Abstract
Charcot-Marie-Tooth disease type 2D (CMT2D) is an autosomal dominant axonal peripheral neuropathy characterized by impaired motor and sensory function in the distal extremities. Mutations in the glycyl-tRNA synthetase (GARS) gene cause CMT2D. GARS is a member of the ubiquitously expressed aminoacyl-tRNA synthetase (ARS) family and is responsible for charging tRNA with glycine. To date, thirteen GARS mutations have been identified in patients with CMT disease. While functional studies have revealed loss-of-function characteristics, only four GARS mutations have been rigorously studied. Here, we report the functional evaluation of nine CMT-associated GARS mutations in tRNA charging, yeast complementation, and subcellular localization assays. Our results demonstrate that impaired function is a common characteristic of CMT-associated GARS mutations. Additionally, one mutation previously associated with CMT disease (p.Ser581Leu) does not demonstrate impaired function, was identified in the general population, and failed to segregate with disease in two newly identified families with CMT disease. Thus, we propose that this variant is not a disease-causing mutation. Together, our data indicate that impaired function is a key component of GARS-mediated CMT disease and emphasize the need for careful genetic and functional evaluation before implicating a variant in disease onset.
Keywords: Charcot-Marie-Tooth disease, aminoacyl-tRNA synthetase, peripheral neuropathy, glycyl-tRNA synthetase, GARS
INTRODUCTION
Charcot-Marie-Tooth (CMT) disease encompasses a clinically and genetically heterogeneous group of distal, symmetric polyneuropathies characterized by muscle weakness and loss of sensation in the extremities (Reilly et al., 2011; Millecamps and Julien, 2013). CMT disease is the most common inherited peripheral neuropathy and is estimated to affect one in 2,500 individuals worldwide (Skre, 1974). To date, over 80 loci have been associated with CMT disease (Timmerman et al., 2014), which is divided into two major subclasses based on electrophysiological studies. Patients with demyelinating CMT disease (CMT1) have decreased motor nerve conduction velocities (MNCVs) due to defects in myelinating Schwann cells (Dyck and Lambert, 1968). Not surprisingly, genes implicated in CMT1 have important roles in myelination including peripheral myelin protein 22 (PMP22) (Patel et al., 1992) and myelin protein zero (MPZ) (Hayasaka et al., 1993). Patients with axonal CMT disease (CMT2) have normal MNCVs but decreased amplitudes of evoked muscle action potentials due to axonal dysfunction (Dyck and Lambert, 1968). Also consistent with electrophysiological studies, the genes associated with CMT2 encode proteins critical for axon function including mitofusin 2 (MFN2) (Züchner et al., 2004) and neurofilament light chain (NEFL) (Mersiyanova et al., 2000). Interestingly, mutations in six genes encoding enzymes indispensable for protein translation have been implicated in CMT disease with an axonal pathology (Antonellis et al., 2003; Jordanova et al., 2006; Latour et al., 2010; McLaughlin et al., 2010; Vester et al., 2012; Gonzalez et al., 2013).
Aminoacyl-tRNA synthetases (ARSs) are ubiquitously expressed, essential enzymes responsible for charging tRNA molecules with cognate amino acids—the first step of protein translation—in the cytoplasm and mitochondria (Delarue, 1995). There are 37 ARS genes in the human nuclear genome that code for 17 cytoplasmic, 17 mitochondrial, and 3 bi-functional enzymes; the latter group charge tRNA in both cellular compartments (Antonellis and Green, 2008). Glycyl-tRNA synthetase (GARS; MIM# 600287) mutations have been identified in patients with CMT2D or distal spinal muscular atrophy type-V (dSMA-V), two autosomal dominant forms of axonal peripheral neuropathy characterized by distal muscle atrophy, which is typically more severe in the hands (Christodoulou et al., 1995; Ionasescu et al., 1996; Pericak-Vance et al., 1997; Sambuughin et al., 1998; Ellsworth et al., 1999; Antonellis et al., 2003). Subsequently, mutations in the tyrosyl- (YARS; MIM# 603623), alanyl- (AARS; MIM# 601065), histidyl- (HARS; MIM# 142810) and methionyl- (MARS; MIM# 156560) tRNA synthetase genes have been implicated in dominantly inherited CMT disease with an axonal pathology (Jordanova et al., 2006; Latour et al., 2010; Vester et al., 2012; Gonzalez et al., 2013). Finally, one individual with CMT disease plus multiple neurological and non-neurological sequelae was found to be compound heterozygous for two mutations in lysyl-tRNA synthetase (KARS; MIM# 601421) (McLaughlin et al., 2010). The occurrence of mutations in multiple ARS family members suggests a common pathogenic mechanism of ARS-related axonal CMT disease.
While there is strong genetic evidence linking ARS mutations and CMT disease, the molecular pathology underlying the axonal phenotype remains unknown. A number of hypotheses have been presented regarding the mechanism of ARS-associated CMT disease, including both loss- and gain-of-function effects (Antonellis and Green, 2008; Motley et al., 2010; Wallen and Antonellis, 2013). Since ARSs are responsible for charging tRNA molecules with amino acids, one possibility is that ARS-associated CMT disease is caused by the failure to charge sufficient quantities of tRNA; mutations in ARS genes may inhibit tRNA charging by impairing enzyme activity or by altering proper cellular localization (Antonellis and Green, 2008). As a result, the translational machinery may not be able to meet the demand for protein synthesis in highly metabolic, terminally differentiated motor and sensory axons.
To date, functional studies performed on a subset of CMT-associated ARS mutations have revealed loss-of-function characteristics. All nine of the YARS, KARS, AARS, HARS, and MARS mutations tested to date cause a severe decrease in enzyme activity and/or an inability to rescue deletion of the endogenous ARS gene in yeast complementation assays (Jordanova et al., 2006; McLaughlin et al., 2010, 2011; Vester et al., 2012; Gonzalez et al., 2013). However, the majority of CMT-associated ARS mutations have been identified in GARS and there is conflicting data regarding the presence or absence of loss-of-function characteristics. The functional consequences of human GARS mutations have been assessed in enzyme activity assays, yeast and fly complementation assays, and localization studies in cultured neurons. While many of the mutations demonstrate a loss-of-function effect in at least one of these assays (Antonellis et al., 2006; Chihara et al., 2007; Nangle et al., 2007; Xie et al., 2007; Stum et al., 2011), nine of the thirteen identified GARS mutations have not been rigorously tested (Wallen and Antonellis, 2013). Indeed, this includes five mutations (p.Ala57Val, p.Asp146Asn, p.Ser211Phe, p.Pro244Leu and p.Ile280Phe (James et al., 2006; Rohkamm et al., 2007; Abe and Hayasaka 2009; Lee et al., 2012)) that have not been evaluated in any assay. Characterizing the full panel of GARS mutations will be necessary to determine if a loss-of-function mechanism is a component of CMT2D and dSMA-V. These data will be critical for a better understanding of the molecular pathology of ARS mutations and for developing effective therapies for patients with ARS-related CMT disease.
Here we describe genetic, enzyme kinetic, yeast complementation, and protein localization studies that reveal the functional consequences of disease-associated GARS mutations. Combined with previously reported data, our findings support the hypothesis that impaired GARS function plays a central role in CMT2D and dSMA-V. Additionally, further screening of patients for disease-associated GARS mutations revealed that a variant previously associated with neuropathy (p.Ser581Leu) does not segregate with disease. These findings illustrate the need for careful genetic and functional evaluation of variants identified in patient populations before reporting them as causative factors in human disease.
MATERIALS AND METHODS
Computational assessment of GARS mutations
GARS protein sequences were collected from the NCBI Protein Database (http://www.ncbi.nlm.nih.gov/protein/) for the indicated species using the following accession numbers: human (Homo sapiens, accession number AAA57001.1), mouse (Mus musculus, accession number AAH21747.1), zebrafish (Danio rerio, accession number XP_692410.4), roundworm (Caenorhabditis elegans, accession number NP_498093.1), and baker’s yeast (Saccharomyces cerevisiae, accession number NP_009679.2). Multiple-species amino acid sequence alignments were then generated using ClustalW2 software (Larkin et al., 2007).
GARS and GRS1 expression constructs
DNA constructs for aminoacylation, yeast complementation and localization studies where generated using Gateway cloning technology (Invitrogen, Carlsberg, CA). Briefly, the human GARS open reading frame was amplified from a human cDNA sample, and the S. cerevisiae GRS1 locus was amplified from S. cerevisiae genomic DNA. Primers were designed with flanking Gateway sequences attB1 (forward) and attB2 (reverse; primer sequences available on request). Entry clones were generated by recombining PCR purified amplicons into the pDONR221 vector using BP clonase and the manufacturer’s specifications. After transformation into E. coli, DNA from individual entry clones was isolated and subjected to DNA sequencing analysis to confirm the presence of the appropriate wild-type gene. For each mutation studied here, mutation-containing oligonucleotides were generated, and the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA) was used per the manufacturer’s instructions. After transformation into E. coli, DNA from individual clones was purified and sequenced to confirm the presence of each mutation and the absence of any cycle-induced errors. Subsequently, validated entry clones were purified and recombined into the appropriate Gateway-compatible vector using LR clonase and the manufacturer’s specifications. The vectors included pET-21a(+) (aminoacylation assays), pEGFP-N2 (for expressing GARS with a C-terminal EGFP tag), and pRS315 (yeast complementation assays). DNA from the resulting expression constructs was purified and digested with the restriction enzyme BsrGI (New England Biosystems, Ipswich, MA) to confirm the presence of the appropriate insert.
ARS Aminoacylation Assays
Wild-type and mutant human GARS proteins (see above) were expressed in E. coli Rosetta 2 (DE3) pLys cells with a C-terminal in-frame His tag and purified with nickel affinity resin according to the manufacturer’s protocol (Novagen, Rockland, MA). Based on the wild-type enzyme, the active fraction of human GARS in our preparations was ~20%. The T7 transcript of human tRNAGly/UCU (UCU, anticodon) was prepared and purified as previously described (Hou et al., 1993) and was heat denatured at 85°C for 3 min and annealed at 37°C for 20 min before use. Steady-state aminoacylation assays were monitored at 37°C in 50 mM HEPES (pH 7.5), 20 mM KCl, 10 mM MgCl2, 4 mM DTT, 2 mM ATP, and 50 μM 3H-glycine (Perkin Elmer, Waltham, MA) at a specific activity of 16,500 dpm/pmole. The reaction was initiated by mixing a GARS enzyme (20–600 nM) with varying concentrations of tRNA (0.3–20 μM). Aliquots of a reaction mixture were spotted on filter papers, quenched by 5% trichloroacetic acid (TCA), washed, dried, and measured for radioactivity by a liquid scintillation counter (LS6000SC, Beckman Coulter Inc., Fullerton, CA). The amount of radioactivity retained on filter pads was corrected for quenching effects to determine the amount of synthesis of Gly-tRNAGly. Steady-state kinetics were determined by fitting the initial rate of aminoacylation as a function of tRNA concentration to the Michaelis–Menten equation (Schreier and Schimmel, 1972).
Yeast complementation assays
The RJT3/II-1 haploid yeast strain (MATα grs1::HIS3, his3Δ200, leu2Δ1, lys2Δ202, trpΔ63, ura3–52, pTsscII-maint (cen, GRS1, URA3)) carrying a deleted endogenous GRS1 allele and wild-type GRS1 on a URA3-bearing pRS316 maintenance vector was previously reported (Turner et al., 2000; Antonellis et al., 2006). RJT3/II-1 was transformed with a LEU2-bearing pRS315 vector containing wild-type or mutant GRS1 (described above) or pRS315 with no insert as previously reported (Antonellis et al., 2006). Each transformation was performed at least four times with at least three independent plasmid DNA preparations. Four colonies were selected from each transformation for additional analysis. Each colony was grown to saturation in –leu -ura selection medium for 48 hours. Next, 10 μL of undiluted and diluted (1:10 and 1:100) samples from each culture were spotted on plates containing 0.1% 5-Fluoroorotic Acid (5-FOA) (Boeke et al., 1987) complete medium, yeast extract-peptone-glycerol (YPG) plus 0.1% 5-FOA medium, or SD -leu -ura growth medium (Teknova, Hollister, CA) and incubated at 30°C or 37°C for 72 hours. Yeast cell growth was determined by visual inspection.
Cell culture and protein localization studies
The mouse motor neuron, neuroblastoma fusion cell line (MN-1) was cultured and transfected with constructs to express wild-type or mutant GARS in-frame with a C-terminal enhanced green fluorescent protein (EGFP) tag as previously described (Salazar-Grueso et al., 1991; Antonellis et al., 2006). After 48 hours, growth medium was removed, and cells were washed in 1X PBS and then incubated in 1X PBS/0.4% paraformaldehyde for 10 min at room temperature. Cells were washed in 1X PBS, co-stained with 300 nM DAPI for 5 min, washed again in 1X PBS, and finally coated with Pro-Long antifade reagent (Invitrogen). Images were obtained with an IX71 Inverted Microscope using cellSens Standard image software (Olympus, Center Valley, PA).
Population Screening and Segregation analysis
The presence of each GARS variant in the general population was assessed using the Exome Variant Server (snp.gs.washington.edu/EVS/), the 1000 Genomes Project database (www.1000genomes.org/), and dbSNP (www.ncbi.nlm.nih.gov/snp). Additionally, a search of the GEM.app exome-sequencing database was performed for GARS variants in patients with CMT disease (genomics.med.miami.edu). For p.Ser581Leu GARS segregation studies, oligonucleotide primers were designed to amplify GARS exon 16, which harbors the 2nd and 3rd nucleotides of the serine 581 codon. Patient DNA samples were subjected to PCR amplification under standard conditions along with a ‘no DNA’ PCR-negative control reaction. Subsequently, PCR products were purified on Mini Spin Columns (Epoch Life Science, Inc., Missouri city, TX), eluted, and then subjected to Sanger DNA sequencing analysis using the ‘forward’ and ‘reverse’ amplification primers in separate reactions (University of Michigan DNA Sequencing Core). The appropriate review boards of each participating institution approved these studies.
RESULTS
Localization and conservation of human GARS mutations
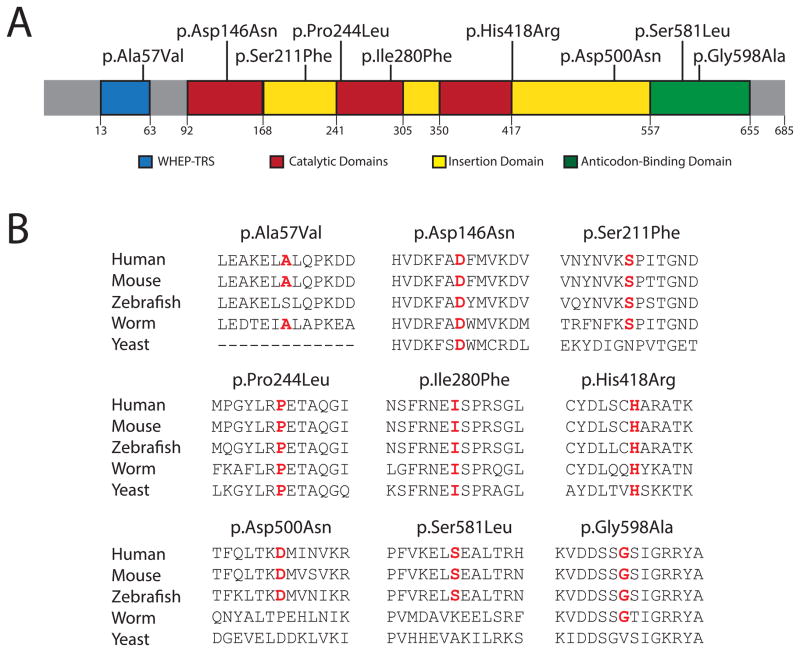
GARS mutations found in patients with CMT disease are distributed throughout the primary structure of the protein. Notably, the mutations affect conserved residues, which are often conserved between human and yeast, suggesting a critical role in protein function (Antonellis et al., 2003). To determine if newly reported GARS mutations demonstrate a similar level of conservation, we mapped the residues onto the human protein and generated an alignment with evolutionarily diverse organisms (Fig. 1A and B). Mutations were evenly distributed throughout the major functional domains of the protein (Xie et al., 2007)(Fig. 1A): (1) p.Ala57Val affects a residue in the disordered WHEP-TRS domain (residues 13–63; pfam00458 in the NCBI Conserved Domain Database), which is putatively involved in tRNA binding and interacting with other aminoacyl-tRNA synthetases in enzyme complexes (Ray et al., 2011), but that is not critical for GARS function (Xie et al., 2007); (2) p.Asp146Asn, p.Pro244Leu, and p.Ile280Phe affect amino acids in the catalytic core (residues 92–168, 241–305, and 350–418; pfam 00587) responsible for ligation of the amino acid to tRNA; (3) p.Ser211Phe, p.His418Arg and p.Asp500Asn affect amino acids within insertion domains of uncertain function; and (4) p.Ser581Leu and p.Gly598Ala reside in the anticodon-binding domain (residues 557–655; pfam03129) that binds to glycine-specific tRNAs.
Figure 1.
Localization and conservation of GARS variants. (A) Mutations are depicted on a cartoon of the known functional and structural domains of GARS in blue (WHEP-TRS domain), red (catalytic domain), yellow (insertion domains), and green (anticodon-binding domain). The amino-acid positions for each domain are indicated below the cartoon. (B) GARS protein sequence alignments from multiple, evolutionarily diverse species are depicted. The amino acid change is listed at the top of each protein fragment. The affected amino acid is highlighted in red.
The evolutionary conservation of mutated GARS residues varies greatly (Fig. 1B). Asp146, Pro244, Ile280, and His418 are identical between human and yeast, while Ser211 and Gly598 are conserved, but to a lesser degree (e.g., between human and worm). In contrast, Ala57 is conserved among mammals and worm, but not zebrafish, and Asp500 and Ser581 are conserved only among vertebrates. Thus Asp146, Ser211, Pro244, Ile280, His418, and Gly598 are predicted to be critical for enzyme function, while Ala57, Asp500, and Ser581 are less likely to be essential for GARS activity.
The majority of GARS mutations reduce aminoacylation activity
The primary function of ARS enzymes is to charge tRNA molecules with cognate amino acids (Delarue, 1995) in a two-step aminoacylation reaction: (i) binding and activation of the amino acid with ATP and (ii) conjugation of the activated amino acid to the 3′ end of the appropriate tRNA (Antonellis and Green, 2008). The aminoacylation assay evaluates the combined reactions of both steps and has been used to establish if CMT-associated ARS mutations disrupt this critical canonical function (Nangle et al., 2007; McLaughlin et al., 2010, 2011; Froelich and First 2011). Here, we assessed previously untested GARS variants (p.Ala57Val, p.Asp146Asn, p.Ser211Phe, p.Pro244Leu, p.Ile280Phe, p.His418Arg and p.Gly598Ala) for the ability to charge human cytoplasmic tRNAGly with tritium-labeled glycine. For comparison purposes, we included two GARS mutations previously shown to impair enzyme kinetics: p.Gly240Arg and p.Gly526Arg (Nangle et al., 2007). With regard to the untested variants, p.Asp146Asn, p.Ser211Phe, p.Pro244Leu, p.Ile280Phe, p.His418Arg, and p.Gly598Ala GARS demonstrate less than 10% aminoacylation activity compared to wild-type GARS (Table 1). In contrast, p.Ala57Val displayed the ability to charge tRNAGly molecules with a much higher efficiency than other mutant proteins, albeit with a 50% reduction relative to wild-type GARS.
Table 1.
Aminoacylation kinetics of GARS protein variants
| Variant | Km (μM) | kcat (s−1) | kcat/Km (μM−1s−1) | Relative to WT |
|---|---|---|---|---|
| Wild-type | 1.03 ± 0.03 | 0.14 ± 0.02 | 0.14 ± 0.02 | 1 |
| p.Ala57Val | 0.92 ± 0.22 | 0.07 ± 0.01 | 0.07 ± 0.02 | 1/2 |
| p.Asp146Asn | 0.6 ± 0.1 | 0.006 ± 0.002 | 0.01 ± 0.01 | 1/14 |
| p.Ser211Phe | - | - | (3.2 ± 1.5) × 10−5 | 1/4,400 |
| p.Gly240Arg | 3.2 ± 0.6 | (4.2 ± 0.3) × 10−3 | (1.3 ± 0.4) × 10−3 | 1/110 |
| p.Pro244Leu | - | - | undetectable | undetectable |
| p.Ile280Phe | - | - | (7.9 ± 2.0) × 10−5 | 1/1,700 |
| p.His418Arg | 0.004 ± 0.001 | (3.9 ± 0.1) × 10−5 | 0.01 ± 0.01 | 1/16 |
| p.Gly526Arg | 7.1 ± 1.3 | (2.3 ± 0.4) × 10−5 | (3.2 ± 2.0) × 10−6 | 1/44,000 |
| p.Gly598Ala | 1.4 ± 0.3 | (1.1 ± 0.1) × 10−3 | (7.9 ± 3.0) × 10−4 | 1/180 |
Amino-acid coordinates correspond to GenBank accession number AAA57001.1
± indicates standard deviations
- indicates that kcat and km are not directly measured; the value for kcat/Km (where applicable) was measured under an approximate condition where the substrate concentration is below an estimated Km
“Undetectable” indicates that values are below the limits of detection
The majority of testable GARS mutations dramatically reduce yeast cell viability
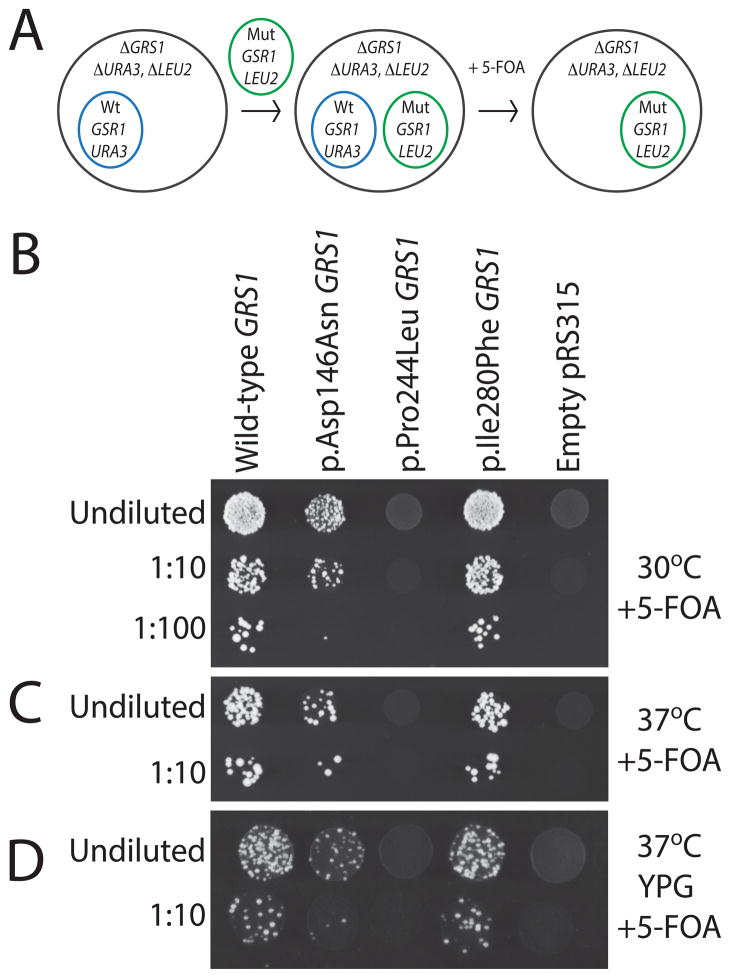
Yeast complementation assays have been performed to determine the functional consequences of CMT-associated ARS mutations in the context of a living cell (Antonellis et al., 2006; Jordanova et al., 2006; McLaughlin et al., 2010, 2011; Stum et al., 2011; Vester et al., 2012; Gonzalez et al., 2013). Here, mutations are modeled in the S. cerevisiae GARS ortholog GRS1, and cellular growth is used as a proxy for enzyme function; this model employs a haploid yeast strain with the endogenous GRS1 locus deleted and viability maintained via expression of wild-type GRS1 on a URA3-bearing maintenance vector (Turner et al., 2000; Antonellis et al., 2006; Stum et al., 2011) (Fig. 2A). Based upon ClustalW alignments (Fig. 1B), we identified the orthologous GRS1 amino acid of each previously untested GARS mutation and performed site-directed mutagenesis to model the human mutations in yeast. We were able to confidently model p.Asp146Asn, p.Pro244Leu, and p.Ile280Phe GARS in GRS1 based on homology; p.Ala57Val, p.Ser211Phe, p.Asp500Asn and p.Ser581Leu are not conserved between human and yeast and thus could not be modeled in this assay. Based on protein alignments, GARS p.Asp146Asn, p p.Pro244Leu, and p.Ile280Phe (GenBank accession number AAA57001.1) correspond to GRS1 residues p.Asp96Asn, p.Pro227Leu, and p.Ile255Phe (GenBank accession number NP_009679.2), respectively. GRS1 mutations are referred to in Figure 2 based on the human mutation they model. Yeast cells were transformed with the experimental alleles on a LEU2-harboring vector and plated on solid growth medium containing 0.1% 5-Fluoroorotic Acid (5-FOA; Fig. 2A). 5-FOA is toxic to yeast cells expressing URA3 and thus selects for cells that have spontaneously lost the maintenance vector (Boeke et al., 1987). Only yeast cells containing a functional GRS1 allele on the LEU2-bearing experimental plasmid are able to complement the deleted endogenous GRS1 allele and survive on 5-FOA plates. Our wild-type GRS1 expression construct allowed robust yeast cell growth and our experimental vector without a GRS1 gene (‘Empty’ in Fig. 2B) was unable to complement the null allele in yeast cells (Fig. 2B). These results are consistent with the construction of a functional GRS1 experimental system and with GRS1 being an essential gene, respectively (Turner et al., 2000; Antonellis et al., 2006). With regard to the modeled mutations, p.Ile280Phe GRS1 was able to maintain yeast viability (Fig. 2B) in a comparable manner to wild-type GRS1. In contrast, p.Asp146Asn and p.Pro244Leu GRS1 resulted in a severe reduction of yeast cell growth compared to wild-type GRS1 (Fig. 2B).
Figure 2.
Characterization of yeast expressing wild-type and mutant GRS1. Cartoon illustrates yeast complementation strategy (A). Each yeast strain was transformed with a LEU2-bearing pRS315 vector containing wild-type GRS1 (Wt), the indicated mutant form of GRS1 (Mut), or no insert (‘Empty’). Cultures for each strain (labeled along the top for each panel in B, C, and D) were grown for two days in liquid media and spotted on selective solid growth medium directly or after dilution as indicated. Strains were plated on medium containing 5-FOA to determine if the GRS1 alleles complement loss of endogenous GRS1 gene at 30°C (B) and 37°C (C). (D) Strains were plated on solid medium containing 5-FOA and glycerol (YPG) to assess for an effect on mitochondrial function.
We also evaluated yeast cells incubated at 37°C to assess for temperature-sensitive effects of the studied mutations. These analyses did not reveal any further depletion of growth associated with the modeled mutations compared to wild-type GRS1 (Fig. 2C). Finally, since GARS is a bi-functional ARS enzyme responsible for charging tRNA in both the cytoplasm and mitochondria (Antonellis and Green, 2008), yeast were grown on medium containing glycerol and 5-FOA at 30°C to require yeast to rely solely on mitochondrial respiration, which allows the detection of impaired mitochondrial function. Similar to the studies at 37°C, the relative pattern of viability remained consistent when comparing mutant and wild-type GRS1 and therefore did not reveal any further mutation-dependent depletion in yeast cell growth (Fig. 2D). Thus, two of the three human GARS mutations modeled in GRS1 dramatically decrease cell viability in vivo when expressed in isolation, consistent with a loss-of-function effect.
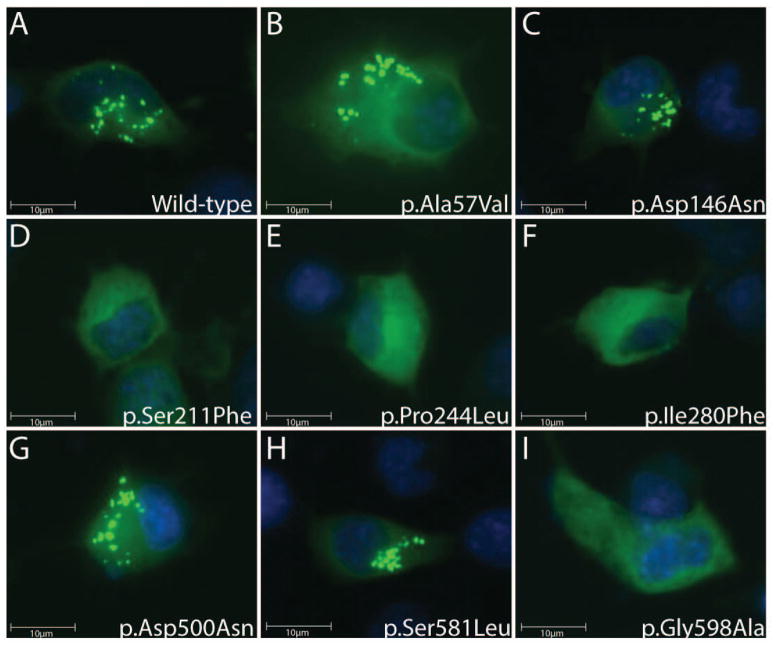
The majority of GARS mutations reduce punctate localization in cultured neurons
The wild-type GARS protein localizes to ‘puncta’ within axons in the human peripheral nerve where it likely functions in local protein translation (Antonellis et al., 2006). Certain GARS mutations impair the punctate localization in cultured neurons, suggesting that reduced localization to axons is a component of CMT2D and dSMA-V disease pathogenesis (Antonellis et al., 2006). However, not all GARS mutations have been tested for an effect on puncta formation in neuronal cells. We therefore expressed wild-type and eight untested mutant GARS-EGFP fusion proteins in a mouse motor neuron, neuroblastoma fusion cell line (MN-1 cells) (Salazar-Grueso et al., 1991; Antonellis et al., 2006). Fluorescence microscopy revealed that wild-type, p.Ala57Val, p.Asp146Asn, p.Asp500Asn, and p.Ser581Leu GARS-EGFP all demonstrated a punctate localization pattern in MN-1 cells (Fig. 3A, B, C, G and H). In contrast, p.Ser211Phe, p.Pro244Leu, p.Ile280Phe, and p.Gly598Ala GARS-EGFP proteins did not associate with puncta in MN-1 cells (Fig. 3D, E, F, I). Importantly, for the mutant GARS-EGFP proteins that did form puncta, these structures were also observed in MN-1 neurite projections, consistent with previous observations of wild-type GARS localization (data not shown) (Antonellis et al., 2006; Fallini et al., 2011).
Figure 3.
Expression of wild-type and mutant GARS in mouse motor neuron-derived cells. MN-1 cells expressing EGFP-tagged wild-type (A), p.Ala57Val (B), p.Asp146Asn (C), p.Ser211Phe (D), p.Pro244Leu (E), p.Ile280Phe (F), p.Asp500Asn (G), p.Ser581Leu (H), or p.Gly598Ala GARS (I) were evaluated for the presence or absence of puncta by fluorescence microscopy.
p.Ser581Leu GARS does not segregate with disease in newly discovered pedigrees
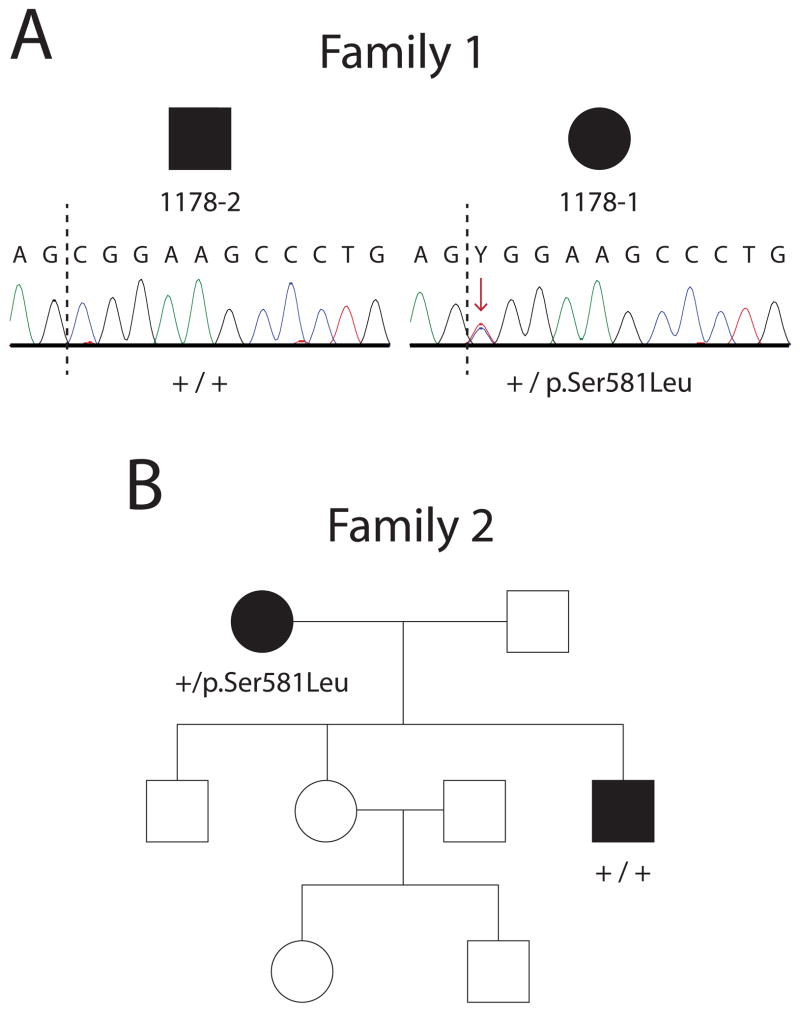
The p.Ala57Val variant was identified in a single individual with weakness and muscle wasting in the upper extremities (Rohkamm et al., 2007) and p.Ser581Leu was identified in a single proband with an extensive family history of lower limb predominant axonal CMT disease (James et al., 2006). No further segregation analyses were performed on either mutation. Furthermore, ethnically-matched control populations were not screened for the absence of p.Ala57Val or p.Ser581Leu. Therefore, we searched genome variant databases for the occurrence of these two mutations. p.Ala57Val and p.Ser581Leu were both identified by the NHLBI GO Exome Sequencing Project at a low frequency of 1/11,842 (rs370531212:C>T) and 2/11,940 (rs201358272:C>T), respectively. The other 11 GARS mutations associated with disease have not been reported in control or general populations.
Our on-going mutation analyses revealed p.Ser581Leu GARS in two unrelated probands with dominant axonal CMT disease. To determine if this mutation is associated with disease in the two respective families, we performed segregation studies. DNA sequence analysis of additional affected individuals revealed that p.Ser581Leu GARS does not segregate with disease in either family (Fig. 4A and B). Specifically, clinically affected individuals in these two families do not carry p.Ser581Leu GARS. Combined with the identification of this variant in the general population, these data indicate that p.Ser581Leu GARS should not be considered a disease-causing mutation.
Figure 4.
p.Ser581Leu GARS does not segregate with CMT disease. DNA samples from affected individuals from two presumably unrelated families with autosomal dominant, axonal CMT disease were genotyped for p.Ser581Leu GARS. Female patients are indicated with circles and male patients are indicated with squares. Filled symbols represent affected individuals with a diagnosis of dominant axonal CMT disease and empty symbols indicate unaffected individuals. Where applicable, an individual’s genotype is indicted with + (for the wild-type allele) or p.Ser581Leu (for the variant allele). (A) Samples from individuals in Family 1 were anonymized resulting in loss of the pedigree structure for this family; however, individuals 1178–1 and 1178–2 are directly related, both affected, and reside in a pedigree originally described as dominant axonal CMT disease. Representative chromatographs from sequencing analysis of genomic DNA from affected individuals are shown. The sequence includes the AG splice site acceptor at the 3′ end of intron 15 followed by the first 12 nucleotides of exon 16 (intron/exon junction marked by dashed line). The p.Ser581 codon (TCG) is encoded by the last nucleotide of exon 15 (T; not shown) and the first two nucleotides (CG) of exon 16. Individual 1178–1 is heterozygous for a C>T mutation (red arrow) in the first base of exon 16 resulting in the p.Ser581Leu allele. 1178–2 is homozygous for the wild-type genotype at this nucleotide (C). (B) The pedigree for Family 2 illustrates the affected status of each family member and the p.Ser581Leu genotypes of two affected individuals with dominant axonal CMT disease.
DISCUSSION
Aminoacyl-tRNA synthetases (ARSs) are essential enzymes that charge tRNA molecules with cognate amino acids (Delarue, 1995). Interestingly, genes encoding ARSs have been implicated in a phenotypically diverse panel of dominant and recessive diseases (Wallen and Antonellis, 2013). While the majority of disease-associated alleles demonstrate a primary loss of function, only four of the 13 reported GARS mutations have been rigorously tested. In this study, we evaluated nine GARS mutations, and our results indicate that impaired function, either by reduced enzyme function or altered cellular localization, is an important component of CMT disease pathogenesis. We determined that p.Asp146Asn, p.Ile280Phe, p.Ser211Phe, p.Pro244Leu, p.His418Arg, and p.Gly598Ala GARS encode a protein product with a severe defect in tRNA charging. Additionally, p.Ser211Phe, p.Pro244Leu, p.Ile280Phe and p.Gly598Ala GARS proteins fail to form puncta characteristic of wild-type GARS (Antonellis et al., 2006; Fallini et al., 2011). Finally, our genetic analyses rule out p.Ser581Leu GARS as a pathogenic GARS mutation. Combined with previous reports, the data presented here reveal that 10 of the 12 disease-associated GARS mutations identified to date demonstrate loss-of-function characteristics (Antonellis et al., 2006; Chihara et al., 2007; Nangle et al., 2007; Xie et al., 2007; Stum et al., 2011). This study furthers our understanding of the allelic spectrum of GARS-associated CMT disease and suggests that impaired GARS function is an important component of the axonal peripheral neuropathy.
Aminoacylation and yeast growth assays are established systems for testing the function of ARS enzymes in vitro and in vivo, respectively. Importantly, all GARS mutations that fail to complement yeast cell growth also have a severe reduction in aminoacylation activity. However, certain mutations, such as p.Ile280Phe, show reduced aminoacylation activity in vitro but complement yeast cell growth in a manner similar to the wild-type enzyme. One possible explanation for this discrepancy is that bacterially produced proteins may lack post-translational modifications important for enzyme function in human cells or for proper activity in specific cellular contexts. Additionally, other cellular components may stabilize the mutant protein product in the yeast system, allowing for improved function and viability. The yeast assay could not be used to evaluate all GARS mutations due to a lack of conservation with human residues. Alternative assays (see below) should be explored to determine the in vivo consequences of such mutations.
While yeast offers insight into the functional consequences of GARS mutations in vivo, it does not reveal how GARS mutations affect axons. The function of GARS and other ARS enzymes in axons remains unclear; however, there is strong evidence that they provide tRNA charging for local protein translation (Ingoglia et al., 1983; Giuditta et al., 2002). Interestingly, wild-type endogenous GARS forms discrete puncta in peripheral neurons and axons from healthy human subjects (Antonellis et al., 2006). Data collected to date show that seven of the 12 disease-associated GARS mutations give rise to proteins that are unable to localize to puncta in cultured neurons (Table 2). Another study showed that mutant GARS proteins mislocalize in neurite projections (Nangle et al., 2007) but did not reveal discrete puncta, which is likely due to differential placement of the epitope tag. The loss of a punctate localization pattern observed for certain GARS mutant proteins may be due to altered protein structure and/or reduced binding to specific protein partners or RNA molecules required for puncta formation. It will be critical to identify proteins and/or RNAs that GARS interacts with in these structures to further understand puncta function and how disruption of these structures may lead to CMT disease.
Table 2.
Summary of functional studies on GARS variants
| Variant | Enzyme Activity | Yeast Viability | Puncta Formation | References |
|---|---|---|---|---|
| Wild-type | Normal | Viable | Yes | Antonellis et al. 2006, and this study |
| p.Ala57Val | Normal1 | NA | Yes | This study |
| p.Glu71Gly | Normal | Viable | Yes | Antonellis et al. 2006, Nangle et al. 2006 |
| p.Leu129Pro | Reduced | Reduced | No | Antonellis et al. 2006, Nangle et al. 2006 |
| p.Asp146Asn | Reduced | Reduced | Yes | This study |
| p.Ser211Phe | Reduced | NA | No | This study |
| p.Gly240Arg | Reduced | Viable | No | Antonellis et al. 2006, Nangle et al. 2006 |
| p.Pro244Leu | Reduced | Lethal | No | This study |
| p.Ile280Phe | Reduced | Viable | No | This study |
| p.His418Arg | Reduced | Lethal | No | Antonellis et al. 2006, Nangle et al. 2006, and this study |
| p.Asp500Asn | Normal | NA | Yes | Nangle et al. 2006, and this study |
| p.Gly526Arg | Reduced | Lethal | Yes | Antonellis et al. 2006, Nangle et al. 2006, and Xie et al. 2007 |
| p.Ser581Leu | Normal | NA | Yes | Nangle et al. 2006, Cader et al. 2007, and this study |
| p.Gly598Ala | Reduced | Viable | No | Stum et al. 2011, and this study |
Amino-acid coordinates correspond to GenBank accession number AAA57001.1
Red text indicates a result consistent with a loss-of-function effect.
This variant was associated with ~50% activity compared to wild-type GARS and we considered a >90% reduction in activity as dramatically ‘Reduced’.
NA – not applicable; mutation could not be modeled in yeast ortholog
Although impaired function is associated with GARS-mediated CMT disease, three disease-associated mutations have not demonstrated impaired function in any of the assays used in this study: p.Ala57Val, p.Glu71Gly, and p.Asp500Asn (Table 2). The p.Glu71Gly GARS mutation segregates with disease in a large pedigree with upper limb predominant axonal neuropathy characteristic of CMT2D (Antonellis et al., 2003). Analysis of human p.Glu71Gly GARS in a fly model revealed that it cannot complement the loss of gars in neurons, and the authors conclude that p.Glu71Gly represents a loss-of-function allele (Chihara et al., 2007). Similar to p.Glu71Gly, p.Asp500Asn GARS segregates with disease in a large pedigree with dominantly inherited, upper limb predominate axonal neuropathy (Del Bo et al., 2006), providing strong genetic and phenotypic evidence supporting a role in CMT2D disease pathogenesis. In contrast, p.Ala57Val has weak genetic evidence for a role in CMT disease. First, this variant was identified in a single individual with upper limb predominant CMT disease and no additional family members were available for segregation studies (Rohkamm et al., 2007). Second, ethnically matched controls were not tested to determine the presence of this allele in appropriate populations. Finally, p.Ala57Val was identified by the NHLBI GO Exome Sequencing Project (rs370531212:C>T) at a low frequency; however, since individuals in the NHLBI study were not subjected to neurological exam and since CMT is often a late onset disease, inclusion in this database does not exclude p.Ala57Val from having a causative role in CMT disease. Evaluation of p.Asp500Asn and p.Ala57Val in the fly model will be important to determine if these variants have impaired function that cannot be detected by other assays.
To date, loss-of-function mutations at five ARS loci have been implicated in dominantly inherited CMT disease (Antonellis et al., 2003; Jordanova et al., 2006; Latour et al., 2010; Vester et al., 2012; Gonzalez et al., 2013). Thus, mechanisms of impaired function in dominant disease should be carefully evaluated, including haploinsufficiency, a dominant-negative effect, and a toxic gain of function. First, while mice heterozygous for loss-of-function missense Gars mutations (C201R and P234KY) demonstrate a dominant axonal neuropathy (Seburn et al., 2006; Achilli et al., 2009; Motley et al., 2011), mice heterozygous for a Gars null (gene-trap) allele have a wild-type phenotype (Seburn et al., 2006). In addition, GARS null alleles have not been identified in patients with CMT2D and dSMA-V. Combined, these findings argue against haploinsufficiency. Second, transgenic overexpression of wild-type human GARS in the two mouse models restored viability but did not rescue the neuropathy phenotype (Motley et al., 2011), arguing against a dominant-negative effect. However, overexpression of human GARS was unable to rescue the lethality associated with homozygosity for the Gars gene trap allele, indicating that the cDNA transgene may not function in all tissues and during all developmental periods (Motley et al., 2011). Thus, it will be important to assess the activity of wild-type:mutant GARS dimers and the effect of wild-type GARS overexpression in the context of mutations identified in human patients before excluding a dominant-negative mechanism (Wallen and Antonellis, 2013). Finally, impaired GARS function may be a prerequisite for a toxic gain-of-function effect. While mutant protein aggregates have not been reported (Antonellis et al., 2006; Seburn et al., 2006; Nangle et al., 2007; Achilli et al., 2009; Motley et al., 2011; Stum et al., 2011), conformational changes of human mutant GARS proteins have been observed (He et al., 2011). These proteins should be assessed for mutant-specific binding to neuronal RNAs and/or proteins. If these experiments are informative, then therapies should be designed to reduce the expression of mutant GARS alleles in patients with CMT2D (Motley et al., 2011). Importantly, the three pathogenic mechanisms outlined here are not mutually exclusive; mutation-specific utilization of haploinsufficiency, dominant-negative, and/or toxic gain-of-function effects may explain the observed clinical spectrum of GARS-associated peripheral neuropathy.
The development of rapid, affordable technologies has allowed the use of DNA sequencing for research and diagnostic purposes. Importantly, once a gene is implicated in disease, patients with a similar phenotype can be rapidly screened for variants at that locus. However, rare variants identified in individual patients or small pedigrees are difficult to implicate in disease phenotypes due to the absence of strong genetic evidence (MacArthur et al., 2014). Here, we demonstrate that p.Ser581Leu GARS, a rare variant previously identified in patients with CMT disease, does not segregate with neuropathy in two newly identified families with dominant axonal CMT disease. This variant was originally identified in individuals with lower limb predominant CMT disease (James et al., 2006), which is uncharacteristic of GARS-associated CMT2D. Interestingly, p.Ser581Leu has normal aminoacylation activity (Nangle et al., 2007) and localization in neurons (Fig. 3). Thus, our data indicate that p.Ser581Leu is not a primary disease-causing mutation and should be removed from further studies to determine the pathogenic mechanism of GARS mutations.
In conclusion, our data indicate that impaired function is a common characteristic of CMT-associated GARS mutations and that this may be a necessary first step for disease pathogenesis. However, additional research is needed to implicate or refute a causative link between impaired tRNA charging and CMT2D disease. Importantly, determining the precise mechanism of mutant GARS toxicity in axons will be essential for designing effective therapies for patients with CMT2D.
Acknowledgments
Contract Grant Sponsor: Muscular Dystrophy Association (grant MDA294479 to A.A.); the National Institute of Neurological Diseases and Stroke (grants NS075764 and NS065712 to S.Z.); Charcot-Marie-Tooth Association; the National Institutes of Health Cellular and Molecular Biology Training Grant (T32 GM007315 to L.B.G); the National Institutes of Health Medical Scientist Training Grant (T32 GM07863 to L.B.G).
We are indebted to the patients and their families for their participation in this study. The authors would like to thank Victor Ionasescu (University of Iowa) for anonymized patient DNA samples, Paul Schimmel (The Scripps Research Institute) for the GRS1 yeast strain, Kurt Fischbeck (NIH/NINDS) for the MN-1 cells, William Law for assistance with counting cells, the laboratory of Miriam Meisler for helpful discussions, and John Moran for critical evaluation of the manuscript.
References
- Abe A, Hayasaka K. The GARS gene is rarely mutated in Japanese patients with Charcot-Marie-Tooth neuropathy. Journal of human genetics. 2009;54:310–312. doi: 10.1038/jhg.2009.25. [DOI] [PubMed] [Google Scholar]
- Achilli F, Bros-Facer V, Williams HP, Banks GT, AlQatari M, Chia R, Tucci V, Groves M, Nickols CD, Seburn KL, Kendall R, Cader MZ, et al. An ENU-induced mutation in mouse glycyl-tRNA synthetase (GARS) causes peripheral sensory and motor phenotypes creating a model of Charcot-Marie-Tooth type 2D peripheral neuropathy. Disease Models & Mechanisms. 2009;2:359–373. doi: 10.1242/dmm.002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin S-Q, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, Sivakumar K, Ionasescu V, et al. Glycyl tRNA Synthetase Mutations in Charcot-Marie-Tooth Disease Type 2D and Distal Spinal Muscular Atrophy Type V. The American Journal of Human Genetics. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Green ED. The Role of Aminoacyl-tRNA Synthetases in Genetic Diseases*. Annual Review of Genomics and Human Genetics. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- Antonellis A, Lee-Lin SQ, Wasterlain A, Leo P, Quezado M, Goldfarb LG, Myung K, Burgess S, Fischbeck KH, Green ED. Functional Analyses of Glycyl-tRNA Synthetase Mutations Suggest a Key Role for tRNA-Charging Enzymes in Peripheral Axons. Journal of Neuroscience. 2006;26:10397–10406. doi: 10.1523/JNEUROSCI.1671-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods in enzymology. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Chihara T, Luginbuhl D, Luo L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nature neuroscience. 2007;10:828–837. doi: 10.1038/nn1910. [DOI] [PubMed] [Google Scholar]
- Christodoulou K, Kyriakides T, Hristova AH, Georgiou DM, Kalaydjieva L, Yshpekova B, Ivanova T, Weber JL, Middleton LT. Mapping of a distal form of spinal muscular atrophy with upper limb predominance to chromosome 7p. Human molecular genetics. 1995;4:1629–1632. doi: 10.1093/hmg/4.9.1629. [DOI] [PubMed] [Google Scholar]
- Del Bo R, Locatelli F, Corti S, Scarlato M, Ghezzi S, Prelle A, Fagiolari G, Moggio M, Carpo M, Bresolin N, Comi GP. Coexistence of CMT-2D and distal SMA-V phenotypes in an Italian family with a GARS gene mutation. Neurology. 2006;66:752–754. doi: 10.1212/01.wnl.0000201275.18875.ac. [DOI] [PubMed] [Google Scholar]
- Delarue M. Aminoacyl-tRNA synthetases. Current opinion in structural biology. 1995;5:48–55. doi: 10.1016/0959-440x(95)80008-o. [DOI] [PubMed] [Google Scholar]
- Dyck PJ, Lambert EH. Lower motor and primary sensory neuron diseases with peroneal muscular atrophy. I. Neurologic, genetic, and electrophysiologic findings in hereditary polyneuropathies. Archives of neurology. 1968;18:603–618. doi: 10.1001/archneur.1968.00470360025002. [DOI] [PubMed] [Google Scholar]
- Ellsworth RE, Ionasescu V, Searby C, Sheffield VC, Braden VV, Kucaba TA, McPherson JD, Marra MA, Green ED. The CMT2D locus: refined genetic position and construction of a bacterial clone-based physical map. Genome research. 1999;9:568–574. [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Zhang H, Su Y, Silani V, Singer RH, Rossoll W, Bassell GJ. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. Journal of Neuroscience. 2011;31:3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich CA, First EA. Dominant Intermediate Charcot-Marie-Tooth disorder is not due to a catalytic defect in tyrosyl-tRNA synthetase. Biochemistry. 2011;50:7132–7145. doi: 10.1021/bi200989h. [DOI] [PubMed] [Google Scholar]
- Giuditta A, Kaplan BB, van Minnen J, Alvarez J, Koenig E. Axonal and presynaptic protein synthesis: new insights into the biology of the neuron. Trends in neurosciences. 2002;25:400–404. doi: 10.1016/s0166-2236(02)02188-4. [DOI] [PubMed] [Google Scholar]
- Gonzalez M, McLaughlin H, Houlden H, Guo M, Yo-Tsen L, Hadjivassilious M, Speziani F, Yang X-L, Antonellis A, Reilly MM, Züchner S Inherited Neuropathy Consortium (INC) Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. Journal of neurology, neurosurgery, and psychiatry. 2013 doi: 10.1136/jnnp-2013-305049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka K, Himoro M, Sato W, Takada G, Uyemura K, Shimizu N, Bird TD, Conneally PM, Chance PF. Charcot-Marie-Tooth neuropathy type 1B is associated with mutations of the myelin P0 gene. Nature genetics. 1993;5:31–34. doi: 10.1038/ng0993-31. [DOI] [PubMed] [Google Scholar]
- He W, Zhang H-M, Chong YE, Guo M, Marshall AG, Yang X-L. Dispersed disease-causing neomorphic mutations on a single protein promote the same localized conformational opening. Proceedings of the National Academy of Sciences. 2011;108:12307–12312. doi: 10.1073/pnas.1104293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YM, Westhof E, Giegé R. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6776–6780. doi: 10.1073/pnas.90.14.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingoglia NA, Giuditta A, Zanakis MF, Babigian A, Tasaki I, Chakraborty G, Sturman JA. Incorporation of 3H-amino acids into proteins in a partially purified fraction of axoplasm: evidence for transfer RNA-mediated, post-translational protein modification in squid giant axons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1983;3:2463–2473. doi: 10.1523/JNEUROSCI.03-12-02463.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionasescu V, Searby C, Sheffield VC, Roklina T, Nishimura D, Ionasescu R. Autosomal dominant Charcot-Marie-Tooth axonal neuropathy mapped on chromosome 7p (CMT2D) Human molecular genetics. 1996;5:1373–1375. doi: 10.1093/hmg/5.9.1373. [DOI] [PubMed] [Google Scholar]
- James PA, Cader MZ, Muntoni F, Childs A-M, Crow YJ, Talbot K. Severe childhood SMA and axonal CMT due to anticodon binding domain mutations in the GARS gene. Neurology. 2006;67:1710–1712. doi: 10.1212/01.wnl.0000242619.52335.bc. [DOI] [PubMed] [Google Scholar]
- Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, Rao CV, Tournev I, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nature genetics. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Latour P, Thauvin-Robinet C, Baudelet-Méry C, Soichot P, Cusin V, Faivre L, Locatelli M-C, Mayençon M, Sarcey A, Broussolle E, Camu W, David A, et al. A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. American journal of human genetics. 2010;86:77–82. doi: 10.1016/j.ajhg.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Park J, Nakhro K, Park JM, Hur Y-M, Choi B-O, Chung KW. Two novel mutations of GARS in Korean families with distal hereditary motor neuropathy type V. Journal of the Peripheral Nervous System. 2012;17:418–421. doi: 10.1111/j.1529-8027.2012.00442.x. [DOI] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin HM, Sakaguchi R, Giblin W, Wilson TE, Biesecker L, Lupski JR, Talbot K, Vance JM, Züchner S, Lee Y-C, Kennerson M, et al. NIH Intramural Sequencing Center. A Recurrent loss-of-function alanyl-tRNA synthetase (AARS) mutation in patients with charcot-marie-tooth disease type 2N (CMT2N) Human Mutation. 2011;33:244–253. doi: 10.1002/humu.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin HM, Sakaguchi R, Liu C, Igarashi T, Pehlivan D, Chu K, Iyer R, Cruz P, Cherukuri PF, Hansen NF, Mullikin JC, Biesecker LG, et al. Compound Heterozygosity for Loss-of-Function Lysyl-tRNA Synthetase Mutations in a Patient with Peripheral Neuropathy. The American Journal of Human Genetics. 2010;87:560–566. doi: 10.1016/j.ajhg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersiyanova IV, Perepelov AV, Polyakov AV, Sitnikov VF, Dadali EL, Oparin RB, Petrin AN, Evgrafov OV. A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. The American Journal of Human Genetics. 2000;67:37–46. doi: 10.1086/302962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S, Julien J-P. Axonal transport deficits and neurodegenerative diseases. Nature reviews Neuroscience. 2013;14:161–176. doi: 10.1038/nrn3380. [DOI] [PubMed] [Google Scholar]
- Motley WW, Seburn KL, Nawaz MH, Miers KE, Cheng J, Antonellis A, Green ED, Talbot K, Yang X-L, Fischbeck KH, Burgess RW. Charcot-Marie-Tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. PLoS genetics. 2011;7:e1002399. doi: 10.1371/journal.pgen.1002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley WW, Talbot K, Fischbeck KH. GARS axonopathy: not every neuron’s cup of tRNA. Trends in neurosciences. 2010;33:59–66. doi: 10.1016/j.tins.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangle LA, Zhang W, Xie W, Yang X-L, Schimmel P. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11239–11244. doi: 10.1073/pnas.0705055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PI, Roa BB, Welcher AA, Schoener-Scott R, Trask BJ, Pentao L, Snipes GJ, Garcia CA, Francke U, Shooter EM, Lupski JR, Suter U. The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nature genetics. 1992;1:159–165. doi: 10.1038/ng0692-159. [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Speer MC, Lennon F, West SG, Menold MM, Stajich JM, Wolpert CM, Slotterbeck BD, Saito M, Tim RW, Rozear MP, Middleton LT, et al. Confirmation of a second locus for CMT2 and evidence for additional genetic heterogeneity. Neurogenetics. 1997;1:89–93. doi: 10.1007/s100480050013. [DOI] [PubMed] [Google Scholar]
- Ray PS, Sullivan JC, Jia J, Francis J, Finnerty JR, Fox PL. Evolution of function of a fused metazoan tRNA synthetase. Molecular biology and evolution. 2011;28:437–447. doi: 10.1093/molbev/msq246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MM, Murphy SM, Laurà M. Charcot-Marie-Tooth disease. Journal of the peripheral nervous system : JPNS. 2011;16:1–14. doi: 10.1111/j.1529-8027.2011.00324.x. [DOI] [PubMed] [Google Scholar]
- Rohkamm B, Reilly MM, Lochmüller H, Schlotter-Weigel B, Barisic N, Schöls L, Nicholson G, Pareyson D, Laurà M, Janecke AR, Miltenberger-Miltenyi G, John E, et al. Further evidence for genetic heterogeneity of distal HMN type V, CMT2 with predominant hand involvement and Silver syndrome. Journal of the neurological sciences. 2007;263:100–106. doi: 10.1016/j.jns.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Grueso EF, Kim S, Kim H. Embryonic mouse spinal cord motor neuron hybrid cells. Neuroreport. 1991;2:505–508. doi: 10.1097/00001756-199109000-00002. [DOI] [PubMed] [Google Scholar]
- Sambuughin N, Sivakumar K, Selenge B, Lee HS, Friedlich D, Baasanjav D, Dalakas MC, Goldfarb LG. Autosomal dominant distal spinal muscular atrophy type V (dSMA-V) and Charcot-Marie-Tooth disease type 2D (CMT2D) segregate within a single large kindred and map to a refined region on chromosome 7p15. Journal of the neurological sciences. 1998;161:23–28. doi: 10.1016/s0022-510x(98)00264-0. [DOI] [PubMed] [Google Scholar]
- Schreier AA, Schimmel PR. Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry. 1972;11:1582–1589. doi: 10.1021/bi00759a006. [DOI] [PubMed] [Google Scholar]
- Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron. 2006;51:715–726. doi: 10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- Skre H. Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clinical genetics. 1974;6:98–118. doi: 10.1111/j.1399-0004.1974.tb00638.x. [DOI] [PubMed] [Google Scholar]
- Stum M, McLaughlin HM, Kleinbrink EL, Miers KE, Ackerman SL, Seburn KL, Antonellis A, Burgess RW. An assessment of mechanisms underlying peripheral axonal degeneration caused by aminoacyl-tRNA synthetase mutations. Molecular and cellular neurosciences. 2011;46:432–443. doi: 10.1016/j.mcn.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman V, Strickland AV, Züchner S. Genetics of Charcot-Marie-Tooth (CMT) Disease within the Frame of the Human Genome Project Success. Genes. 2014;5:13–32. doi: 10.3390/genes5010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RJ, Lovato M, Schimmel P. One of two genes encoding glycyl-tRNA synthetase in Saccharomyces cerevisiae provides mitochondrial and cytoplasmic functions. The Journal of biological chemistry. 2000;275:27681–27688. doi: 10.1074/jbc.M003416200. [DOI] [PubMed] [Google Scholar]
- Vester A, Velez-Ruiz G, McLaughlin HM, Lupski JR, Talbot K, Vance JM, Züchner S, Roda RH, Fischbeck KH, Biesecker LG, Nicholson G, et al. NISC Comparative Sequencing Program. A Loss-of-Function Variant in the Human Histidyl-tRNA Synthetase (HARS) Gene is Neurotoxic In Vivo. Human Mutation. 2012 doi: 10.1002/humu.22210. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallen RC, Antonellis A. To Charge or Not to Charge: Mechanistic Insights into Neuropathy-Associated tRNA Synthetase Mutations. Current opinion in genetics & development. 2013;23:302–309. doi: 10.1016/j.gde.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Nangle LA, Zhang W, Schimmel P, Yang X-L. Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9976–9981. doi: 10.1073/pnas.0703908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nature genetics. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]