Abstract
The recent discovery of conditions that induce nanoribbon structures of amelogenin protein in vitro raises questions about their role in enamel formation. Nanoribbons of recombinant human full-length amelogenin (rH174) are about 17 nm wide and self-align into parallel bundles; thus, they could act as templates for crystallization of nanofibrous apatite comprising dental enamel. Here we analyzed the secondary structures of nanoribbon amelogenin by x-ray diffraction (XRD) and Fourier transform infrared spectroscopy (FTIR) and tested if the structural motif matches previous data on the organic matrix of enamel. XRD analysis showed that a peak corresponding to 4.7 Å is present in nanoribbons of amelogenin. In addition, FTIR analysis showed that amelogenin in the form of nanoribbons was comprised of β-sheets by up to 75%, while amelogenin nanospheres had predominantly random-coil structure. The observation of a 4.7-Å XRD spacing confirms the presence of β-sheets and illustrates structural parallels between the in vitro assemblies and structural motifs in developing enamel.
Keywords: self-assembly, structure, development, powder diffraction, Fourier transform infrared spectroscopy, secondary protein structure
Introduction
Amelogenin has been classified as an intrinsically disordered protein lacking considerable secondary structure, but, at the same time, several in vitro studies have reported the presence of a broad range of α-helices, poly-proline, b-sheets, and b-spirals (Renugopalakrishnan et al., 1989; Delak et al., 2009; Jin et al., 2009; Zhang et al., 2011). Amelogenin, which makes up about 90% of all enamel matrix protein, is critical to enamel formation – the most intricate architecture of a mineralized tissue in the human body.
Dental enamel is comprised of fibrous apatite nanocrystals to 95% by weight. These crystals measure about 50 nanometers in diameter and are hundreds of micrometers long, most likely forming a continuous mineral fiber that runs from the surface of the dentin to the occlusal surface of the enamel. The mineral fibers are well-aligned and packed into rods that develop in association with the movement of the Tomes’ process at the distal ends of the ameloblasts. It is therefore surprising that the highly organized architecture of enamel, with its structural control of mineralization at the nanometer level, has not been associated with an organic matrix that is able to assemble into similar higher order structures. Amelogenin is a highly hydrophobic protein and forms nanospheres in an aqueous environment in vitro (Fincham et al., 1999). Detailed structural evaluations of recombinant amelogenin and native extracted amelogenin reported in the literature have been performed exclusively on nanospherical amelogenin or molecular amelogenin (Delak et al., 2009; Jin et al., 2009; Zhang et al., 2011).
Only recently have elongated and fibrous structures of this protein, exhibiting a structural similarity with the fibrillar morphology of apatite crystals in enamel, been reported (He et al., 2011). Amelogenin is also an amphiphilic molecule, since it contains a short, about 15 residues long, hydrophilic C-terminus. On the basis of the amphiphilic character, an emulsion system was developed that facilitated assembly of amelogenins into nanoribbons at the water-oil interface (He et al., 2011; Martinez-Avila et al., 2011). In these studies, amelogenin showed the ability to form nanoribbons of 16.7 (± 1) nm in width and several micrometers in length. Most importantly, these initial studies revealed that 2 ions, calcium and phosphate, were essential for the formation of ribbons from recombinant amelogenin, suggesting that they are required for proper folding of the protein. Both ions are the main components of apatite mineral and are available in abundance in developing enamel. The ions appear to stabilize protein dimers, which act as building blocks for assembly into nanoribbons. In a subsequent study, identical amelogenin nanoribbons were reproduced in calcium- and phosphate-rich solutions in the absence of an oil phase (Martinez-Avila et al., 2012). Such ribbons form with similar kinetics over a period of days. Ribbon assembly requires a slightly acidic pH between 4 and 6 and is attributed to the importance of histidine protonation for proper folding of amelogenin molecules and possible phosphate interactions (Martinez-Avila et al., 2012).
The structure of developing enamel and its insoluble portion of the organic matrix has been extensively analyzed, and evidence of aligned fibrillar protein structures has been provided by electron microscopy (Perdok and Gustafson, 1961; Travis and Glimcher, 1964; Nanci et al., 1985). In addition, x-ray diffraction (XRD) studies on fixed and demineralized enamel matrices were performed to investigate the presence of secondary structures of the enamel proteins. A sharply oriented reflection at 4.7 Å was observed by several investigators between 1958 and 1964 in human, bovine, and rodent enamel (Little, 1958; Pautard, 1961; Perdok and Gustafson, 1961; Travis and Glimcher, 1964; Glimcher et al., 1965). A detailed study that carefully aimed to preserve the protein native structure by a series of different fixation protocols confirmed the 4.7-Å reflection in 1986 and also reported a weaker reflection at 4.2 Å (Jodaikin et al., 1986). The d-spacing at 4.7 Å has been associated with β-sheet structures of many proteins and peptides, while the 4.2-Å reflection is not as common but has been associated with the distance between aromatic residues in β-sheets or the presence of certain lipids (Jodaikin et al., 1986; Aggeli et al., 1997; Kubelka and Keiderling, 2001; Xu, 2009). Structural Fourier transform infrared spectroscopy (FTIR) analysis of the enamel matrix has not yet been reported. Extracted and recombinant amelogenins have been studied by FTIR, but there seems to be no agreement on what secondary structure predominates in amelogenin assemblies of the nanosphere type, since many major motifs have been observed (Lakshminarayanan et al., 2007; Beniash et al., 2012).
In this study, we used XRD and FTIR to analyze the structure of nanoribbons of recombinant human amelogenin protein rH174 and compared the data with previous findings on the enamel matrix. Our analysis suggests that the nanoribbons are predominantly (75%) comprised of antiparallel beta-sheets, as proposed in a previous model (He et al., 2011; Martinez-Avila et al., 2012). The diffraction pattern at 4.7 Å corresponds very well with observations made by others on the enamel matrix, indicating that this form of assembled structure may also be adopted by amelogenin during amelogenesis.
Materials & Methods
Recombinant Amelogenin Protein and Assembly
The full-length human amelogenin protein rH174 was expressed in BL21DE3 pLys Escherichia coli and purified on C4 hydrophobic beads as previously described (Li et al., 2003). Compared with the native protein (H175), rH174 lacks the first residue methionine and is not phosphorylated at serine-16. The purity of rH174 was above 95% as assessed by HPLC. The protein was self-assembled into nanoribbons in an oil-free environment by the mixing of 2 mg/mL with aqueous solutions containing 33.4 mM CaCl2 and 20.9 mM KH2PO4 at pH between 4.5 and 5.0, as described in detail previously (Martinez-Avila et al., 2012). Nanoribbon-free protein control samples were prepared in aqueous solutions at pH 5 containing 33 mM KCl, without calcium and phosphate. Protein-free controls consist of aqueous solutions containing 33.4 mM CaCl2 and 20.9 mM KH2PO4 at pH between 4.5 and 5.0, without amelogenin. The presence of nanoribbons and nanospheres, respectively, was confirmed by atomic force microscopy.
Atomic Force Microscopy (AFM)
We performed AFM analysis by pipetting 25 μL of the respective solution onto a silica glass slide. The droplet was left for 1 hr in a wet cell to facilitate immobilization of the protein to the glass surface. Subsequently, the droplet was rinsed off with a few drops of deionized water. Canned air was used to blow off any residual water (Martinez-Avila et al., 2012). All imaging was performed under dry conditions in tapping mode with silicon cantilevers at about 300 kHz on a Multi-Mode AFM with Nanoscope III controller (Digital Instruments, Goleta, CA, USA). Width and height measurements were performed without corrections for tip-broadening from height-mode image data with Nanoscope software 5.37.
Fourier Transform Infrared Spectroscopy
Samples studied by AFM were used for FTIR analysis. Specimens with nanospheres or nanoribbons were directly placed in contact with a diamond crystal for Attenuated Total Reflection Analysis (ATR) by infrared spectroscopy with a Nicolet 6700 spectrometer from Thermo Fisher Scientific (Waltham, MA, USA). The ATR spectra were collected between 500 and 4,000 cm−1 at a resolution of 4 cm−1 with 256 accumulations per run. Spectra were analyzed with FTIR software OMNIC 8.2.0.387 (Thermo Fisher Scientific). Spectral deconvolution of the Amide-I peak was performed with Origin 7.5 software, as described in the Appendix.
X-ray Diffraction
Powder XRD crystallography was performed with 0.11159-nm synchrotron radiation at beam line 8.3.1 of the Advanced Light Source at Lawrence Berkeley National Laboratory. XRD experiments were performed on nanoribbons immobilized on a thin Kapton grid, from a 20-µL extract of protein solution (2 mg/mL) which was left to dry at room temperature. XRD patterns were recorded for: (i) the dehydrated samples of self-assembled rH174 from calcium phosphate solution, (ii) precipitates from calcium phosphate solution without protein, and (iii) the lyophilized protein only. Reflections were identified and assigned according to the International Centre for Diffraction Data (ICDD) databank for inorganic minerals. XRD sample images were initially processed with the FIT2D data analysis program (Hammersley, 1997). Images from the XRD beam line were background-subtracted using a blank sample with a comparable exposure time and detector distance, taken within a few hours of the measurement. They were radially integrated, and the profiles were processed in GNU Octave. Peaks were assigned manually based on the following criteria: d-spacing within 0.03 Å of previously published data, and presence of the previously published first- or second-highest-intensity peak in our dataset. Several low-intensity peaks remained unassigned.
Results
Self-assembly experiments of rH174 were performed for 7 days in potassium chloride solutions with and without calcium phosphate. AFM analysis of the solutions without calcium phosphate showed the presence of nanospheres with diameters between 14 and 63 nm (Fig. 1A). In contrast, in the presence of calcium and phosphate, nanoribbons developed, as described previously (Fig. 1B) (Martinez-Avila et al., 2012). The specimen surface revealed zones of protein aggregations, with large numbers of ribbons stacked on top of each other (Fig. 1C).
Figure 1.
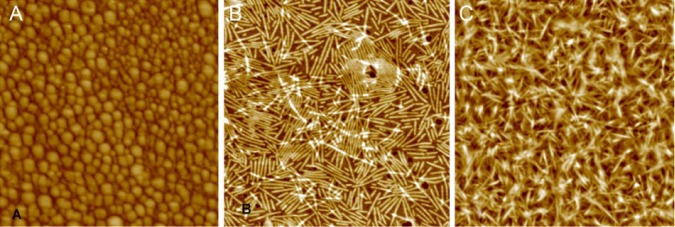
AFM analysis of self-assembled structures of amelogenin. After 7 days of incubation, rH174 assembled into (A) nanospheres in the absence of calcium phosphate (scan size, 850 nm); (B) self-aligning nanoribbons in the presence of calcium phosphate (scan size, 1,200 nm); and (C) ribbon agglomerates, observed in several zones (scan size, 1,300 nm).
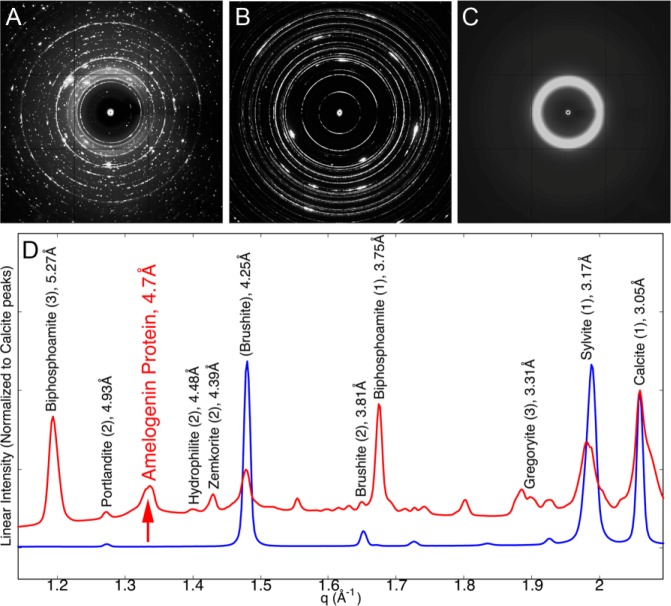
Fig. 2 shows characteristic powder XRD patterns obtained on rH174 nanoribbons in calcium phosphate solutions (Fig. 2A) in comparison with protein-free calcium phosphate solutions (Fig. 2B) and the lyophilized protein (Fig. 2C). The radially averaged XRD data for samples with and without amelogenin are plotted as a function of the q-spacing. Numerous sharp peaks are present in both the protein-free control and the sample with nanoribbons (e.g., 2.0 Å, 2.6 Å, 3.0 Å) (Fig. 2D and Appendix Fig. 1). These peaks are associated with regular salt structures that precipitated upon dehydration of the samples. According to the ICDD database, intense reflections were assigned to phosphate, carbonate, and chloride minerals, as listed in the Appendix Table. Brushite [CaHPO4• 2(H2O)], calcite (CaCO3) biphosphoamite (KH2PO4), and sylvite (KCl) are the main components of the precipitates observed after complete evaporation of the solvent. In addition to the reflections associated with mineral, a broader peak at 4.7 Å (Fig. 2D) was observed only in the sample containing rH174 nanoribbons and not in the protein-free control. This reflection could not be assigned to any mineral phase listed in the ICDD database for elements selected in these experiments. Furthermore, the sample containing rH174 shows a broad zone of elevated intensity between d-spacings of 2 to 6 Å, which is most likely associated with the presence of an amorphous phase (Fig. 2A, Appendix Fig. 1). The pure lyophilized protein sample produced a single, very broad peak centered around 5.1 Å and is shown in Fig. 2C (see Appendix Fig. 3 for comparison).
Figure 2.
X-ray diffraction analysis. (A) Amelogenin nanoribbon with buffer, (B) buffer only, and (C) pure lyophilized protein. Images obtained from a 1.1159-Å source, at a detector distance of 200 mm, and scale to 313 mm per side. (D) Radial averages of the x-ray diffraction of the nanoribbon+buffer (red) and buffer-only samples (blue). Assigned peaks are labeled with their mineral names. Full assignments can be found in Appendix Fig. 1 and the Appendix Table.
FTIR analyses of rH174 in the form of nanoribbons, nanospheres, and as lyophilized powder are shown in Fig. 3A from 900 to 1,800 cm−1. All samples show strong bands for amide-I (1,600-1,700 cm−1), amide-II (1,500-1,600 cm−1), and CH modes (1,450 cm−1). Because of the high salt content, several minerals were detected in the spectra from rH174 ribbons. Carbonates are associated with peaks at 1,409 cm−1 and at 871 cm−1, while phosphate vibrational modes were detected between 980 and 1,115 cm−1. A significant difference in the amide-I peak maxima was observed in comparison of amelogenin in the form of nanosphere with nanoribbon assemblies. The amide-I peak maximum was 1,660 and 1,629 cm−1 for rH174 in the form of nanospheres and nanoribbons, respectively. In addition, a shoulder at 1,715 cm−1 was observed for the ribbon-like assemblies of rH174 (see Fig. 3B). Deconvolution of the amide-I peak was used to identify secondary structural components of the protein and to calculate their proportional content (Table). According to the FTIR analysis, amelogenin nanospheres are composed of about 53% of random coil structure, while nanoribbons have obtained b-sheet configuration to about 75%, with only 21% of random coil structure remaining.
Figure 3.
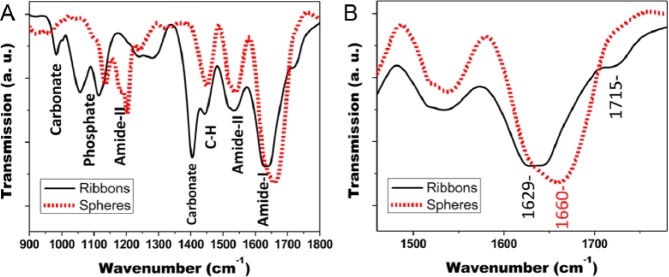
FTIR analysis of amelogenin nanoribbons and nanospheres. (A) Spectral range from 900 to 1,800 cm−1 shows mineral precipitates as phosphates and carbonates in nanoribbon sample. (B) Sections of amide-I and amide-II show the strong shift of the amide-I peak to lower wavelength in nanoribbons and a high-frequency peak at 1,715 cm−1, indicating increased b-sheet content. Amide-I peak deconvolution is shown in the Appendix.
Table.
Calculated Relative Areas for Each Deconvoluted Peak Associated with Amide-I in the Range from 1,600 to 1,700 cm−1 as Determined by FTIR, Comparing Amelogenin rH174 as Nanosphere vs. Nanoribbon
| Nanospheres |
Nanoribbons |
||
|---|---|---|---|
| Assignment | Peak-Max | Peak Area (%) | Peak Area (%) |
| Extended | 1,612 cm−1 | 3 | 0 |
| β-sheet | 1,625 cm−1 | 16 | 75 |
| Random coil | 1,650 cm−1 | 53 | 21 |
| α-helix/β-turn | 1,665 cm−1 | 24 | 4 |
| β-sheet (secondary) | 1,685 cm−1 | 4 | Outside range |
Peak assignments are based on literature listed in the Appendix.
Discussion
Amelogenin is the main organic component of the enamel matrix and has a significant role in controlling enamel mineralization, as shown by knock-out and transgenic mice models (Gibson et al., 2001; Snead et al., 2011). Self-assembly of matrix molecules is nature’s tool to build tissues, and collagen-based mineralized tissues are a prime example of a strategy to use fibrous assemblies for the synthesis of larger structures. In the case of enamel, the current hypothesis assumes the formation of nanospheres to be the structural component of the developing enamel matrix (Fincham et al., 1999). This theory is mainly derived from the observation of the amelogenin nanospheres that form in vitro when the protein is suspended in aqueous solutions. Very few studies have actually shown spherical features of about 20 nm in enamel, leaving little evidence that such nanospheres are actually present in enamel (Fincham et al., 1995). In contrast, there have been TEM studies that indicate the presence of filamentous structures in the enamel matrix, and these were listed in a previous publication (Martinez-Avila et al., 2011).
In vitro self-assembly of full-length amelogenin into nanoribbons requires the presence of calcium and phosphate ions (Martinez-Avila et al., 2012). In the presence of these ions, salt crystals precipitate during the evaporation of the aqueous solvent, resulting in a large number of crystal diffraction patterns (Fig. 2). We have identified intensive reflections of the XRD data, as listed in the Appendix Table. In addition to these mineral phases, a relatively strong reflection with a d-spacing of 4.7 Å was detected. In 1965, Glimcher and co-workers performed x-ray diffraction on a calcium-depleted incisor of an embryonic calf, and found a 4.77-Å peak that they associated with the organic matrix (Glimcher et al., 1965). Numerous studies on proteins and peptides have also shown that XRD spacings around 4.7 Å are characteristic of b-sheets (Jodaikin et al., 1986; Symmons et al., 1997; von Bergen et al., 2005).
FTIR analysis further supports this finding, since amide-I peaks between 1,615 and 1,640 cm−1 have been associated with b-sheet formation in proteins (Haris and Chapman, 1995). Deconvolution indicates that 75% of the proteins have adopted b-sheet structure in nanoribbons, vs. only 16% in nanospheres (Table). An amide-I peak around 1,650 cm−1 is associated with random-coil structure and may be the main component of amelogenin nanospheres (Table), as also reported by others (Beniash et al., 2012). The random-coil configuration may allow for more flexibility and explain the large distribution of nanosphere diameters between 10 and 100 nm that have been reported (Moradian-Oldak, 2001). The nanoribbon configuration of amelogenin is quite different. Nanoribbons of rH174 have a precise width of 16.7 (± 1) nm, possibly related to the width of an antiparallel amelogenin dimer which may constitute the building block of the ribbons (He et al., 2011; Martinez-Avila et al., 2012). There is evidence in the literature that antiparallel b-sheets have a secondary amide-I peak at increased wavenumbers between 1,680 and 1,715 cm−1 (Haris and Chapman, 1995). Similar FTIR absorption peaks have been observed in amyloids and ascribed to extended conformation of the b-sheet domains (Shivu et al., 2013).
This study provided additional evidence that assemblies of the full-length amelogenin protein differ strongly in their secondary structure when assembled into nanospheres or nanoribbons. The presence of b-sheets in amelogenin nanoribbons which match the XRD diffraction pattern of matrices in developing enamel suggests that nanoribbons may also be the functional supramolecular structure of amelogenin that is able to control mineralization in amelogenesis.
Supplementary Material
Acknowledgments
We thank Drs. Feroz Khan and Wu Li (UCSF) for providing us with amelogenin protein rH174, Dr. Sebnem Inceoglu (UCB) for support of FTIR analysis, and James Holton for support of XRD analysis.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This study was funded by National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) grant R21-023422. Portions of this work were performed at the Molecular Foundry and the Advanced Light Source, both of which are supported by the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Aggeli A, Bell M, Boden N, Keen JN, Knowles PF, McLeish TC, et al. (1997). Responsive gels formed by the spontaneous self-assembly of peptides into polymeric beta-sheet tapes. Nature 386:259-262. [DOI] [PubMed] [Google Scholar]
- Beniash E, Simmer JP, Margolis HC. (2012). Structural changes in amelogenin upon self-assembly and mineral interactions. J Dent Res 91:967-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan Y, Moradian-Oldak J, et al. (2009). The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry 48:2272-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Diekwisch TG, Lyaruu DM, Wright JT, Bringas P, Jr, et al. (1995). Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. J Struct Biol 115:50-59. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Simmer JP. (1999). The structural biology of the developing dental enamel matrix. J Struct Biol 126:270-299. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, et al. (2001). Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem 276:31871-31875. [DOI] [PubMed] [Google Scholar]
- Glimcher MJ, Daniel EJ, Travis DF, Kamhi S. (1965). Electron optical and x-ray diffraction studies of organization of inorganic crystals in embryonic bovine enamel. J Ultrastruct Res 50(Suppl 7):1-77. [PubMed] [Google Scholar]
- Hammersley AP. (1997). FIT2D: an introduction and overview. ESRF Internal Report ESRF97HA02T. [Google Scholar]
- Haris PI, Chapman D. (1995). The conformational-analysis of peptides using Fourier-transform IR spectroscopy. Biopolymers 37:251-263. [DOI] [PubMed] [Google Scholar]
- He X, Wu S, Martinez-Avila O, Cheng Y, Habelitz S. (2011). Self-aligning amelogenin nanoribbons in oil-water system. J Struct Biol 174:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Ito Y, Luan X, Dangaria S, Walker C, Allen M, et al. (2009). Elongated polyproline motifs facilitate enamel evolution through matrix subunit compaction. PLoS Biol 7:e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodaikin A, Traub W, Weiner S. (1986). Protein conformation in rat tooth enamel. Arch Oral Biol 31:685-689. [DOI] [PubMed] [Google Scholar]
- Kubelka J, Keiderling TA. (2001). Differentiation of beta-sheet-forming structures: ab initio-based simulations of IR absorption and vibrational CD for model peptide and protein beta-sheets. J Am Chem Soc 123:12048-12058. [DOI] [PubMed] [Google Scholar]
- Lakshminarayanan R, Fan D, Du C, Moradian-Oldak J. (2007). The role of secondary structure in the entropically driven amelogenin self-assembly. Biophys J 93:3664-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Gao C, Yan Y, DenBesten PK. (2003). X-linked amelogenesis imperfecta May result from decreased formation of tyrosine rich amelogenin peptide (TRAP). Arch Oral Biol 48:177-183. [DOI] [PubMed] [Google Scholar]
- Little K. (1958). Electron microscope studies on human dental enamel. J R Microsc Soc 78:58-66. [DOI] [PubMed] [Google Scholar]
- Martinez-Avila OM, Wu S, Cheng Y, Lee R, Khan F, Habelitz S. (2011). Self-assembly of amelogenin proteins at the water-oil interface. Eur J Oral Sci 119(Suppl 1):75-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Avila O, Wu S, Kim SJ, Cheng Y, Khan F, Samudrala R, et al. (2012). Self-assembly of filamentous amelogenin requires calcium and phosphate: from dimers via nanoribbons to fibrils. Biomacromolecules 13:3494-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradian-Oldak J. (2001). Amelogenins: assembly, processing and control of crystal morphology. Matrix Biol 20:293-305. [DOI] [PubMed] [Google Scholar]
- Nanci A, Bendayan M, Slavkin HC. (1985). Enamel protein biosynthesis and secretion in mouse incisor secretory ameloblasts as revealed by high-resolution immunocytochemistry. J Histochem Cytochem 33:1153-1160. [DOI] [PubMed] [Google Scholar]
- Pautard FG. (1961). An x-ray diffraction pattern from human enamel matrix. Arch Oral Biol 3:217-220. [DOI] [PubMed] [Google Scholar]
- Perdok WG, Gustafson G. (1961). X-ray diffraction studies of the insoluble protein in mature human enamel. Arch Oral Biol 4:70-75. [DOI] [PubMed] [Google Scholar]
- Renugopalakrishnan V, Prabhakaran M, Huang SG, Balasubramaniam A, Strawich E, Glimcher MJ. (1989). Secondary structure and limited three-dimensional structure of bovine amelogenin. Connect Tissue Res 22:131-138. [PubMed] [Google Scholar]
- Shivu B, Seshadri S, Li J, Oberg KA, Uversky VN, Fink AL. (2013). Distinct beta-sheet structure in protein aggregates determined by ATR-FTIR spectroscopy. Biochemistry 52:5176-5183. [DOI] [PubMed] [Google Scholar]
- Snead ML, Zhu DH, Lei Y, Luo W, Bringas PO, Jr, Sucov HM, et al. (2011). A simplified genetic design for mammalian enamel. Biomaterials 32:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons MF, Buchanan SGS, Clarke DT, Jones G, Gay NJ. (1997). X-ray diffraction and far-UV CD studies of filaments formed by a leucine-rich repeat peptide: structural similarity to the amyloid fibrils of prions and Alzheimer’s disease beta-protein. FEBS Lett 412:397-403. [DOI] [PubMed] [Google Scholar]
- Travis DF, Glimcher MJ. (1964). The structure and organization of, and the relationship between the organic matrix and the inorganic crystals of embryonic bovine enamel. J Cell Biol 23:447-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergen M, Barghorn S, Biernat J, Mandelkow EM, Mandelkow E. (2005). Tau aggregation is driven by a transition from random coil to beta sheet structure. Biochim Biophys Acta 1739:158-166. [DOI] [PubMed] [Google Scholar]
- Xu S. (2009). Cross-beta-sheet structure in amyloid fiber formation. J Phys Chem B 113:12447-12455. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ramirez BE, Liao X, Diekwisch TG. (2011). Amelogenin supramolecular assembly in nanospheres defined by a complex helix-coil-PPII helix 3D-structure. PLoS One 6:e24952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.