Abstract
Background and Purpose
Common variants have been identified using genome-wide association studies which contribute to intracranial aneurysms (IA) susceptibility. However, it is clear that the variants identified to date do not account for the estimated genetic contribution to disease risk.
Methods
Initial analysis was performed in a discovery sample of 2,617 IA cases and 2,548 controls of Caucasian ancestry. Novel chromosomal regions meeting genome-wide significance were further tested for association in two independent replication samples: Dutch (717 cases; 3,004 controls) and Finnish (799 cases; 2,317 controls). A meta-analysis was performed to combine the results from the three studies for key chromosomal regions of interest.
Results
Genome-wide evidence of association was detected in the discovery sample on chromosome 9 (CDKN2BAS; rs10733376: p< 1.0 × 10−11), in a gene previously associated with IA. A novel region on chromosome 7, near HDAC9, was associated with IA (rs10230207; p= 4.14 × 10−8). This association replicated in the Dutch sample (p=0.01), but failed to show association in the Finnish sample (p=0.25). Meta-analysis results of the three cohorts reached statistical significant (p=9.91 × 10−10).
Conclusions
We detected a novel region associated with IA susceptibility that was replicated in an independent Dutch sample. This region on chromosome 7 has been previously associated with ischemic stroke and the large vessel stroke occlusive subtype (including HDAC9), suggesting a possible genetic link between this stroke subtype and IA.
Keywords: intracranial aneurysm, genomewide association study, chromosome 7
INTRODUCTION
Subarachnoid hemorrhage (SAH) due to the rupture of an intracranial aneurysm (IA) occurs in 16,000 to 17,000 persons in the U.S. annually, over 40% die within 30 days.1, 2 There is evidence that aneurysmal SAH has a genetic contribution. First degree relatives of an SAH patient have 2–6 times greater frequency of an SAH as compared with age-matched controls.3–6 First and second degree relatives of an SAH or IA patient also have a greater risk of an unruptured IA (8.7% – 13.9%), compared with the general population (1%).7–9 These data support a genetic contribution but could also be due to common life-style related risk factors.
Genome-wide association studies (GWAS) identified and replicated significant associations at chromosome 4q31.23 (EDNRA), 5q31.3, 6q24.2, 8q12.1 (SOX17), 9p21.3 (CDKN2A/CDKN2B/CDKN2BAS), 10q24.32 (CNNM2), 12q22, 13q13.1 (KL/STARD13), 18q11.2 (RBBP8), and 20p12.1.10–15 We utilized a case-control design to identify additional loci associated with IA in a large sample of European ancestry. IA cases were enriched for a positive family history of IA, which may increase the contribution of genetic factors. Two independent replication samples were examined to confirm the most promising findings in the discovery sample.
SUBJECTS AND METHODS
Discovery Sample
The discovery sample involved 2,617 Caucasian IA cases and 2,548 Caucasian controls identified through different studies. Some of these samples were reported as part of a previous analysis.12 IA cases were ascertained through 5 studies and controls through 5 studies. All studies obtained appropriate institutional ethics approvals (Supplemental Materials).
Genotyping and Quality Review
The discovery sample, with the exception of the Atherosclerosis Risk in Communities (ARIC) samples, was genotyped on the Affymetric Axiom array. The ARIC samples were genotyped on the Affymetric SNP array 6.0. All released Axiom genotypes underwent a common quality review pipeline that included identification of sample duplicates, related individuals, and gender discrepancies. Prior to performing imputation, SNPs were excluded if there were: (i) improper mapping to Genome Reference Consortium GRCh37; (ii) a minor allele frequency (MAF) <0.03; (iii) SNP genotype call rate <95%; or (iv) Hardy Weinberg Equilibrium (HWE) p-value <10−4 in controls. MAF and call rates were calculated by combining all Axiom array data. From the 597,320 SNPs on the Axiom array, 464,632 were retained following this quality review. The ARIC samples genotyped on the Affymetric SNP array 6.0 underwent the same quality review as the Axiom genotypes. From the 793,799 autosome SNPs on the Affymetric SNP array 6.0 that were provided by ARIC following their initial data review, a total of 626,645 were retained (Supplemental Materials).
Principal component analysis (PCA) was performed using the SNPs genotyped on both the Axiom and Affymetric SNP array 6.0 platforms. Analyses were performed using Eigenstrat16 using data from 11 HapMap phase III populations. To ensure that the discovery sample included only Caucasian subjects, all samples clustering outside the CEU and TSI samples were excluded from further analysis. (Supplemental Materials, Supplemental Figure I).
Imputation and Statistical Analysis
Imputation was performed for all autosomes using IMPUTE2 (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html). All samples genotyped on the Axiom array that passed quality review (n=4,060) were imputed together using the 1000Genomes haplotypes (n=1092) as the phased reference panel. Only variants with more than one minor copy across all 1000Genome populations were imputed. Original genotypes were not overwritten. ARIC samples that passed quality review (n=1,132) were imputed separately using the same reference panel.
All ARIC samples genotyped on the Affymetrix SNP array 6.0 were controls, whereas all cases and the remaining controls were genotyped on the Axiom array. Therefore, extensive and detailed quality review was performed to ensure that spurious association was not detected based on platform effects. SNPs imputed in all datasets were not included in the analysis sample; only SNPs genotyped on at least one of the arrays (1,195,878 SNPs) were considered for inclusion in the analysis. We used the approach described by Sinnott and Kraft17 which implements an aggressive filtering approach to remove SNPs having the potential for systematic platform differences (Supplemental Materials). The final dataset retained 672,210 autosome SNPs for analysis.
The genotyped and imputed SNPs were tested for association with IA susceptibility using a logistic regression model. No additional covariates such as principal components for ancestry were necessary. Analysis was performed using the SNPTEST v2 software using an additive model of SNP effect. Imputed genotypes were encoded in the logistic model as the expected allele count.18 For autosomal SNPs, all samples were analyzed together. For chromosome X, only SNPs that were on the Axiom array were included because the ARIC study samples only had genotypes for autosomal SNPs. Using the same QC as above, 13,071 chromosome X SNPs were used. Genomic control was applied to correct for inflation (initial lambda = 1.104; after genomic control, = 1.0). Between the autosomes and X chromosome SNPs, the dataset included 685,281 SNPs for the association analysis. We applied a Bonferroni correction to obtain the appropriate genome-wide threshold for significance (0.05/685,281= p<7.3 × 10−8).
To test whether there might be more than one risk variant in a particular gene or gene region contributing to the association, conditional analyses were performed. A logistic regression model was used to include the genotype at the most significant SNP from the meta-analysis and to test for association with other SNPs in the region.
Replication and Meta-Analysis
Two independent case-control cohorts were used as replication samples. The first was a Dutch case-control sample. IA patients (n=786) admitted to the Utrecht University Medical Center (in the Netherlands) between 1997 and 2007 were included. This included both patients with ruptured and unruptured IA (Supplemental Materials). Controls were 3110 Dutch subjects recruited as part of the Nijmegen Biomedical Study and the Nijmegen Bladder Cancer Study.19, 20
The second replication cohort was a case-control Finnish sample. The IA patients (n=851), included both ruptured and unruptured IA, treated at the Helsinki and Kuopio University Hospitals (Supplemental Materials). Controls were 2,317 Finnish individuals from three samples (Supplemental Materials): 1) Anonymous Finnish patients at the same hospitals as Finnish cases who gave blood samples for unrelated causes in consecutive days; 2) the Helsinki Birth Cohort Study (HBCS);21 and 3) The Health 2000 cohort.22, 23
The replication cohorts were genotyped on the Illumina CNV370-duo chips (Illumina Inc., San Diego, CA, USA) and results were previously reported.10, 15 Extensive quality control was performed in both subject and SNP data. Imputation was also performed in these samples (Supplemental Materials). The final Replication Samples included 717 Dutch cases, 3004 Dutch controls, 799 Finnish cases, and 2317 Finnish controls.
Replication was performed initially by reviewing results in each of the replication studies for the SNP on chromosome 7 that was found to be significantly associated with IA in the discovery sample. Replication was defined as p≤0.01 in either of the replication samples, with the direction of the SNP effect the same as in the Discovery Sample.
Subsequently, meta-analysis was performed in the chromosome 7 region by combining individual SNP results from the discovery sample and the replication sample. METAL24 was employed using an inverse-variance weighting scheme. We evaluated results to identify findings meeting genome-wide significance criteria.
RESULTS
Discovery Sample
The final sample included 2,617 IA cases and 2,548 controls (Table 1). The cases across the studies had similar age of onset and all had a preponderance of females. The controls across the studies also had a similar age at the time of recruitment.
Table 1.
Sample Demographics
| Cases | Number | Mean Age of Onset (SD) |
% Male |
|---|---|---|---|
| FIA Study multiplex families (1/family) | 388 | 50.7 (11.9) | 31.4 |
| FIA Study (general recruitment) | 1441 | 54.1 (11.7) | 20.1 |
| GERFHS | 44 | 54.7 (12.9) | 40.9 |
| Australia | 118 | 53.4 (16.3) | 34.8 |
| UCSF | 128 | 55.8 (12.0) | 32.8 |
| Poland | 498 | 52.1 (12.9) | 41.2 |
| TOTAL number of Cases | 2,617 | 53.2 (12.3) | 27.4 |
| Controls |
Mean Age at Recruitment (SD) |
||
| CCC | 294 | 64.0 (14.8) | 46.9 |
| GERFHS | 484 | 66.3 (13.0) | 52.7 |
| Australia | 154 | 50.9 (16.1) | 39.0 |
| Poland | 484 | 56.2 (15.8) | 39.7 |
| ARIC | 1,132 | 54.3 (7.5) | 27.9 |
| TOTAL number of Controls | 2,548 | 57.9 (13.1) | 37.7 |
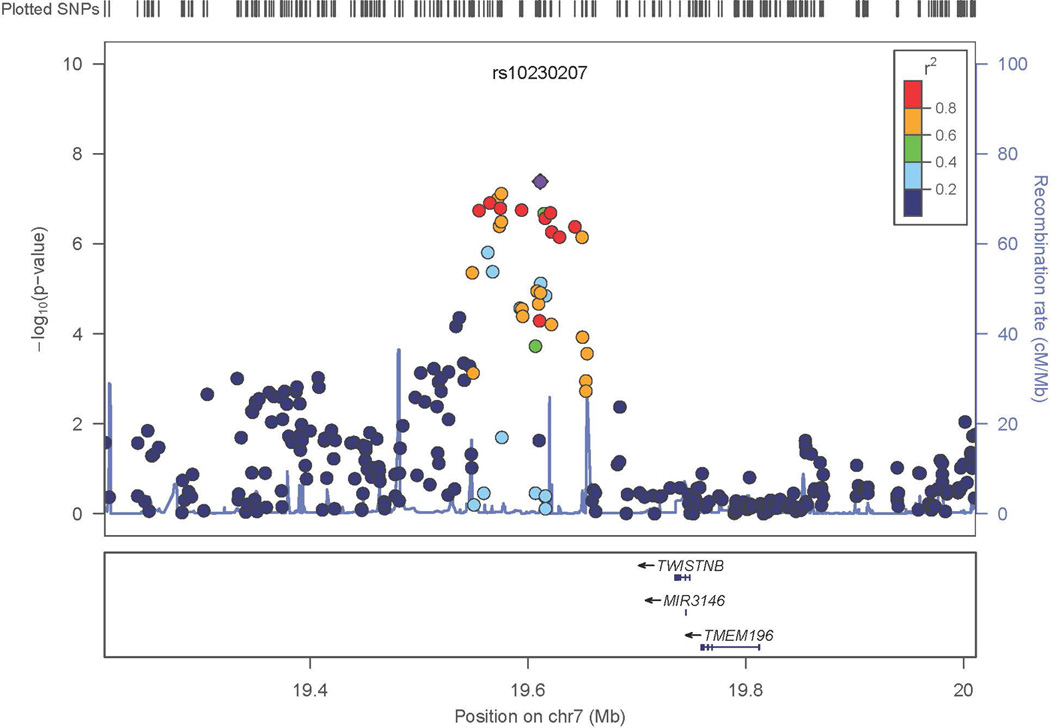
The most significant genome-wide results included SNPs in CDKN2BAS, also known as ANRIL (rs10733376; p=4.07 × 10−12; OR=1.34, 95% CI: 1.23 – 1.45) on chromosome 9 (Figures 1, 2A) previously associated with IA, as well as with other phenotypes.12–15 A SNP on chromosome 7 also met genome-wide significance (rs10230207; p=4.14 × 10−8) (Figures 1, 2B). This SNP is in an intergenic region 3’ of TWISTNB, MIR3146, and TMEM196 and near SNPs previously associated with ischemic stroke and large vessel occlusive ischemic subtype.25 On chromosome 5, rs9687972 also reached genome-wide significance; however, no other SNPs in the same region provided similar evidence of association. This SNP was genotyped in the Axiom samples and imputed in the ARIC samples. The imputation information metric is 0.966, which indicates a good imputation; however, the genotype call rate in the Axiom samples was 95.5%, just above our threshold of 95%. The MAFs are 0.130 (in ARIC controls) and 0.115 (in Axiom controls) (p value is 0.12, just above our threshold of 0.1). Therefore, this finding is likely a false positive association.
Figure 1. Genome-wide Association Analysis in the Discovery Sample.
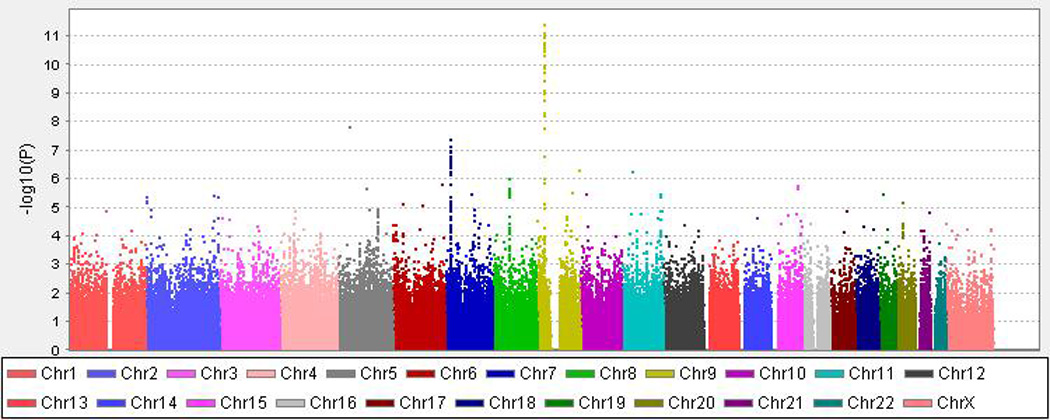
X-axis is the physical position along the genome. Y-axis denotes the −log10 (p-value) for association.
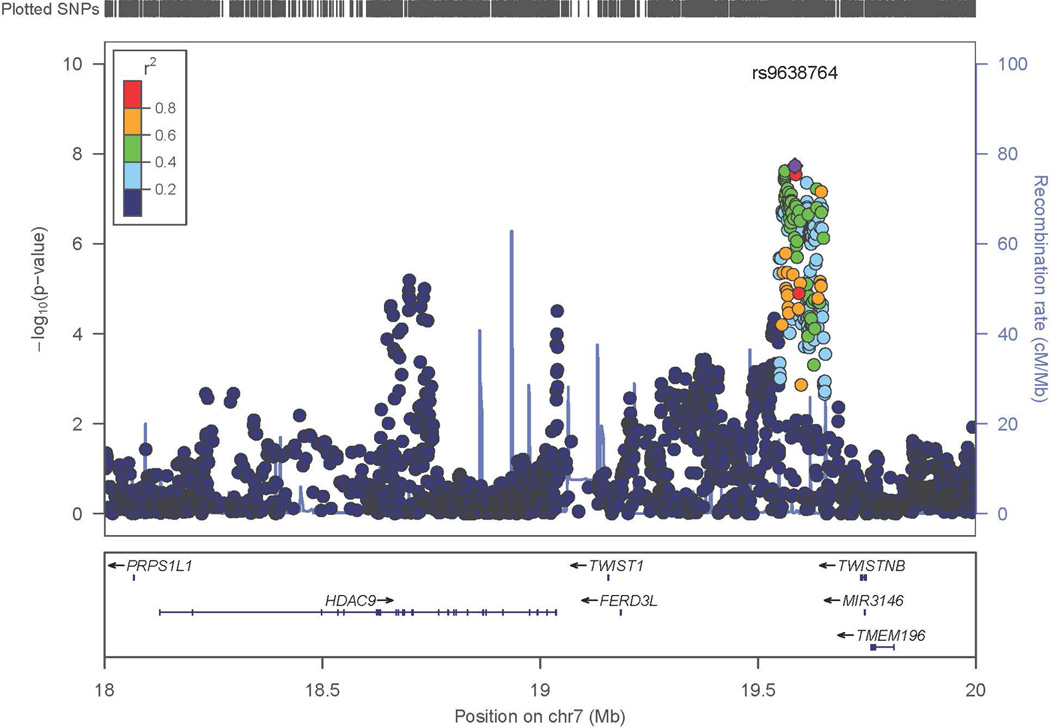
Figure 2. Regional Association Results in the Discovery Sample.
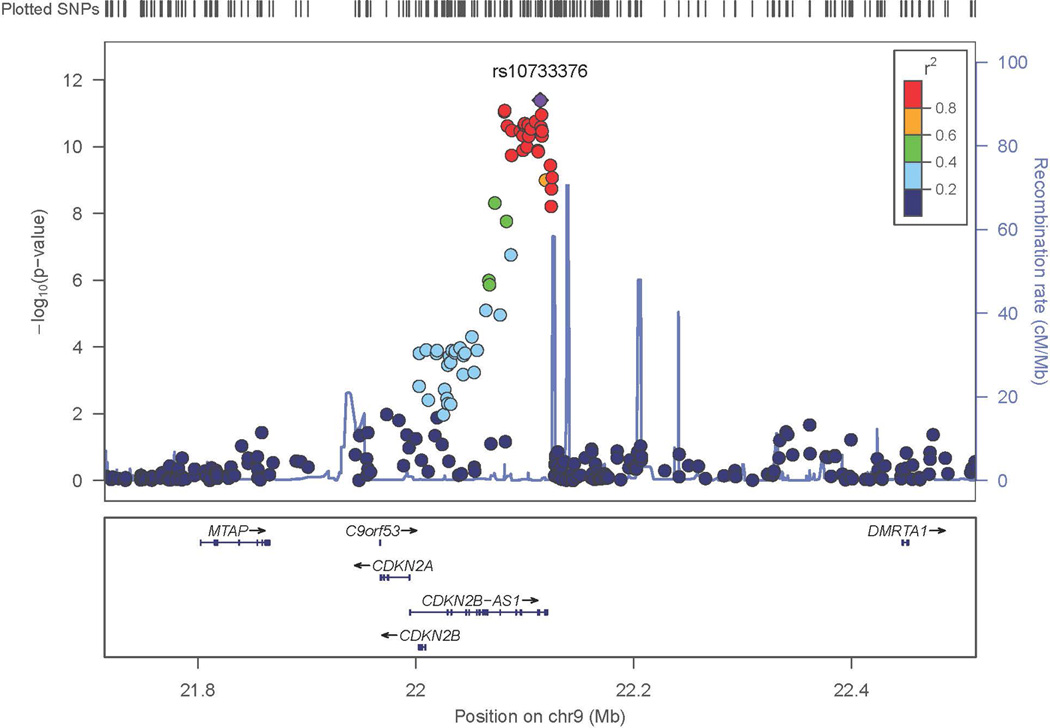
- Chromosome 9
- Chromosome 7
- Chromosome 7 (Expanded view)
On chromosome 7 imputed SNPs were analyzed to further evaluate the evidence of association. As expected, the imputed SNPs in high linkage disequilibrium (LD) with the most significant SNPs initially analyzed further supported the association with IA. The three neighboring regions on chromosome 7 which were examined (Figure 2C) were independent of each other, suggesting that there might be independent evidence of association to IA susceptibility factors within this chromosomal region. Conditional analysis was performed and as expected, based on the lack of LD between the primary region and the two more distant regions, conditioning on the most significant SNP (rs10230207) did not significantly reduce the evidence of association in the other regions. However, none of these other regions on chromosome 7 attained genome-wide criteria, and therefore these results remained tentative.
Results did not exceed the genome-wide threshold in any other chromosomal region. In the chromosomal regions nominated by previous studies, p-values < 0.01 were attained with the key SNPs nominated at chromosomes 4q31.23 and 8q12.1. In addition, although top SNPs previously reported on chromosomes 13q13.1 and 18q11.2 were not available in our sample, SNPs in LD with these SNPs attained p values < 0.01 (Supplemental Figure II).
Replication Sample
Two independent replication samples were analyzed for evidence of association the SNPs on chromosome 7 identified in the discovery sample. The sample demographics are in Table 2.
Table 2.
Replication Sample Demographics
| Cases | Number | Mean Age of Onset | % Male |
|---|---|---|---|
| Dutch | 717 | 54.4 | 31.2 |
| Finnish | 799 | 49.6 | 42.2 |
| Controls |
Mean Age at Recruitment |
||
| Dutch | 3,004 | 61.7 | 62.8 |
| Finnish | 2,317 | 60.9 | 53.7 |
Initially, only the single SNP (rs10230207) which had attained genome-wide evidence of association in the discovery sample was tested for association. When analyzing this SNP, the Dutch sample provided replication (p=0.01), whereas the Finnish sample failed to do so (p= 0.25). Subsequently, meta-analysis was performed across the discovery sample and the two replication samples (Table 3).
Table 3.
Results of Replication Analysis in the Chromosome 7 region
| Discovery Sample | Dutch | Finnish | Meta-analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Position | Effect Allele |
p-value | OR 95%CI |
p-value | OR 95%CI |
p-value | OR 95%CI |
p-value | OR 95%CI |
| rs12669789 | 18,734,065 | C | 9.52 × 10−8 | 1.42 (1.22 –1.67) | 0.714 | 1.05 (0.83– 1.32) | 0.02 | 1.29 (1.04– 1.60) | 1.73×10−6 | 1.30(1.17– 1.45) |
| rs7798197 | 19,037,661 | A | 9.52 × 10−4 | 1.21(1.09–1.34) | 0.13 | 1.13(0.97– 1.32) | 0.02 | 1.18 (1.03– 1.35) | 4.67 × 10−6 | 1.18 (1.10– 1.27) |
| rs10230207* | 19,611,307 | T | 4.14×10−8 | 1.27 (1.17– 1.38) | 0.011 | 1.18 (1.04– 1.33) | 0.25 | 1.08 (0.95– 1.23) | 9.91 × 10−10 | 1.21 (1.14– 1.28) |
| rs2192476 | 19,612,305 | T | 4.53 × 10−7 | 1.26 (1.16– 1.36) | 0.010 | 1.18 (1.04– 1.34) | 0.26 | 1.08 (0.95–1.23) | 3.51 × 10−9 | 1.20 (1.13– 1.27) |
rs10230207 achieved genome-wide significance in the discovery sample and the SNP tested initially for replication. OR – odds ratio 95% CI – 95% confidence interval for the odds ratio rs7798197 and rs12669789 are top SNPs in second and third region respectively.
DISCUSSION
We detected genome-wide evidence of association to a novel region on chromosome 7. The evidence of association was replicated in an independent Dutch sample with the same SNP (rs10230207) and in the same direction of effect. This region failed to show replication in the Finnish cohort, although the direction of effect was the same as in the other 2 samples and may be due to the different genetic architecture of the Finnish population. In addition, the incidence of SAH is higher in Finland as compared to most other parts of the world,26 suggesting that unique genetic factors may segregate in the Finnish population. Another possible explanation is that the size of the replication samples did not have sufficient power to replicate the association. Of note, the association on chromosome 9 with ANRIL has been replicated across many European populations, including the Finns.13
The region on chromosome 7 identified in this study was associated with other stroke phenotypes in previous GWAS studies. Matarin et al (2007)25 reported association to chromosome 7p21.1 for ischemic stroke, although the finding did not meet genome-wide significance. Support for the association of ischemic stroke to this region was also found in a Han Chinese sample.27 Large-vessel stroke was associated with SNPs in HDAC9 on chromosome 7p21.128 ~600kb away from the initial reports for the more general ischemic stroke phenotype. Two other genes, TWIST1 and FERD3L, downstream of HDAC9 could not be excluded as underlying the association. Interestingly, unlike the earlier reports from Matarin and Ding, the SNPs in this region were not associated with the general ischemic stroke phenotype or other subtypes, such as small-vessel stroke or cardioembolic stroke. The METASTROKE Collaboration confirmed the association of large vessel stroke the region around HDAC9.29
Our genome-wide significant association is closest to the finding by Matarin. There is LD between our top SNP (rs10230207) and theirs (r2=0.921). In our study, SNPs in and around HDAC9 did not attain genome-wide significance, but they provided modest evidence of association (p= 10−6, Table 3) that was independent of the association with rs10230207. The Dutch and Finnish sample did not have significant evidence of linkage with the SNPs in and around HDAC9. We also did not find extensive LD (r2=0.117) between our top SNP in this region and the SNP reported by the International Stroke Genetics Consortium (rs11984041). We performed a similar comparison with the METASTROKE association. We had modest evidence of association with the SNPs in the METASTROKE-associated region (p<10−5); however, our most significant SNP was not in LD with the SNP identified in the METASTROKE study (rs2107595).
Our results along with those of other stroke consortium suggest that this region on chromosome 7 is associated with both IA and large vessel ischemic stroke subtype. Combined with our observation, one could speculate that IA, a disease of large intracranial arteries, and ischemic stroke due to large artery disease, may share similar gene pathways and overlapping pathophysiology. Interestingly, the strongest GWAS evidence for IA to this point is for the region on chromosome 9p21.3, which has also been linked to large vessel ischemic stroke, myocardial infarction, and aortic aneurysm. Thus, genetic risk factors for large vessel vasculopathy may lead to different phenotypes depending on coexisting risk factors such as smoking, hypertension, hyperlipidemia, and other genetic risk factors.
A recent meta-analysis combined results from 61 candidate gene and GWAS studies and employed association analysis to identify genes contributing to IA susceptibility.30 The overall number of samples was 32,887 IA cases and 83,683 controls. The strongest evidence of association was on chromosome 9 in CDKN2B, chromosome 8 near SOX17 and chromosome 4 near EDNRA. These results were influenced by the available data, which typically did not include results for all SNPs, making it difficult to detect association to SNPs that were not identified in the initial report. In addition, by including candidate gene papers, the amount of available data was also limited, reducing the ability to detect novel associations in regions that have not yet been studied in the context of IA.
In summary, we identified and replicated in one sample a novel association with IA on chromosome 7 in a region previously reported to be associated with stroke. The associated region is near, although not in LD with HDAC9, which has been associated with large vessel stroke. Studies to replicate this association in additional cohorts of European or other ancestry are necessary and may suggest some overlapping etiology across stroke types and would provide important new insights into the causes and potential targets for stroke prevention.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the staff, participants and the families of the participants for their important contributions in this research study.
SOURCES OF FUNDING
Infrastructure was partly supported by Grant Number UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. This study was also supported by R01NS39512 (JPB), R03NS083468 (TF), the National Health and Medical Research Council (NHMRC) of Australia and the Health Research Council of New Zealand. Additional funding was provided by an NHMRC Senior Principal Research Fellowship (CA), a Veni grant by the Netherlands Organization for Scientific Research (NOW) (project no. 91610016) (YMR), a grant from the Dutch Heart Foundation (project no. 2008B004) (FNGvH) and a grant from Jagiellonian University Medical College K/ZDS/001456.
ARIC is carried out as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; NHGRI contract U01HG004402; and NIH contract HHSN268200625226C. Neurocognitive data is collected by U01 HL096812, HL096814, HL096899, HL096902, and HL096917 with previous brain MRI examinations funded by R01-HL70825.
Footnotes
DISCLOSURES:
Aki Laakso has received consulting fees from Orion Pharma Ltd. Bradford Worrall is Associate Editor of AAN/Neurology. The other authors report no conflict.
REFERENCES
- 1.Broderick JP, Brott T, Tomsick T, Huster G, Miller R. The risk of subarachnoid and intracerebral hemorrhages in blacks as compared with whites. N Engl J Med. 1992;326:733–736. doi: 10.1056/NEJM199203123261103. [DOI] [PubMed] [Google Scholar]
- 2.Nieuwkamp DJ, Setz LE, Algra A, Linn FH, de Rooij NK, Rinkel GJ. Changes in case fatality of aneurysmal subarachnoid haemorrhage over time, according to age, sex, and region: a meta-analysis. Lancet Neurol. 2009;8:635–642. doi: 10.1016/S1474-4422(09)70126-7. [DOI] [PubMed] [Google Scholar]
- 3.Bromberg JE, Rinkel GJ, Algra A, Greebe P, van Duyn CM, Hasan D, et al. Subarachnoid haemorrhage in first and second degree relatives of patients with subarachnoid haemorrhage. BMJ. 1995;311:288–289. doi: 10.1136/bmj.311.7000.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Braekeleer M, Perusse L, Cantin L, Bouchard JM, Mathieu J. A study of inbreeding and kinship in intracranial aneurysms in the Saguenay Lac-Saint-Jean region (Quebec, Canada) Ann Hum Genet. 1996;60:99–104. doi: 10.1111/j.1469-1809.1996.tb01181.x. [DOI] [PubMed] [Google Scholar]
- 5.Schievink WI, Schaid DJ, Michels VV, Piepgras DG. Familial aneurysmal subarachnoid hemorrhage: a community-based study. J Neurosurg. 1995;83:426–429. doi: 10.3171/jns.1995.83.3.0426. [DOI] [PubMed] [Google Scholar]
- 6.Vlak MH, Algra A, Brandenburg R, Rinkel GJ. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. Lancet Neurol. 2011;10:626–636. doi: 10.1016/S1474-4422(11)70109-0. [DOI] [PubMed] [Google Scholar]
- 7.Brown BM, Soldevilla F. MR angiography and surgery for unruptured familial intracranial aneurysms in persons with a family history of cerebral aneurysms. AJR Am J Roentgenol. 1999;173:133–138. doi: 10.2214/ajr.173.1.10397113. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa T, Hashi K, Kurokawa Y, Yamamura A. Family history of subarachnoid hemorrhage and the incidence of asymptomatic, unruptured cerebral aneurysms. J Neurosurg. 1999;91:391–395. doi: 10.3171/jns.1999.91.3.0391. [DOI] [PubMed] [Google Scholar]
- 9.Ronkainen A, Hernesniemi J, Puranen M, Niemitukia L, Vanninen R, Ryynanen M, et al. Familial intracranial aneurysms. Lancet. 1997;349:380–384. doi: 10.1016/S0140-6736(97)80009-8. [DOI] [PubMed] [Google Scholar]
- 10.Bilguvar K, Yasuno K, Niemela M, Ruigrok YM, von Und Zu Fraunberg M, van Duijn CM, et al. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat Genet. 2008;40:1472–1477. doi: 10.1038/ng.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deka R, Koller DL, Lai D, Indugula SR, Sun G, Woo D, et al. The relationship between smoking and replicated sequence variants on chromosomes 8 and 9 with familial intracranial aneurysm. Stroke. 2010;41:1132–1137. doi: 10.1161/STROKEAHA.109.574640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foroud T, Koller DL, Lai D, Sauerbeck L, Anderson C, Ko N, et al. Genome-Wide Association Study of Intracranial Aneurysms Confirms Role of Anril and SOX17 in Disease Risk. Stroke. 2012;43:2846–2852. doi: 10.1161/STROKEAHA.112.656397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 14.Yasuno K, Bakircioglu M, Low SK, Bilguvar K, Gaal E, Ruigrok YM, et al. Common variant near the endothelin receptor type A (EDNRA) gene is associated with intracranial aneurysm risk. Proc Natl Acad Sci U S A. 2011;108:19707–19712. doi: 10.1073/pnas.1117137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasuno K, Bilguvar K, Bijlenga P, Low SK, Krischek B, Auburger G, et al. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat Genet. 2010;42:420–425. doi: 10.1038/ng.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 17.Sinnott JA, Kraft P. Artifact due to differential error when cases and controls are imputed from different platforms. Hum Genet. 2012;131:111–119. doi: 10.1007/s00439-011-1054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wetzels JF, Kiemeney LA, Swinkels DW, Willems HL, den Heijer M. Age- and gender-specific reference values of estimated GFR in Caucasians: the Nijmegen Biomedical Study. Kidney Int. 2007;72:632–637. doi: 10.1038/sj.ki.5002374. [DOI] [PubMed] [Google Scholar]
- 21.Barker DJ, Osmond C, Forsen TJ, Kajantie E, Eriksson JG. Trajectories of growth among children who have coronary events as adults. N Engl J Med. 2005;353:1802–1809. doi: 10.1056/NEJMoa044160. [DOI] [PubMed] [Google Scholar]
- 22.Aromaa A, Koskinen S. Health and functional capacity in Finland : baseline results of the Health 2000 health examination survey. Helsinki: National Public Health Institute; 2004. [Google Scholar]
- 23.Health 2000. Helsinki: THL - National Institute for Health and Welfare; 2000. [Google Scholar]
- 24.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matarin M, Brown WM, Scholz S, Simon-Sanchez J, Fung HC, Hernandez D, et al. A genome-wide genotyping study in patients with ischaemic stroke: initial analysis and data release. Lancet Neurol. 2007;6:414–420. doi: 10.1016/S1474-4422(07)70081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Rooij NK, Linn FH, van der Plas JA, Algra A, Rinkel GJ. Incidence of subarachnoid haemorrhage: a systematic review with emphasis on region age, gender and time trends. J Neurol Neurosurg Psychiatry. 2007;78:1365–1372. doi: 10.1136/jnnp.2007.117655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding H, Xu Y, Bao X, Wang X, Cui G, Wang W, et al. Confirmation of genomewide association signals in Chinese Han population reveals risk loci for ischemic stroke. Stroke. 2010;41:177–180. doi: 10.1161/STROKEAHA.109.567099. [DOI] [PubMed] [Google Scholar]
- 28.Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, Burgess AI, Pirinen M, et al. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE Collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alg VS, Sofat R, Houlden H, Werring DJ. Genetic risk factors for intracranial aneurysms: a meta-analysis in more than 116,000 individuals. Neurology. 2013;80:2154–2165. doi: 10.1212/WNL.0b013e318295d751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


