Abstract
Aim
To determine microbial profiles that discriminate periodontal health from different forms of periodontal diseases.
Methods
Subgingival biofilm was obtained from patients with periodontal health (27), gingivitis (11), chronic periodontitis (35) and aggressive periodontitis (24), and analyzed for the presence of >250 species/phylotypes using HOMIM. Microbial differences among groups were examined by Mann-Whitney. Regression analyses were performed to determine microbial risk indicators of disease.
Results
Putative and potential new periodontal pathogens were more prevalent in subjects with periodontal diseases than periodontal health. Detection of Porphyromonas endodontalis/Porphyromonas spp. (OR 9.5 [1.2–73.1]) and Tannerella forsythia (OR 38.2 [3.2–450.6]), and absence of Neisseria polysaccharea (OR 0.004 [0–0.15]) and Prevotella denticola (OR 0.014 [0–0.49], p<0.05) were risk indicators of periodontal disease. Presence of Aggregatibacter actinomycetemcomitans (OR 29.4 [3.4–176.5]), Cardiobacterium hominis (OR 14.9 [2.3–98.7]), Peptostreptococcaceae sp. (OR 35.9 [2.7–483.9]), P. alactolyticus (OR 31.3 [2.1–477.2]), and absence of Fretibacterium spp. (OR 0.024 [0.002–0.357]), Fusobacterium naviforme/Fusobacterium nucleatum ss vincentii (OR 0.015 [0.001–0.223]), Granulicatella adiacens/Granulicatella elegans (OR 0.013 [0.001–0.233], p<0.05) were associated with aggressive periodontitis.
Conclusion
There were specific microbial signatures of the subgingival biofilm that were able to distinguish between microbiomes of periodontal health and diseases. Such profiles may be used to establish risk of disease.
Keywords: microbiota, biofilm, gingivitis, aggressive periodontitis, chronic periodontitis, microarray analysis
Introduction
The human microbiome is important in the establishment and maintenance of human health (Cho & Blaser 2012). Due to many ecological determinants, the oral cavity comprises a very complex and diverse microbiota in the human body (Paster et al. 2001, Aas et al. 2005, Dewhirst et al. 2010). For most part, the oral microbiota presents a symbiotic relationship with the host; however, major disturbances in this microbial community (dysbiosis) may lead to the development of several oral diseases, such as gingivitis, different forms of periodontitis, caries, and endodontic infections (Darveau 2010). Periodontal diseases are among the most common infectious oral diseases associated with a pathogenic biofilm. The resultant inflammatory process adds to further destruction of the periodontal apparatus and eventual tooth loss (Armitage 1999). Over the years, specific bacteria have been associated with different forms of periodontal disease and disease severity (Socransky & Haffajee 2005). Furthermore, the application of molecular methods of microbial identification has allowed for the detection and association of numerous novel potential commensal and pathogenic species, including not-yet-cultivated taxa (Kumar et al. 2005). Although periodontal pathogens have been recognized, increasing evidence indicates that periodontal diseases are polymicrobial infections related to distinct microbial consortia (Darveu 2010, Wade 2013). Nevertheless, little is known about the mechanisms involved in shifts from a health-related commensal microbiota to a pathogenic complex microbiome associated with periodontal disease. This holistic concept of bacterial communities in contrast to the single-pathogen concept as the unit of pathogenicity has provided a more ecological view with respect to the etiology of oral diseases (Siqueira & Roças 2009). Thus, understanding the composition of the periodontal microbiota and its interaction with the host and environmental factors will give new insights into the role these microbial communities play in health and disease, leading to novel therapeutic strategies aimed at correcting dysbiosis and restoring the beneficial periodontal microbiome. The goal of this study was to determine microbial signature profiles that could discriminate periodontal health from different forms of periodontal diseases.
Material and Methods
Subject Population
In this observational study, 97 patients diagnosed as having periodontal health (H), gingivitis (G), generalized aggressive (AgP) or chronic periodontitis (CP) were recruited between 2009 and 2012, from a pool of first-time patients referred to the Division of Graduate Periodontics of the School of Dentistry at the Federal University of Rio de Janeiro (UFRJ), Brazil. All subjects were ≥18 years of age, had ≥14 teeth, and were diagnosed according to criteria described by the American Academy of Periodontology (Armitage 1999), with modifications. Briefly, H patients presented ≤ 10% of sites with BOP, no PD or CAL > 3 mm, although PD or CAL = 4 mm in up to 5% of the sites without BOP was allowed; G patients had > 10% of sites with BOP, no PD or CAL > 3 mm, although PD or CAL = 4 mm in up to 5% of the sites without BOP was allowed; CP patients presented > 10% of teeth with PD and/or CAL ≥ 5 mm and BOP; AgP presented ≥ 30% of teeth with PD and/or CAL ≥ 5 mm with BOP, including at least one incisor and one first molar, and ≤ 39 years of age (da Silva-Boghossian et al. 2011).
Exclusion criteria included systemic conditions that could affect the progression or treatment of periodontal diseases, long-term administration of anti-inflammatory medication, periodontal treatment and/or use of antibiotics in the last 6 months; pregnancy, and nursing. Research was conducted according to the principles outlined in the Declaration of Helsinki on experimentation involving human subject. All subjects were informed about the nature of the study and a signed consent form was obtained from each individual. The study protocol was approved by the Ethics in Human Research Committee of the Clementino Fraga Filho University Hospital-UFRJ (#1361/2003).
Clinical examination
Subjects were submitted to a medical/dental anamnesis, and information regarding age, gender, ethnicity/color, and smoking status was obtained. Clinical examination was performed by 2 trained and calibrated examiners (D. H. and C.M.S.B). In a group of 10 individuals who did not participate in this study, pairs of examinations were conducted in each individual with a 1-hour interval between them. Intraclass correlation coefficients for pocket depth (PD) and clinical attachment level (CAL) were calculated at the sites level. Intra- and inter-examiner coefficient s for CAL ranged between 0.90 and 0.97, and for PD, between 0.80 and 0.94. Full-mouth clinical measurements included presence/absence of visible supragingival biofilm, suppuration, bleeding on probing (BOP), PD and CAL recorded using a North Carolina periodontal probe (Hu-Friedy, Chicago, IL, USA).
Biofilm Sampling
Subgingival biofilm samples were obtained from 7 healthy sites (PD and/or CAL < 4 mm, no BOP) and 7 sites with the greatest PD (PD and/or CAL > 4 mm with BOP) from periodontitis patients; 7 sites with gingivitis (PD and/or CAL < 4 mm with BOP) from G patients, and 7 healthy sites from H patients. Subgingival samples were collected using sterile Gracey curettes (Hu-Friedy), pooled and placed into microtubes containing TE buffer. DNA was isolated from samples using a commercial kit (MasterPure DNA Purification Kit, Epicentre, Madison, WI, USA). Each DNA sample was examined for its quality and quantity on a 1.5% agarose gel (MassRuler DNA Ladder Mix, Thermo Fisher Scientific Inc., Waltham, MA, USA).
Microbiological Assessment
Microbial analysis of DNA samples was carried out using the Human Oral Microbe Identification Microarray (HOMIM) as described in previously published papers (Colombo et al. 2009; 2012). Briefly, 16S rRNA-based, reverse-capture oligonucleotide probes targeting >250 bacterial taxa (http://mim.forsyth.org/bacteria.html) were printed on aldehyde-coated glass slides. 16S rRNA genes were PCR amplified using 16S rRNA universal primers (NF1: 5’- CCA GRG TTY GAT YMT GGC -3’; 1541R: 5’- RAA GGA GGT GWT CCA DCC -3’; 1492R: 5’- GDT AYC GGT GWT CCA DCC -3’), and labeled via incorporation of Cy3-dCTP in a second nested PCR (9F: 5'- GRG TTY GAT YMT GGC TCA G -3’, and 1492R). The labeled amplicons were hybridized to probes on the slides. After washing, microarray slides were scanned and crude data was extracted using a program for microarray analysis (GenePix Pro 6.0, MDS Analytical Technologies, Sunnyvale, CA, USA).
Statistical Analysis
Full-mouth clinical measurements were computed for each subject and averaged across subjects within groups. Microbial data were generated from scanned arrays using an online analysis tool (at http://bioinformatics.forsyth.org/homim). Signals were normalized by comparing individual signal intensities to the average of signals for the universal probes. Any original signal <2 times the background value was reset to 1 and was assigned to the level 0. All the values >1 were categorized into scores 1 to 5, corresponding to different signal levels. The frequency of scores was computed for each species/phylotype within groups, using patients as units of analysis. Significant differences among groups were sought by Kruskal-Wallis, Mann-Whitney, Chi-square and Fisher’s exact tests. Logistic regression analyses (forward stepwise Wald) were carried out to determine possible microbial indicators of risk for periodontal diseases. Only variables that were significant indicators (p<0.05) in the univariate model were included in the multivariate analyses. These analyses were carried out using the SPSS program v. 19.0 (IBM, Armonk, New York, NY, USA). Cluster analyses using Pearson’s coefficient and UPGMA were performed in order to classify patients based on their microbial profiles. Likewise, to determine variation of bacterial community compositions across groups, total hybridization HOMIM profiles for each sample were compared using correspondence analysis (CoA) in MeV v. 4.8 (Saeed et al. 2006). For comparisons among groups, regression and cluster analyses, only the diseased sites of G, CP and AgP individuals were considered. Samples from healthy sites of diseased individuals were used for comparisons between healthy sites of H and diseased subjects. Adjustments for multiple comparisons of the 380 probes among groups were carried out (adjusted p = 0.00013; Socransky et al. 1991). For all the other analyses the significance level was set at 5%.
Results
Clinical data
The demographic and clinical characteristics of the study population are presented in Table S1. A greater proportion of smokers/former-smokers was observed in the CP as compared to AgP (p<0.05). Caucasians were more prevalent in the H than G and AgP groups. In contrast, the AgP group presented a greater percent of African-Americans than CP patients (p<0.05). CP patients were older than patients in the other groups, and H patients were significantly younger than AgP individuals (p<0.001). Patients with periodontal diseases presented significantly higher values for all clinical parameters in relation to H patients (<0.001). Among periodontitis patients, the AgP group showed significantly greater disease severity than CP patients (p<0.001).
Microbiological data
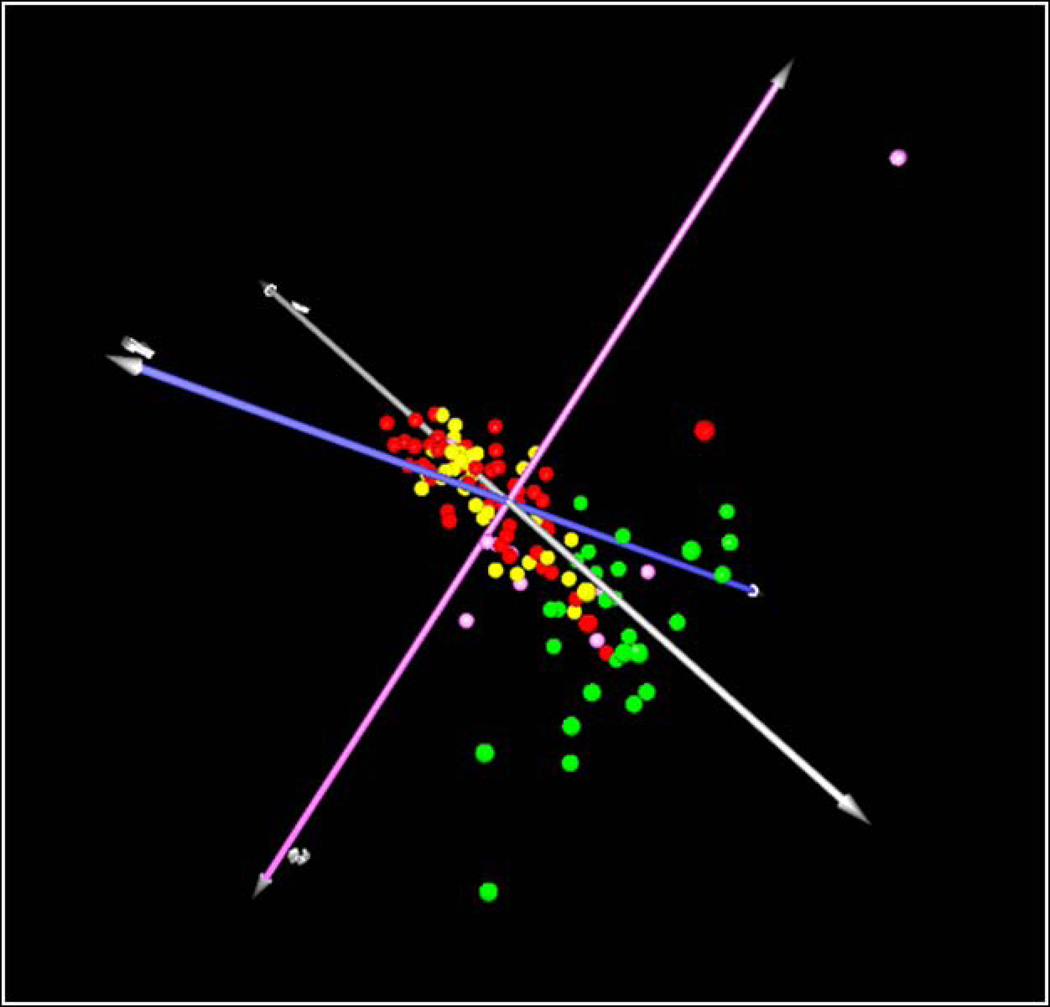
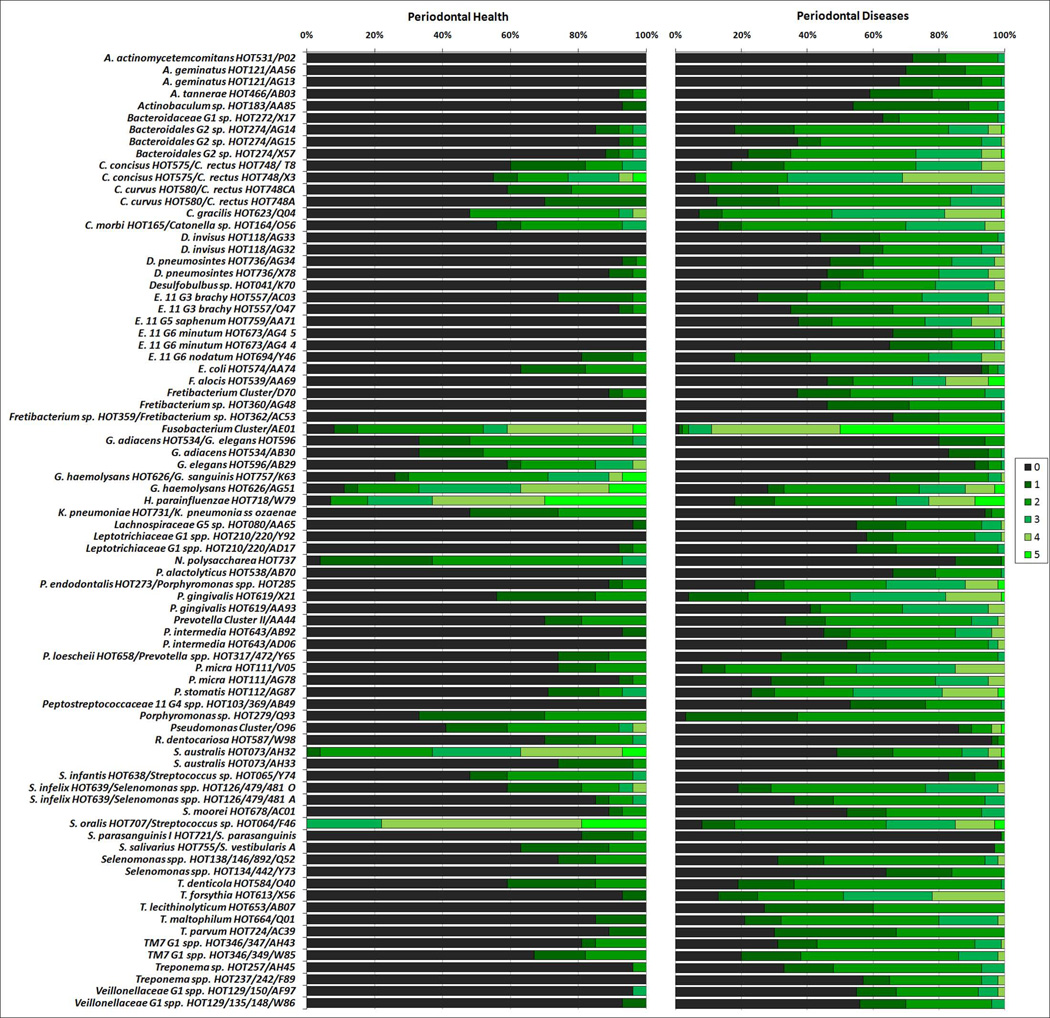
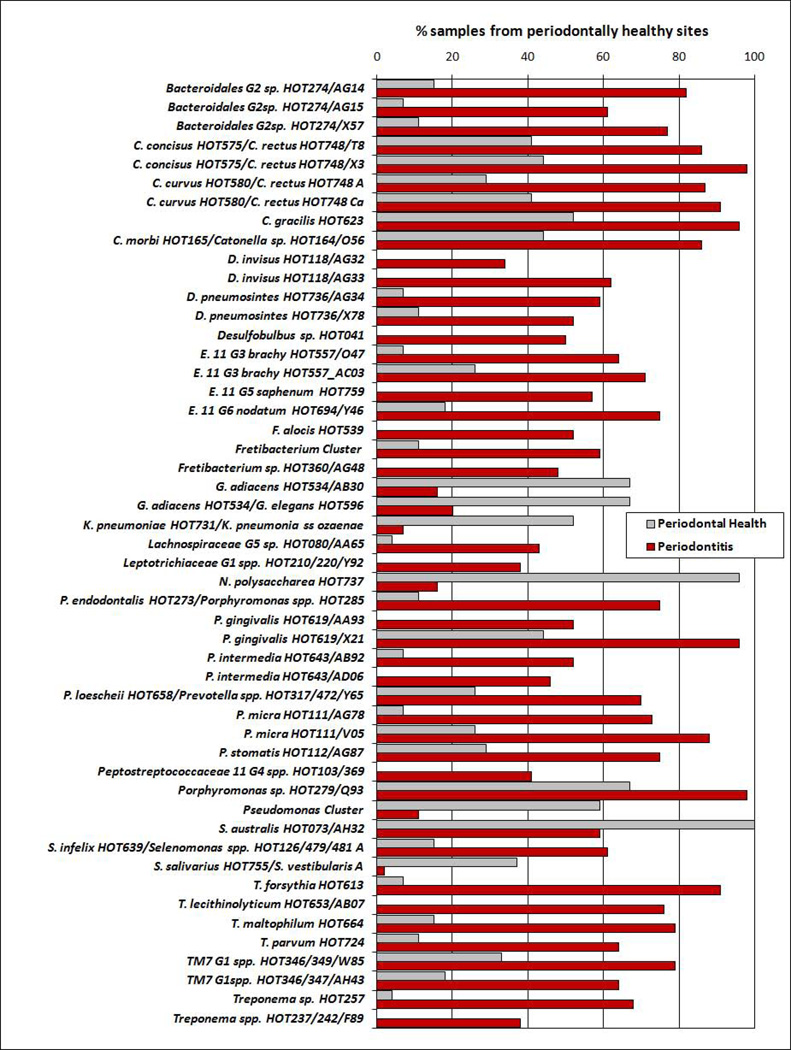
Classification of patients based on microbial signatures of the subgingival microbiota was obtained by cluster analysis (Figure 1) and CoA (Figure 2). Two major separated groups can be seen, one comprising of the majority of periodontitis patients, and the other consisting of H patients. However, a high microbial heterogeneity is noticed within these two groups, as demonstrated by the presence of several clusters within them (Figure 1). Of interest, G patients seemed to present an intermediate microbial profile between H and disease. No specific profiles to distinguish CP from AgP patients may be observed. Comparisons of all species/phylotypes among groups showed that very few microorganisms differed significantly between H and G (Figure S1) or G and periodontitis (Figure S2). Likewise, significant differences between CP and AgP were detected only for 4 species (Aggregatibacter actinomycetemcomitans [Aa], Neisseria elongata, Pseudoramibacter alactolyticus, Prevotella intermedia) which were more prevalent in AgP than in CP patients (p<0.00013, Figure S3). In contrast, several species/phylotypes had significantly different frequencies between all diseased (G, CP and AgP) and H patients (Figure 3, p<0.00013). Most of those microorganisms were detected in greater prevalence in diseased subjects, including known periodontal pathogens (Aa, Campylobacter spp., Eubacterium spp., Fusobacterium spp., Porphyromonas gingivalis, Parvimonas micra, Prevotella spp., Selenomonas spp., Tannerella forsythia, Treponema denticola), as well as possible novel pathogens such as Actinobaculum sp., Alloprevotella tannerae, Anaeroglobus geminatus, Bacteroidales spp., Catonella morbi, Desulfobulbus sp., Dialister spp., Filifactor alocis, Fretibacterium spp., Peptostreptococcus stomatis, Pseudoramibacter alactolyticus, Solobacterium moorei, and TM7 spp. The species/phylotypes that predominated in H compared to diseased individuals included Escherichia coli, Gemella spp., Granulicatella spp., Haemophilus parainfluenzae, Klebsiella pneumoniae, Neisseria polysaccharea, Pseudomonas spp., Rothia dentocariosa and Streptococcus spp. (p<0.00013). These differences were maintained even when controlling for smoking. Of interest, significant differences between healthy sites from H patients and healthy sites from periodontitis patients were also observed (Figure 4). Overall, healthy sites from periodontitis patients harbored several pathogenic species, while Granulicatella spp., K. pneumoniae, N. polysaccharea, Pseudomonas spp., Streptococcus australis and Streptococcus salivarius/Streptococcus vestibularis were more predominant in healthy sites of H individuals (p<0.00013). Of all 380 probes tested as microbial discriminators, only 4 species were found to be risk indicators of disease (Table 1). Presence of Porphyromonas endodontalis/Porphyromonas spp. and T. forsythia, and absence of Prevotella denticola and N. polysaccharea in the subgingival plaque increased significantly the likelihood of a patient to have periodontal disease (p<0.05). To discriminate individuals with CP from AgP, 17 variables were entered in the multivariate model (Table 2). Detection of Aa, Cardiobacterium hominis, Peptostreptococcaceae sp., P. alactolyticus, and absence of Fretibacterium spp., Fusobacterium naviforme/Fusobacterium nucleatum ss vincentii and Granulicatella adiacens/Granulicatella elegans were associated with a higher risk for AgP in relation to CP.
Figure 1.
Microbial profiles of subgingival plaque samples from individual patients (columns) of the four clinical groups: health (green boxes), gingivitis (pink boxes), chronic (red boxes) and aggressive periodontitis (yellow boxes). Patients were grouped by cluster analysis based on the frequency of scores of 380 probes (lanes) grouped by genera or species. The different intensities of green correspond to signal intensities of the arrays (scores 0 to 5). The vertical dashed lane separates the two major clinical groups (periodontal health, on the left; and periodontal disease, on the right).
Figure 2.
Correspondence analysis 3D plot for clustering individuals with different clinical status based on their microbial profiles (frequency of species/phylotypes) determined using HOMIM. Green circles: periodontally healthy individuals. Pink circles: individuals with gingivitis. Red circles: chronic periodontitis individuals. Yellow circles: aggressive periodontitis individuals.
Figure 3.
Stacked bar chart of the frequency of scores (0 a 5) of the species/phylotypes detected by HOMIM in subgingival plaque samples of patients with periodontal health (n=27) and periodontal diseases (gingivitis, 11, chronic, 35, and aggressive periodontitis, 24). These microorganisms represent the ones that differed significantly in prevalence between groups, after adjusting for multiple comparisons (p<0.00013, Mann-Whitney test). The green shades of the bars correspond to the scores (0 to 5) of fluorescence intensities obtained in the arrays.
Figure 4.
Stacked bar chart of the frequency of the species/phylotypes detected by HOMIM in subgingival plaque samples obtained from periodontally healthy sites of patients with periodontal health (n=27) and periodontitis (n=59). These microorganisms represent the ones that differed significantly in prevalence between groups, after adjusting for multiple comparisons (p<0.00013, Chi-square test).
Table 1.
Multivariate logistic regression analysis (stepwise forward Wald) employed to determine microbial indicators of risk for periodontal diseases (gingivitis and periodontitis).
| Predictors | β | SE | Wald | p | OR* | 95% CI | |
|---|---|---|---|---|---|---|---|
| Constant | 3.55 | 1.75 | 4.12 | 0.042 | 34.99 | ||
| N. polysaccharea*** | −5.44 | 1.82 | 8.98 | 0.003 | 0.004 | 0 | 0.15 |
| P. endodontalis/Porphyromonas spp.# | 2.25 | 1.04 | 4.65 | 0.031 | 9.47 | 1.23 | 73.08 |
| P. denticola† | −4.27 | 1.81 | 5.57 | 0.018 | 0.014 | 0 | 0.49 |
| T. forsythia** | 3.64 | 1.26 | 8.38 | 0.004 | 38.23 | 3.24 | 450.60 |
Reference: patients with periodontal health;
Variable entered in the first step;
Variable entered in the second step;
Variable entered in the third step;
Variable entered in the fourth and final model.
β: regression coefficient for the predictor variable; SE: standard error of the coefficient; Wald: Wald chi-square test; p: 2-tailed p-value (significance) OR: odds ratio for the predictors; CI: confidence interval.
Table 2.
Multivariate logistic regression analysis (stepwise forward Wald) employed to determine microbial indicators of risk for aggressive periodontitis.
| Predictors | β | SE | Wald | p | OR* | 95% CI | |
|---|---|---|---|---|---|---|---|
| Constant | −1.866 | 0.935 | 3.980 | 0.460 | 0.155 | ||
| Aa** | 3.381 | 1.096 | 9.521 | 0.002 | 29.399 | 3.433 | 176.47 |
| C. hominis† | 2.703 | 0.964 | 7.873 | 0.005 | 14.931 | 2.259 | 98.686 |
| Fretibacterium spp.§ | −3.722 | 1.373 | 7.347 | 0.007 | 0.024 | 0.002 | 0.357 |
| F. naviforme/F. nucleatum ss vincentii*** | −4.173 | 1.363 | 9.375 | 0.002 | 0.015 | 0.001 | 0.223 |
| G. adiacens/G. elegans# | −4.312 | 1.457 | 8.755 | 0.003 | 0.013 | 0.001 | 0.233 |
| Peptostreptococcaceae sp.¶ | 3.582 | 1.327 | 7.288 | 0.007 | 35.932 | 2.668 | 483.977 |
| P. alactolyticus£ | 3.444 | 1.390 | 6.140 | 0.013 | 31.310 | 2.054 | 477.198 |
Reference: patients with chronic periodontitis;
Variable entered in the first step;
Variable entered in the second step; P. intermedia: variable entered in the third step;
Variable entered in the fourth step;
Variable entered in the fifth step;
Variable entered in the sixtieth step;
Variable entered in the seventh step;
Variable entered in the eighth step;
Aa: Aggregatibacter actinomycetemcomitans. β: regression coefficient for the predictor variable; SE: standard error of the coefficient; Wald: Wald chi-square test; p: 2-tailed p-value (significance) OR: odds ratio for the predictors; CI: confidence interval.
Discussion
A better comprehension of the etiology and pathogenesis of periodontal diseases is essential to develop more effective diagnostic tools and classification systems, as well as more efficacious and affordable periodontal therapies (Armitage 2013). Researchers have been struggling for years to develop reliable diagnostic tests capable of defining and identifying etiological and risk factors for periodontal diseases, particularly at the earliest phases of periodontal infection. In this context important progress in the understanding of the complex interactions between periodontal microbiota and host in health and disease has been made. In polymicrobial periodontal infections, determination of the microbial taxa is the first step to comprehend the dynamic interactions among microorganisms, host and environment. In this investigation, we used this “first step” approach in order to define microbial signatures that could discriminate periodontal health and disease, and disease severity. The data showed that most patients with periodontal health and disease were separated into two major clusters based on their microbial profiles. Between these two clusters, a somewhat intermediate microbial profile including mainly G patients was observed. In fact, G patients shared most of their subgingival microbiota with periodontitis and H individuals, and only very few species/phylotypes differed in frequency among these groups. Regardless of the clinical status, the large majority of the bacterial taxa were comprised of species commonly found in the periodontal microbiota, including Fusobacterium spp., Gemella spp. and some streptococci. Fusobacterium spp., in particular, is a major co-aggregating microorganism within the periodontal biofilm (Jakubovics & Kolenbrander 2010), and it is present in high proportions in the subgingival biofilm associated to various periodontal clinical conditions (Loozen et al. 2014). Oral streptococci are established primary colonizers of the dental biofilm, comprising about 80% of the biofilm (Jakubovics & Kolenbrander 2010). These major groups of microorganisms could be considered part of the core microbiome of the periodontal microbiota (Zaura et al. 2009, Abusleme et al. 2013). Despite the high diversity of the subgingival biofilm, a reduced number of species was able to discriminate between health and disease. In addition to putative periodontal pathogens, new candidate pathogens were detected in significantly high frequency in diseased individuals. In contrast, Gemella spp., Granulicatella spp., Haemophilus spp., Klebsiella spp., Neisseria spp., Pseudomonas spp., Rothia spp. and Streptococcus spp. were more prevalent in H, corroborating the data reported by other studies (Kumar et al. 2005; Aas et al. 2005; Keijser et al. 2008; Colombo et al. 2009, 2012; Bik et al. 2010, Huang et al. 2011; Griffen et al. 2012; Liu et al.,2012; Abusleme et al. 2013; Ge et al. 2013; Klister et al. 2013). More recently,Belstrom et al. (2014) showed by using HOMIM that a relatively small number of bacterial taxa differed significantly in prevalence in saliva samples between patients with periodontitis and periodontal health. These authors have also shown that these differences were independent of the individuals’ smoking status. In addition to the limited differences in the prevalence of species/phylotypes between H and periodontitis individuals observed in the current investigation, diseased-associated taxa were present in the subgingival biofilm of H patients, although in lower proportions, and health-related species were detected in individuals with periodontitis. These findings support the concept that the marked diversity of the oral microbiota provides functional redundancy, and therefore versatility to the microbial community to cope with environmental disturbances (Wade 2013). Thus, periodontitis seems to be associated with ecological shifts in community structure rather than shifts in members of this microbial community (Darveau 2010; Abusleme et al. 2013). In other words, changes from health to periodontitis do not necessarily result from the replacement of health-associated species, but from the rise of new dominant species/phylotypes present previously in low frequency and/or levels (Abusleme et al. 2013). This may explain, in part, the significantly higher prevalence of classical and novel pathogens in periodontally healthy sites of periodontitis patients compared to healthy sites of H patients, also reported in other studies (Riviere et al. 1996, Haffajee et al. 1998). It is speculated that the continuous intra-oral dissemination of periodontal pathogens from periodontal pockets to healthy sulcus in individuals with periodontitis may lead to a greater colonization of the periodontally healthy sites by these pathogens. Whether the elevated proportion of these pathogenic species in these healthy sites will result in a dysbiosis of the periodontal microbiota, and consequently lead to destructive periodontal disease is unknown. In the periodontal ecosystem, the dynamic interactions among numerous microorganisms involve very complex and sophisticated mechanisms (Kolenbrander et al. 2002, Loosen et al. 2014), many of which we are just beginning to comprehend. Pathogenic species do not usually play a role in the periodontal microbiota as a single pathogenic entity (Socransky et al. 1998), and antagonic and/or synergistic relationships among several species will in fact determine the pathogenic role of that microbiota. Due to this strong correlation/dependence among many members of the periodontal microbiota, comparing individual species/phylotypes (pathogenic or host-compatible species) may not be suitable to determine microbial signatures capable of discriminating between health and disease. Using multivariate regression analyses, our findings demonstrated that a consortium composed of high prevalence of P. endodontalis/Porphyromonas spp. and T. forsythia, and low/no detection of P. denticola and N. polysaccharea was associated with a greater probability for having periodontal disease. Porphyromonas spp. (especially P. gingivalis) and T. forsythia have been strongly associated with periodontitis (Socransky et al. 1998, Socransky & Haffajee 2005). However, the negative association between P. denticola and periodontitis was quite surprising since this species has been related to disease (Griffen et al. 2012). Conceivably, methodological differences, and the fact that bacterial taxa should be analyzed not as single beneficial or pathogenic entities but within a consortium (Siqueira & Roças 2009) may explain the discrepancies among studies. N. polysaccharea is a nonpathogenic species that has been isolated from the throats of healthy children (Riou et al., 1983). The role of N. polysaccharea as a potential beneficial species has not been determined (Aas et al. 2005), but some species of Neisseria have been considered to be first colonizers of the supragingival biofilm, and are often related to periodontal health (Diaz et al. 2006; Teles et al. 2012; Ge et al. 2013). CP and AgP are diseases difficult to differentiate based only on clinical parameters (Armitage & Cullinan 2010). In the current study, a multivariate regression model including high prevalence of Aa, C. hominis, N. elongata, Peptostreptococcaceae sp. HOT113, P. alactolyticus, and low prevalence of Fretibacterium spp., F. naviforme/F. nucleatum ss vincentii and G. adiacens/G. elegans was associated with a greater risk for AgP. Except for species of Cardiobacterium and Granulicatella, all the other species have been associated with periodontitis (Paster et al. 2001; Kumar et al. 2005, 2006; Aas et al. 2005; Colombo et al. 2009, 2012; Abusleme et al. 2013; Ge et al. 2013). Unfortunately, there are no available studies comparing the microbiota of CP and AgP by using high-throughput sequencing or microarray techniques. In a longitudinal study, Fine and co-workers (2013) showed by HOMIM that Aa-positive adolescents who presented bone loss had also high prevalence of P. micra, F. alocis, and Peptostreptococcus sp. (HOT113). At the site level, the presence of Aa, S. parasanguinis, and F. alocis together was associated with further bone loss. Likewise,Shaddox et al. (2012) reported that in addition to Aa, the species P. micra, S. moorei, Tannerella sp., F. alocis, and Capnocytophaga sp. were more prevalent in localized AgP than in healthy children. This same group has recently demonstrated that the presence or absence of Aa in the subgingival biofilm of localized AgP adolescents was associated with distinct microbial compositions (Gonçalves et al. 2013). These data suggest that a microbial consortium combining Aa and other potential pathogens may be helpful to discriminate between AgP and CP or H. Periodontal diseases are likely syndromes of complex etiopathogeny and diagnosis, and defining the microbiota associated with these infections is just a piece of the puzzle. Further studies should also look at the functional and metabolic features of the periodontal microbiome to obtain a full understanding of the health-associated status that should be achieved after treatment. Nevertheless, this study showed that there are indeed specific microbial signatures of the periodontal biofilm that were able to distinguish between the microbiomes of periodontal health and diseases, as well as disease severity. These profiles were more complex than previously believed. Such profiles may be used to help establish risk of disease.
Supplementary Material
Clinical relevance.
Scientific rationale for the study
Analysis of different periodontal microbial profiles may provide an additional tool to discriminate periodontal health from various forms of periodontal diseases.
Principal findings
Considering the high diversity of the subgingival microbiota, a relatively small subset of species/phylotypes differed between periodontally healthy and diseased individuals. Specific microbial consortia were able to discriminate periodontal health from different types of periodontal diseases.
Practical implications
Specific microbial signatures of the subgingival biofilm may help to distinguish periodontal health from periodontal diseases, as well as to establish risk of disease.
Acknowledgments
This study was supported by National Council for Scientific and Technological Development (CNPq); Coordination of Improvement of Higher Education Personnel (CAPES); Foundation for Research Financial Support in the State of Rio de Janeiro (FAPERJ), Brazil, and a grant from The National Institutes of Health, DE021565 (BP).
Footnotes
Conflict of interest The authors declare that they have no conflict of interests regarding this study.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacteria flora of the oral cavity. Journal of Clinical Microbiology. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME Journal. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage GC. Development of a classification system for periodontal diseases and conditions. Annals of Periodontology. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- Armitage GC. Learned and unlearned concepts in periodontal diagnostics: a 50-year perspective. Periodontology 2000. 2013;62:20–36. doi: 10.1111/prd.12006. [DOI] [PubMed] [Google Scholar]
- Armitage GC, Cullinan MP. Comparison of the clinical features of chronic and aggressive periodontitis. Periodontology 2000. 2010;53:12–27. doi: 10.1111/j.1600-0757.2010.00353.x. [DOI] [PubMed] [Google Scholar]
- Belstrøm D, Fiehn NE, Nielsen CH, Kirkby N, Twetman S, Klepac-Ceraj V, Paster BJ, Holmstrup P. Differences in bacterial saliva profile between periodontitis patients and a control cohort. Journal of Clinical Periodontology. 2014;41:104–112. doi: 10.1111/jcpe.12190. [DOI] [PubMed] [Google Scholar]
- Bik EM, Long CD, Armitage GC, Loomer PM, Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM, Relman DA. Bacterial diversity in the oral cavity of 10 healthy individuals. The ISME Journal. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. Journal of Periodontology. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AP, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst FE, Paster BJ. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. Journal of Periodontology. 2012;83:1279–1287. doi: 10.1902/jop.2012.110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva-Boghossian CM, do Souto RM, Luiz RR, Colombo AP. Association of red complex,A. actinomycetemcomitans and non-oral bacteria with periodontal diseases. Archives of Oral Biology. 2011;56:899–906. doi: 10.1016/j.archoralbio.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nature Reviews Microbiology. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W, Lakshmanan A, Wade WG. The Human Oral Microbiome. Journal of Bacteriology. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ, Jr, Kolenbrander PE. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Applied and Environmental Microbiology. 2006;72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine DH, Markowitz K, Fairlie K, Tischio-Bereski D, Ferrendiz J, Furgang D, Paster BJ, Dewhirst FE. A consortium of Aggregatibacter actinomycetemcomitans(Aa),Streptococcus parasanguinis and Filifactor alocis are present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. Journal of Clinical Microbiology. 2013;51:2850–2861. doi: 10.1128/JCM.00729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Rodriguez R, Trinh M, Gunsolley J, Xu P. Oral Microbiome of Deep and Shallow Dental Pockets In Chronic Periodontitis. PLoS ONE. 2013;8:e65520. doi: 10.1371/journal.pone.0065520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves PF, Klepac-Ceraj V, Huang H, Paster BJ, Aukhil I, Wallet SM, Shaddox L. Correlation of Aggregatibacter actinomycetemcomitans detection with Clinical/Immunoinflammatory Profile of Localized Aggressive Periodontitis Using a 16S rRNA Microarray Method: A Cross-Sectional Study. PLoS ONE. 2013;8:e85066. doi: 10.1371/journal.pone.0085066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. The ISME Journal. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffajee AD, Cugini MA, Tanner A, Pollack RP, Smith C, Kent RL, Jr, Socransky SS. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. Journal of Clinical Periodontology. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- Huang S, Yang F, Zeng X, Chen J, Li R, Wen T, Li C, Wei W, Liu J, Chen L, Davis C, Xu J. Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC Oral Health. 2011;11:33. doi: 10.1186/1472-6831-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics NS, Kolenbrander PE. The road to ruin: the formation of disease-associated oral biofilms. Oral Disease. 2010;16:729–739. doi: 10.1111/j.1601-0825.2010.01701.x. [DOI] [PubMed] [Google Scholar]
- Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC, ten Cate JM, Crielaard W. Pyrosequencing analysis of the oral microflora of healthy adults. Journal of Dental Research. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- Kistler JO, Booth V, Bradshaw DJ, Wade WG. Bacterial Community Development in Experimental Gingivitis. PLoS ONE. 2013;8:e71227. doi: 10.1371/journal.pone.0071227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr Communication among oral bacteria. Microbiology and Molecular Biology Reviews. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of Candidate Periodontal Pathogens and Beneficial Species by Quantitative 16S Clonal Analysis. Journal of Clinical Microbiology. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. Journal of Clinical Microbiology. 2006;44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang Y-C, Li S, Stine OC, Hasturk H, Kasif S, Segrè D, Pop M, Amar S. Deep Sequencing of the Oral Microbiome Reveals Signatures of Periodontal Disease. PLoS ONE. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loozen G, Ozcelik O, Boon N, De Mol A, Schoen C, Quirynen M, Teughels W. Inter-bacterial correlations in subgingival biofilms: a large-scale survey. Journal of Clinical Periodontology. 2014;41:1–10. doi: 10.1111/jcpe.12167. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. Journal of Bacteriology. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou JY, Guibourdenche M, Popoff MY. A new taxon in the Genus Neisseria. Annals of Microbiology. 1983;134:257–267. doi: 10.1016/s0769-2609(83)80038-6. [DOI] [PubMed] [Google Scholar]
- Riviere GR, Smith KS, Tzagaroulaki E, Kay SL, Zhu X, DeRouen TA, Adams DF. Periodontal status and detection frequency of bacteria at sites of periodontal health and gingivitis. Journal of Periodontology. 1996;67:109–115. doi: 10.1902/jop.1996.67.2.109. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods in Enzymology. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Shaddox LM, Huang H, Lin T, Hou W, Harrison PL, Aukhil I, Walker CB, Klepac-Ceraj V, Paster BJ. Microbiological characterization in children with aggressive periodontitis. Journal of Dental Research. 2012;91:927–933. doi: 10.1177/0022034512456039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira JF, Jr, Rôças IN. Community as the unit of pathogenicity: An emerging concept as to the microbial pathogenesis of apical periodontitis. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontics. 2009;107:870–878. doi: 10.1016/j.tripleo.2009.01.044. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Smith C, Dibart S. Relation of counts of microbial species to clinical status at the sampled site. Journal of Clinical Periodontology. 1991;18:766–775. doi: 10.1111/j.1600-051x.1991.tb00070.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontology 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. Journal of Clinical Periodontology. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Teles FR, Teles RP, Uzel NG, Song XQ, Torresyap G, Socransky SS, Haffajee AD. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. Journal of Periodontal Research. 2012;47:95–104. doi: 10.1111/j.1600-0765.2011.01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade WG. The oral microbiome in health and disease. Pharmacological Research. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiology. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.