Abstract
Neuronal production in the mammalian cortex depends on extensive mitoses of radial glial progenitors (RGPs) residing in the ventricular zone (VZ). Here, we examine the function of centrioles in RGPs during cortical neurogenesis in mice by conditional removal of SAS-4, a protein required for centriole biogenesis. SAS-4 deletion leads to a progressive loss of centrioles, accompanied by RGP detachment from the VZ. Delocalized RGPs do not become outer subventricular zone RGPs (oRGs). While remaining proliferative, ectopic RGPs, as well as those in the VZ with a centrosomal deficit exhibit prolonged mitosis, p53 up-regulation and apoptosis, resulting in neuronal loss and microcephaly. Simultaneous removal of p53 fully rescues RGP death and microcephaly, but not RGP delocalization and randomized mitotic spindle orientation. Our findings define centriolar functions in anchoring RGPs in the VZ and ensuring their efficient mitoses, and also reveal the remarkable adaptability of RGPs in the developing cortex.
INTRODUCTION
Radial glial progenitors (RGPs) are responsible for generating nearly all cortical excitatory neurons during embryonic development 1. They reside in the ventricular zone (VZ), a germinal zone of the developing cortex, and possess a short apical process that anchors at the VZ surface (ventricular endfoot) and a long basal process that reaches the pia (radial glial process). During embryonic cortical development, RGPs undergo extensive divisions at the VZ surface to produce neurons either directly or indirectly via transient amplifying progenitors, such as intermediate progenitors (IPs, also called basal progenitors) 2–4, outer subventricular zone radial glial progenitors (oRGs; also called outer SVZ progenitors, OSVZ progenitor, intermediate RGs, or translocating RGs, tRGs) 5–10 and short neural precursors 11. Successive waves of neurogenesis and coordinated neuronal migration lead to a stereotypic “inside-out” sequence of lamina formation in the cortex 12, the largest brain structure that supports all high-order brain functions.
The centrosome is a cellular organelle in animals that serves as the main microtubule-organizing center (MTOC) and provides the basal body that acts as a template for cilia and flagella assembly 13–15. It is composed of a core of two centrioles surrounded by a mass of proteins termed the pericentriolar material (PCM). Mutations in centrosomal proteins have been linked to human autosomal recessive primary microcephaly (MCPH), a neurodevelopmental disorder characterized by small brain size and deficient neuronal production in the cortex 16, 17. While centrosomal proteins are ubiquitously expressed, the selective vulnerability of cortical development demonstrates the importance of an intact centrosome in cortical neurogenesis. A number of mouse models with mutations in centrosomal proteins have been established and characterized 18–20. However, the precise functions of the centriole, along with the centrosome in actively dividing RGPs and the centrosome-related cell biological roots of MCPH remain elusive.
To define the functions of centrioles and centrosomes in RGPs, we used a conditional null allele to delete Sas-4 (also called CENPJ, CPAP or MCPH6 in humans) 21, which encodes a protein required for centriole biogenesis 22–25, in the developing mouse brain. As expected, removal of SAS-4 in RGPs led to a progressive loss of centrioles and centrosomes. This caused p53 up-regulation and widespread cell death of progenitors, resulting in a substantial loss of neurons and microcephaly. Simultaneous removal of p53 fully rescued the Sas-4 conditional mutant from microcephaly. Further analysis showed that the microcephaly arose largely from the preferential apoptosis of RGPs delocalized from the VZ due to the loss of centrosomes. Although RGPs in the Sas-4−/− p53−/− double mutant brain became ectopically localized and divided with randomly oriented mitotic spindles, they retained full neurogenic potential and generated the normal number of neurons. These results suggest that the key functions of the centrosome in cortical neurogenesis are to anchor RGPs in the VZ, and to facilitate the formation of the mitotic spindle and prevent activation of p53 and cell death. Our data reveal surprising plasticity of RGPs in the developing cortex in vivo and challenge prevailing views of the importance of mitotic spindle orientation and progenitor localization to the VZ in cortical neurogenesis.
RESULTS
Sas-4 deletion causes p53-dependent microcephaly
Mutations in the SAS-4/CENPJ gene cause microcephaly in humans 26. Mouse embryos that completely lack Sas-4 die at mid-gestation around embryonic (E) day 8.5, due to massive p53-dependent cell death 21. To define the function of centrioles in cortical neurogenesis and the cell biological basis of centrosome-associated microcephaly, we selectively removed Sas-4 from neural progenitors beginning at ~E10–11 by crossing mice carrying a conditional null allele of Sas-4 with mice carrying Nestin-Cre27 (Supplementary Fig. 1a). This led to a dramatic decrease in the amount of SAS-4 in the developing brain at E13.5 (Supplementary Fig. 2a). The conditional Sas-4 mutants showed a strong microcephalic phenotype that was obvious by E15.5 (Fig. 1a and Supplementary Fig. 2b middle). Compared to wild-type (WT) littermate controls, the telencephalic area estimated by circumference measurement of Sas-4−/− mice was reduced by ~38 ± 5% at this stage (n = 4; p<0.01).
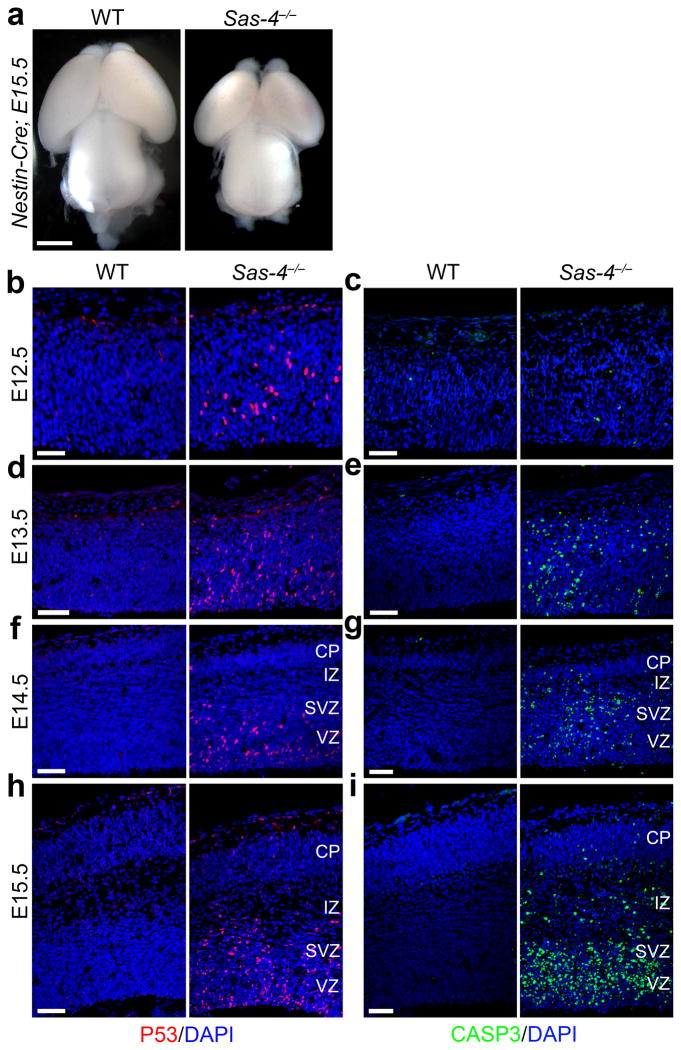
Fig. 1. Sas-4 deletion causes microcephaly with widespread p53 up-regulation and apoptosis.
(a) Representative whole mount images of E15.5 brains from WT and Sas-4−/− embryos. Scale bar: 0.5 mm. (b–i) Representative images of WT and Sas-4−/− cortices stained for p53 (red) and cleaved-caspase 3 (CASP3, green), and DAPI (blue) at E12.5 (b, c), E13.5 (d, e), E14.5 (f, g) and E15.5 (h, i). Note the obvious up-regulation of p53 (b) but no significant activation of CASP3 (c) in the Sas-4−/− cortex at E12.5, and the progressive increase in p53 up-regulation and CASP3 expression as time proceeds, primarily outside the cortical plate (CP) that harbors post-mitotic neurons. VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone. Scale bars: 25 μm (b, c) and 50 μm (d–i).
To pinpoint the origins of microcephaly, we examined cortical organization and cell death by assessing cleaved caspase 3 (CASP3) and p53 expression. Although there was no apparent difference in cortical organization or cell death between WT and Sas-4−/− mice at E12.5 (Fig. 1c), p53 was clearly up-regulated in the Sas-4−/− but not WT cortex (Fig. 1b), suggesting that an apoptosis cascade is initiated in the Sas-4−/− cortex at E12.5. Indeed, apoptosis was evident one day later at E13.5 and progressively increased as time proceeded (Fig. 1d–g). By E15.5, extensive p53 up-regulation and apoptosis were observed in the VZ, the subventricular zone (SVZ) and the intermediate zone (IZ), but not in the cortical plate (CP), of the mutant cortex (Fig. 1h,i).
To test whether the up-regulation of p53 was responsible for the cell death and microcephaly, we analyzed cortical development in mice with simultaneous conditional deletion of both p53 and Sas-4 in neural progenitors using Nestin-Cre. The telencephalic area of Sas-4−/− p53−/− mice was indistinguishable from that of WT mice at E15.5, postnatal (P) day 1 and P14 (Fig. 2a,b,f and Supplementary Fig. 2b right) and no obvious apoptosis was detected in the Sas-4−/− p53−/− cortex (Supplementary Fig. 2c right), indicating that removal of p53 suppressed apoptosis and fully rescued the microcephaly caused by Sas-4 deletion. Conditional deletion of p53 alone using Nestin-Cre did not cause any obvious defects in progenitor behavior and neurogenesis in the developing cortex (Supplementary Fig. 3), as suggested previously 20.
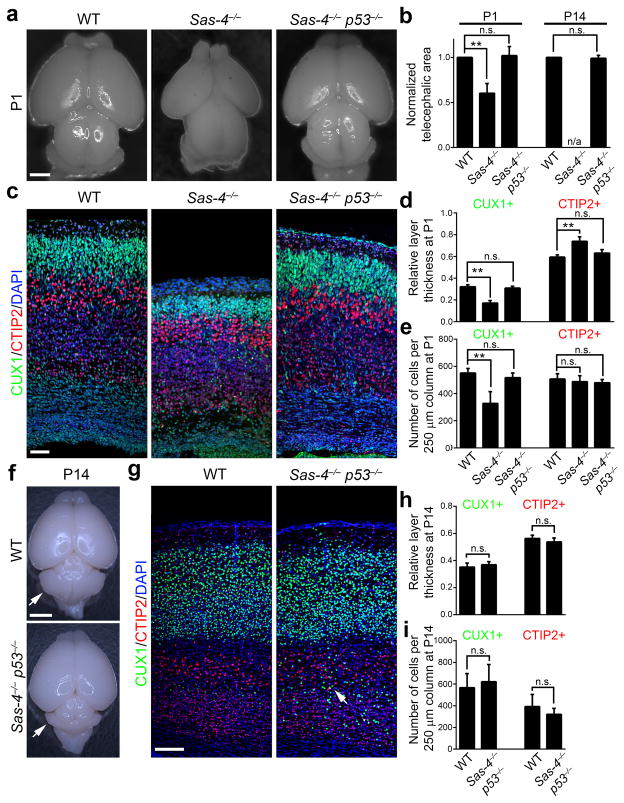
Fig. 2. Simultaneous removal of p53 rescues microcephaly caused by SAS-4 ablation.
(a) Representative whole mount images of WT, Sas-4−/−, and Sas-4−/− p53−/− brains at P1. Note the severe microcephaly in the Sas-4−/− brain, which is rescued by concurrent p53 deletion. Scale bar: 1 mm. (b) Quantification of the telencephalic area of mutant brains relative to WT littermates. (P1: WT, n=4; Sas-4−/−, n=4; Sas-4−/− p53−/−, n=4; P14: WT, n=3; Sas-4−/− p53−/−, n=4). **, p<0.01; n.s., not significant; n/a, not applicable. (c) Representative images of WT, Sas-4−/−, and Sas-4−/− p53−/− cortices at P1 stained for the deep layer marker CTIP2 (red), the superficial layer maker CUX1 (green) and DAPI (blue). Scale bar: 50 μm. (d, e) Quantification of the thickness (d) and cell number (e) per unit area (i.e. a 250 μm radial column) of the deep (CTIP2+) and superficial (CUX1+) layers relative to the total cortical thickness at P1 (n=6). **, p<0.01; n.s., not significant. (f) Representative whole mount images of WT and Sas-4−/− p53−/− brains at P14. Arrows indicate the cerebellar hypoplasia due to the lack of primary cilia in the Sas-4−/− p53−/− brain. Scale bar: 2.5 mm. (g) Representative images of WT and Sas-4−/− p53−/− cortices at P14 stained for CTIP2 (red), CUX1 (green) and DAPI (blue). The arrow indicates a small number of CUX1+ neurons in the deep layers of the Sas-4−/− p53−/− cortex. Scale bar: 150 μm. (h, i) Quantification of the thickness (h) and cell number (i) per unit area (i.e. 250 μm radial column) of the deep (CTIP2+) and superficial (Cux1+) layers relative to the total cortical thickness at P14 (n=4). n.s., not significant. Data are presented as mean ± SD. Individual p values and degrees of freedom are available in the Supplementary Methods Checklist.
Because the cortex develops in a birth date-dependent inside-out sequence, in which neurons born at later times migrate past earlier-born neurons and occupy superficial positions 12, we examined the identity of neurons that were lost in the Sas-4−/− cortex. Compared with the WT control, the number of early-born CTIP2-expressing deep layer neurons 28 was not significantly changed, but the number of late-born CUX1-expressing superficial layer neurons 28 was reduced by ~41% in the Sas-4−/− cortex (Fig. 2c–e). The preferential loss of the superficial, but not deep, layer neurons most probably reflects the progressive loss of centrioles and appearance of apoptotic cells upon SAS-4 removal (Fig. 1c–j); that is, apoptosis mostly occurs when the superficial layer neurons are produced. Moreover, the neuronal loss and microcephaly in Sas-4−/− mice were not due to a loss of primary cilia, as there was no detectable apoptosis, neuronal loss or microcephaly in the conditional Ift88−/− brain that lacked primary cilia (Supplementary Fig. 4).
Remarkably, removal of p53 completely restored the superficial layer neurons lost in the Sas-4−/− cortex. The thickness and cell number of the superficial layers marked by CUX1 in the Sas-4−/− p53−/− cortex were not significantly different from those of the WT control at P1 (Fig. 2c–e) and P14 (Fig. 2g–i). These results suggest that Sas-4−/− cells in the developing cortex retain the ability to generate the correct number and type of neurons if they are rescued from apoptosis by removal of p53. Notably, we observed a minor but significant population of CUX1-expressing neurons located in the deep layers (Fig. 2g arrow and Supplementary Fig. 5a) as well as a small heterotopia in the posterior dorsomedial region of the Sas-4−/− p53−/− cortex (Supplementary Fig. 6), indicating subtle defects in cortical neuronal migration. There was no obvious change in the number or distribution of neurons expressing FOXP2 and SATB2 (Supplementary Fig. 5b,c), two additional cortical neuronal markers 28. In addition, similar to the Ift88−/− brain (Supplementary Fig. 7 right), the Sas-4−/− p53−/− brain showed hydrocephalus (Supplementary Fig. 7 middle) and cerebellar hypoplasia (Fig. 2f arrows), likely due to the loss of cilia (see below) 29.
Microcephaly arises from a loss of displaced progenitors
Cell death in the embryonic cortex of Sas-4−/− mice was most prominent in the regions where progenitors reside (Fig. 1e–j). There are two major populations of progenitors in the developing mouse cortex: PAX6-expressing RGPs located in the VZ and TBR2-expressing IPs in the SVZ 4. We found that in the Sas-4−/− cortex, p53 expression was detected exclusively in proliferating cells expressing Ki67 (Supplementary Fig. 8a,b), and mostly in PAX6-expressing cells (Supplementary Fig. 8c,d). Compared with the WT control at E15.5, the total number of PAX6-expressing cells was drastically reduced in the Sas-4−/− cortex (Fig. 3a,b, left and middle). In addition, some PAX6-expressing cells were located in the SVZ and the IZ, instead of their normal position in the VZ. TBR2-expressing IPs exhibited a similar reduction in number in the Sas-4−/− cortex within the SVZ (Fig. 3c,d, left and middle).
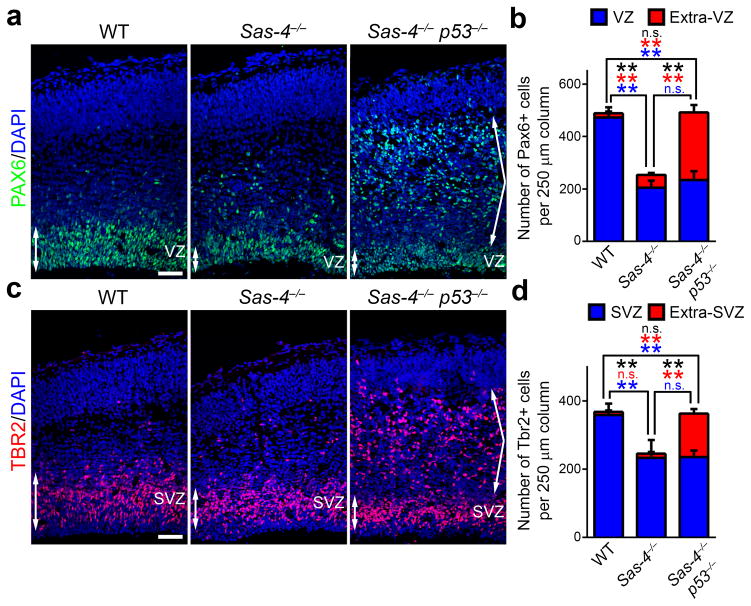
Fig. 3. Sas-4 deletion leads to RGP delocalization and loss.
(a) Representative images of E15.5 WT, Sas-4−/−, and Sas-4−/− p53−/− cortices stained with the antibody against PAX6 (green), an RGP marker, and DAPI (blue). Double-headed arrows indicate RGPs in the VZ and angled arrows indicate displaced RGPs outside the VZ. Scale bar: 50 μm. (b) Quantification of the number of Pax6+ cells per unit area (i.e. 250 μm radial column) within the VZ (blue) and outside the VZ (Extra-VZ, red) (WT, n=5; Sas-4−/−, n=4; Sas-4−/− p53−/−, n=5). Asterisks indicate the statistical significance of the differences in the number of total PAX6+ cells (black), the Extra-VZ PAX6+ cells (red) and the VZ PAX6+ cells (blue). **, p<0.01; n.s., not significant. (c) Representative images of E15.5 WT, Sas-4−/−, and Sas-4−/− p53−/− cortices stained with the antibody against TBR2 (red), an IP marker, and DAPI (blue). Double-headed arrows indicate IPs in the SVZ and angled arrows indicate displaced IPs outside the SVZ. Scale bar: 50 μm. (d) Quantification of the number of TBR2+ cells per unit area (i.e. 250 μm radial column) within the SVZ (blue) and outside the SVZ (Extra-SVZ, red) (WT, n=5; Sas-4−/−, n=4; Sas-4−/− p53−/−, n=5). Asterisks indicate the statistical significance of the differences in the total TBR2+ cells (black), the Extra-SVZ TBR2+ cells (red) and the SVZ TBR2+ cells (blue). **, p<0.01; n.s., not significant. Data are presented as mean ± SD. Individual p values and degrees of freedom are available in the Supplementary Methods Checklist.
We then examined progenitors in the Sas-4−/− p53−/− brain, which did not show microcephaly. Although the number of PAX6-expressing cells in the Sas-4−/− p53−/− VZ was comparable to that in the Sas-4−/− cortex (Fig. 3a,b, middle and right), there was an additional large population of ectopic PAX6-expressing cells, mostly located in the IZ (Fig. 3a, right, arrows). Consequently, the total number of PAX6-expressing cells in the Sas-4−/− p53−/− cortex was indistinguishable from that in the WT control (Fig. 3a,b, left and right). Moreover, similar to those located in the VZ, virtually all ectopic PAX6-expressing cells were positive for SOX2 (Supplementary Fig. 9a,b), another transcription factor expressed in RGPs 30, as well as brain lipid-binding protein (BLBP) (Supplementary Fig. 9c,d), a bone fide RGP marker 31, further suggesting that they are ‘RGP’ in nature.
Similarly, the total number of TBR2-expressing IPs in the Sas-4−/− p53−/− cortex was comparable to that in the WT control, but significantly more than that in the Sas-4−/− cortex. Most of the additional IPs were located ectopically in the IZ (Fig. 3c,d). Given that there was no significant displacement of IPs away from the SVZ in the Sas-4−/− single mutant cortex, the ectopic IPs in the Sas-4−/− p53−/− cortex most probably originated from displaced RGPs that were rescued from apoptosis. Notably, we obtained similar results in progenitor delocalization and loss, and microcephaly when an Emx1-Cre allele 32 (Supplementary Fig. 1b) was used to delete Sas-4 alone or Sas-4 together with p53 in the cortex (Supplementary Fig. 10). Together, these results strongly suggest that neuronal loss and microcephaly in the Sas-4−/− brain arise primarily from the loss of ectopically localized ‘RGPs’ and their IP progeny.
Centrosome loss leads to delocalization of RGPs
Consistent with the essential role of SAS-4 in centriole biogenesis 22–25, we observed a progressive loss of centrosomes, as well as a depletion of the primary cilia that grow from the functional centrioles, in the Sas-4−/− cortex, revealed by the staining for pericentrin (PCNT), a PCM protein, and ARL13B, a marker of the primary cilia 33. The loss of centrosomes and primary cilia outside the VZ surface (extra-VZ) was clear by E12.5 (Supplementary Fig. 11a,b, bottom). In contrast, the loss of centrosomes and primary cilia at the VZ surface, where the centrosomes of RGPs normally reside 34, 35, was not obvious until E14.5 (Supplementary Fig. 11a,b, top). These results suggest that RGPs having a centriole are selectively maintained in the VZ, whereas those lacking the centriole/centrosome are progressively delocalized from the VZ.
Consistent with this notion, while there was a significant number of ARL13B-labeled primary cilia at the VZ surface associated with RGPs in the VZ in the Sas-4−/− p53−/− cortex at E1.5.5, there were virtually no primary cilia detected in ectopically localized PAX6-expressing cells (Fig. 4a), indicating a lack of the centriole/centrosome in displaced RGPs. Together, these results suggest that centriole/centrosome loss causes RGPs to delocalize from the VZ.
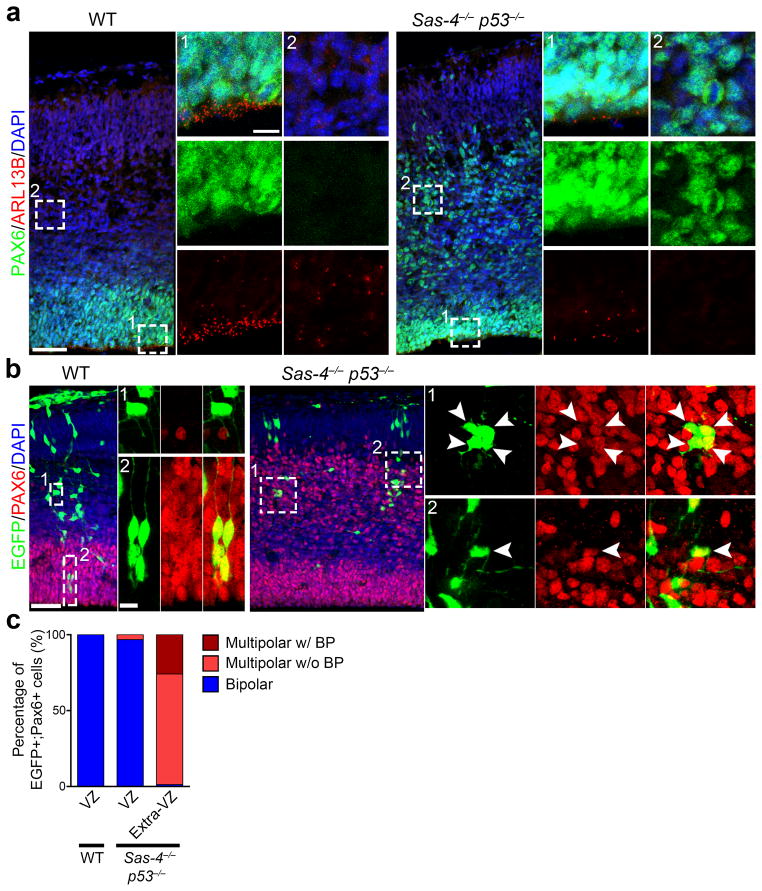
Fig. 4. Displaced RGPs lack centrioles/centrosomes and are not oRGs.
(a) Representative images of E15.5 WT (left) and Sas-4−/− p53−/− (right) cortices stained with the antibodies against PAX6 (green) and ARL13B (red), a primary cilium marker, and DAPI (blue). High magnification images of a region in the VZ and outside the VZ (Extra-VZ) (broken lines, 1 and 2) are shown to the right. Scale bars: 50 μm and 10 μm. (b) Representative images of E15.5 WT (left) and Sas-4−/− p53−/− (right) cortices that received in utero intraventricular injection of EGFP-expressing retroviruses (green) at E11.5 and stained with the antibody against PAX6 (red) and DAPI (blue). High magnification images of individual PAX6-expressing cells in the VZ and Extra-VZ (broken lines) are shown to the right. Note that Extra-VZ PAX6-expressing cells in Sas-4−/− p53−/− cortices possess multiple short processes, but not a prominent pia-directed radial glial process, indicating that they are not oRGs. Scale bars: 50 μm and 10 μm. (c) Quantification of the percentage of PAX6-expressing cells that are bipolar, multipolar without a basal process (w/o BP) or multipolar with a basal process (w/BP). Note that the PAX6-expressing cells in the extra-VZ of the Sas-4−/− p53−/− cortex are predominantly multipolar in morphology, whereas those in the VZ of the WT or Sas-4−/− p53−/− cortex are bipolar. (WT: VZ, n=105 cells; Sas-4−/− p53−/−: VZ, n=91 cells; Extra-VZ, n=143 cells).
Delocalized RGPs are not oRGs and remain proliferative
A small number of RGP-like progenitors have been found outside the VZ of the rodent cortex 5–7. They retain the basal process to the pia but lack the apical process to the ventricular surface. To test whether delocalized RGPs become oRGs, we examined the morphology of delocalized RGPs in the Sas-4−/− p53−/− cortex. We injected retroviruses expressing enhanced green fluorescent protein (EGFP) into the lateral ventricle of the WT and Sas-4−/− p53−/− embryos at E11.5 to label dividing RGPs at the VZ surface and their progeny. Brains were recovered at E15.5 for analysis. While EGFP-expressing cells positive for PAX6 were frequently observed in the VZ of the WT cortex (Fig. 4b left), they were rarely found in the VZ of the Sas-4−/− p53−/− cortex; instead, they were predominantly located outside the VZ (Fig. 4b right), consistent with a delocalization of RGPs in the Sas-4−/− p53−/− cortex.
As expected, the PAX6-expressing RGPs in the VZ of the WT cortex displayed a characteristic bipolar morphology (Fig. 4b left and 4c). In contrast, most of the PAX6-expressing cells outside the VZ in the Sas-4−/− p53−/− cortex exhibited a multipolar morphology with several short processes emanating from the cell body (Fig. 4b right and 4c). Similar morphological features were revealed by BLBP labeling (Supplementary Fig. 9e). Moreover, while PAX6-expressing cells in the VZ were readily labeled by the lipophilic dye DiI applied at the pia, those in the extra-VZ were rarely labeled (Supplementary Fig. 12a,b), suggesting that the majority of ectopic PAX6-expressing cells in the Sas-4−/− p53−/− cortex do not possess a pia-reaching basal process. Consistent with a lack of the long basal process, we observed mitotic EGFP- and PAX6-expressing cells outside the VZ labeled by phosphorylated VIMENTIN (P-VIMENTIN) 36 as rounded cells with no discernible pia-directed process (Supplementary Fig. 12c). Together, these results suggest that delocalized RGPs originate from dividing RGPs in the VZ; however, they are largely multipolar in morphology without a prominent basal process and are thus not oRGs.
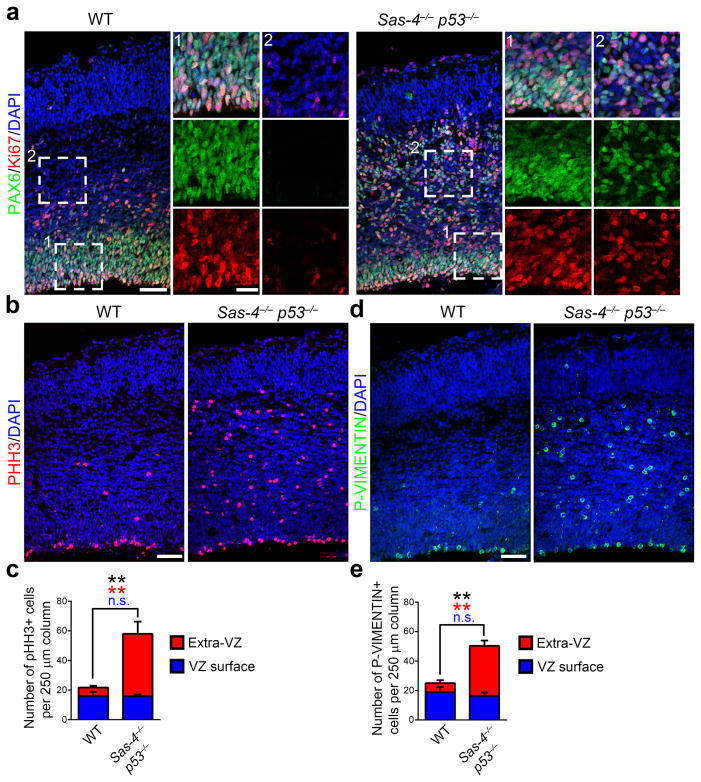
The VZ is considered to be a niche for RGPs 37 and detachment of RGPs from the VZ has been associated with premature differentiation 35, 38. However, we found that, similar to control RGPs in the VZ of the WT cortex, PAX6-expressing RGPs in the Sas-4−/− p53−/− cortex, regardless of their localization in or out of the VZ, were positive for the proliferation marker Ki67 (Fig. 5a). Moreover, we observed numerous mitotic cells labeled with phosphorylated histone H3 (PHH3) or P-VIMENTIN both at and away from the VZ surface (Fig. 5b,d). In fact, unexpectedly, there was a substantial increase in the number of M-phase cells in the Sas-4−/− p53−/− cortex compared with the WT control (Fig. 5b–e). These results clearly suggest that delocalized RGPs lacking the centriole/centrosome remain proliferative, and do not prematurely exit the cell cycle and differentiate.
Fig. 5. Displaced RGPs remain proliferative.
(a) Representative images of E15.5 WT and Sas-4−/− p53−/− cortices stained with the antibodies against PAX6 (green) and Ki67 (red), a cell proliferation marker, and DAPI (blue). Note that similar to RGPs in the VZ of the WT cortex, nearly all RGPs in the Sas-4−/− p53−/− cortex, regardless of their localization in the VZ (area 1) or outside the VZ (area 2), are positive for Ki67, indicating that they remain proliferative. Scale bars: 50 μm and 20 μm. (b, d) Representative images of E15.5 WT and Sas-4−/− p53−/− cortices stained with the antibodies against PHH3 (red, b) or P-VIMENTIN (green, d) that labels mitotic cells and DAPI (blue). Note the increase in the number of mitotic cells in the Sas-4−/− p53−/− cortex. Scale bar: 50 μm. (c, e) Quantification of the number of PHH3+ (c) or P-VIMENTIN+ (e) cells per unit area (WT, n=5; Sas-4−/− p53−/−, n=5). **, p<0.01. Data are presented as mean ± SD. Individual p values and degrees of freedom are available in the Supplementary Methods Checklist.
Centrosome loss results in mitotic delay
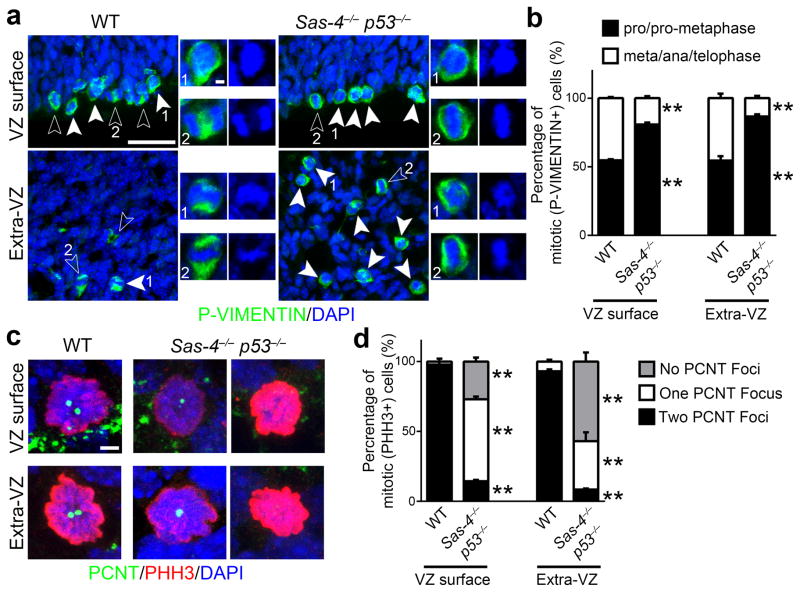
Loss of centrioles/centrosomes has been reported to cause mitotic spindle assembly defects and delays in mitotic progression 39. Given that there was no increase in the total number of progenitors in the Sas-4−/− p53−/− cortex (Fig. 3), the increase in mitotic cells (Fig. 5b–e) indicates that the mitotic progression of Sas-4−/− p53−/− progenitors is affected. To test this, we analyzed the fraction of dividing cells in metaphase, anaphase or telophase (meta/ana/telophase) versus those in prophase or pro-metaphase (pro/pro-metaphase), which reflects mitotic progression. Dividing cells were identified by their expression of P-VIMENTIN and their mitotic phase was inferred from the arrangement of chromosomes (Fig. 6a). We found that in the WT cortex, the fraction of mitotic cells in pro/pro-metaphase was ~55% both at the VZ surface and away from the VZ surface (Fig. 6a,b). In contrast, this fraction increased to ~80% for cells of the Sas-4−/− p53−/− cortex that divided at the VZ surface and to ~86% for cells that divided away from the VZ surface (Fig. 6a,b). These results suggest that the duration of pro/pro-metaphase is prolonged in centriole-deficient mitotic progenitors lacking SAS-4. Consistent with a delay in mitotic progression, we observed a significant increase in the number of proliferative cells labeled by Ki67 (Supplementary Fig. 13a,b), but no obvious change in the cell cycle exit (Supplementary Fig. 13a,c), in the Sas-4−/− p53−/− cortex.
Fig. 6. Centrosome loss causes mitotic delay.
(a) Representative images of mitotic cells at the VZ surface (top) and Extra-VZ (bottom) in E15.5 WT and Sas-4−/− p53−/− cortices stained for P-VIMENTIN (green) and DAPI (blue). Closed arrowheads indicate cells in pro/pro-metaphase, and open arrowheads indicate cells in meta/ana/telophase. High magnification images of pro/pro-metaphase (labeled as 1) and meta/ana/telophase (labeled as 2) are shown to the right. Note that the Sas-4−/− p53−/− cortex contains more cells in pro/pro-metaphase compared to WT control. Scale bars: 25 μm and 2 μm. (b) Quantification of the fraction of mitotic cells in pro/pro-metaphase (black) versus in meta/ana/telophase (white), which reflects mitotic progression (WT, 461 cells from n=3 brains; Sas-4−/− p53−/−, 1,366 cells from n=3 brains). **, p<0.01. (c) Representative images of mitotic cells labeled by PHH3 (red) at the VZ surface and away from the VZ surface (Extra-VZ) in E15.5 WT and Sas-4−/− p53−/− cortices stained for PCNT (green), a centrosomal marker, and DAPI (blue). Note that while WT mitotic cells possess two PCNT foci, mitotic cells at the VZ surface (top) and in the Extra-VZ (bottom) of the Sas-4−/− p53−/− cortex largely contain either none or one PCNT focus. Scale bar: 2 μm. (d) Quantification of the number of PCNT foci in mitotic cells at the VZ surface or Extra-VZ (WT, 226 cells from n=3 brains; Sas-4−/− p53−/−, 690 cells from n=3 brains). **, p<0.01. Data are presented as mean ± SD of individual brains. Individual p values and degrees of freedom are available in the Supplementary Methods Checklist.
We then examined the spindle pole status in mitotic cells by staining them with the antibody against PCNT. While the mitotic cells in the WT cortex contained two prominent PCNT foci (Fig. 6c left, d), those in the Sas-4−/− p53−/− cortex exhibited a clear deficit of the PCNT foci (Fig. 6c right, d). Specifically, the mitotic cells at the VZ surface mostly contained only one PCNT focus, whereas those away from the VZ surface possessed either none or only one PCNT focus. These results suggest that mitotic progenitors lacking SAS-4 exhibit defects in spindle pole assembly, which likely trigger mitotic delay, and subsequent p53 up-regulation and apoptosis. It is important to note that there is no neuronal loss or microcephaly in the Sas-4−/− p53−/− cortex, suggesting that the mitosis of centriole-deficient progenitors lacking SAS-4 does proceed to completion, should the apoptosis be prevented. Moreover, analysis of the chromosome number by fluorescence in situ hybridization (FISH) showed that there was no substantial increase in aneuploidy in the Sas-4−/− p53−/− cortex, when compared with the control (Supplementary Fig. 14a,b). In line with this, we did not observe any dividing cells with multipolar spindles or lagging chromosomes in the Sas-4−/− p53−/− cortex (Supplementary Fig. 14c,d). In addition, no substantial increase in DNA damage was observed, as revealed by γ-H2AX expression (Supplementary Fig. 14e,f). Similarly, no significant increase in aneuploidy or DNA damage was detected in Sas-4 null embryos at mid-gestation 21.
Proliferation of delocalized RGPs
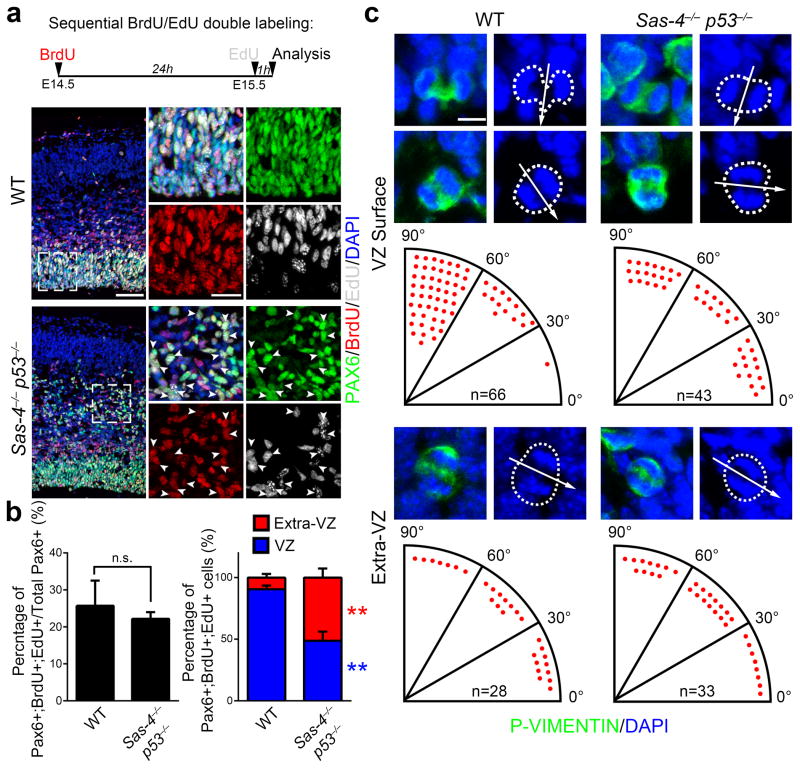
An important feature of RGPs during cortical neurogenesis is their ability to undergo consecutive rounds of mitosis. This is achieved through asymmetric cell division that renews the RGPs while producing neurons or transient amplifying progenitors 2, 3. To test whether ectopically localized RGPs are capable of self-renewing, we performed sequential pulse-chase experiments using 5-bromo-2′-deoxyuridine (BrdU) and 5-ethynyl-2′-deoxyuridine (EdU), two thymidine analogues, spaced 24 hours apart (Fig. 7a, top). If a progenitor goes through S-phase more than once during this period, it should incorporate both BrdU and EdU. Interestingly, similar to PAX6-expressing RGPs in the VZ of the control cortex (Fig. 7a,b), ectopically localized BLBP- and PAX6-expressing cells in the Sas-4−/− p53−/− cortex were positive for both BrdU and EdU (Fig. 7a arrowheads, 7b and Supplementary Fig. 15), suggesting that these delocalized ‘RGPs’ actively divide and self-renew outside the VZ niche.
Fig. 7. VZ niche and cleavage plane orientation are not essential for consecutive rounds of RGP division and cortical neurogenesis.
(a) Representative images of E15.5 WT and Sas-4−/− p53−/− cortices subjected to dual pulse-chase labeling of BrdU (red) and EdU (white) and stained for PAX6 (green), an RGP marker, and DAPI (blue). The pulse-chase protocol is shown at the top. High magnification images of RGPs in the VZ of WT cortex and delocalized RGPs in the IZ of the Sas-4−/− p53−/− cortex are shown to the right. Note that similar to RGPs in the VZ of WT control, many ectopically located RGPs in the Sas-4−/− p53−/− cortex (arrowheads) are positive for both BrdU and EdU, indicating that they undergo consecutive rounds of divisions. Scale bars, 50 μm and 20 μm. (b) Quantification of the percentage of PAX6-expressing cells that are positive for both BrdU and EdU (left) and the relative distribution of Pax6+;BrdU+;EdU+ triple positive cells in the VZ (blue) or Extra-VZ (red) between WT and Sas-4−/− p53−/− cortices (right) (WT, 1,397 PAX6+ cells and 361 PAX6+;BrdU+;EdU+ cells from 3 brains; Sas-4−/− p53−/−, 2,027 PAX6+ cells and 449 PAX6+;BrdU+;EdU+ cells from 4 brains). Data are presented as mean ± SD of individual brains. **, p<0.01; n.s., not significant. (c) Representative images of mitotic cells labeled by P-VIMENTIN (green) in ana/telophase revealed by DAPI staining (blue) at the VZ surface (top) or away from the VZ surface (bottom) in E15.5 WT and Sas-4−/− p53−/− cortices. Broken lines indicate the contours of dividing cells. Arrows indicate the cleavage plane. Determination of the cleavage plane orientation as the angle between the cleavage plane (arrows) and the VZ surface is shown at the bottom of images. Each red dot represents one dividing cell. Scale bar: 5 μm. Individual p values and degrees of freedom are available in the Supplementary Methods Checklist.
The centrosomes at the mitotic spindle poles are likely important in controlling the position of the mitotic spindle and thus the cleavage plane orientation 40–42. We therefore analyzed the cleavage plane orientation of dividing progenitors both at and away from the VZ surface in the Sas-4−/− p53−/− cortex. Because the metaphase plate of dividing progenitors in the cortex rocks extensively and is not set until anaphase 43, 44, we focused on progenitors at ana/telophase, revealed by P-VIMENTIN staining and chromosomal arrangement (Fig. 7c). We assessed the cleavage plane orientation based on three-dimensional analysis of the entire dividing progenitors. As shown previously 45, 46, the majority of WT RGPs at E15.5 divided with axes that were vertical relative to the VZ surface (the angle between the cleavage plane and the VZ surface was between 60° and 90°) (Fig. 7c, left, top), while IPs divided with a random cleavage plane orientation (Fig. 7c, left, bottom). In contrast, the cleavage plane orientation of progenitors including RGPs, regardless of their localization, in the Sas-4−/− p53−/− cortex appeared to be random (Fig. 7c, right). Together, these results suggest that the centrosome is required for normal orientation of the mitotic spindle and the cleavage plane in dividing RGPs. However, these results also argue that the cleavage plane orientation is not essential for RGP asymmetric division and self-renewal. Furthermore, given that neuron number and corticogenesis appeared grossly normal in the Sas-4−/− p53−/− cortex, these results suggest that the cleavage plane orientation of RGPs is not an essential determinant of neurogenesis in the developing cortex.
DISCUSSION
In this study, we used a conditional null allele of Sas-4 to compare RGP behavior and cortical neurogenesis in the presence and absence of centrioles. We found that deletion of Sas-4 in RGPs leads to loss of centrioles, which triggers delocalization of RGPs from the VZ, progenitor mitotic delay, p53-dependent cell death, neuronal loss and microcephaly.
The underlying cause of microcephaly has been investigated in a number of mouse models, including partial loss-of-function of Cdk5rap2 (MCPH3/centrosomin) 18 and Sas-4/Cenpj 19 mutants. For example, the Sas-4 hypomorphic mice exhibit dwarfism from birth as well as mild microcephaly due to cell death associated with genomic instability and DNA damage 19. A similar link between aneuploidy-linked cell death and microcephaly phenotype was observed in a mouse model that overexpressed the centriole duplication protein Polo-like kinase 4 (PLK4) 20. In comparison, we observed no substantial increase in aneuploidy or DNA damage in the Sas-4−/− p53−/− brain. Instead, our data suggested that mitotic delay in progenitors that lack the centrosome(s) leads to wide scale p53 activation, cell death and eventually microcephaly.
While some apoptosis during embryonic brain development has also been observed in other mouse models of microcephaly 18–20, the microcephaly has generally been attributed to an increased rate of cell cycle exit and premature differentiation 18, 38, 47. It has also been suggested that the depletion of neural progenitors is caused by spindle abnormalities and changes in the cleavage plane orientation 18, 47. Even though the cleavage plane orientation was abnormal in Sas-4−/− p53−/− brains, they were not microcephalic, arguing that the cleavage plane orientation defect arising from the absence of centrosomes is not the main cause of microcephaly. While it is possible that mutations of different centrosomal proteins result in distinct cellular defects and microcephaly 18–20, our study suggested that p53-dependent cell death is likely to be a key cause of centrosome-associated microcephaly. Future characterization of this cell death pathway in vivo will provide crucial new information about p53 regulation and its relationship to microcephaly.
The VZ has long been thought of as a proliferative niche in the developing cortex 37, in which the centrosome helps to anchor the RGPs 35, 38. Our results supported the hypothesis that the centrosome is required to maintain RGPs in the VZ. However, in contrast to earlier reports of RGPs losing their proliferative capacity and differentiating when they exit the VZ 35, 38, we found that ~50% of RGPs outside the VZ in the Sas-4−/− cortex were eliminated through p53-dependent apoptosis. When apoptosis was prevented by simultaneous removal of p53, these ectopic RGPs continued to divide and produced a normal number of IPs and neurons, despite having lost both the direct contact with the ventricular cerebrospinal fluid (CSF) and the orientation of their cleavage plane relative to the VZ surface. Although ectopic RGPs arise from RGPs in the VZ and retain asymmetric division and self-renewal capability, they are not oRGs; instead, based on their morphology, mitotic behavior and marker expression, they are displaced RGPs with a unique morphology. Therefore, our data suggested that the VZ, the progenitor morphology, or the cleavage plane orientation is not essential for RGP asymmetric division and self-renewal. The displaced RGPs have an intrinsic program of proliferation and differentiation that can be executed outside the VZ with non-characteristic morphology and mitotic spindle orientation, as suggested by previous studies of isolated cortical progenitors in vitro 48 and embryonic stem cells in culture 49, 50. While it is likely that there are additional aspects of neuronal differentiation and cortical organization such as neuronal migration and precise lamination that depend on the centriole/centrosome, our study defined the principal functions of the centriole in RGPs during neocortical neurogenesis and revealed the robust plasticity of RGPs in the developing cortex in vivo.
ONLINE METHODS
Animals
The Sas-4 conditional allele was recently described 21. In brief, knockout-first ES cell line (EPD0028_7_G05) was obtained from the International Knockout Mouse Consortium (Cenpjtm1a(EUCOMM)Wtsi) (http://www.knockoutmouse.org). Actin-FLP transgenic mice (JAX stock #005703) were used to excise the gene trap and obtain Sas-4+/fl (fl, floxed allele). Nestin-Cre (JAX stock #003771), Emx1-Cre (JAX stock #005628), Ai9/tdTomato (JAX stock #007909) mice were obtained from the Jackson Laboratory. Trp53ftm1Brn mice were a kind gift from David Solit at SKI (JAX stock #008462). The analysis was performed in a mixed FVB/NJ and C57BL6 mouse background. Genotyping was carried out using standard PCR protocols. All experiments were conducted in accordance with animal protocols approved by the Institutional Animal Care and Use Committee (IACUC).
In utero retrovirus Injection
Moloney murine leukemia retroviruses were produced as previously described 51. In utero intraventricular injection was performed as previously described 51. In brief, uterine horns of pregnant mice at E11.5 gestation stages were exposed in a clean environment. Retrovirus solution (~1.0 μL) mixed with fast green (2.5 mg/mL, Sigma) was injected into the lateral ventricle through a beveled, calibrated glass micropipette (Drummond Scientific). During the procedure, the embryos were constantly bathed with warm phosphate buffered saline (PBS, pH 7.4). After injection, the uterus was placed back in the abdominal cavity and the wound was surgically sutured. After surgery, the animal was placed in a 28°C recovery incubator under close monitoring until it recovered and resumed normal activity.
Brain sections, immunohistochemistry, DiI labeling and confocal imaging
For analysis of brain sections, timed pregnant females that carried the Sas-4 and p53 conditional alleles and the Nestin-Cre transgene were anesthetized, embryos were removed and perfused with ice-cold phosphate buffered saline (PBS), followed by 4% PFA and prepared for cryosectioning or vibratrome sectioning and immunohistochemistry as previously described 52. The following antibodies were used: rabbit anti-ARL13B as previously described 45, rabbit anti-SAS-4 (a gift from Pierre Gönczy Institut Suisse de Recherches Experimentales sur le Cancer, Lausanne, Switzerland, 1:1000), rabbit anti-CUX1 (Santa Cruz, sc-13024, 1:100), rat anti-CTIP2 (Abcam, ab18465, 1:500), rabbit anti-PHH3 (Millipore, 06-570, 1:500), goat anti-FOXP2 (Santa Cruz, sc-21069, 1:100), rabbit anti-SATB2 (Abcam, ab92446, 1:500), rabbit anti-TBR2 (Abcam, ab23345, 1:500), rabbit anti-PCNT (BioLegend, prb-432c, 1:500), mouse anti-PCNT (BD Transduction Labs, 611814, 1:500), rabbit anti-PAX6 (Biolegend, prb-278p, 1:500), ), mouse anti-PAX6 (Developmental Hybridoma Bank, 1:100)), rabbit anti-CASP3 (Promega, G7481, 1:200), rabbit anti-p53 (Leica Microsystems, P52-CM5P, 1:200), mouse anti-Ki67 (BD Transduction Laboratories, 610968, 1:200, ), rabbit anti-Ki67 (Abcam, ab16667, 1:500), rabbit anti-SOX2 (Millipore, AB5603, 1:500), rabbit anti-BLBP (Abcam, ab32423, 1:500), mouse anti-P-VIMENTIN (Abcam, ab22651, 1:500), rat anti-BrdU (Accurate, YSRTMCA2060GA, 1:500), rabbit anti-γ-H2AX (Cell Signaling, 9718, 1:200), mouse anti-tyrosinated α-TUBULIN (Sigma, T9028, 1:1000) and rat anti-EGFP (Nacalai Tesque, GF090R, 1:1000). Alexa fluor 488-, 546-, or 647-conjugated secondary antibodies (Life Technologies) were used. BrdU injection and staining were performed as described previously 53, and EdU staining was performed using the manufacturer’s protocol (Life Technologies). DiI labeling was carried out by sprinkling small DiI crystals (Life Technologies) onto fixed E15.5 brains with the meninges removed. After incubation at 37° for 24 hours, brains were sectioned on a vibratome, and stained/washed as previously described 52, but only with solutions containing 0.3% Tween-20 (Sigma). FISH staining of brain sections was performed according to standard protocols 54. Images were obtained using a FV1000 (Olympus), or Leica SP5, and SP8 confocal microscopes (Leica microsystems). Confocal images were analyzed using Volocity software package (Improvision), Photoshop (Adobe) and ImageJ (NIH).
Quantification and statistical analysis
For the measurement of telencephalic area, a single cerebral hemisphere was traced from a dorsally oriented whole mount image. For relative thickness of cortical layers, the thickness of the major layer (i.e. CUX1+ or CTIP2+) was measured relative to the overall thickness of the cortex (from the pial surface to the white matter). For cell counts in cortical layers, all cells within a 250 μm width columnar area from the white matter to the pial surface were counted. For cell counts of embryonic progenitors, all cells within a 250 μm width columnar area from the ventricular surface to the pial surface were counted. For centrosomal and primary cilia to nuclei ratio quantification, the total PCNT or ARL13B puncta along 50 μm of the VZ surface of the ventricle was counted in relation to the overlying nuclei (DAPI) within the VZ. For extra-VZ quantification, the total PCNT or ARL13B puncta within a 50 μm2 area were counted in relation to total nuclei in the area. The cleavage plane orientation was defined by the angle between the ventricular surface and the cleavage plane. Morphology of cells was defined as the following: bipolar - two processes in the direction of the pial and apical surface; unipolar - a single process in the direction of the pial surface; multipolar - more than two visible processes (with or without a prominent process oriented towards the pial surface). Any cell that the morphology was not clearly visible was excluded from analysis.
Animals were assigned to groups on the basis of genotype. Age-matched littermates were used as controls in all experiments. No randomization was used and no animals were excluded from analyses. No statistical methods were used to predetermine sample sizes. Each experiment was done by analyzing at least three different brains. Our sample sizes are similar to those reported in previous publication 21. For all experiments, data distribution was assumed to be normal, but this was not formally tested. Two groups of data were assessed for significant differences using a two-tailed unpaired Student’s t-test with a cutoff for significance of p<0.05 (Prism software, GraphPad). The data are presented as the mean ± SD (standard deviation).
A Supplementary Methods Checklist with individual p values and degrees of freedom is available.
Supplementary Material
Acknowledgments
We thank Bryan M.-F. Tsou for reagents and advice; Pierre Gönczy (Institut Suisse de Recherches Experimentales sur le Cancer, Lausanne, Switzerland) for reagents; David Solit for p53fl/fl mice; and Vitaly Boyko and Yevgeniy Romin at the Molecular Cytology Core Facility for their technical support. We thank the Anderson and Shi lab members for their input and comments on the manuscript. This work was supported by grants from the National Institutes of Health (R01DA024681 and R01NS085004 to S.-H.S, and R01NS044385 to K.V.A.). H.B. was supported by NKF and NRSA postdoctoral fellowships.
Footnotes
AUTHOR CONTRIBUTIONS
R.I., H.B., K.V.A., and S.-H.S. conceived the project and designed the experiments. R.I. and W.S. analyzed brain phenotype, and H.B. generated mutant animals. All authors discussed the results and contributed to aspects for preparing the manuscript.
References
- 1.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 3.Miyata T, et al. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 4.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci. 2011;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Cerdeno V, et al. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PLoS One. 2012;7:e30178. doi: 10.1371/journal.pone.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reillo I, de Juan Romero C, Garcia-Cabezas MA, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex. 2011;21:1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- 9.Fietz SA, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 10.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 11.Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayer SA, Altman J. Neocortical development. Ravens; New York: 1991. [Google Scholar]
- 13.Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193:435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- 15.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 16.Gilmore EC, Walsh CA. Genetic causes of microcephaly and lessons for neuronal development. Wiley interdisciplinary reviews Developmental biology. 2013;2:461–478. doi: 10.1002/wdev.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woods CG, Bond J, Enard W. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. American journal of human genetics. 2005;76:717–728. doi: 10.1086/429930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lizarraga SB, et al. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development. 2010;137:1907–1917. doi: 10.1242/dev.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McIntyre RE, et al. Disruption of mouse Cenpj, a regulator of centriole biogenesis, phenocopies Seckel syndrome. PLoS Genet. 2012;8:e1003022. doi: 10.1371/journal.pgen.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marthiens V, et al. Centrosome amplification causes microcephaly. Nature cell biology. 2013;15:731–740. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- 21.Bazzi H, Anderson KV. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci U S A. 2014;111:E1491–1500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt TI, et al. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 23.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. CPAP is a cell-cycle regulated protein that controls centriole length. Nature cell biology. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 24.Basto R, et al. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 25.Leidel S, Gonczy P. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Developmental cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 26.Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 27.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 28.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 29.Chizhikov VV, et al. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. J Neurosci. 2007;27:9780–9789. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutton SR, Pevny LH. SOX2 expression levels distinguish between neural progenitor populations of the developing dorsal telencephalon. Dev Biol. 2011;352:40–47. doi: 10.1016/j.ydbio.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- 32.Gorski JA, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Chenn A, Zhang YA, Chang BT, McConnell SK. Intrinsic polarity of mammalian neuroepithelial cells. Molecular and cellular neurosciences. 1998;11:183–193. doi: 10.1006/mcne.1998.0680. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamei Y, et al. Visualization of mitotic radial glial lineage cells in the developing rat brain by Cdc2 kinase-phosphorylated vimentin. Glia. 1998;23:191–199. doi: 10.1002/(sici)1098-1136(199807)23:3<191::aid-glia2>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 37.Lehtinen MK, et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchman JJ, et al. Cdk5rap2 interacts with pericentrin to maintain the neural progenitor pool in the developing neocortex. Neuron. 2010;66:386–402. doi: 10.1016/j.neuron.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 39.Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–1292. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 40.Buchman JJ, Tsai LH. Spindle regulation in neural precursors of flies and mammals. Nat Rev Neurosci. 2007;8:89–100. doi: 10.1038/nrn2058. [DOI] [PubMed] [Google Scholar]
- 41.Lancaster MA, Knoblich JA. Spindle orientation in mammalian cerebral cortical development. Curr Opin Neurobiol. 2012;22:737–746. doi: 10.1016/j.conb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nature cell biology. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 43.Haydar TF, Ang E, Jr, Rakic P. Mitotic spindle rotation and mode of cell division in the developing telencephalon. Proc Natl Acad Sci U S A. 2003;100:2890–2895. doi: 10.1073/pnas.0437969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanada K, Tsai LH. G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Yingling J, et al. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konno D, et al. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nature cell biology. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- 47.Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci U S A. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Q, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 49.Gaspard N, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 50.Eiraku M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell stem cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 51.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 52.Bultje RS, et al. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown KN, et al. Clonal production and organization of inhibitory interneurons in the neocortex. Science. 2011;334:480–486. doi: 10.1126/science.1208884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leversha MA. Mapping of genomic clones by fluorescence in situ hybridization. Methods in molecular biology. 2001;175:109–127. doi: 10.1385/1-59259-235-X:109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.