or collection (v). Because of internal circulation inside the droplets, the injected materials are rapidly mixed with the original contents [21]. Note that the picoinjection interface is well maintained during the whole injection process due to the reduced pressure fluctuation. In Figure 3B, droplets containing GFP expressing E. coli cells flow by the picoinjector where 400 mM KOH solution is injected into the droplet for cell lysis. The injected KOH rapidly mixes within the droplet and immediately lyses the cells and denatures the protein and nucleic acid of the cells inside. This can be visualized by disappearance of discrete spots of GFP fluorescence (v). Figure 3C shows the results of on-chip droplet MDA of E. coli cells. E. coli cells are thermally or chemically lysed and denatured prior to encapsulation in droplets, and an MDA reagent mix is injected into the droplets via a single picoinjector. The droplets with lysed cells and amplification reagents are collected in a separate “reservoir” chip for incubation. After an 18-hour-long incubation on chip at 30°C, very bright fluorescent signals are observed in droplets, which are condensed (likely hyperbranched) structures typical of randomly primed multiple displacement amplification of genomic DNA [22]. The successful MDA results imply that the pressure stabilizer enables robust and precise controls of the amounts of MDA reagents during picoinjection for thousands of droplets in sample.
Figure 3.
(A) Operation of the stabilized injection (dye injection), using a pressure stabilizer and a nozzle picoinjection. Electrodes (not shown) apply an electric field near the junction to trigger injection; i) An incoming droplet. ii) Initiation of injection. iii) Continued injection. iv) Pinching off the neck of the droplet. v) The separated droplet with progressed mixing. (B) Demonstration of chemical cell lysis by Injecting potassium hydroxide into droplets containing GFP expressing E. coli cells (bright dots). Upon the injection, the cells are immediately lysed and lose their fluorescence thereafter. Image sequences in (A) and (B) have an interval of 250ms. (C) Results of on-chip droplet MDA of lysed E. coli cells enabled by a single picoinjector with a pressure stabilizer. Thermally or chemically lysed E. coli cells are encapsulated in droplets, followed by picoinjection of an MDA reagent mix; (top) droplets collected in a reservoir chip before incubation (0 hour), (bottom) droplets after on-chip incubation at 30°C (18 hours). In the white box inlet, a zoomed-in view of droplets shows a multi-branched structure of amplified genomic DNA. Scale bars indicate 100 μm.
The stability and reproducibility of picoinjection is investigated during on-demand operation, which renders picoinjection uncontrollable without a pressure stabilizer (Figure 4). When the pressure actively varies during droplet generation, the polydispersity of droplets increases. Reproducible picoinjection requires achieving the same final concentration of reagents for droplets of different sizes. As the size of the incoming droplet increases, the volume added to the droplet by picoinjection increases (Figure 4A), because longer droplets stay in contact with the injection nozzle longer. The resultant concentration of injection reagents remains almost constant for incoming droplet sizes ranging from 250-650 pL (Figure 4B). Incoming droplets smaller than 250pL in volume show larger variations in the resultant dye concentrations (Figure 4B, blue bars). part to the inherent fluctuation in injected volume, and also due The variance in concentration is due in part to the inherent fluctuation in injected volume, and also due to the short length of the droplets relative to the size of the nozzle. The reagent concentration increases for incoming droplets larger than 700 pL. A possible explanation is that an elongated large droplet increases flow resistance in the nozzle, which slows the droplet and prolongs contact time with the injection fluid. The width of the channel in the vicinity of the picoinjection nozzle can be made narrower or wider to cause greater or less droplet elongation, thereby tuning the size range over which reproducible picoinjection is achieved.
Figure 4.
Reproducibility of picoinjection over a wide range of incoming droplet sizes at a fixed injection pressure. (A) Bar plot of the amount of injected volume by picoinjection as a function of the incoming droplet size. (B) Relative dye concentrations of droplets after picoinjection (the left blue axis and blue bars) and the percent increase of volume after picoinjection (the right red axis and red bars). The error bars refer to ±1 standard deviation of 7-24 droplets for each size range.
The pressure stabilizer also facilitates use of two-injector systems, which are otherwise challenging to implement. In Figure S1, we demonstrate a droplet-based bioassay system with two separate picoinjectors stabilized with a single pressure stabilizer upstream of the first picoinjector. In this example, two different color dyes were used for injection fluids at different positions. As an example of a multi-step bioassay, this system can be used for droplet-based single cell genomic amplification, combining the individual operations of cell lysis and genomic amplification shown in Figure 3 into a single device. Single cell-containing droplets generated “on-demand” flow past the pressure stabilizer to a series of two picoinjectors. The first injector will inject an alkaline (potassium hydroxide) cell lysis solution, resulting in immediate cell lysis and DNA denaturation during the short translocation. The second picoinjector then supplies the droplets with a neutralizing solution to reduce the pH and also provides MDA reagents including random hexamer primers, 29 DNA polymerase, nucleotides, and cofactors.
Conclusions
We have presented a pressure stabilizer that greatly improves the reproducibility of picoinjection. This unique fork-like structure retains droplets in the main channel while venting a large portion of continuous oil phase to an alternate outlet, resulting in significant reduction of internal pressure at the injection junction. The pressure stabilizer thus makes it possible to use the picoinjection in applications that require a wide range of flow rates and internal pressure variation. We have shown that stable picoinjection is achieved in aid of the pressure stabilizer during on-demand droplet operation where picoinjection would not work. We have also demonstrated that a single pressure stabilizer can enable both single and sequential picoinjection to implement multi-step bioassays. This highly reproducible injection capability with our pressure stabilizer can facilitate various on-chip droplet applications engaged with multiple reaction steps [23], including single cell studies [24], enzymatic assays [25], and genetic sequencing.
Supplementary Material
Acknowledgements
We thank David Brekke for his guidance and advice for experiments. We are also grateful to Victoria VanderNoot, David Heredia, Matthew Piccini, and Ben Schudel for helpful conversations and extensive help on laboratory work. Financial support for the work was provided by the grants: R01 DE020891, funded by the NIDCR; ENIGMA, a LBNL Scientific Focus Area Program supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research; and the DOE Joint BioEnergy Institute supported by the US DOE, Office of Science, Office of Biological and Environmental Research through contract DEAC02-05 CH11231 (LBNL). Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin Company, for US DOE's Nuclear Security Administration under contract DE-AC04-94AL85000.
Footnotes
† Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Seemann R, Brinkmann M, Pfohl T, Herminghaus S. Reports on Progress in Physics. 2012;75:016601. doi: 10.1088/0034-4885/75/1/016601. [DOI] [PubMed] [Google Scholar]
- 2.Huebner A, Sharma S, Srisa-Art M, Hollfelder F, Edel JB, Demello AJ. Lab on a Chip. 2008;8:1244–1254. doi: 10.1039/b806405a. [DOI] [PubMed] [Google Scholar]
- 3.Teh SY, Lin R, Hung LH, Lee AP. Lab on a Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- 4.Simon MG, Lin R, Fisher JS, Lee AP. Biomicrofluidics. 2012;6:014110. doi: 10.1063/1.3687400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baret JC. Lab on a Chip. 2012;12:422–433. doi: 10.1039/c1lc20582j. [DOI] [PubMed] [Google Scholar]
- 6.Lorenz RM, Edgar JS, Jeffries GDM, Chiu DT. Analytical Chemistry. 2006;78:6433–6439. doi: 10.1021/ac060748l. [DOI] [PubMed] [Google Scholar]
- 7.Baroud CN, de Saint Vincent MR, Delville JP. Lab on a Chip. 2007;7:1029–1033. doi: 10.1039/b702472j. [DOI] [PubMed] [Google Scholar]
- 8.Mazutis L, Baret JC, Treacy P, Skhiri Y, Araghi AF, Ryckelynck M, Taly V, Griffiths AD. Lab on a Chip. 2009;9:2902–2908. doi: 10.1039/b907753g. [DOI] [PubMed] [Google Scholar]
- 9.Lee M, Collins JW, Aubrecht DM, Sperling RA, Solomon L, Ha JW, Yi GR, Weitz DA, Manoharan VN. Lab on a Chip. 2014;14:509–513. doi: 10.1039/c3lc51214b. [DOI] [PubMed] [Google Scholar]
- 10.Mazutis L, Griffiths AD. Lab on a Chip. 2012;12:1800–1806. doi: 10.1039/c2lc40121e. [DOI] [PubMed] [Google Scholar]
- 11.Schoeman RM, Kemna EWM, Wolbers F, van den Berg A. Electrophoresis. 2014;35:385–392. doi: 10.1002/elps.201300179. [DOI] [PubMed] [Google Scholar]
- 12.Abate AR, Hung T, Mary P, Agresti JJ, Weitz DA. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19163–19166. doi: 10.1073/pnas.1006888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eastburn DJ, Sciambi A, Abate AR. PLoS One. 2013;8:e62961. doi: 10.1371/journal.pone.0062961. doi: 10.1371/journal.pone.0062961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beneyton T, Coldren F, Baret J-C, Griffiths AD, Taly V. Analyst. 2014;139:3314. doi: 10.1039/c4an00228h. [DOI] [PubMed] [Google Scholar]
- 15.Rhee M, Liu P, Meagher RJ, Light YK, Singh AK. Biomicrofluidics. 2014;8:034112. doi: 10.1063/1.4874715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanapalli SA, Banpurkar AG, van den Ende D, Duits MHG, Mugele F. Lab on a Chip. 2009;9:982–990. doi: 10.1039/b815002h. [DOI] [PubMed] [Google Scholar]
- 17.Zeng SJ, Li BW, Su XO, Qin JH, Lin BC. Lab on a Chip. 2009;9:1340–1343. doi: 10.1039/b821803j. [DOI] [PubMed] [Google Scholar]
- 18.Whitesides GM, Ostuni E, Takayama S, Jiang XY, Ingber DE. Annual Review of Biomedical Engineering. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 19.Blainey PC, Quake SR. Nucleic Acids Research. 2011;39:e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priest C, Herminghaus S, Seemann R. Applied Physics Letters. 2006;89:134101. [Google Scholar]
- 21.Rhee M, Burns MA. Langmuir. 2008;24:590–601. doi: 10.1021/la702575j. [DOI] [PubMed] [Google Scholar]
- 22.Dean FB, Nelson JR, Giesler TL, Lasken RS. Genome Research. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo MT, Rotem A, Heyman JA, Weitz DA. Lab on a Chip. 2012;12:2146–2155. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 24.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang C, Sustarich J, Bharadwaj R, Chandrasekaran A, Adams PD, Singh AK. Lab Chip. 2013;13:1817–1822. doi: 10.1039/c3lc41418c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.